Abstract
Ribosome assembly in eukaryotes requires approximately 200 essential assembly factors (AFs), and occurs via ordered events that initiate in the nucleolus and culminate in the cytoplasm. Here we present the cryo-electron microscopy (cryo-EM) structure of a late cytoplasmic 40S ribosome assembly intermediate from Saccharomyces cerevisiae. The positions of bound AFs were defined using cryo-EM reconstructions of pre-ribosomal complexes lacking individual components. All seven AFs are positioned to prevent each step in the translation initiation pathway by obstructing the binding sites for initiation factors, by preventing the opening of the mRNA channel, by blocking 60S subunit joining, and by disrupting the decoding site. We suggest that these highly redundant mechanisms ensure that pre-40S particles do not enter the translation pathway, which would result in their rapid degradation. Implications for the regulation of 40S maturation are also discussed.
In eukaryotes, the assembly of the ribosomal subunits from the four rRNAs (18S, 5.8S, 25S and 5S) and 78 ribosomal proteins is facilitated by a conserved macromolecular machinery comprising ~200 assembly factors (AFs). These proteins, mostly essential, catalyze the modification, cleavage from precursor transcripts and folding of the rRNA, and facilitate the binding of ribosomal proteins (1). While the components of this assembly line have been identified, their functions remain largely unknown. Pioneering cryo-EM studies (2–7) followed by the crystal structures of prokaryotic (8–11) and very recently of eukaryotic mature ribosomes (12, 13) have provided functional insight to the complex architecture. However, little is known about the structure of assembly intermediates, or the binding sites for AFs.
While most ribosome assembly steps take place in the nucleolus, where rRNA is transcribed, final maturation of both subunits occurs in the cytoplasm. There, assembling ribosomal subunits encounter large pools of mature subunits, mRNA, and translation factors, representing a unique challenge: the prevention of premature translation initiation on immature subunits. This is of particular concern for assembling 40S subunits, as translation initiation factors, mRNAs and the 60S subunit all bind to 40S subunits to initiate translation, and furthermore, because premature 40S ribosomes, when incorporated into 80S ribosomes, are rapidly degraded (14). These considerations suggest the existence of mechanisms to prevent 80S formation before 40S maturation is complete. Here we report the structure of a late cytoplasmic pre-40S assembly intermediate. Our results show that bound AFs are positioned to cooperatively prevent each step in the course of translation initiation: the binding of initiation factors, the opening of the mRNA channel, the joining of 60S subunits, and finally the formation of the decoding site.
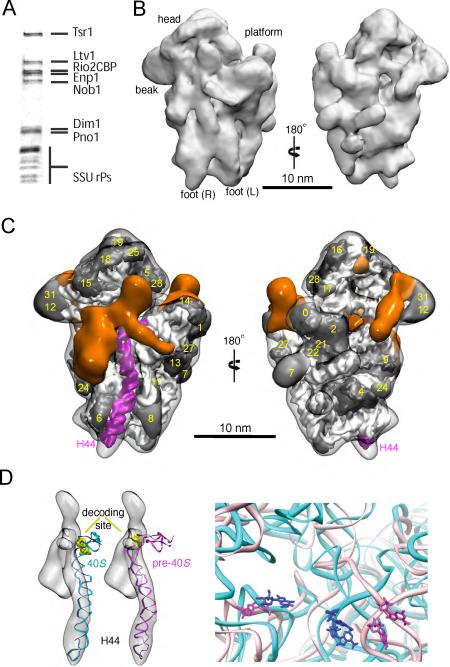
Using cryo-electron microscopy (cryo-EM), we have determined, at 18 Å resolution, the structure of a late pre-40S ribosome assembly intermediate, purified from S. cerevisiae via TAP-tagged Rio2, an essential kinase required for 18S rRNA production (Fig. 1, S1). As expected (15), this particle includes seven stably bound AFs: the methylase Dim1, the endonuclease Nob1, which produces the mature 3'-end, its regulator Pno1 (also referred to as Dim2), as well as the kinase Rio2, the GTPase-like protein Tsr1, the export adaptor Ltv1, and Enp1, a protein of unknown function (Fig. 1A). Furthermore, all small subunit ribosomal proteins (Rps), except Rps10 and