Abstract
MicroRNA-200c (miR-200c) through repression of specific target genes has been associated with cellular transition, tumorigenesis, and tissue fibrosis. We explored the expression and functional aspects of miR-200c in genesis of leiomyomas (LYO), benign uterine tumors with fibrotic characteristic. Using LYO and matched myometrium (MYO; n=76) from untreated and from patients exposed to hormonal therapies (GNRH agonist (GNRHa), Depo-Provera, and oral contraceptives), we found that miR-200c was expressed at significantly lower levels (P<0.05) in LYO as compared with MYO. These levels were lower in LYO from African Americans as compared with Caucasians, patients experiencing abnormal uterine bleeding and those exposed to GNRHa therapy. Gain-of-function of miR-200c in isolated leiomyoma smooth muscle cells (LSMCs), myometrial smooth muscle cells (MSMCs), and leiomyosarcoma cell line (SKLM-S1) repressed ZEB1/ZEB2 mRNAs and proteins, with concurrent increase in E-cadherin (CDH1) and reduction in vimentin expression, phenotypic alteration, and inhibition of MSMC and LSMC proliferations. We further validated TIMP2, FBLN5, and VEGFA as direct targets of miR-200c through interaction with their respective 3′ UTRs, and other genes as determined by microarray analysis. At tissue levels, LYO expressed lower levels of TIMP2 and FBLN5 mRNAs but increased protein expressions, which to some extent altered due to hormonal exposure. Given the regulatory functions of ZEBs, VEGFA, FBLN5, and TIMP2 on cellular activities that promote cellular transition, angiogenesis, and matrix remodeling, we concluded that altered expression of miR-200c may have a significant impact on the outcome of LYO growth, maintenance of their mesenchymal and fibrotic characteristics, and possibly their associated symptoms.
Introduction
Leiomyomas (LYO) are the most common benign uterine tumors that develop during reproductive age with a higher prevalence among African Americans (Parker 2007, Peddada et al. 2008). Although their etiology is unknown, LYO are considered to develop from myometrial cellular transition into a fibrotic tissue with myofibroblastic characteristic, which is a hallmark of tissues undergoing fibrosis (Hinz et al. 2007, Ono et al. 2007, Parker 2007, Chang et al. 2010, Chegini 2010). Ischemia, hypoxia, inflammation, and cellular injury, resulting in altered expression of local angiogenic, proinflammatory, and profibrotic mediators, are among the direct causes of development and progression of tissue fibrosis (Wallez et al. 2006, Hinz 2007, Wynn 2008, Chegini 2010, Fragiadaki & Mason 2011). Although some of the above conditions and mediators have been considered to account for genesis of LYO (Walker & Stewart 2005, Leppert et al. 2006, Spoelstra et al. 2006, Tanwar et al. 2009, Chegini 2010), the identity and mechanism of actions of the molecule(s) that initiates myometrial cellular transition into LYO remain elusive.
Accumulated evidence has provided strong support for the expression and regulatory function of microRNAs (miRNAs) on protein-coding gene expression through interacting with their 3′ UTR resulting in their posttranscriptional repression (Djuranovic et al. 2011). Through this mechanism, miRNAs regulate various aspects of normal biological processes and their aberrant expression has been closely associated with many disorders, including cellular transition, tumorigenesis, and tissue fibrosis (Thiery et al. 2009, Dykxhoorn 2010, Fabian et al. 2010, Jiang et al. 2010, Djuranovic et al. 2011, Small & Olson 2011). The miR-200 family, including miR-200c, through repression of specific number of genes such as transcription factors, ZEBs have been recognized to play central roles in epithelial to mesenchymal transition (EMT) and mesenchymal to epithelial transition (Korpal & Kang 2008, Davalos & Esteller 2009, Bendoraite et al. 2010, Dykxhoorn 2010, Oba et al. 2010, Chan et al. 2011, Roybal et al. 2011). Differential expression of ZEBs resulting in altered expression of their downstream target genes, including transcriptional repression of E-cadherin (CDH1), has been well documented as a mechanism of cellular transition that results in tumorigenesis and tissue fibrosis (Kalluri & Weinberg 2009, Katoh & Katoh 2009, Thiery et al. 2009, Chai et al. 2010, Fragiadaki & Mason 2011, Gregory et al. 2011). In the uterus, expression profiling and next generation sequencing have identified many miRNAs, including several members of miR-200 family whose expression altered in LYO, ectopic endometrium, endometrial cancer, and myometrium (MYO) during parturition (Luo & Chegini 2008, Renthal et al. 2010, Hawkins et al. 2011a,b, Lee et al. 2011).
In LYO, miRNA profiling has indicated a menstrual cycle-, tumor size-, and ethnic-dependence of their expression, including a lower miR-200a as compared with MYO (Wang et al. 2007), however, only the expression of a few miRNAs and their regulatory function on specific target genes have been confirmed and/or validated (Luo & Chegini 2008, Peng et al. 2008). Progesterone treatments have also been shown to alter the expression of miR-200b/429 and ZEBs in mice uterus during gestation and in isolated myometrial smooth muscle cells (MSMCs) where miR-200b/429 overexpression repressed ZEBs, oxytocin receptor, and connexin-43 (Renthal et al. 2010). In human MYO and LYO, immunohistochemical localization indicated a similar staining of ZEB1 with elevated intensity in leiomyosarcomas and MYO of ovariectomized mice treated with progesterone or estrogen (Spoelstra et al. 2006). As such, altered expression of miR-200 in LYO and their specific target genes, including ZEBs, may serve to maintain their mesenchymal and promote their fibrotic characteristics. However, evidence suggests that miRNA expression and regulatory functions occur in cell- and tissue-dependent manners (Dahiya et al. 2008, Inomata et al. 2009). As such, in the present study we explored the expression and regulatory function of miR-200c on specific target genes, including ZEBs, in LYO and matched MYO from follicular and luteal phases of the menstrual cycle and women experiencing abnormal uterine bleeding (AUB), and those who were exposed to GNRH agonist (GNRHa), Depo-Provera (Depo), or oral contraceptives (OCPs). In addition, using isolated MSMC, leiomyoma smooth muscle cell (LSMC), and SKLM-S1, a leiomyosarcoma cell line as in vitro models, we assessed the regulatory function of miR-200c on specific target gene expression, their cellular phenotypes and growth.
Our results indicated that miR-200c expression is downregulated in LYO as compared with MYO in an ethnic-dependent manner, which to some extent is influenced by hormonal exposure. We confirmed ZEB1/2, validated VEGFA, FBLN5, and TIMP2 as direct targets of miR-200c in MSMC, LSMC, and SKLM-S1, and showed that overexpression of miR-200c caused phenotypic modification of MSMC and LSMC, but not SKLM-S1, and inhibited their proliferation. The results suggest that low expression of miR-200c and CDH1 may serve to maintain myometrial and LYO mesenchymal characteristic, and miR-200c through direct and/or indirect mechanism regulate the expression of specific target genes which promotes LYO fibrotic characteristic and possibly their associated symptoms.
Materials and methods
Tissue collection
Portions of LYO and matched MYO were collected from patients (n=76) who were scheduled to undergo hysterectomy. The patients' age ranged from 20 to 59 years (median=42±7.968) and 32 were African Americans, 38 were Caucasians, and six were unknown. Of these patients, 43 were not taking any hormonal medications 3 months before surgery (17 were African Americans, 23 were Caucasians, and three were unknown); and based on their last menstrual periods they were from follicular (Follicular, n=12) and luteal (Luteal, n=19) phases of the menstrual cycle, or experiencing AUB (n=12). The remaining patients were exposed to either GNRHa (n=9), Depo (n=17), or OCP (n=7) for medical management of their symptomatic LYO. All LYO used in this study were 3–5 cm in diameter and were collected at the University of Florida affiliated Shands Hospital with prior approval from the Institutional Review Board. Tissues were snap-frozen and stored in liquid nitrogen for further analysis, or used for the isolation of LSMC and MSMC.
Isolation of MSMC and LSMC
Small portions of LYO and matched MYO from untreated patients (n=15) were used for isolation of MSMC and LSMC, as previously described in detail (Chegini et al. 2002). The isolated MSMC and LSMC as well as a human leiomyosarcoma cell line, SKLM-S1 (ATCC, Manassas, VA, USA), were cultured in DMEM supplemented with 10% FBS until reaching the confluence (Chegini et al. 2002). All supplies for isolation and culturing of these cells were purchased from Sigma–Aldrich, Invitrogen, and Fisher Scientific (Atlanta, GA, USA).
RNA isolation and analysis
Total RNA was extracted from paired tissues and cell cultures using TRIzol (Invitrogen). The quantity and quality of the isolated RNAs were determined (ND-1000 Spectrophotometer, NanoDrop Technologies, Wilmington, DE, USA), and 10 ng (for miRNA) or 2 μg were reverse transcribed using specific stem–loop primer for miR-200c or random primers for ZEBs, VEGFA, TIMP2, FBLN5, KLF9, and FLT1 according to the manufacturer's guidelines (Applied Biosystems, Foster city, CA, USA). Quantitative RT-PCR (qRT-PCR) was carried out using TaqMan or SYBR gene expression master mix, TaqMan miRNA or TaqMan gene expression assays (Applied Biosystems). Reactions were incubated for 10 min at 95 °C followed by 40 cycles for 15 s at 95 °C and for 1 min at 60 °C, and the level of mRNA and miRNA expression was determined using Applied Biosystems 7300 Detection System with 18S and RNU6B used for normalization respectively. All reactions were run in triplicate, and relative expression was analyzed with the comparative cycle threshold method (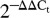 ) according to the manufacturer (Applied Biosystems). The primer sequences used in SYBR system for amplification of VEGFA and 18S were sense, 5′-GATCCGCAGACGTGTAAATGTTC-3′; antisense, 5′-GCTGCCTCGCCTTGCA-3′ and sense, 5′-GACGGACCAGAGCGAAAGC-3′; antisense, 5′-CCTCCGACTTTCGTTCTTGATT-3′ respectively.
