Abstract
We reported previously that 10 older men (66.4±4.6 years) with premutation alleles (55–200 CGG repeats) of the FMR1 gene, with or without FXTAS, had decreased telomere length when compared to sex- and age-matched controls. Extending our use of light intensity measurements from a telomere probe hybridized to interphase preparations, we have now found shortened telomeres in 9 younger male premutation carriers (31.7±17.6 years). We have also shown decreased telomere length in T lymphocytes from 6 male individuals (12.0±1.8 years) with full mutation FMR1 alleles (>200 CGG repeats). These findings support our hypothesis that reduced telomere length is a component of the sub-cellular pathology of FMR1-associated disorders. The experimental approach involved pair-wise comparisons of light intensity values of 20 cells from an individual with either premutation or full mutation CGG-repeat expansions relative to an equivalent number of cells from a sex- and age-matched control. In addition, we demonstrated reduced telomere size in T-lymphocyte cultures from eight individuals with the FMR1 premutation by using six different measures. Four relied on detection of light intensity differences, and two involved measuring the whole chromosome, including the telomere, in microns. This new approach confirmed our findings with light intensity measurements and demonstrated the feasibility of direct linear measurements for detecting reductions in telomere size. We have thus confirmed our hypothesis that reduced telomere length is associated with both premutation and full mutation-FMR1 alleles and have demonstrated that direct measurements of telomere length can reliably detect such reductions.
INTRODUCTION
Telomeres or chromosome ends consist of highly conserved TTAGGG repeats that become reduced in length with each subsequent cell cycle [Samassekou et al., 2010]. Reduced telomere length is found in a number of conditions. They include replicative cellular senescence and apoptosis [Allsopp et al., 1992; Hao et al., 2004], tumorigenesis [Plentz et al., 2004; Oeseburg et al., 2010; Martinez-Delgado et al., 2011], in vivo cellular aging [Hastie et al., 1990; Lindsey et al., 1991; Ahmed et al., 2001; Flanary et al., 2003], heart disease [Samani et al., 2001; Benetos et al., 2004; Epel et al., 2009; Wong et al., 2010], stress [Epel et al., 2004; Damjanovic et al., 2007; Puterman et al., 2010; Wolkowitz et al., 2011; Drury et al., 2011; Entringer et al., 2011], dyskeratosis congenita [Vulliamy et al., 2007; Nelson & Bertuch, 2011], Alzheimer disease [Panossian et al., 2004], and dementia in Down syndrome (Jenkins et al., 2006, 2010).
We reported previously that older men (56–73 years) with premutation alleles (55–200 CGG repeats) of the fragile X mental retardation (FMR1) gene, including seven individuals with fragile X-associated tremor/ataxia syndrome (FXTAS) and three without, exhibited reductions in telomere length relative to controls [Jenkins et al., 2008a]. We have now observed similar reductions in younger male carriers (8.6–56 years) of either a premutation or a full mutation. We hypothesize that increased telomere shortening may be intrinsic to the nature of the premutation and the full mutation. Data presented in this paper support this hypothesis. Here we show that shorter telomeres may be detected in FMR1 premutation male individuals by using six different measures including the novel approach of measuring chromosome 1 telomeres in microns.
MATERIALS AND METHODS
Subjects, Molecular Analysis
Twenty-nine males with either the FMR1 premutation or full mutation allele, as well as controls, age 8.6 to 62 years, were recruited through the Fragile X Research and Treatment Center at the University of California Davis MIND Institute. This research was conducted under an IRB-reviewed/approved protocol.
FMR1 genotyping was performed using both PCR and Southern blot analyses as previously described [Tassone et al., 2008]. Analyses of repeat sizes involved the use of an Alpha Innotch FluorChem 880-0 Image Detection System.
Telomere Analysis
Anonymous buffy coat samples, obtained by gradient centrifugation with a Ficoll Paque protocol, were cultured at 37°C for four days at an initial concentration of 200,000–400,000 viable mononuclear cells per ml of PHA-containing medium. Metaphase preparations were hybridized with both an FITC(fluorescein isothiocyanate)-labeled PNA (peptide nucleic acid) telomere probe (DAKO, North America) and a centromere 2 (cen 2) probe (a gift for investigational use from DAKO, Glostrup, Denmark), and DAPI(4',6-diamidino-2-phenylindole)-counterstained. Light intensity differences, as well as linear differences in microns, were detected by an image analyzer (MetaSystems Inc., Waltham, MA) as previously described [Jenkins et al., 2008]. Light intensity data from the cen 2 PNA probe provided a non-telomere standard that allowed us to generate light intensity ratios from telomere and cen 2 light intensities [Perner et al., 2003], thus providing a means for normalization of intensities. For telomere length analysis, a ratio was calculated by dividing total telomere length for chromosome 1 by total chromosome 1 length minus total telomere length (i.e., the non-telomere portion of chromosome 1).