Rps26 are present (Table S1). The 3D cryo-EM reconstruction, as well as 2D class averages of negative stained particles (Fig. S2) clearly show all features of the mature small subunit (SSU): the head, beak, platform, left and right foot (16). For a better interpretation of the EM densities, we applied explicit-solvent molecular dynamics flexible fitting (MDFF) (17–20) to fit the structure of the mature 40S ribosome into the cryo-EM map (Fig. S3&4, see Materials and Methods), and subsequently subtracted the resulting model from our reconstruction. This strategy allowed us to clearly reveal the densities corresponding to AFs, which are located in three major regions (orange in Fig. 1C): on the subunit interface, at the back of the beak, as well as on the back of the platform. All of these regions are important for translation initiation as discussed below. In addition, MDFF allowed us to characterize a major shift in conformation between mature and pre-40S particles at the upper part of H44, which affects the decoding site region, as discussed below (Fig. 1D).
Figure 1.
Molecular architecture of late pre-40S ribosomes. (A) SDS-PAGE analysis of pre-40S particles purified via Rio2-TAP. (B) 18Å cryo-EM reconstruction of pre-40S particles purified via Rio2-TAP. (C) Densities of AFs (in orange) revealed after explicit-solvent MDFF of the structure of mature 40S ribosomes into the cryo-EM map. rRNA is shown in white, Rps are shown in graphite and are annotated as in (13). H44 and H45 are highlighted in magenta. Subunit (left) and solvent (right) interface views are shown. (D) H44 is distorted in pre-40S particles. Rigid-body docking of the mature H44 structure (cyan) does not fit the corresponding density in the cryo-EM map (gray). MDFF allows for improved fit of H44 (magenta), accompanied by a change in the positioning of the decoding site residues (yellow). Close-up view reveals that the distance between G577 and A1755 is increased from 3 to ~4.5 nm in the pre-40S (magenta) or mature 40S (cyan) ribosomes.
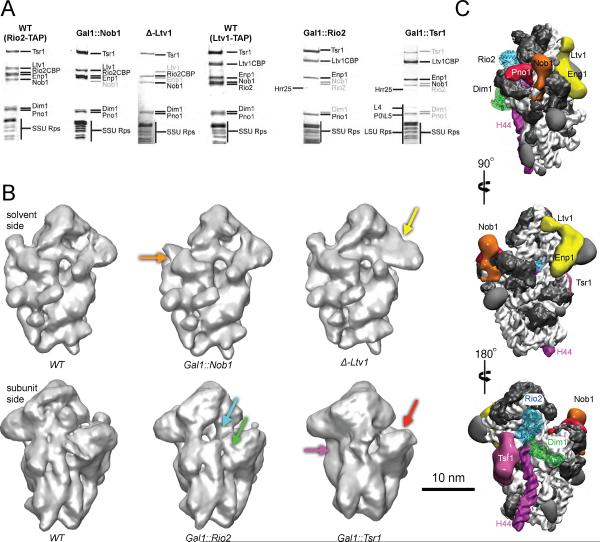
To assign the extra densities to individual AFs, we individually depleted (or deleted) the assembly factors Nob1, Rio2, Tsr1 and Ltv1, and determined the cryo-EM structures of the resulting particles to resolutions of 20 Å, 22 Å, 26 Å and 20 Å, respectively (Fig. 2A,B, S5–10). Comparisons, including t-test difference mapping, of the wild type cryo-EM map with maps of partial particles lacking individual AFs allows for localization of factors. E.g. in the Nob1 depletion, Nob1 was the only missing protein, allowing us to unambiguously assign its density at the platform. In contrast, in the Rio2 depletion, Nob1 and Dim1 were also missing. In that case, additional information from previous footprinting data was used to assign the two possible densities to Dim1 and Rio2. This assignment was also aided by the available crystal structures for Dim1 and Rio2, which fit the assigned densities well with cross correlation values of 0.854 and 0.904, respectively (Fig. 2C and S8). In addition, we obtained a 30 Å 3D reconstruction of negative stained recombinant Tsr1 (Fig. S11–12), and employed antibody labeling against Ltv1 and Enp1 (Fig. S13&14) to substantiate our assignments for Tsr1, Ltv1 and Enp1 (see Supplementary Materials for a detailed discussion on AF assignment). These results are summarized in Figure 2C. Importantly, these placements are largely consistent with previous crosslinking data, as well as a systematic analysis of protein-protein interactions [(21–24), Fig. S15&16, see supplemental discussion].