) according to the manufacturer (Applied Biosystems). The primer sequences used in SYBR system for amplification of VEGFA and 18S were sense, 5′-GATCCGCAGACGTGTAAATGTTC-3′; antisense, 5′-GCTGCCTCGCCTTGCA-3′ and sense, 5′-GACGGACCAGAGCGAAAGC-3′; antisense, 5′-CCTCCGACTTTCGTTCTTGATT-3′ respectively.
Gain- or lose-of-function of miR-200c
MSMC, LSMC, and SKLM-S1 were seeded at a cell density of 3.5×104/well in six-well plates and at sub-confluence transfected with 50 nM of pre-miR-200c, anti-miR-200c, pre-miR negative control (preNC), or anti-miR negative control (Applied Biosystems) for 96 h using PureFection Transfection Reagent (System Biosciences, Inc., Mountain View, CA, USA) according to the manufacturer's protocol.
Western blot analysis
Total protein isolated from tissues and/or cells was subjected to immunoblotting as previously described (Chuang et al. 2012). Briefly, 30–50 μg total protein were subjected to SDS–PAGE, transferred onto polyvinylidene difluoride membranes, blocked, and incubated with antibody against ZEB2 (1:1000), VEGFA (1:1000), KLF9 (1:1000) (Santa Cruz Biotechnology, Santa Cruz, CA, USA), E-cadherin (1:1000), TIMP2 (1:1000) (Cell Signaling Technology, Inc., Danvers, MA, USA), ZEB1 (1:1000) (Abcam, Inc., Cambridge, MA, USA), FBLN5 (1:500), and FLT1 (1:1000) (R&D Systems, Minneapolis, MN, USA). The membranes were also stripped and probed with antibody generated against β-actin (Sigma) or α-tubulin (Abcam, Inc.) serving as loading control. The membranes were exposed to HRP-labeled secondary antiserum, and immunoreactive proteins were detected by chemiluminescence (ECL, Amersham). The band densities were determined using Image J program (http://imagej.nih.gov/ij/) and normalized to α-tubulin accordingly, and their ratios were calculated using the values in control groups as 1.
Luciferase reporter assay
MCMC, LSMC, and SKLM-S1 were seeded at a cell density of 3.5×104/well in six-well plates and cultured until reaching a sub-confluence. The cells were then transiently cotransfected with pre-miR-200c or preNC at the concentration mentioned earlier and with luciferase reporter plasmid (1 μg/well) containing 3′ UTR sequences for ZEB1, ZEB2, VEGFA, TIMP2, or FBLN5, respectively (GeneCopoeia, Inc., Rockville, MD, USA), using PureFection transfection reagent. Firefly and Renilla luciferase activities were measured after 48 h of transfection using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer's instruction. Firefly luciferase activity was normalized to Renilla luciferase activity, and the level of induction was reported as the mean±s.e.m. of three experiments performed in duplicates and compared with a ratio of cells transfected with preNC independently set at 1.
ELISA
Culture-conditioned media collected from the above experiments were centrifuged, supernatants were collected, and their total protein content was determined by standard method. The TIMP2 and VEGF contents of the supernatants were determined using Quantikine human TIMP2 and VEGF ELISA kit (R&D Systems) with limited detection of 11 pg/ml and sensitivity of 5 pg/ml. The levels of VEGF and TIMP2 were calculated as picogram per milligram of protein and reported as fold change compared with control experiments.
Phenotypic cellular assessment and cell proliferation assay
MSMC and LSMC were plated at a cell density of 3.5×104/well in six-well plates for 48 h and subjected to gain-of-function of miR-200c (transfected with pre-miR-200c or preNC) as described earlier. The cell shapes were monitored after 96 h of transfection and images were captured using an Olympus IX70 microscope equipped with digital camera (Olympus, Inc., Melville, NY, USA). Cell survival and proliferation were assessed using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and crystal violet assays. Briefly, cells were cultured in 96-well plates at a density of 103 cells/well for 48 h and transfected with miR-200c or preNC. For MTT assay after 96 h of incubation, MTT (Sigma) was added to the culture medium at a final concentration of 1 mg/ml for 2 h at 37 °C and the media were removed. The formazan product was solubilized in dimethyl sulfoxide and the absorbance at 490 nm was determined by subtracting from absorbance at 630 nm (background) for each well using multi-plate reader (Molecular Device, Inc., Sunnyvale, CA, USA). For crystal violet assay, the medium was removed and cells were washed twice with PBS, fixed with 100 μl of acetone: methanol (1:1) at 4 °C, and stained with 1% crystal violet (Sigma) in 20% methanol for 10 min. The cells were washed and crystal violet was solubilized in 200 μl of 1% SDS solution, and the absorbance was measured using a multiplate reader at 595 nm. The result was expressed as fold change relative to negative control (preNC) and the assay was performed in six replicates per condition and repeated four times using two independent cell preparations.
Microarray and bioinformatic analysis
Total RNA isolated from MSMC and LSMC following gain-of-function of miR-200c was subjected to gene expression profiling using Human HT-12 v4 Expression BeadChip (Illumina, Inc., San Diego, CA, USA) and was carried out at gene array core facility at the University of Florida Interdisciplinary Center for Biotechnology Research, according to manufacturer's protocol. The expression values were background-subtracted, globally normalized (BeadStudio version 1.5.1.3, Illumina), and subjected to statistical analysis as previously described (Luo et al. 2005). The differentially expressed genes were selected based on P≤0.05 (two-way ANOVA and Tukey's test) and a 1.5-fold change cutoff and subjected to cluster and tree-view and functional analyses using Ingenuity Pathways Software (Ingenuity Systems, Redwood City, CA, USA).
For bioinformatic analysis, the list of miR-200c predicted target genes was selected from TargetScan (http://www.targetscan.org) and their expression values were retrieved from the list of genes expressed in MSMC and LSMC with miR-200c gain-of-function using Microsoft Access. The gene list was sorted based on a 1.5-fold change cutoff (Supplementary Table 2, see section on supplementary data given at the end of this article) and analyzed by cluster and tree-view analyses and subjected to functional analysis using Ingenuity Pathways Software.
Statistical analysis
Whenever appropriate, the results were reported as mean±s.e.m. All in vitro experiments were performed using paired MSMC and LSMC preparations from at least three different patients. Comparisons between two or among groups were made using unpaired nonparametric Student's t-test and ANOVA followed by a Tukey's HSD post hoc multiple comparison respectively. A P<0.05 was considered statistically significant.
Results
miR-200c is aberrantly expressed in LYO
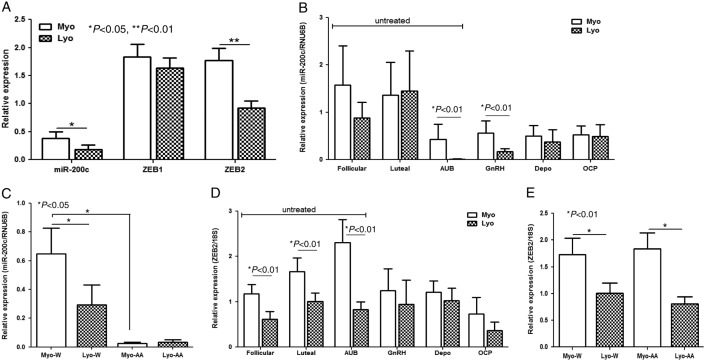
Using qRT-PCR, we found that all LYO and paired MYO from follicular and secretory phases of the menstrual cycle, women experiencing AUB, and those who were exposed to GNRHa, Depo, and OCPs, expressed low but variable levels of miR-200c. The relative mean expression of miR-200c was lower in LYO as compared with matched MYO from untreated group (Fig. 1A, P<0.05). The relative expression of miR-200c declined in LYO from patients experiencing AUB (P<0.01) with a slight increase in tissues from luteal phase as compared with MYO (Fig. 1B). Further analysis of miR-200c expression in paired tissues from untreated group indicated that both LYO and MYO from African Americans expressed lower levels as compared with Caucasians (P<0.05, Fig. 1C). In LYO from cohorts exposed to hormonal therapies, the mean expression of miR-200c was declined in patients who received GNRHa therapy (P<0.01) as compared with Depo and OCPs (Fig. 1B).
Figure 1.
(A) The relative mean expression of miR-200c, ZEB1, and ZEB2 in leiomyoma (LYO) and myometrium (MYO) from untreated group including follicular (n=12) and luteal (n=19) phases of the menstrual cycle and women experiencing AUB (n=12). Relative (mean±s.e.m.) expression of miR-200c (B) and ZEB2 (D) in LYO and matched MYO from untreated group corresponding to follicular (Follicular) and luteal (Luteal) phases of the menstrual cycle and AUB, and patients who received GNRHa, Depo, and those taking OCP. (C) miR-200c and (E) ZEB2 expression of untreated group based on ethnicity in Caucasians (W, n=23) and African Americans (AA, n=17). The relative expression was determined by setting the expression value of each gene in a myometrium from follicular phase as 1. The data were analyzed using nonparametric Student's t-test with P values indicated by corresponding lines for each group.