We had observed previously that interphase preparations, whole metaphase preparations, and individual chromosomes (21, 1, 2, and 16) could each be used separately to detect telomere length differences in people with Down syndrome, with and without dementia or MCI (mild cognitive impairment) [Jenkins et al., 2006; 2010]. The current study therefore used both interphase and metaphase preparations. In addition, the metaphases were used to determine the mean number of chromosome arms with no telomere probe signal, since we have shown this measure to be effective in detecting telomere changes in people with Down syndrome and dementia or MCI [Jenkins et al., 2008b].
Finally, each analysis was performed in a pairwise fashion, such that 20 cells from a person with an expanded CGG repeat were compared to 20 cells from an age- and sex-matched control. t tests were conducted on each paired analysis. All light intensity analyses were blinded by the image analyzer whereas absolute physical length in microns and determination of the mean number of chromosome arms with no signals utilized preparations that had already been analyzed blindly for light intensity differences, where metaphases were relocated and analyzed for absolute length determination. The lengths themselves were blind as the computer (using MetaSystems Image Analyzer software called “Isis”) determined the length of each telomere in pixels which converted those values to micron lengths that were then tabulated by the microscopist for final statistical analysis. The same relocation system was used for actual signal number counts, but the counts were done by the microscopist who did not know whether the material came from an individual with the premutation or full mutation or a control until the analysis was completed.
Results
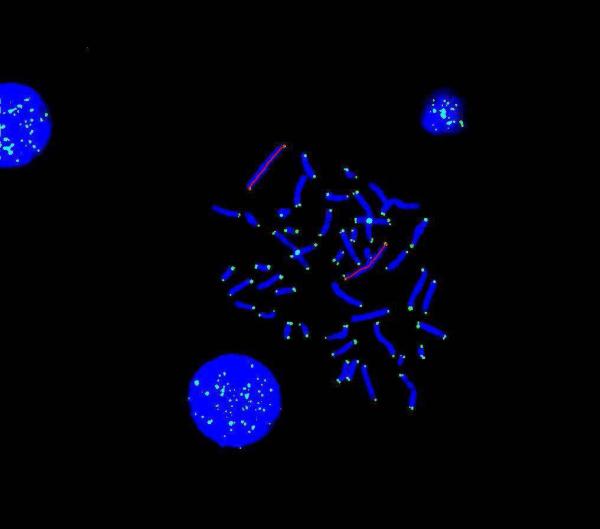
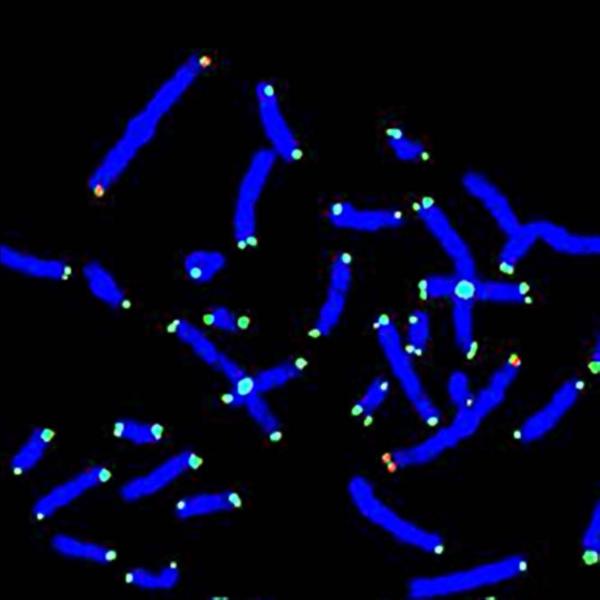
Reduced telomere size was found in metaphase chromosome 1 telomeres in cells from all 8 of the FMR1 premutation males, aged 8.6 to 56.2 years (33.±6.2), as shown in Table 1 (Cases 1P-8P). PNA telomere probe light intensity values were compared to those of age-matched controls (p < 0.0001). Analysis of cen 2 light intensity ratios gave the same comparative results except that all comparisons resulted in greater statistical significance (p < 10−6). Similarly, when telomere length was measured directly and comparisons were made on the same 8 premutation males, all telomeres were shorter than for age-matched controls (p < 10−6). Fig. 1a shows a DAPI-stained metaphase preparation where telomeres and chromosome 2 centromeres were hybridized by FITC-labeled PNA probes and a red line indicates the length of the longitudinal axis of the whole chromosome1 (both telomere and “inter-telomere” chromosomal material) while Fig. 1b shows the lengths of the chromosome 1 telomeres.