Figure 2.
(A) SDS-PAGE analysis of wild-type Rio2TAP, Gal1∷Nob1, Δ-Ltv1, wild-type Ltv1TAP, Gal1∷Rio2 and Gal1∷Tsr1 used in the cryo-EM shows depleted and co-depleted proteins. (B) cryo-EM maps for the wild type, Gal1∷Nob1, Δ-Ltv1 (solvent view) and wild type, Gal1∷Rio2, Gal1∷Tsr1 (subunit view) pre-40S particles identify densities belonging to individual assembly factors (See also Fig. S5–S14). Colored arrows point to the missing densities with color-coding as in (C). (C) Positioning of AFs on pre-40S particles. The structures of archeal Rio2 (blue) and human Dim1 (green) are docked within the corresponding cryo-EM densities.
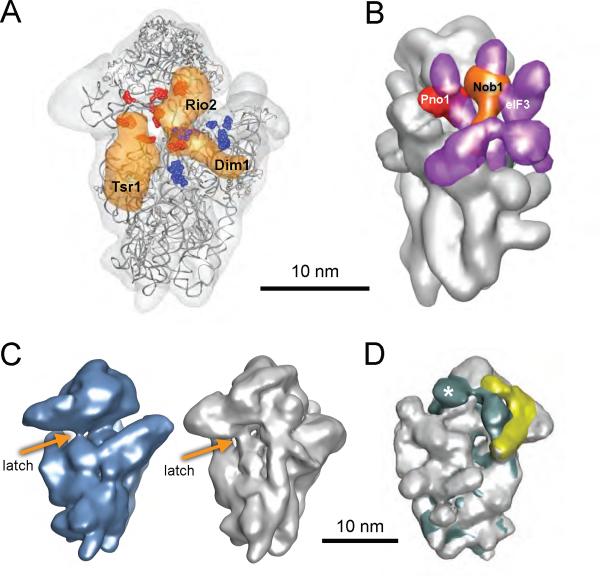
The cryo-EM maps show that the AFs on the subunit interface, Rio2, Tsr1 and Dim1 overlap the binding sites of translation initiation factors eIF1, and eIF1A (Fig. 3A), as previously shown for the bacterial homologs of Dim1 and eIF1 (25). Furthermore, the binding sites for the nuclease Nob1 and its regulator Pno1 overlap the binding site for eIF3 at the platform (Fig. 3B). Thus, recruitment of initiation factors that directly bind to the 40S subunit is prevented by the joint activity of Nob1, Pno1, Tsr1, Dim1 and Rio2.
Figure 3.
AFs obstruct translation initiation factor binding sites, and prevent mRNA binding. (A) Tsr1, Rio2 and Dim1 block binding of eIF1 and eIF1A. Sites of RNA footprints from Fe-labeled eIF1 and eIFA are shown in red and blue, respectively (44, 45). (B) Nob1 and Pno1 block binding of eIF3. Nob1 and Pno1 densities are shown in orange and red, respectively. The density for eIF3 (46) is shown in purple. (C) The latch between H18 and H34 closes the mRNA channel in mature (26) (blue) and pre-40S ribosomes (grey). The Tsr1 density was removed here to allow for better visualization of the latch. (D) Overlay of the structure of mature 40S ribosomes with eIF1 and eIF1A bound (26) (in aqua) and pre-40S particles (grey) shows that the hinge on the back of the beak (aqua) and the density for the Enp1/Ltv1/Rps3 complex (yellow) partially overlap. RACK1 is present only in mature 40S and its density is indicated with an asterisk.