The expression of ZEBs and their downstream target gene, CDH1, in the above tissues also indicated considerable variations in the level of their expression. Overall, the mean expression of ZEB2, but not ZEB1, was lower in LYO as compared with MYO from untreated group (Fig. 1A, P<0.01) with lower ZEB1 expression in LYO from luteal phase (P<0.05, Supplementary Figure 1A, see section on supplementary data given at the end of this article) and lower ZEB2 expression from follicular and luteal phases and AUB (Fig. 1D, P<0.01). Analysis of ZEBs expression in paired tissues from untreated group also indicated a lower ZEB2, but not ZEB1, expression in LYO from African Americans and Caucasians as compared with their corresponding paired MYO (P<0.01, Fig. 1E and Supplementary Figure 1B). Although LYO and paired MYO from patients exposed to GNRHa and OCPs expressed lower levels of ZEB1, the mean expression of ZEBs did not significantly alter as a result of hormonal exposure (Supplementary Figure 1A and B). The level of CDH1 was very low in all the tissues examined (data not shown).
Gain-of-function of miR-200c alters the expression of ZEBs, CDH1, and cell morphology
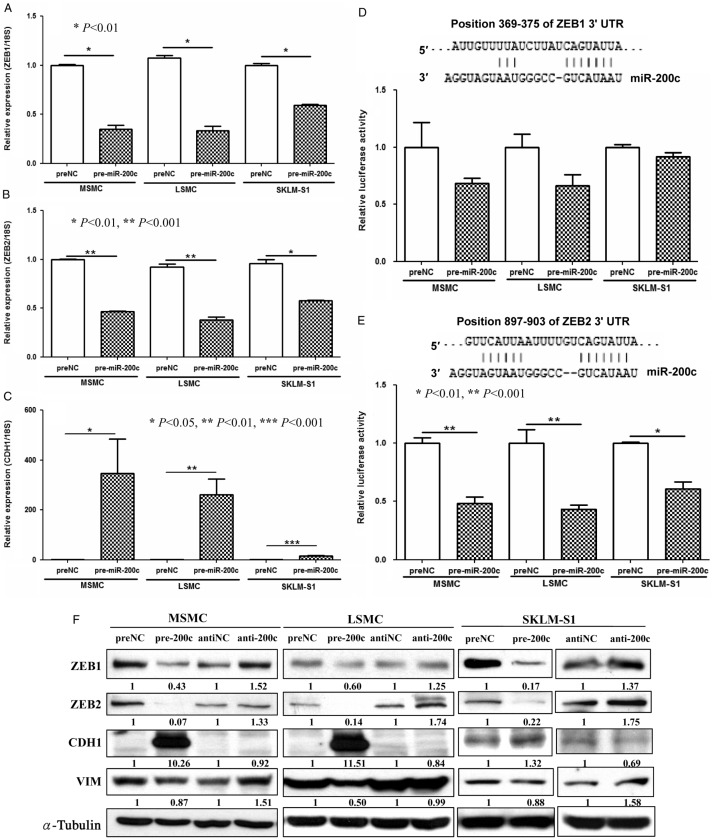
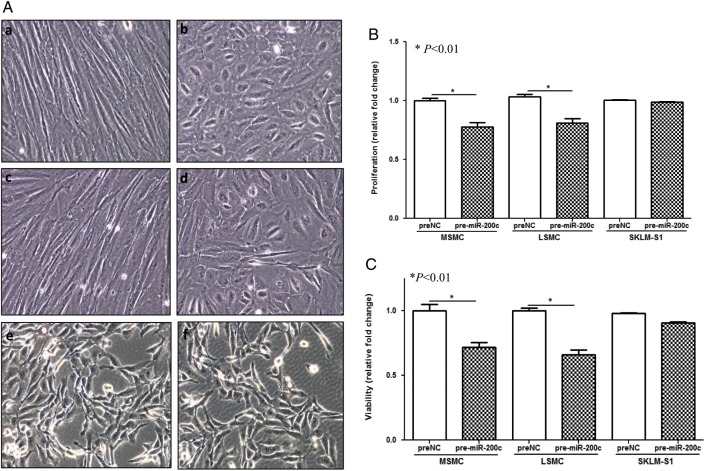
As miR-200c was expressed at lower levels in LYO, we next examined the influence of gain-of-function of miR-200c on ZEB expression, which are well-established targets of miR-200 family, including miR-200c. We confirmed that miR-200c repressed ZEB1 and ZEB2 in MSMC, LSMC, and SKLM-S1 at mRNA and protein levels (Fig. 2A, B, C, D, E and F), through direct interaction with their respective 3′ UTRs as indicated by luciferase reporter assay (Fig. 2D and E). ZEB repression (pre-miR-200c transfected cells) was accompanied by a significant increase in CDH1 in MSMC and LSMC, while a slight decrease in vimentin was observed in LSMC (Fig. 2C and F) in contrast to anti-miR-200c transfected cells. As compared with SKLM-S1, the basal expression of CDH1 was very low in MSMC and LSMC. We also found that gain-of-function of miR-200c in MSMC and LSMC resulted in an alteration of their elongated smooth muscle cell characteristics into round shapes (Fig. 3A) with limited effect on SKLM-S1. Using crystal violet staining (Fig. 3B) and MTT assay (Fig. 3C), we demonstrated that overexpression of miR-200c reduced the rate of proliferation and cellular viability of MSMC and LSMC but not SKLM-S1.
Figure 2.
The influence of overexpression of miR-200c (pre-miR-200c transfected) on the expression of ZEB1 (A), ZEB2 (B), and CDH1 (C) in MSMC, LSMC, and SKLM-S1 as determined by qRT-PCR (A, B and C) as compared with pre-miR control (preNC). Firefly luciferase assay with pZEX-MT01 constructs carrying 3′ UTR fragments of ZEB1 (D) and ZEB2 (E) respectively. The cells were cotransfected with firefly luciferase reporters, pre-miR-200c, or preNC and the ratio of firefly:Renilla was determined after 48 h and reported as relative luciferase activity as compared with preNC which was independently set at 1 for each cell. The results presented from three sets of independent experiments performed in duplicates. Sequence alignment of miR-200c seed regions and ZEB1 and ZEB2 mRNA target sits at their 3′ UTRs with the coordinated positions are shown at the top of each graph. The results are presented as mean±s.e.m. and analyzed using nonparametric Student's t-test with P values indicated by corresponding lines. (F) Western blot analysis shows the effects of pre-miR-200c or anti-miR-200c on ZEB1, ZEB2, CDH1, and vimentin expression. The assays were performed using three to five sets of independent cell preparations.
Figure 3.
(A) Photomicrographs showing the morphological appearance of MSMC (a and b), LSMC (c and d), and SKLM-S1 (e and f) following gain-of-function of miR-200c (b, d and f) as compared with control (preNC) (a, c and e) for 96 h. The influence of miR-200c and corresponding control (preNC) on cell proliferation (B) and cellular viability (C) were determined by crystal violet and MTT assay after 4 days of incubation with culture media changed every 2 days. The assays were performed using three to five sets of independent cell preparation and the results were presented as mean±s.e.m. and analyzed by nonparametric Student's t-test with P values shown as corresponding lines. A full colour version of this figure is available via http://dx.doi.org/10.1530/ERC-12-0007.