TABLE 1.
Shorter telomeres, determined by six different measures including micron (micrometer) linear length, in 15 male individuals with the FMR1 premutation (n=9) and full mutation (n=6)
| Case#1Premutation (P) Control (C) | Age (yrs) | cgg2 | Mean (sem) Chr. 1 tel light inten.3 ×103 | Cen 2 Ratio4 | Interph Light Inten5 (sem) | mncans6 (sem) | Chr. 1 μm tel length7 | Premut chr. 1 tel Length ratio8 |
|---|---|---|---|---|---|---|---|---|
| 1P (20-08) | 56.2 | 121 | 3.3 (0.2) | 0.7 (0.02) | 58.8 (4.3) | 19.9 (1.1) | 2.2 (0.13) | 0.11 (0.006) |
| 1C (295-08) | 62 | 19 | 5.8( 0.3) | 1.5 (0.04) | 140.7 (14.8) | 7.3 (0.4) | 4.9 (0.13) | 0.2 (0.006) |
| 2P (361-08) | 56 | 90 | 3.0 (0.2) | 0.7 (0.02) | 53.9 (3.9) | 24.3 (0.9) | 2.4 (0.13) | 0.11(0.006) |
| 1C (295-08) | 62 | 19 | 5.8 (0.3) | 1.5 (0.04) | 140.7 (14.8) | 7.3 (0.4) | 4.9 (0.13) | 0.2 (0.006) |
| 3P (231-08) | 38.5 | 98 | 3.4 (0.2) | 0.9 (0.02) | 61.4 (9.9) | 15.4 (0.8) | 2.5 (0.13) | 0.11(0.006) |
| 2C (246.08) | 39.1 | 28 | 5.3 (0.3) | 1.8 (0.09) | 137.1 (13.4) | 6.9 (0.4) | 4.6 (0.14) | 0.2 (0.006) |
| 4P (203-08) | 38.9 | 140 | 2.6 (0.2) | 0.9 (0.02) | 47.1 (4.9) | 17.6 (1.0) | 2.5 (0.13) | 0.11 (0.006) |
| 2C (246.08) | 39.1 | 28 | 5.3 (0.3) | 1.8 (0.09) | 137.1 (13.4) | 6.9 (0.4) | 4.6 (0.14) | 0.2 (0.006) |
| 5P (216-08) | 34.6 | 92 | 3.4 (0.2) | 0.8 (0.04) | 107.3 (7.5) | 16.3 (0.8) | 2.8 (0.13) | 0.11(0.006) |
| 3C (25-08) | 35.2 | 20 | 5.6 (0.4) | 1.5 (0.07) | 214.7 (18.7) | 6.2 (0.6) | 4.6 (0.14) | 0.21 (0.006) |
| 6P (354-08) | 23.1 | 74 | 3.3 (0.2) | 0.7 (0.02) | 82.7 (6.6) | 22.0 (1.2) | 2.7 (0.14) | 0.13 (0.01) |
| 4C (352-08) | 23 | 29 | NA10 | NA | 140.3 (9.0) | NA | - | - |
| 5C (299-08) | 34.3 | 31 | 5.9 (0.3) | 1.5 (0.04) | 144.1 (13.4) | 10.0 (0.5) | 5.6 (0.13) | 0.25 (0.01) |
| 7P (199-08) | 14.2 | 92 | 3.6 (0.2) | 0.8 (0.02) | 89.9 (9.2) | 20.2 (1.1) | 2.1 (0.1) | 0.13 (0.006) |
| 6C (212-08) | 15.3 | 27 | 6.5 (0.6) | 1.6 (0.07) | 145.0 (6.6) | 8.5 (0.6) | 4.5 (0.14) | 0.22 (0.01) |
| 8P (177-08) | 8.6 | 124 | 3.2 (0.2) | 0.7 (0.04) | 69.3 (6.0) | 18.9 (1.1) | 2.5 (0.13) | 0.13 (0.006) |
| 7C (178-08) | 9.1 | 36 | 5.7 (0.4) | 1.6 (0.09) | 125.0 (8.9) | 8.5 (0.7) | 4.3 (0.14) | 0.22 (0.01) |
| 9P (197-09)n | 15.3 | 104 | - | - | 53.9 (6.1) | - | - | - |
| 8C (142-09) | 15.2 | 30 | - | - | 136.7(9.3) | - | - | - |