During translation initiation, recruitment of the mRNA to the 40S subunit is facilitated by the activity of eIF1 and eIF1A (26). Their joint binding promotes a conformational switch in the 40S subunits, whereby the mRNA channel opens up, breaking the latch between helix 18 and helix 34 (Fig. 3C). This opening is stabilized on the solvent side by a hinge formed between the beak and shoulder region (26) (Fig. 3D). The Enp1/Ltv1/Rps3 complex at the back of the beak overlaps the bridge formed upon mRNA channel opening (Fig. 3D). Furthermore, the latch on the front site is closed in pre-40S particles (Fig. 3C). Thus, the structure suggests that opening of the mRNA channel is inhibited by the Enp1/Ltv1/Rps3 complex.
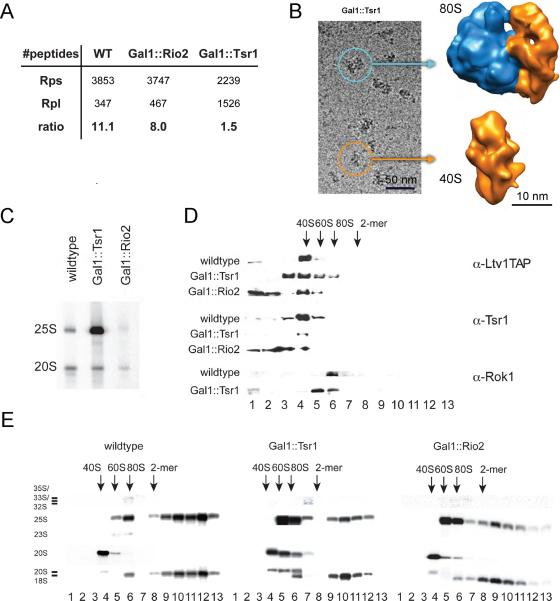
Despite the abundance of mRNAs and 60S subunits, 80S ribosomes are rarely (<10% of particles) observed in pre-40S purifications. Depletion of Tsr1 from the pre-40S particle leads to a >7-fold increase in peptides of ribosomal proteins from the 60S subunit (Rpl) that copurify with pre-40S subunits, such that the number of Rpl peptides is similar to that of Rps peptides, and copurifying Rpl4, Rpl5 and P0 are identified by SDS-PAGE (Fig. 2A, 4A). Furthermore, the amount of 25S rRNA is increased 20-fold (Fig. 4C), and 80S particles are frequently observed in cryo EM images upon Tsr1 depletion (Fig. 4B). To confirm that depletion of Tsr1 leads to formation of 80S ribosomes containing pre-40S-bound Ltv1 in vivo, we carried out sucrose gradient analysis to separate free proteins from 40S, 80S and polysome fractions, and used the TAP-tag on Ltv1 to probe for its localization. As expected, depletion of Tsr1, but not Rio2, shifts Ltv1 from being exclusively bound to a 40S-like particle to an 80S particle, indicative of 60S subunit binding (Fig. 4D). Northern analysis confirms that this 80S particle contains 20S and 25S rRNAs, indicating that it is not a 90S pre-ribosome, which would contain 35S rRNA, but not 25S or 20S rRNAs (Fig. 4E). This is also confirmed by Northern analysis of the TAP purification, which shows no 35S pre-rRNA in any strain (data not shown). Tsr1 and Pno1 are both depleted in the Tsr1 depletion but present in all other purifications, and therefore could both contribute to this effect. Alternatively, the loss of all interface-binding AFs together could also be responsible for this effect. Pno1, but not Tsr1, can be found in polysome fractions (data not shown, (27, 28)), indicating that the presence of Pno1 does not antagonize 80S formation. Furthermore, depletion of human Tsr1 leads to increased turnover of cytoplasmic 40S pre-ribosomes, as expected when pre-40S ribosomes form inactive 80S ribosomes, which are degraded by the no-go pathway (29). These data indicate that Tsr1 inhibits the premature association of 60S subunits with assembling 40S subunits in the cytoplasm. This anti-subunit-association activity of Tsr1 can be readily explained by the large surface it occupies on the subunit interface, and is analogous to that performed by eIF6 and Nmd3 for late cytoplasmic 60S precursors (30–32). We also note however that Tsr1 does not provide a complete block to 60S joining, as low amounts of 60S subunits can be found copurifying with wild type Rio2TAP pre-40S subunits, consistent with the previous observation that low amounts of Nob1 and 20S rRNA can be found in polysomes (14).