TIMP2, FBLN5, and VEGFA are direct targets of miR-200c
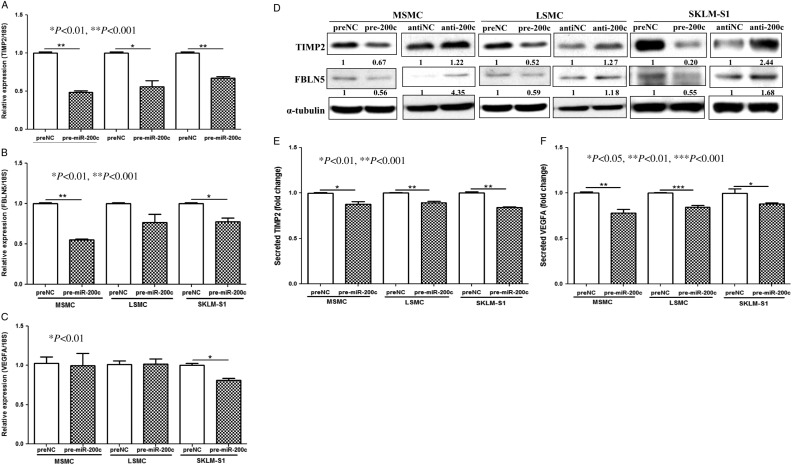
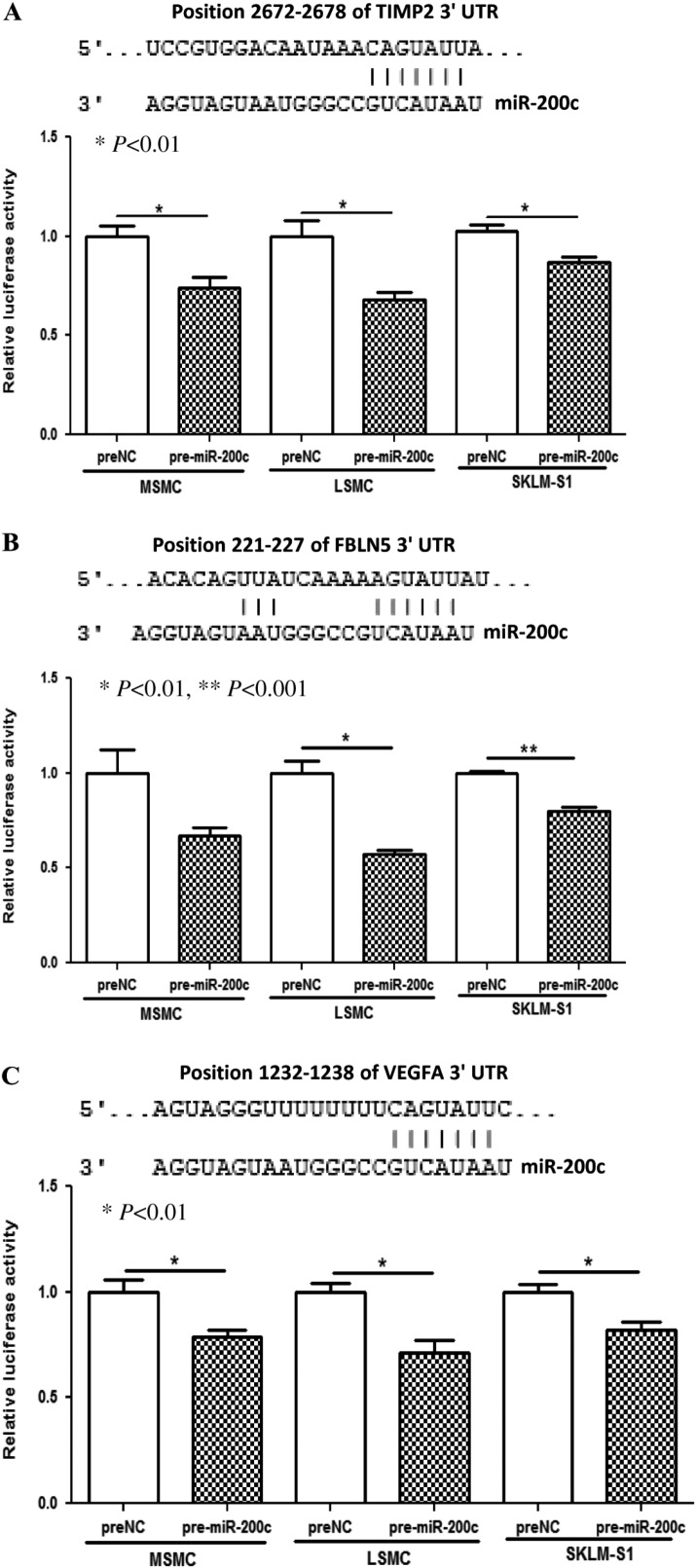
VEGFA, FLT1, FBLN5, TIMP2, and KLF9 are among several hundred genes predicted as targets of miR-200. As the expression of VEGFA, FLT1, FBLN5, TIMP2, and KLF9 and to some extent their biological actions have been documented in uterine tissue and cells, we next examined possible regulatory function of miR-200c on their expression. We found that gain-of-function of miR-200c in MSMC, LSMC, and SKLM-S1 in a cell-specific manner repressed TIMP2, FBLN5, and VEGFA at mRNA (VEGFA only in SKLM-S1) and protein levels (Fig. 4A, B, C and D), and at the levels of VEGFA and TIMP2 secreted into their culture-conditioned media (Fig. 4E and F). However, gain-of-function of miR-200c had a limited effect on FLT1 and KLF9 expression (data not shown). The regulatory function of miR-200c on TIMP2, FBLN5, and VEGFA expression occurred through direct interactions with their respective 3′ UTRs as demonstrated by luciferase reporter assay (Fig. 5A, B and C).
Figure 4.
The influence of overexpression of miR-200c (pre-miR-200c transfected) on the expression of TIMP2 (A), FBLN5 (B), and VEGFA (C) in MSMC, LSMC, and SKLM-S1 as determined by qRT-PCR (A, B and C). The assays were performed using three to five sets of independent cell preparation in triplicates. Western blot analysis of TIMP2, FBLN5 (D), and ELISA (E and F) of VEGF-A and TIMP2 in MSMC, LSMC, and SKLM-S1 following transfection with pre-miR-200c and control (preNC) with α-tubulin was used as loading control. The assays were performed using three to five sets of independent cell preparation and in duplicated in ELISA. The data are presented as mean±s.e.m. and analyzed using nonparametric Student's t-test with P values presented as indicated by corresponding lines.
Figure 5.
Firefly luciferase assay with pZEX-MT01 constructs carrying 3′ UTR fragments of TIMP2 (A), FBLN5 (B), and VEGFA (C) respectively. MSMC, LSMC, and SKLM-S1 were cotransfected with firefly luciferase reporters, pre-miR-200c or preNC. The ratio of firefly:Renilla was determined after 48 h and reported as relative luciferase activity as compared with preNC which was independently set at 1 for each cell. The results presented from three sets of independent experiments performed in duplicates and analyzed using unpaired t-test. Sequence alignment of miR-200c seed regions and TIMP2, FBLN5, and VEGFA mRNAs target sites at their 3′ UTRs with the coordinated positions are shown at the top of each graph.
Expression of TIMP2, FBLN5, and VEGFA is altered in LYO
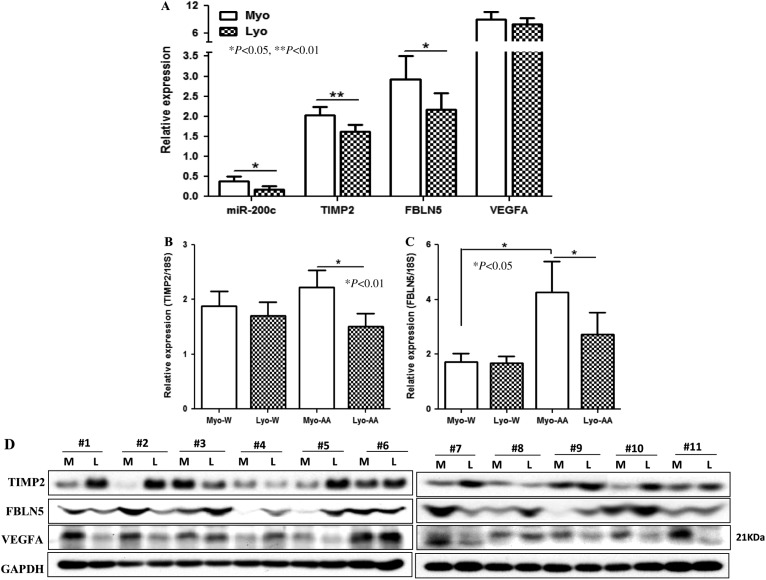
We next examined the expression of TIMP2, FBLN5, and VEGFA in the same tissues used for the analysis of miR-200c expression. The mean expression of TIMP2 and FBLN5, but not VEGFA, was lower in LYO as compared with MYO from untreated group (P<0.01 and P<0.05, Fig. 6A). Further analysis also indicated a lower TIMP2 expression in LYO from follicular and luteal phases of the menstrual cycle (P<0.05), with a trend toward lower levels in GNRHa-treated group (P=0.062; Supplementary Figure 2A, see section on supplementary data given at the end of this article). The expression of FBLN5 was also lower in LYO from luteal phase (P<0.05), and GNRHa treated (P=0.063), and those taking OCP (P<0.05; Supplementary Figure 2B). VEGFA displayed a trend toward an increased expression in LYO from follicular (P=0.078) and lower expression in luteal phase (P<0.05), without significant difference among other groups (Supplementary Figure 2C). In addition, the levels of TIMP2 and FBLN5 expression were lower in LYO from African Americans, but not Caucasians, as compared with MYO (P<0.01 and P<0.05, Fig. 6B and C), with significantly higher FBLN5 expression in MYO from African Americans as compared with Caucasians (P<0.05, Fig. 6C). The summary of the ratio of miR-200c, ZEBs, TIMP2, FBLN5, and VEGFA expression at 1.3-fold change cutoff in LYO over its own paired MYO is shown in Table 1. On western blotting using paired LYO and MYO from 11 untreated patients, TIMP2, VEGFA, and FBLN5 were detected at variable levels with increased TIMP2 and FBLN5 and lower VEGFA productions, at least in seven out of 11 LYO as compared with paired MYO (Fig. 6D).
Figure 6.
Relative (mean±s.e.m.) expression of miR-200c, TIMP2, FBLN5, and VFGFA (A) in LYO and matched MYO from untreated including follicular (n=12) and luteal (n=19) phases of the menstrual cycle and women experiencing AUB (n=12). (B) TIMP2 and (C) FBLN5 expression of untreated group based on ethnicity in Caucasians (W, n=23) and African Americans (AA, n=17). (D) Western blot analysis shows the expression of TIMP2, FBLN5, and VEGFA of tissue extracts from 11 leiomyomas (L) and paired myometrium (M) from untreated groups. GAPDH was used as loading control. The mean values were analyzed using nonparametric t-test with P values indicated by corresponding lines for each group.
Table 1.