| Case#1 Full mutation (F) Control (C) | Age (yrs) | cgg2 | Mean (sem) Chr. 1 tel light inten.3 ×103 | Cen 2 Ratio4 | InterphLight Inten5 (sem) | mncans6 (sem) | Chr. 1 μm tel length7 | Full mut chr. 1 tel length ratio8 |
|---|---|---|---|---|---|---|---|---|
| Full mut. (F) 10F 12 (21-10) | 19.4 | Fm12 | - | - | 50.1 (6.3) | - | - | - |
| 9C (333-09) | 20.8 | 26 | - | - | 148.4 (10.0) | - | - | - |
| 11F (424-09) | 8.5 | Fm | - | - | 38.7 (4.2) | - | - | - |
| 10C (421-09) | 8.8 | 30 | - | - | 99.0 (6.7) | - | - | - |
| 12F (143-09) | 14 | Fs12 | - | - | 55.0 (6.5) | - | - | - |
| 11C (346-09) | 14.8 | 30 | - | - | 150.0 (9.0) | - | - | - |
| 13F (177-09) | 12.4 | Fm | - | - | 47.0 (3.9) | - | - | - |
| 12C (305-09) | 12.6 | 20 | - | - | 122.7 (6.7) | - | - | - |
| 14F (22-10) | 7.1 | Fs | - | - | 38.8 (3.7) | - | - | - |
| 13C (504-09) | 7.4 | 32 | - | - | 118.7(4.9) | - | - | - |
| 15F (46-10) | 10.7 | Fs | - | - | 45.5 (3.0) | - | - | - |
| 14C (349-08) | 11.4 | 24 | - | - | 115.2 (4.7) | - | - | - |
Case number 1P and partial log number (20-08);
CGG repeat number;
LI = mean light intensity from chromosome 1 telomeres (note: all numbers in parentheses are SEM values);
ratio of Chr. 1 telomere light intensity/centromere 2 light intensity;
interphase preparations; pair-wise comparisons of interphase light intensity values [e.g., 1P (20-08 - premutation) vs. 1C (295-08 – control; control values are immediately under each Premutation or Full Mutation case and are all in “shaded” rows.)] resulted in p values ranging from < 0.0003 - < 1×10−6;
mean number of chromosome arms with no signal (mncans);
chromosome 1 telomere length in microns from premutation (8 individuals) – Case 9P (197-09) had insufficient metaphase material; only interphase could be used) versus age- and sex-matched controls (6) resulted in p values of < 1×10−6;
ratio of chromosome 1 telomere length/(chromosome 1 length – telomere length) from a premutation individual versus control resulted in p values < 1×10−6;
similar to 8 except the person (control) is age- and sex-matched with no premutation as indicated by cgg value;
Insufficient metaphase material but older control (299-08) used for metaphase comparisons;
Only interphase available;
full mutation, methylation mosaic; fs = full mutation, size mosaic; pair-wise comparisons of light intensity values from interphase preparations of 6 individuals with full mutations versus controls resulted in p values of < 1×10−6. The six full mutation cases are labeled 10F-15F.