Figure 4.
Tsr1 blocks 60S subunit joining. (A) Peptides from ribosomal proteins belonging to the large subunit (Rpl; identified by mass spectrometry) copurify heavily with pre-40S particles purified from Gal1∷Tsr1 strains, but not from the wild type control, or the Gal1∷Rio2 strain. (B) 3D cryo-EM reconstructions confirm that larger particles in Ltv1TAP purifications from Gal1∷Tsr1 cells are 80S particles. (C) Northern blotting shows that 25S rRNA from the large subunit copurifies with pre-40S particles in Gal1∷Tsr1 cells relative to wild type or Gal1∷Rio2 cells. (D) Western blot of fractions from 10–50% sucrose gradients demonstrates that Ltv1TAP shifts from being bound only to 40S-preribosomes to 80S fractions when Tsr1 is depleted, but not when Rio2 is depleted. (E) Northern probing on gradient fractions demonstrates that 80S fractions in the Tsr1 depleted strain contain 20S and 25S rRNA, but not 35S or 23S pre-rRNA. The position of 40S, 60S and 80S fractions determined by absorbance is indicated. Fig. S19 shows the absorbance profiles from these gradients.
During decoding, A1755/A1756 flip out of H44 and G577 rotates around the glycosidic bond to interrogate the mRNA/tRNA base pair by hydrogen-bonding with the minor groove (33). The cryo-EM map of the pre-40S particle shows that H44 is kinked in the lower part, which drives the upper portion outwards and to the left (from the perspective of the 60S subunit). As a result, the nucleotides that make up the decoding site are displaced from their location in mature 40S subunits as indicated by the cryo-EM density (Fig. 1D). Importantly, A1755/A1756 and G577 move into opposite directions (Fig.1D) so that their distance increases, thereby precluding interactions of all three nucleotides with the mRNA/tRNA duplex. Thus, the shift in H44 disrupts the decoding site and prevents the subsequent steps leading to peptide bond formation.
Collectively these data indicate that in a highly redundant and multi-pronged approach all seven late-binding cytoplasmic ribosome AFs contribute to chaperoning pre-40S subunits to prevent them from prematurely engaging the translational apparatus. In this model, binding of translation factors eIF1, eIF1A, and eIF3 is precluded by binding of Dim1, Tsr1, Rio2, Pno1 and Nob1, while opening of the mRNA channel is prevented by binding of Enp1 and Ltv1 (Fig. 3). Tsr1 blocks the premature binding of 60S subunits (Fig. 4), and the decoding site is disrupted as a result of a prominent kink in H44 (Fig. 1). Bacteria lack all of these factors, with the exception of Dim1, which blocks binding of IF3, the homolog of eIF1 (25). However, during 30S assembly in bacteria the GTPase Era binds to the anti-Shine-Dalgarno sequence (34). Since bacterial mRNA recruitment occurs via base pairing between Shine-Dalgarno and anti-Shine Dalgarno sequences, binding of Era might prevent mRNA recruitment (34) analogous to Enp1 and Ltv1, although by entirely different mechanisms. Overlapping the function of Era is RbfA, whose binding to the bacterial SSU leads to a deformation in H44 similar to the one observed here. Interestingly, the binding site for RbfA is similar to the Pno1 binding site (35). Thus, it seems possible that the function of AFs is conserved between kingdoms, but different proteins have evolved to maintain these functions, reflecting the differences in the translation initiation pathways. The lack of redundancy in prokaryotes might reflect the fact that SSU assembly intermediates in polysomes are not as efficiently degraded in bacteria (36), as they are in eukaryotes (14).