The ratio of the expression of miR-200c, ZEB1, ZEB2, TIMP2, FBLN5, and VEGFA in each leiomyoma (L) over its own paired myometrium (M) from untreated group (n=43) representing to follicular (Follicular; n=12) and luteal (Luteal; n=19) phases of the menstrual cycle and women experiencing AUB (n=12) and those who received GNRHa (n=9), Depo (n=17), and those taking OCPs (n=7). The ratio represents the number of leiomyoma over its own paired myometrium expressing elevated (up) or lower (down) levels of miR-200c and its target genes at 1.3-fold change cutoff with percentage shown in parentheses for each group and total number of paired tissues analyzed (n value)
| L/M | Follicular | Luteal | AUB | GNRH | Depo | OCP |
|---|---|---|---|---|---|---|
| miR-200c | ||||||
| Up | 3/12 (0.25) | 6/19 (0.316) | 1/12 (0.083) | 0 | 6/17 (0.353) | 3/7 (0.429) |
| Down | 8/12 (0.667) | 11/19 (0.579) | 10/12 (0.833) | 7/9 (0.778) | 8/17 (0.471) | 2/7 (0.286) |
| n value | 12 | 19 | 12 | 9 | 17 | 7 |
| ZEB1 | ||||||
| Up | 2/11 (0.182) | 3/19 (0.158) | 5/12 (0.417) | 1/9 (0.111) | 3/14 (0.214) | 0 |
| Down | 4/11 (0.364) | 6/19 (0.316) | 3/12 (0.25) | 7/9 (0.778) | 4/14 (0.286) | 4/5 (0.8) |
| n value | 11 | 19 | 12 | 9 | 14 | 5 |
| ZEB2 | ||||||
| Up | 1/9 (0.111) | 3/19 (0.158) | 1/12 (0.083) | 2/9 (0.222) | 2/14 (0.143) | 0 |
| Down | 7/9 (0.778) | 9/19 (0.474) | 10/12 (0.833) | 5/9 (0.556) | 8/14 (0.571) | 4/4 (1) |
| n value | 9 | 19 | 12 | 9 | 14 | 4 |
| TIMP2 | ||||||
| Up | 1/9 (0.111) | 1/16 (0.063) | 3/12 (0.25) | 0 | 3/11 (0.273) | 0 |
| Down | 5/9 (0.556) | 8/16 (0.5) | 4/12 (0.333) | 4/6 (0.667) | 2/11 (0.182) | 3/3 (1) |
| n value | 9 | 16 | 12 | 6 | 11 | 3 |
| FBLN5 | ||||||
| Up | 1/7 (0.143) | 1/16 (0.063) | 3/12 (0.25) | 0 | 3/11 (0.273) | 0 |
| Down | 2/7 (0.286) | 7/16 (0.438) | 8/12 (0.667) | 5/6 (0.833) | 6/11 (0.545) | 3/3 (1) |
| n value | 7 | 16 | 12 | 6 | 11 | 3 |
| VEGFA | ||||||
| Up | 5/9 (0.556) | 3/16 (0.188) | 8/12 (0.667) | 1/6 (0.167) | 0 | 2/3 (0.667) |
| Down | 2/9 (0.222) | 6/16 (0.375) | 3/12 (0.25) | 4/6 (0.667) | 5/11 (0.455) | 1/3 (0.333) |
| n value | 9 | 16 | 12 | 6 | 11 | 3 |
Gain-of-function of miR-200c targets the expression of many genes
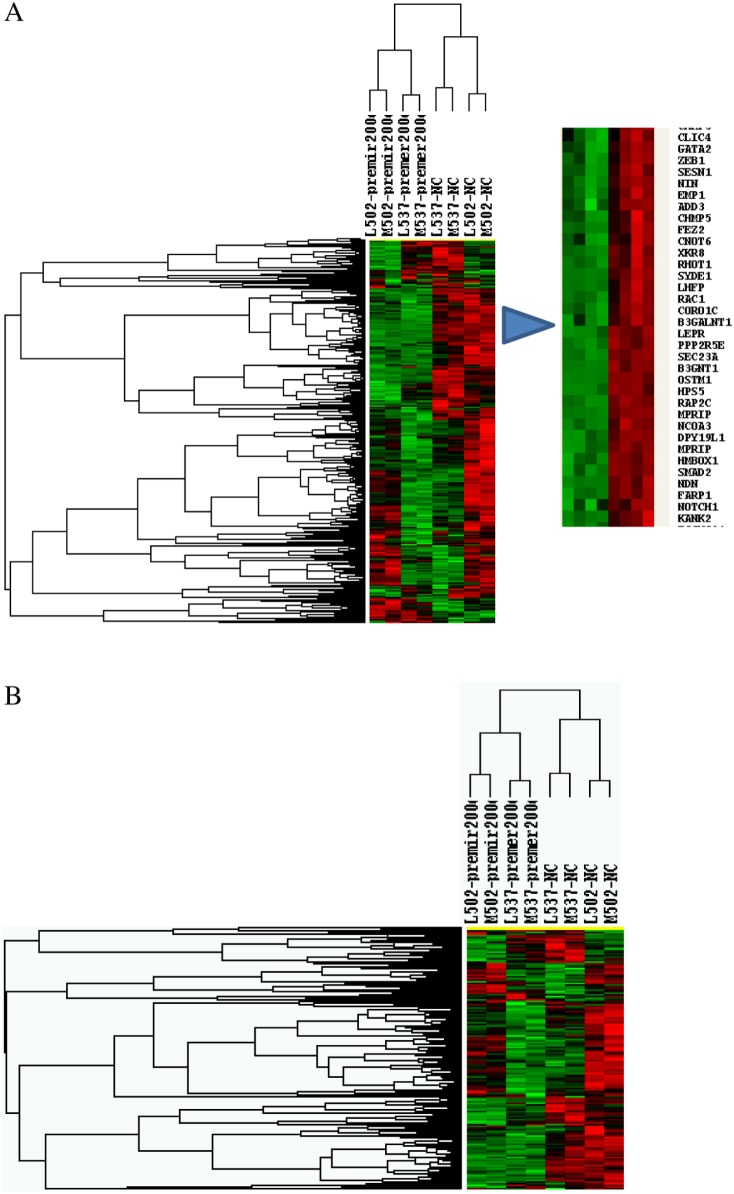
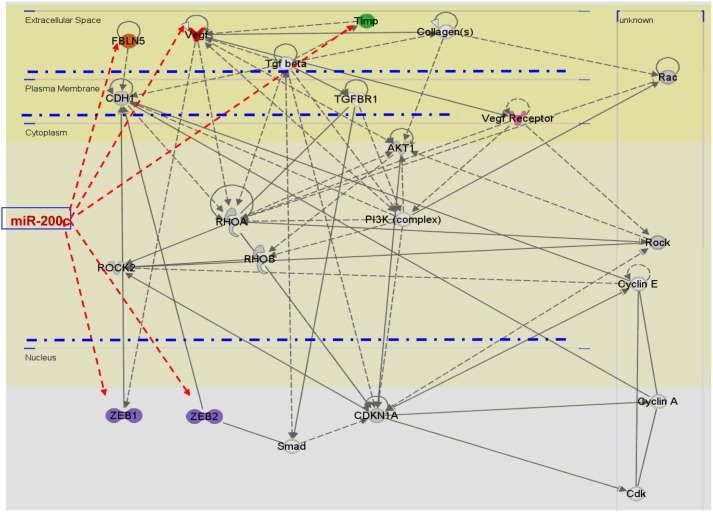
Using microarray, we further assessed the influence of miR-200c gain-of-function on overall gene expression in MSMC and LSMC. Our microarray GEO accession number is GSE38817. The analysis (P≤0.05) indicated that the expression of 350 genes either directly and/or indirectly altered following gain-of-function of miR-200c as compared with control (preNC; tree-view analysis; Fig. 7A and Supplementary Table 1, see section on supplementary data given at the end of this article). Although the expression of a low number of genes was altered (1.5-fold change cutoff) following gain-of-function of miR-200c in MSMC and LSMC, miRNAs repress their target gene expression mainly at translational levels with some target genes repressed without detectable changes in their mRNA levels (Baek et al. 2008, Guo et al. 2010). In addition, bioinformatic analysis of 798 genes predicted as miR-200c targets (TargetScan), including ZEBs, VEGRA, TIMP2, and FBLN5, indicated that 553 of these genes were expressed in MSMC and LSMC (tree-view analysis; Fig. 7B and Supplementary Table 2) of which 18 genes were downregulated and 26 were upregulated in LSMC (1.3-fold change cutoff) following gain-of-function of miR-200c as compared with control (preNC). Ingenuity pathway analysis functionally mapped these genes into several networks corresponding to transcriptional, translational, and signal transduction mediators, cell cycle, extracellular matrix (ECM) turnover, and metabolic activities etc. (Fig. 8 and Supplementary Table 3, see section on supplementary data given at the end of this article).
Figure 7.
Tree-view analysis of differentially expressed genes in MSMC and LSMC following pre-miR-200c transfection (gain-of-function of miR-200c) and pre-miR-control (A) and genes predicted as target of miR-200c (B). A full colour version of this figure is available via http://dx.doi.org/10.1530/ERC-12-0007.
Figure 8.
Ingenuity pathway analysis of selected number of genes differentially expressed in MSMC and LSMC with gain-of-function of miR-200c showing (arrows) the expression of ZEB1, ZEB2, VEGFA, TIMP2, and FBLN5 as direct targets of miR-200c. A full colour version of this figure is available via http://dx.doi.org/10.1530/ERC-12-0007.
Discussion
In the present study, we demonstrated the expression of miR-200c and specific target genes, ZEB1, ZEB2, VEGFA, TIMP2, and FBLN5, in LYO and paired MYO from untreated and from women exposed to different hormonal therapies and assessed miR-200c regulatory functions on the expression of these genes in their isolated smooth muscle cells and a leiomyosarcoma cell line. The level of expression of miR-200c and target genes varied considerably among LYO and paired MYO from the above cohorts, a common phenomenon often observed in studies involving human tissues; however, the mean expression of miR-200c was significantly lower in LYO, specifically in tumors from African Americans and women experiencing AUB. Although differences among individual paired tissues were observed (Table 1) regarding the influence of hormonal milieu on miR-200c expression, its mean expression did not correlate with the phases of the menstrual cycle; however, it was reduced in both tissues from women exposed to GNRHa, Depo, and OCPs. Assessing the overall hormonal influence on miR-200c expression at tissue level is rather complex, specifically in LYO that display variable growth patterns during the menstrual cycle and responses to hormonal therapies such as GNRHa, which often causes hypo-estrogenic condition and regresses LYO growth. However, lower miR-200c expression in tissues from patients experiencing AUB and those exposed to GNRHa suggests possible hormonal regulation of miR-200c. Our finding with miR-200c expression to some extent is supported by a previous microarray study which reported a menstrual cycle-, tumor size-, and ethnic-dependent expression of miRNAs, including a lower miR-200a expression in LYO (Wang et al. 2007) and correlate with aberrant expression of miR-200 family in other disorders (Kalluri & Weinberg 2009, Katoh & Katoh 2009, Thiery et al. 2009, Chai et al. 2010, Fragiadaki & Mason 2011, Gregory et al. 2011).