Chrom 1 LI: 20 vs 295 p < 1×10−6; 361 vs 295 p < 1×10−6; 231 vs 246 p < 9×10−6; 203 vs 246 p < 1×10−6; 216 vs 25 p < 6×10−5; 354 vs 352 P NA; 354 vs 299 p < 1×10−6; 199 vs 212 p < 9×10−5; 177 vs 178 p < 2×10−5
Cen2 Chrom 1: 20 vs295 p < 1×10−6; 361 vs 295 p < 1×10−6; 231 vs 246 p 1×10−6; 203 vs 246 p < 1×10−6; 216 vs 25 p < 1×10−6; 352 vs354 p NA; 299 vs 354 p < 1×10−6;
199 vs 212 p < 1×10−6; 177 vs 178 p < 1×10−6
Signal no: mncans 20 vs 295 p < 1×10−6; 361 vs 295 p < 1×10−6; 231 vs 246 P < 1×10−6; 203 vs 246 p < 1×10−6; 216 vs 25 p < 1×10−6; 354 vs 299 p < 1×10−6; 199 vs 212 p < 1×10−6; 177 vs178p < 1×10−6
Individual outcome measures include light intensities of chromosome 1 telomeres, interphase telomeres, “loss” of telomeres shown by number of chromosome arms with no signals (mncans) from short-term cultured T lymphocytes. 8 of 8 premutation individuals exhibited shorter chromosome 1 telomeres measured in microns (μm).
Fig. 1a.
Method for determining length. Telomeres in metaphase and interphase, from a male individual with the FMRI premutation, marked by an FITC-labeled PNA probe with DAPI counter staining. PNA cen 2 probe also present. Chromosome 1 lengths marked in red: left at 11:00 = 18.9 μm and right at 3:00 = 19.0 μm.
Fig. 1b.
same as Fig. 1a except the chromosome 1 telomere length is marked in red and the now partial metaphase has been enlarged: Chromosome 1 at 11:00 – upper 1ptel = 0.8 μm and 1qtel = 1.0 μm; Chromosome 1 to right at 3:00 – upper 1ptel = 1.0 μm and 1qtel on left = 0.7 μm and 1qtel on the right = 0.8 μm.
The same results were also obtained when the mean number of chromosome arms with no PNA telomere probe signals from FMR1 premutation individuals was compared to age-matched control values (p < 10−6). When interphase preparations were used to compare PNA telomere probe light intensity values among these same 8 premutation individuals versus controls, all premutation individuals exhibited reduced light intensities (p < 0.0003). Finally, when interphase preparations were analyzed from 6 FMR1 full mutation male individuals from 8.5 – 19.4 (12.0±1.8) years old, versus age-matched male controls, telomeres were shorter in all 6 full mutation male individuals (p < 10−6). The same observation was made for FMR1 premutation case #9P. Thus 9 of 9 FMR1 premutation male individuals from 8.6 years to 56.2 years (31.7±5.9 - Cases 1P-9P, Table 1), exhibited shorter telomeres than controls. Only interphase preparations were used for the full mutation studies, since we had demonstrated the reliability of such preparations for the premutation cases.
Discussion
In conclusion, we have extended our earlier observations of shortened telomeres in older premutation adults [Jenkins et al.., 2008a] to a cohort of younger carriers (mean, 33.8 years; range, 8.6 to 56.2 years) as well as younger males (mean, 12.0 yrs; range 7.1 to 19.4) with full mutation alleles.
Six measures were used to demonstrate shorter telomeres in 8 of 8 male individuals with the FMR1 premutation:
-
1)
PNA telomere probe light intensities on chromosome 1 alone were determined.
-
2)
Using the cen 2 ratio data provided a non-telomere standard that allowed us to generate light intensity ratios: telomere intensities/cen 2 intensities [Perner et al., 2003]. Although we obtained nearly the same results with and without the use of the cen 2 control, we recognize that light intensities can vary from preparation to preparation or from lot-to- lot of PNA probe, so that using the cen 2 probe, which is not expected to be affected by factors that are associated with telomere shortening - such as Alzheimer Disease [Panossian et la., 2003], degenerative defects of aging [Armanios et al., 2009], progerin production during cell senescence [Cao et al., 2011], and cellular aging [Mather et al., 2011] - allowed us to normalize the results.
-
3)
Measuring the absolute telomere length in microns (micrometers) was done.
-
4)
The same rationale for using cen 2 ratios was applied for using the absolute telomere length in microns/inter-telomere chromosome 1 length for analysis of telomere length of chromosome 1 in microns. Its application was even more theoretical than cen 2, because the t test p values for comparing telomere lengths alone or analyzing the ratio values were the same (p < 0.000001).