Rps10 and Rps26 are the only ribosomal proteins that are not yet bound to the late pre-40S particles examined here (Table S1), consistent with the observation that their depletion does not stall processing of the 18S precursor, 20S rRNA (37). Furthermore, binding of Rps26 to 20S rRNA is negligible and does not increase when 20S rRNA is accumulated, indicating that Rps26 binds after formation of 18S rRNA (38). Rps26 binds on the platform (13), where it overlaps the binding sites of Pno1 in the pre-40S particle (Fig. S17A), indicating that Rps26 cannot bind before Pno1 dissociates. Similarly, Rps10 binds to the beak, overlapping the binding sites for Ltv1/Enp1 (Fig. S17B). Incorporation of these Rps might be a final regulated step in 40S assembly.
Furthermore, although Rps3 is present in pre-40S particles (Table S1), the cryo-EM map shows that it is not occupying its final position (Fig. S17C). This finding is consistent with previous data suggesting that in this pre-40S particle, but not mature 40S ribosomes, Rps3 is salt labile (15). Additionally, Rps14 is repositioned on the platform, consistent with previous findings that the interactions of its C-terminal extension with rRNA form late during ribosome assembly (39). The late assembly of both the platform region, where Rps26 and Rps14 bind, as well as the head/beak region, where Rps10 and Rps3 bind, is paralleled in the in vitro assembly of the bacterial SSU, where the head and platform region are the last to form, and intermediates lack the homologs for Rps14, Rps26, and Rps3 (40).
Nob1, the nuclease for cleavage at the 3'-end of 18S rRNA, is present in the late pre-40S particle purified here, yet, the particle only contains the 20S rRNA (Fig. 4B), and Nob1 does not cleave even after addition of Mn2+, a known cofactor (unpublished results). These findings suggest that either an external signal is required to activate Nob1, or that Nob1 is mispositioned in the presence of these assembly factors and thereby inactive. Previous in vivo and in vitro footprinting show that Nob1 protects the active site (41), and the EM-map shows that Nob1 is positioned in proximity to the cleavage site (Fig. 2), suggesting that any rearrangement, if it occurs, is limited. The C-terminal extensions of Rps14 and Rps5 are important for D-site cleavage and engage rRNA immediately prior to that step (39, 42).
The data presented here provide important clues about triggers for rearrangements that could reposition Nob1 and its essential effector Pno1, as well as Rps5 and/or Rps14. Previous work has shown that the bacterial homolog for Dim1 is inhibited by ribosomal protein S21. Although bacterial S21 has no eukaryotic sequence homolog, its binding site is almost identical to that of Rps26, suggesting that Rps26 is a functional homolog of S21 (13). We show that Pno1 occupies the Rps26 binding site in pre-40S particles (Fig. S17A). Because Pno1 binds directly to Nob1, and affects its interaction with rRNA (24), reverse communication from the Dim1 site could reposition Pno1, thereby regulating Nob1.
Interestingly Dim1 itself is positioned to receive signals regarding assembly of the head/beak region, which is shown here to be incomplete, as Rps10 is absent and Rps3 is not fully incorporated: Binding of the bacterial homologs for Rps3, and its direct binding partners Rps20 and Rps29, to the head of the SSU stabilizes an RNA conformation that inhibits the activity of Dim1's bacterial homolog at the subunit interface (43). Thus, completion of head assembly could trigger changes at the Dim1 site, which could be relayed to reposition Pno1 and Nob1 for final pre-rRNA cleavage.