miR-200 family, miR-200b/200a/429 and miR-200c/141 are expressed as a single polycistronic transcript from chromosomes 1 and 12 respectively; however, they share the same common targets genes. However, based on their seed regions, they are divided into miR-200bc/429 and miR-200a/141, differing only in their fourth nucleotide, which is considered critical in specificity of their target genes regulation (Lewis et al. 2005). Among several hundred genes predicted as targets of miR-200 family, ZEB1 and ZEB2 are validated as their direct targets whose aberrant expression and repression of CDH1 expression have been closely associated with tumorigenesis and fibrotic disorders (Kalluri & Weinberg 2009, Katoh & Katoh 2009, Thiery et al. 2009, Chai et al. 2010, Fragiadaki & Mason 2011, Gregory et al. 2011). Using the same set of LYO and paired MYO utilized for miR-200c analysis, we found that LYO expressed a low to undetectable levels of CDH1, with higher ZEBs and vimentin expression, a molecular environment reflecting the mesenchymal cellular characteristic of these tissues. Our results with ZEB1 expression corroborate an immunohistochemical study which reported an equal expression of ZEB1 in MYO and LYO, with increased staining in leiomyosarcomas and in MYO of ovariectomized mice treated with progesterone and estrogen (Spoelstra et al. 2006). In contrast, MYO from follicular and luteal phases of the menstrual cycle, and patients experiencing AUB expressed a higher level of ZEB2 which decreased in tissues on exposure to GNRHa, Depo, and OCPs, implying hormonal regulation of ZEB2. In mice, the uterine expression of ZEBs has been reported to increase throughout pregnancy, and daily injections with P4 during late gestation increased ZEB1 but not ZEB2 expression with modest inhibition of miR-200b/429, which was inhibited in P4-injected ovariectomized animals (Renthal et al. 2010). Overexpression of ZEB1 in primary cultures of mouse myometrial cells through miR-200b/429 suppression has also been reported to increase ZEB2, and P4-induced ZEB2 expression was considered to be due to this mechanism (Renthal et al. 2010). Although regulatory function of ovarian steroids on gene expression in uterus and other steroid-sensitive tissues and cells is well established, their influence and molecular mechanism(s) of actions on miRNA expression are beginning to emerge (Luo & Chegini 2008, Bhat-Nakshatri et al. 2009, Maillot et al. 2009, Nothnick & Healy 2010, Renthal et al. 2010, Yoshimoto et al. 2011).
As miR-200c expressed at relatively low levels in LYO and MYO, we overexpressed miR-200c in MSMC, LSMC, and SKLM-S1, a leiomyosarcoma cell line, and as expected found ZEB1 and ZEB2 as its direct targets at both transcriptional and translational levels. However, as indicated by luciferase reporter assay, miR-200c interacted with ZEB1 3′ UTR less efficiently in MSMC and LSMC and was limited in SKLM-S1 as compared with its interaction with ZEB2 3′ UTR. The reasons for less efficient interaction of miR-200c with ZEB1 3′ UTR in MSMC, LSMC, and SKLM-S1 is unclear, although miR-200b/miR-429 has been reported to interact with both ZEB1 and ZEB2 3′ UTRs in an immortalized human myometrial cell line (Renthal et al. 2010). Overexpression of miR-200c in MSMC and LSMC, but not in SKLM-S1, was also accompanied by increased CDH1 and reduced vimentin expression, which expressed at low/undetectable and very high levels, respectively, in MSMC and LSMC and altered phenotypic characteristic of MSMC and LSMC, with only a limited influence on SKLM-S1. Although altered expression of miR-200 family and ZEBs by regulating CDH1 expression has been considered to regulate EMT in tissues undergoing fibrosis (Oba et al. 2010, Chen et al. 2012, Yang et al. 2012), it is unlikely that such mechanism is involved in cellular transition of MYO into LYO. MYO consists of terminally differentiated smooth muscle cells and LYO, which are considered to derive from myometrial cellular transition, have myofibroblastic characteristic that is a common feature of tissues undergoing fibrosis. As such, we consider low miR-200c expression to serve in maintaining myometrial and LYO mesenchymal cellular characteristic and LYO fibrotic features with low CDH1 and high vimentin expression, rather than promoting EMT. However, unlike MSMC and LSMC, SKLM-S1 expressed a higher basal level of CDH1 which was not altered following overexpression of miR-200c though it repressed both ZEBs which regulate CDH1 expression. The molecular mechanism(s) that limits CDH1 induction and phenotypic alteration of SKLM-S1 by miR-200c could be due to possible epigenetic regulation of CDH1 as well as involvement of other gene products which requires detailed investigation.
miR-200 has also been predicted to target the expression of FLT1, VEGFA, TIMP2, FBLN5, and KLF9; of which FLT1 has been validated as a direct target of miR-200c in lung adenocarcinomas (Roybal et al. 2011). Using the same cohorts of LYO and paired MYO, we detected higher VEGFA, FBLN5, and TIMP2 mRNA in both tissues that exhibited an inverse relationship with miR-200c; however, considering that miR-200c was expressed at lower levels in LYO, we expected a higher expression of these genes in LYO as compared with MYO. However, differences in the levels of expression of TIMP2 and FBLN5 mRNAs among individual paired tissues reflected such regulatory interactions, although their mean expression was lower in LYO as compared with their protein expression. As for a lower VEGFA protein expression, VEGFA may not be a major target of miR-200c, or possibly regulated by other miRNAs in LYO, although miR-200c directly interacted with VEGFA 3′ UTR in MSMC, LSMC, and SKLM-S1 and reduced its expression at translational level. Our results with VEGFA mRNA expression in MSMC and LSMC further suggest that miR-200c regulatory function on VEGFA expression occurs in a cell-specific manner and through protein degradation rather than mRNA decay, a mechanism well documented for other miRNAs regulation of their target genes (Djuranovic et al. 2011). In addition, correlation of a miRNA expression with target genes expression at tissue level is rather complex as multiple miRNAs can target the expression of one gene, while many genes could be the target of one miRNA, and their functional regulation of the target genes could occur in a cell- and tissue-specific manner (Dahiya et al. 2008, Inomata et al. 2009). As such correlation between miR-200c expression and that of ZEBs, VEGFA, TIMP2, and FBLN5 expression in LYO and MYO, not only reflects the influence of the above parameters but also differs in LYO with respect to their genetics, response to hormonal therapies, tumor size, and ethnicity (Wang et al. 2007, Peddada et al. 2008, Baird et al. 2011). We provided evidence that TIMP2 and FBLN5, but not KLF9 and FLT1, are direct targets of miR-200c in MSMC, LSMC, and SKLM-S1 which to our knowledge is the first of such report. In addition, the reason for the lack of regulatory action of miR-200c on KLF9 and FLT1 expression in MSMC and LSMC could be cell specific as we have recently reported KLF9 and FLT1 as direct targets of miR-200c in Ishikawa cells (Panda et al. 2012).
The expressions of VEGFA, FLT1, and TIMP2 have previously been reported in LYO and MYO (Nakayama et al. 2006, Xu et al. 2006, 2008, Bogusiewicz et al. 2007, Chegini 2010, Ciarmela et al. 2011, Sanci et al. 2011). Our observation with VEGFA expression is consistent with a previous study on LYO and MYO from untreated and GNRHa-treated patients (Harrison-Woolrych et al. 1995), which is in contrast to another study indicating a higher VEGF immunostaining in LYO from luteal phase with elevated levels in tissues from menopausal period (Lewicka et al. 2010). As LYO have been reported to be less vascular compared with MYO (Pollard et al. 2005), lower VEGF expression may not serve as vasculogenic and angiogenic mediators, although VEGFA, as well as TIMP2 and FBLN5 expression declined in LYO of patients exposed to GNRHa or OCPs treatments which are often used to regress LYO growth and associated symptoms, respectively. Interestingly, similar to GNRHa, selective progesterone receptor modulator (SPRM) therapies also regress LYO growth; however, treatment of MSMC and LSMC with SPRM has been reported to increase the expression of VEGFA, while inhibiting TIMP2 expression (Xu et al. 2006, 2008). Although miR-200c directly targets the expression of VEGFA, TIMP2, and FBLN5, their biological significance such as mediators of angiogenesis, extracellular matrix turnover, and elastic fiber assembly (Harper & Bates 2008, Yanagisawa et al. 2009, Chegini 2010, Ciarmela et al. 2011, Devitt et al. 2011, Guadall et al. 2011) in LYO requires detailed investigation. Additionally, TIMP2 and FBLN5 through interaction with MMP9 have been associated with pelvic organ prolapsed (Budatha et al. 2011) and hypoxia-induced FBLN5 through HIF1α signaling which has been shown to regulate tumor growth, angiogenesis, and EMT (Lee et al. 2008, Yanagisawa et al. 2009, Yue et al. 2009, Guadall et al. 2011). As such, TIMP2 and FBLN5 through such activities may play key roles in the pathogenesis of LYO. As FLT1 and KLF9 are validated as targets of miR-200c in lung adenocarcinomas and Ishikawa, endometrial epithelial cancer cell line (Roybal et al. 2011, Panda et al. 2012), lack of their regulation by miR-200c in MSMC and LSMC further supports the cell specificity of miRNAs regulatory functions as previously reported (Dahiya et al. 2008, Inomata et al. 2009).