-
5)
Determination of the mean number of chromosome arms with no signal and finding significantly more signal loss in premutations versus controls.
-
6)
Finally, we utilized and compared reduced light intensities in interphase preparations.
Our interphase findings, in agreement with the others, allowed us to rely upon interphase preparations solely for light intensity comparisons in cells from male individuals with the full FMR1 mutation. This is important because the use of interphase preparations allows the most efficient use of time to complete the analysis. Use of interphase is less labor-intensive than using metaphase preparations and allows for increased productivity. However, we believe that interphase preparations provide results with increased variability due to increased background fluorescence, resulting in more “noise” in the data generated. In regard to reliability, all methods are appropriate. If interphase preparations are not used, then we recommend physical measurement of telomeres in microns as the next most productive and practical methodology since telomere and inter-telomere chromosome lengths may be analyzed without the use of an expensive image analyzer and it is faster than all other methods except interphase. Similarly, signal number loss can also be determined without the use of an image analyzer and accompanying expensive software, but it is the most labor intensive.
Finally, our observations suggest that telomere shortening in the premutation range is not a consequence of the development of FXTAS, since most individuals in the newer cohort are much younger than the age range for development of the neurodegenerative disorder. Telomere shortening could be due to pathogenic mechanisms that are active much earlier in the lives of premutation carriers, like the alterations in lamin A nuclear morphology [Garcia-Arocena et al., 2010] that was also observed by Cao et al. [2011] during the induction of cell senescence. However, when mean light intensity interphase values from premutations were compared to those of the full mutation males, premutation T lymphocytes exhibited longer telomeres (premutation mean, 71.3 × 103; full mutation mean, 45.8 × 103; p <0.013) suggesting that something in full mutation individuals is causing even greater reduction in telomere length. Therefore, we suggest that telomere shortening may not be solely related to the effects of RNA toxicity, that are largely limited to the premutation range.
ACKNOWLEDGMENTS
This work was supported in part by the New York State Office for People with Developmental Disabilities, NICHD grants HD 036071, HD02274, NINDS NS044299, and the NIH Roadmap Initiative (UL1 RR024922, NCRR: RL1 AG032119. We thank Ezzat El-Akkad and Lawrence Black for their assistance.
Grant sponsor: New York State Office for People with Developmental Disabilities; Grant sponsor: NICHD; Grant numbers: HD036071, HD02274, NINDS NS044299; Grant sponsor: NIH Roadmap Initiative; Grant number: UL1 RR024922; Grant sponsor: NCRR; Grant number: RL1 AG032119; Grant sponsor: NIA.
REFERENCES
- Ahmed A, Tollefsbol T. Telomeres and telomerase: basic science implications for aging. J Am Geriatr Soc. 2001;49:1105–1109. doi: 10.1046/j.1532-5415.2001.49217.x. [DOI] [PubMed] [Google Scholar]
- Allsop RC, Caziri H, Patterson C, Boldsten SE, Younglai V, Futcher AB, Greider CW, Harley CG. Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci USA. 1992;89:1014–1018. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios E, Alder JK, Parry EM, Karim B, Strong MA, Greider CW. Short telomeres are sufficient to cause the degenerative defects. Am J Hum Genet. 2009;85:823–832. doi: 10.1016/j.ajhg.2009.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetos A, Gardner JP, Zureik M, Labat C, Xiaobin L, Adamopoulos C, Temmar M, Bean KE, Thomas F, Aviv A. Short telomeres are associated with increased carotid atherosclerosis in hypertensive subjects. Hypertension. 2004;43(20):182–185. doi: 10.1161/01.HYP.0000113081.42868.f4. [DOI] [PubMed] [Google Scholar]