Supplementary Material
Acknowledgements
We thank M. Campbell for the gift of recombinant Tsr1 and (MBP-)Tsr1, J.P. Gélugne for yeast strains, J. Doudna for making eIF3 maps available, H. Remmers (UM) and M. Chalmers (TSRI) for performing mass spectrometry experiments, S. Ludtke, J. Zhang, and L. Passmore for EMAN scripts to generate variance maps, and J. Doudna, J. Cleveland, K. Nettles, T. Walz, and members of the Karbstein and Skiniotis labs for comments. B.S.S. is supported by an NSF pre-doctoral fellowship. This work is supported by NIH grant R01-GM086451 (to K.K.), the NIH-supported resource Multiscale Modeling Tools for Structural Biology (grant RR12255 to C.L.B.), and the University of Michigan Biological Sciences Scholars Program (G.S.). G.S. is a Pew Scholar of Biomedical Sciences. EM maps have been deposited in the EMDB with accession codes EMD-1922, -1923, -1924, -1925, -1926, and EMD-1927.
References
- 1.Strunk BS, Karbstein K. Powering through ribosome assembly. RNA. 15:2083. doi: 10.1261/rna.1792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frank J, et al. A model of protein synthesis based on cryo-electron microscopy of the E. coli ribosome. Nature. 376:441. doi: 10.1038/376441a0. [DOI] [PubMed] [Google Scholar]
- 3.Beckmann R, et al. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science. 278:2123. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- 4.Spahn CM, et al. Structure of the 80S ribosome from Saccharomyces cerevisiae--tRNA-ribosome and subunit-subunit interactions. Cell. 107:373. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 5.Spahn CM, et al. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 118:465. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Taylor DJ, et al. Comprehensive molecular structure of the eukaryotic ribosome. Structure. 17:1591. doi: 10.1016/j.str.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armache JP, et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc Natl Acad Sci U S A. 107:19748. doi: 10.1073/pnas.1009999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF. X-ray crystal structures of 70S ribosome functional complexes. Science. 285:2095. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 9.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 289:905. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 10.Wimberly BT, et al. Structure of the 30S ribosomal subunit. Nature. 407:327. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 11.Harms J, et al. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 107:679. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 12.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 330:1203. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 13.Rabl J, Leibundgut M, Ataide S, Haag A, Ban N. Crystal Structure of the Eukaryotic 40S Ribosomal Subunit in Complex with Initiation Factor 1. Science. 331:730. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 14.Soudet J, Gelugne JP, Belhabich-Baumas K, Caizergues-Ferrer M, Mougin A. Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae. Embo J. 29:80. doi: 10.1038/emboj.2009.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schafer T, et al. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 441:651. doi: 10.1038/nature04840. [DOI] [PubMed] [Google Scholar]
- 16.Verschoor A, et al. Three-dimensional reconstruction of mammalian 40 S ribosomal subunit. J Mol Biol. 209:115. doi: 10.1016/0022-2836(89)90175-7. [DOI] [PubMed] [Google Scholar]
- 17.Trabuco LG, et al. The role of L1 stalk-tRNA interaction in the ribosome elongation cycle. J Mol Biol. 402:741. doi: 10.1016/j.jmb.2010.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trabuco LG, et al. Applications of the molecular dynamics flexible fitting method. J Struct Biol. 173:420. doi: 10.1016/j.jsb.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 16:673. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 49:174. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell MG, Karbstein K. Protein-Protein Interactions within Late Pre-40S Ribosomes. PLoS One. 2011;6:e16194. doi: 10.1371/journal.pone.0016194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Granneman S, Petfalski E, Swiatkowska A, Tollervey D. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking. Embo J. 29:2026. doi: 10.1038/emboj.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamanna AC, Karbstein K. A Conformational Change Regulates Pre-18S Cleavage. J. Mol. Biol. 2011;405:3. doi: 10.1016/j.jmb.2010.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woolls HA, Lamanna AC, Karbstein K. The Roles of Dim2 in Ribosome Assembly. J. Biol. Chem. 2011;286:2578. doi: 10.1074/jbc.M110.191494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Z, O'Farrell HC, Rife JP, Culver GM. A conserved rRNA methyltransferase regulates ribosome biogenesis. Nat. Struct. Mol. Biol. 15:534. doi: 10.1038/nsmb.1408. [DOI] [PubMed] [Google Scholar]