In summary, we demonstrated that miR-200c is aberrantly expressed in LYO with lower levels in tumors from African Americans as compared with Caucasians, which to some extent is influenced by hormonal milieu. We confirmed ZEB1/2 and VEGFA, validated FBLN5 and TIMP2 as direct targets of miR-200c in MSMC, LSMC, and SKLM-S1, and demonstrated that overexpression of miR-200c results in phenotypic alteration of MSMC and LSMC, but not SKLM-S1, possibly through increased CDH1 expression. As the above gene products regulate various cellular activities ranging from cellular transition, cell growth, angiogenesis, and tissue turnover that are central to LYO development, growth, and fibrotic characteristic, our results support a key role of miR-200c in the genesis of LYO. While this study enhanced our understanding of the regulatory function of miR-200c in the pathogenesis of LYO, further investigation is required to address other aspects of miR-200c and other members of this family in LYO.
Supplementary data
This is linked to the online version of the paper at http://dx.doi.org/10.1530/ERC-12-0007.
Author contribution statement
All authors contributed toward the completion of the paper with T-D Chuang performed most of the experiments, with acquisition, analysis, and interpretation of data; X Luo performed the microarray analysis, and H Panda performed some of the real-time PCR assays; N Chegini conceived and designed the experiments, established the primary cell cultures, and interpreted the data. T-D Chuang and N Chegini wrote the paper.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This work was supported by grants HD37432 and HD58664 from the Eunice Kennedy Shriver National institute of Child Health and Human Development, National Institute of Health. The results are presented, in part, at the 67th annual meeting of American Society for Reproductive Medicine, Orlando FL, 2011, and the 59th annual meeting of the Society for Gynecological Investigation, San Diego, CA, 2012.
References
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird DD, Garrett TA, Laughlin SK, Davis B, Semelka RC, Peddada SD. Short-term change in growth of uterine leiomyoma: tumor growth spurts. Fertility and Sterility. 2011;95:242–246. doi: 10.1016/j.fertnstert.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendoraite A, Knouf EC, Garg KS, Parkin RK, Kroh EM, O'Briant KC, Ventura AP, Godwin AK, Karlan BY, Drescher CW, et al. Regulation of miR-200 family microRNAs and ZEB transcription factors in ovarian cancer: evidence supporting a mesothelial-to-epithelial transition. Gynecological Oncology. 2010;116:117–125. doi: 10.1016/j.ygyno.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat-Nakshatri P, Wang G, Collins NR, Thomson MJ, Geistlinger TR, Carroll JS, Brown M, Hammond S, Srour EF, Liu Y, et al. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Research. 2009;37:4850–4861. doi: 10.1093/nar/gkp500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogusiewicz M, Stryjecka-Zimmer M, Postawski K, Jakimiuk AJ, Rechberger T. Activity of matrix metalloproteinase-2 and -9 and contents of their tissue inhibitors in uterine leiomyoma and corresponding myometrium. Gynecological Endocrinology. 2007;23:541–546. doi: 10.1080/09513590701557416. [DOI] [PubMed] [Google Scholar]
- Budatha M, Roshanravan S, Zheng Q, Weislander C, Chapman SL, Davis EC, Starcher B, Word RA, Yanagisawa H. Extracellular matrix proteases contribute to progression of pelvic organ prolapse in mice and humans. Journal of Clinical Investigation. 2011;121:2048–2059. doi: 10.1172/JCI45636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai JY, Modak C, Mouazzen W, Narvaez R, Pham J. Epithelial or mesenchymal: where to draw the line? Bioscience Trends. 2010;4:130–142. [PubMed] [Google Scholar]
- Chan YC, Khanna S, Roy S, Sen CK. miR-200b targets Ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. Journal of Biological Chemistry. 2011;286:2047–2056. doi: 10.1074/jbc.M110.158790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HL, Senaratne TN, Zhang L, Szotek PP, Stewart E, Dombkowski D, Preffer F, Donahoe PK, Teixeira J. Uterine leiomyomas exhibit fewer stem/progenitor cell characteristics when compared with corresponding normal myometrium. Reproductive Sciences. 2010;17:158–167. doi: 10.1177/1933719109348924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chegini N. Proinflammatory and profibrotic mediators: principal effectors of leiomyoma development as a fibrotic disorder. Seminars in Reproductive Medicine. 2010;28:180–203. doi: 10.1055/s-0030-1251476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chegini N, Ma C, Tang XM, Williams RS. Effects of GnRH analogues, ‘add-back’ steroid therapy, antiestrogen and antiprogestins on leiomyoma and myometrial smooth muscle cell growth and transforming growth factor-beta expression. Molecular Human Reproduction. 2002;8:1071–1078. doi: 10.1093/molehr/8.12.1071. [DOI] [PubMed] [Google Scholar]
- Chen Y, Ge W, Xu L, Qu C, Zhu M, Zhang W, Xiao Y. miR-200b is involved in intestinal fibrosis of Crohn's disease. International Journal of Molecular Medicine. 2012;29:601–606. doi: 10.3892/ijmm.2012.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang TD, Luo X, Panda H, Chegini N. miR-93/106b and their host gene, MCM7, are differentially expressed in leiomyomas and functionally target F3 and IL-8. Molecular Endocrinology. 2012;26:1028–1042. doi: 10.1210/me.2012-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarmela P, Islam MS, Reis FM, Gray PC, Bloise E, Petraglia F, Vale W, Castellucci M. Growth factors and myometrium: biological effects in uterine fibroid and possible clinical implications. Human Reproduction Update. 2011;17:772–790. doi: 10.1093/humupd/dmr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya N, Sherman-Baust CA, Wang TL, Davidson B, Shih I, Zhang Y, Wood W, III, Becker KG, Morin PJ. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS ONE. 2008;3:e2436. doi: 10.1371/journal.pone.0002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos V, Esteller M. Opening the treasure chest of miR-200s family members. Cell Cycle. 2009;8:2141–2142. doi: 10.4161/cc.8.14.9201. [DOI] [PubMed] [Google Scholar]
- Devitt MH, Ralfkjaer U, Cremers N, Frankel M, Troelsgaard PR, Klingelhofer J, Yanagisawa H, Grigorian M, Guldberg P, Sleeman J, et al. Role of fibulin-5 in metastatic organ colonization. Molecular Cancer Research. 2011;9:553–563. doi: 10.1158/1541-7786.MCR-11-0093. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331:550–553. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn DM. MicroRNAs and metastasis: little RNAs go a long way. Cancer Research. 2010;70:6401–6406. doi: 10.1158/0008-5472.CAN-10-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annual Review of Biochemistry. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Fragiadaki M, Mason RM. Epithelial–mesenchymal transition in renal fibrosis the evidence for and against. International Journal of Experimental Pathology. 2011;92:143–150. doi: 10.1111/j.1365-2613.2011.00775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bracken CP, Smith E, Bert AG, Wright JA, Roslan S, Morris M, Wyatt L, Farshid G, Lim YY, et al. An autocrine TGF-{beta}/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial–mesenchymal transition. Molecular Biology of the Cell. 2011;22:1686–1698. doi: 10.1091/mbc.E11-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadall A, Orriols M, Rodriguez-Calvo R, Calvayrac O, Crespo J, Aledo R, Martinez-Gonzalez J, Rodriguez C. Fibulin-5 is up-regulated by hypoxia in endothelial cells through a hypoxia-inducible factor-1 (HIF-1alpha)-dependent mechanism. Journal of Biological Chemistry. 2011;286:7093–7103. doi: 10.1074/jbc.M110.162917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper SJ, Bates DO. VEGF-A splicing: the key to anti-angiogenic therapeutics? Nature Reviews. Cancer. 2008;8:880–887. doi: 10.1038/nrc2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison-Woolrych ML, Sharkey AM, Charnock-Jones DS, Smith SK. Localization and quantification of vascular endothelial growth factor messenger ribonucleic acid in human myometrium and leiomyomata. Journal of Clinical Endocrinology and Metabolism. 1995;80:1853–1858. doi: 10.1210/jc.80.6.1853. [DOI] [PubMed] [Google Scholar]
- Hawkins SM, Buchold GM, Matzuk MM. Minireview: the roles of small RNA pathways in reproductive medicine. Molecular Endocrinology. 2011a;25:1257–1279. doi: 10.1210/me.2011-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins SM, Creighton CJ, Han DY, Zariff A, Anderson ML, Gunaratne PH, Matzuk MM. Functional microRNA involved in endometriosis. Molecular Endocrinology. 2011b;25:821–832. doi: 10.1210/me.2010-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz B. Formation and function of the myofibroblast during tissue repair. Journal of Investigative Dermatology. 2007;127:526–537. doi: 10.1038/sj.jid.5700613. [DOI] [PubMed] [Google Scholar]
- Hinz B, Phan SH, Thannickal VJ, Galli A, Bochaton-Piallat ML, Gabbiani G. The myofibroblast: one function, multiple origins. American Journal of Pathology. 2007;170:1807–1816. doi: 10.2353/ajpath.2007.070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomata M, Tagawa H, Guo YM, Kameoka Y, Takahashi N, Sawada K. MicroRNA-17-92 down-regulates expression of distinct targets in different B-cell lymphoma subtypes. Blood. 2009;113:396–402. doi: 10.1182/blood-2008-07-163907. [DOI] [PubMed] [Google Scholar]