- Cao K, Blair ED, Faddah DA, Kleckheefer JE, Oliver M, Erdos MR, Nabel EG, Collins FS. Progerin and telomere dysfunctional collaborate to trigger cellular senescence in normal human fibroblasts. J Clin Invest. 2011;121(7):2833–2844. doi: 10.1172/JCI43578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damjanovic AK, Yang Y, Glaser R, Kiecolt-Glaser JK, Nguyen H, B Laskowski B, Zou U, Beversdorf DQ, Eng NP. J Immunol. 2007;179:4249–4254. doi: 10.4049/jimmunol.179.6.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury SS, Theall K, Gleason MM, Smyke AT, Vivo ID, Wong JYY, Fox NA, Zeanah CH, Nelson CA. Telomere length and early severe social deprivation: linking early adversity and cellular aging. Molecular Psychiatry. 2011;(2011):1–9. doi: 10.1038/mp.2011.53. doi:10.1038/mp.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entinger S, Epel ES, Kumsta R, Hellhammer LJ, Balckburn EH, Wüst S, Wadhwa PD. Stress exposure in intrauterine life is associated with shorter telomere length in young adulthood. Proc Natl Acad Sci USA. 2011 Aug 3; doi: 10.1073/pnas.1107759108. 2011. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epel ES, Blackburn EH, Lin J, Dhabhar FS, Adler NE, Morrow JD, Cawthon RM. Accelerated telomere shortening in response t life stress. PNAS. 2004;101(49):17312–17315. doi: 10.1073/pnas.0407162101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epel ES, Merkin SS, Cawthon R, Blackburn EH, Adler NE, Petcher MJ, Seeman TE. The rate of leukocyte telomere shortening predicts mortality from cardiovascular disease in elderly men. Aging. 2009;1(1):81–88. doi: 10.18632/aging.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanary BE, Streit WJ. Telomeres shorten with age in rat cerebellum and cortex in vivo. J Anti-Aging Med. 2003;6:299–308. doi: 10.1089/109454503323028894. [DOI] [PubMed] [Google Scholar]
- Garcia-Arocena D, Yang JE, Brouwer JR, Tassone F, Iwahashi C, Berry-Kravis EM, Goetz CG, Sumis AM, Zhou L, Nguyen DV, Campos L, Howell E, Ludwig A, Greco C, Willemsen R, Hagerman RJ, Hagerman PJ. Fibroblast phenotype in male carriers of FMR1 premutation alleles. Hum Mol Genet. 2010 Jan 15;19(2):299–312. doi: 10.1093/hmg/ddp497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RJ, Coffey SM, Maselli R, Soontarapornchai K, Brunberg JA, Leehey MA, Zhang L, Gane LW, Fenton-Farrell G, Tassone F, Hagerman PJ. Neuropathy as a presenting feature in Fragile X-Associated Tremor/Ataxia Syndome. Am J Med Genet Part A. 2007;143A:2256–2260. doi: 10.1002/ajmg.a.31920. [DOI] [PubMed] [Google Scholar]
- Hastie ND, Dempster M, Dunlop MG, Thompson AM, Green DK, Allshire RC. Telomere reduction in human colorectal carcinoma and with aging. Nature. 1990;346(6287):866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- Jenkins EC, Velinov MT, Ye L, Gu H, Li S, Jenkins EC, Jr, Sklower Brooks S, Pang D, Devenny DA, Zigman WB, Schupf N, Silverman WP. Telomere shortening in T lymphocytes of older individuals with Down syndrome and dementia. Neurobiol Aging. 2006;27:941–945. doi: 10.1016/j.neurobiolaging.2005.05.021. [DOI] [PubMed] [Google Scholar]
- Jenkins EC, Tassone F, Ye L, Gu H, Xi M, Velinov M, Brown WT, Hagerman RJ, Hagerman PJ. Reduced telomere length in older men with premutation alleles of the Fragile X Mental Retardation 1 Gene. Am J Med Genet Part A. 2008a;146A:1543–1546. doi: 10.1002/ajmg.a.32342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins EC, Ye L, Gu H, Ni SA, Duncan CJ, Velinov M, Pang D, Krinsky-McHale SJ, Zigman WB, Schupf N, Silverman WP. Increased “Absence” of telomeres may indicate AD/dementia status in older individuals with Down syndrome. Neurosci Letters. 2008b Aug 8;440(3):340–3. doi: 10.1016/j.neulet.2008.05.098. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins EC, Ye L, Gu H, Ni SA, Velinov M, Pang D, Krinsky-McHale SJ, Zigman WB, Schupf N, Silverman WP. Shorter telomeres may indicate dementia status in older individuals with Down syndrome. Neurobiol Aging. 2010;31:765–771. doi: 10.1016/j.neurobiolaging.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey J, McGill NI, Lindsey LA, Green DK, Cooke HJ. In vivo loss of telomeric repeats with age in humans. Mutat Res. 1991;256(1):45–48. doi: 10.1016/0921-8734(91)90032-7. [DOI] [PubMed] [Google Scholar]