- 26.Passmore LA, et al. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007 Apr 13;26:41. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 27.Granneman S, Nandineni MR, Baserga SJ. The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14. Mol. Cell Biol. 25:10352. doi: 10.1128/MCB.25.23.10352-10364.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vanrobays E, Gelugne JP, Caizergues-Ferrer M, Lafontaine DL. Dim2p, a KH-domain protein required for small ribosomal subunit synthesis. RNA. 10:645. doi: 10.1261/rna.5162204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carron C, O'Donohue MF, Choesmel V, Faubladier M, Gleizes PE. Analysis of two human pre-ribosomal factors, bystin and hTsr1, highlights differences in evolution of ribosome biogenesis between yeast and mammals. Nucleic Acids Res. 39:280. doi: 10.1093/nar/gkq734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho JH, Johnson AW. NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol Cell Biol. 19:2389. doi: 10.1128/mcb.19.3.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sengupta J, et al. Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J Cell Biol. 189:1079. doi: 10.1083/jcb.201001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Si K, Maitra U. The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor. Mol Cell Biol. 19:1416. doi: 10.1128/mcb.19.2.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogle JM, et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 292:897. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 34.Sharma MR, et al. Interaction of Era with the 30S ribosomal subunit implications for 30S subunit assembly. Mol. Cell. 18:319. doi: 10.1016/j.molcel.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 35.Datta PP, et al. Structural aspects of RbfA action during small ribosomal subunit assembly. Mol Cell. 28:434. doi: 10.1016/j.molcel.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy-Chaudhuri B, Kirthi N, Culver GM. Appropriate maturation and folding of 16S rRNA during 30S subunit biogenesis are critical for translational fidelity. Proc Natl Acad Sci U S A. 107:4567. doi: 10.1073/pnas.0912305107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferreira-Cerca S, Poll G, Gleizes PE, Tschochner H, Milkereit P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell. 20:263. doi: 10.1016/j.molcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 38.Ferreira-Cerca S, et al. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol. Cell. 28:446. doi: 10.1016/j.molcel.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 39.Jakovljevic J, et al. The carboxy-terminal extension of yeast ribosomal protein S14 is necessary for maturation of 43S preribosomes. Mol. Cell. 14:331. doi: 10.1016/s1097-2765(04)00215-1. [DOI] [PubMed] [Google Scholar]
- 40.Mulder AM, et al. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science. 330:673. doi: 10.1126/science.1193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamanna AC, Karbstein K. Nob1 binds the single-stranded cleavage site D at the 3'-end of 18S rRNA with its PIN domain. Proc Natl Acad Sci U S A. 106:14259. doi: 10.1073/pnas.0905403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neueder A, et al. A local role for the small ribosomal subunit primary binder rpS5 in final 18S rRNA processing in yeast. PLoS One. 2010;5:e10194. doi: 10.1371/journal.pone.0010194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desai PM, Rife JP. The adenosine dimethyltransferase KsgA recognizes a specific conformational state of the 30S ribosomal subunit. Arch Biochem Biophys. 449:57. doi: 10.1016/j.abb.2006.02.028. [DOI] [PubMed] [Google Scholar]
- 44.Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 17:2786. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu Y, et al. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res. 37:5167. doi: 10.1093/nar/gkp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 310:1513. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.