- Jiang X, Tsitsiou E, Herrick SE, Lindsay MA. MicroRNAs and the regulation of fibrosis. FEBS Journal. 2010;277:2015–2021. doi: 10.1111/j.1742-4658.2010.07632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial–mesenchymal transition. Journal of Clinical Investigation. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M, Katoh M. Integrative genomic analyses of ZEB2: transcriptional regulation of ZEB2 based on SMADs, ETS1, HIF1alpha, POU/OCT, and NF-kappaB. International Journal of Oncology. 2009;34:1737–1742. doi: 10.3892/ijo_00000304. [DOI] [PubMed] [Google Scholar]
- Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial–mesenchymal transition and cancer metastasis. RNA Biology. 2008;5:115–119. doi: 10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Albig AR, Regner M, Schiemann BJ, Schiemann WP. Fibulin-5 initiates epithelial–mesenchymal transition (EMT) and enhances EMT induced by TGF-beta in mammary epithelial cells via a MMP-dependent mechanism. Carcinogenesis. 2008;29:2243–2251. doi: 10.1093/carcin/bgn199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Park YA, Choi JJ, Lee YY, Kim CJ, Choi C, Kim TJ, Lee NW, Kim BG, Bae DS. The expression of the miRNA-200 family in endometrial endometrioid carcinoma. Gynecological Oncology. 2011;120:56–62. doi: 10.1016/j.ygyno.2010.09.022. [DOI] [PubMed] [Google Scholar]
- Leppert PC, Catherino WH, Segars JH. A new hypothesis about the origin of uterine fibroids based on gene expression profiling with microarrays. American Journal of Obstetrics and Gynecology. 2006;195:415–420. doi: 10.1016/j.ajog.2005.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewicka A, Osuch B, Cendrowski K, Zegarska J, Stelmachow J. Expression of vascular endothelial growth factor mRNA in human leiomyomas. Gynecological Endocrinology. 2010;26:451–455. doi: 10.3109/09513591003632159. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Luo X, Chegini N. The expression and potential regulatory function of microRNAs in the pathogenesis of leiomyoma. Seminars in Reproductive Medicine. 2008;26:500–514. doi: 10.1055/s-0028-1096130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Ding L, Xu J, Williams RS, Chegini N. Leiomyoma and myometrial gene expression profiles and their responses to gonadotropin-releasing hormone analog therapy. Endocrinology. 2005;146:1074–1096. doi: 10.1210/en.2004-1384. [DOI] [PubMed] [Google Scholar]
- Maillot G, Lacroix-Triki M, Pierredon S, Gratadou L, Schmidt S, Benes V, Roche H, Dalenc F, Auboeuf D, Millevoi S, et al. Widespread estrogen-dependent repression of micrornas involved in breast tumor cell growth. Cancer Research. 2009;69:8332–8340. doi: 10.1158/0008-5472.CAN-09-2206. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Cho YC, Mine Y, Yoshizaki A, Naito S, Wen CY, Sekine I. Expression of vascular endothelial growth factor and its receptors VEGFR-1 and 2 in gastrointestinal stromal tumors, leiomyomas and schwannomas. World Journal of Gastroenterology. 2006;12:6182–6187. doi: 10.3748/wjg.v12.i38.6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothnick WB, Healy C. Estrogen induces distinct patterns of microRNA expression within the mouse uterus. Reproductive Sciences. 2010;17:987–994. doi: 10.1177/1933719110377472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oba S, Kumano S, Suzuki E, Nishimatsu H, Takahashi M, Takamori H, Kasuya M, Ogawa Y, Sato K, Kimura K, et al. miR-200b precursor can ameliorate renal tubulointerstitial fibrosis. PLoS ONE. 2010;5:e13614. doi: 10.1371/journal.pone.0013614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Maruyama T, Masuda H, Kajitani T, Nagashima T, Arase T, Ito M, Ohta K, Uchida H, Asada H, et al. Side population in human uterine myometrium displays phenotypic and functional characteristics of myometrial stem cells. PNAS. 2007;104:18700–18705. doi: 10.1073/pnas.0704472104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda H, Pelakh L, Chuang TD, Luo X, Bukulmez O, Chegini N. Endometrial miR-200c is altered during transformation into cancerous states and targets the expression of ZEBs, VEGFA, FLT1, IKKbeta, KLF9, and FBLN5. Reproductive Sciences. 2012 doi: 10.1177/1933719112438448. [in press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker WH. Etiology, symptomatology, and diagnosis of uterine myomas. Fertility and Sterility. 2007;87:725–736. doi: 10.1016/j.fertnstert.2007.01.093. [DOI] [PubMed] [Google Scholar]
- Peddada SD, Laughlin SK, Miner K, Guyon JP, Haneke K, Vahdat HL, Semelka RC, Kowalik A, Armao D, Davis B, et al. Growth of uterine leiomyomata among premenopausal black and white women. PNAS. 2008;105:19887–19892. doi: 10.1073/pnas.0808188105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Laser J, Shi G, Mittal K, Melamed J, Lee P, Wei JJ. Antiproliferative effects by Let-7 repression of high-mobility group A2 in uterine leiomyoma. Molecular Cancer Research. 2008;6:663–673. doi: 10.1158/1541-7786.MCR-07-0370. [DOI] [PubMed] [Google Scholar]
- Pollard P, Wortham N, Barclay E, Alam A, Elia G, Manek S, Poulsom R, Tomlinson I. Evidence of increased microvessel density and activation of the hypoxia pathway in tumours from the hereditary leiomyomatosis and renal cell cancer syndrome. Journal of Pathology. 2005;205:41–49. doi: 10.1002/path.1686. [DOI] [PubMed] [Google Scholar]
- Renthal NE, Chen CC, Williams KC, Gerard RD, Prange-Kiel J, Mendelson CR. miR-200 family and targets, ZEB1 and ZEB2, modulate uterine quiescence and contractility during pregnancy and labor. PNAS. 2010;107:20828–20833. doi: 10.1073/pnas.1008301107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybal JD, Zang Y, Ahn YH, Yang Y, Gibbons DL, Baird BN, Alvarez C, Thilaganathan N, Liu DD, Saintigny P, et al. miR-200 Inhibits lung adenocarcinoma cell invasion and metastasis by targeting Flt1/VEGFR1. Molecular Cancer Research. 2011;9:25–35. doi: 10.1158/1541-7786.MCR-10-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanci M, Dikis C, Inan S, Turkoz E, Dicle N, Ispahi C. Immunolocalization of VEGF, VEGF receptors, EGF-R and Ki-67 in leiomyoma, cellular leiomyoma and leiomyosarcoma. Acta Histochemica. 2011;113:317–325. doi: 10.1016/j.acthis.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoelstra NS, Manning NG, Higashi Y, Darling D, Singh M, Shroyer KR, Broaddus RR, Horwitz KB, Richer JK. The transcription factor ZEB1 is aberrantly expressed in aggressive uterine cancers. Cancer Research. 2006;66:3893–3902. doi: 10.1158/0008-5472.CAN-05-2881. [DOI] [PubMed] [Google Scholar]
- Tanwar PS, Lee HJ, Zhang L, Zukerberg LR, Taketo MM, Rueda BR, Teixeira JM. Constitutive activation of beta-catenin in uterine stroma and smooth muscle leads to the development of mesenchymal tumors in mice. Biology of Reproduction. 2009;81:545–552. doi: 10.1095/biolreprod.108.075648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial–mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Walker CL, Stewart EA. Uterine fibroids: the elephant in the room. Science. 2005;308:1589–1592. doi: 10.1126/science.1112063. [DOI] [PubMed] [Google Scholar]
- Wallez Y, Vilgrain I, Huber P. Angiogenesis: the VE-cadherin switch. Trends in Cardiovascular Medicine. 2006;16:55–59. doi: 10.1016/j.tcm.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Wang T, Zhang X, Obijuru L, Laser J, Aris V, Lee P, Mittal K, Soteropoulos P, Wei JJ. A micro-RNA signature associated with race, tumor size, and target gene activity in human uterine leiomyomas. Genes, Chromosomes & Cancer. 2007;46:336–347. doi: 10.1002/gcc.20415. [DOI] [PubMed] [Google Scholar]
- Wynn TA. Cellular and molecular mechanisms of fibrosis. Journal of Pathology. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Ohara N, Chen W, Liu J, Sasaki H, Morikawa A, Sitruk-Ware R, Johansson ED, Maruo T. Progesterone receptor modulator CDB-2914 down-regulates vascular endothelial growth factor, adrenomedullin and their receptors and modulates progesterone receptor content in cultured human uterine leiomyoma cells. Human Reproduction. 2006;21:2408–2416. doi: 10.1093/humrep/del159. [DOI] [PubMed] [Google Scholar]
- Xu Q, Ohara N, Liu J, Amano M, Sitruk-Ware R, Yoshida S, Maruo T. Progesterone receptor modulator CDB-2914 induces extracellular matrix metalloproteinase inducer in cultured human uterine leiomyoma cells. Molecular Human Reproduction. 2008;14:181–191. doi: 10.1093/molehr/gan004. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Schluterman MK, Brekken RA. Fibulin-5, an integrin-binding matricellular protein: its function in development and disease. Journal of Cell Communication and Signaling. 2009;3:337–347. doi: 10.1007/s12079-009-0065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Banerjee S, de Freitas A, Sanders YY, Ding Q, Matalon S, Thannickal VJ, Abraham E, Liu G. Participation of miR-200 in pulmonary fibrosis. American Journal of Pathology. 2012;180:484–493. doi: 10.1016/j.ajpath.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto N, Toyama T, Takahashi S, Sugiura H, Endo Y, Iwasa M, Fujii Y, Yamashita H. Distinct expressions of microRNAs that directly target estrogen receptor alpha in human breast cancer. Breast Cancer Research and Treatment. 2011;130:331–339. doi: 10.1007/s10549-011-1672-2. [DOI] [PubMed] [Google Scholar]
- Yue W, Sun Q, Landreneau R, Wu C, Siegfried JM, Yu J, Zhang L. Fibulin-5 suppresses lung cancer invasion by inhibiting matrix metalloproteinase-7 expression. Cancer Research. 2009;69:6339–6346. doi: 10.1158/0008-5472.CAN-09-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]