- Martinez-Delgado B, Yanowsky K, Inglada-Perez L, Domingo S, Urioste M, Osorio A, Benitez J. Genetic anticipation is associated with telomere shortening in hereditary breast cancer. PLoS Genet. 2011;7(7):e1002182. doi: 10.1371/journal.pgen.1002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather KA, Jorm AF, Parslow RA, Christensen H. Is telomere length a biomarker of aging? A review. J Gerontol A Biol Sci Med Sci. 2011;66A(2):202–213. doi: 10.1093/gerona/glq180. [DOI] [PubMed] [Google Scholar]
- Nelson ND, Bertuch AA. Dyskeratosis congenital as a disorder of telomere maintenance. Mutat Res. 2011 Jul 2; doi: 10.1016/j.mrfmmm.2011.06.008. 2011. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeseburg H, de Boer RA, van Gilst WH, van der Harst P. Telomere biology in healthy aging and disease. Pflugers Arch. 2010;459(2):259–268. doi: 10.1007/s00424-009-0728-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panossian LA, Porter VR, Valenzuela HG, Zhu X, Reback E, Masterman D, Cummings JL, Effros RB. Telomere shortening in T cells correlates with Alzheimre's disease status. Neurobiol Aging. 2003;24:77–84. doi: 10.1016/s0197-4580(02)00043-x. [DOI] [PubMed] [Google Scholar]
- Perner S, Brüderlein S, Hasel C, Walbel I, Holdenried A, Giloglu N, Chopurian H, Nielsen KV, Plesch A, Högel J, Möller P. Quantifying telomere lengths of human individual chromosome arms by centromere-calibrated fluorescence in situ hybridization and digital imaging. Am J Pathol. 2003;163:1751–1756. doi: 10.1016/S0002-9440(10)63534-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panossian LA, Porter VR, Valenzuela HF, Xnhu X, Reback E, Masterman D, Cummings JL, Effros RB. Telomere shortening in T cells correlates with Alzheime's disease status. Neurobiool. Aging. 2003;24:77–84. doi: 10.1016/s0197-4580(02)00043-x. [DOI] [PubMed] [Google Scholar]
- Plentz RR, Caselitz M, Bleck JS, Gebel M, Flemming P, Kubicka S, Manns MP, Rudolph KL. Hepatocellular telomere shortening correlates with chromosomal insability and development of human hepatoma. Hepatology. 2004;40(1):80–86. doi: 10.1002/hep.20271. [DOI] [PubMed] [Google Scholar]
- Puterman E, Lin J, Blackburn E, O'Donovan A, Adler N, Epel E. The power of exercise: buffering the effect of chronic stress on telomere length. PLoS One. 2010;5(5):e10837. doi: 10.1371/journal.pone.0010837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samani JJ, Boultby R, Butler R, Thompson JR, Goodall AH. Telomere shortening in atherosclerosis. Lancet. 2001;358(9280):472–473. doi: 10.1016/S0140-6736(01)05633-1. [DOI] [PubMed] [Google Scholar]
- Samassekou O, Gadji M, Drouin R, Yan J. Sizing the ends: Normal length of human telomeres. Ann Anat. 2010;192:284–291. doi: 10.1016/j.aanat.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Vulliamy TJ, Dokal I. The diverse clinical presentation of mutations in the telomerase complex. Biochimie. 2007;90(1):122–130. doi: 10.1016/j.biochi.2007.07.017. [DOI] [PubMed] [Google Scholar]
- Wolkowitz OM, Mellon SH, Epel ES, Lin J, Dhabhar FS, Su Y, Reus VI, Rosser R, Burke HM, Kupferman E, Compagnone M, Nelson JC, Balckburn EH. Leukocyte telomere length in major depression: Correlations with chronicity, inflammation and oxidative stress – preliminary findings. PLoS One. 2011;6(3):e17837. doi: 10.1371/journal.pone.0017837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LSM, van der Harst P, de Boer RA, Huzen J, van Gilst WH, van Veldhuisen DJ. Aging, telomeres and heart failure. Heart Fail Rev. 2010;15(5):479–486. doi: 10.1007/s10741-010-9173-7. [DOI] [PMC free article] [PubMed] [Google Scholar]