Abstract
In industrialized and/or agriculturally used landscapes, inhabiting species are exposed to a variety of anthropogenic changes in their environments. Genetic diversity may be reduced if populations encounter founder events, bottlenecks, or isolation. Conversely, genetic diversity may increase if populations adapt to changes in selective regimes in newly created habitats. With the present study, genetic variability of 918 sticklebacks from 43 samplings (21.3 ± 3.8 per sample) at 36 locations from cultivated landscapes in Northwest Germany was analyzed at nine neutral microsatellite loci. To test if differentiation is influenced by habitat alterations, sticklebacks were collected from ancient running waters and adjacent artificial stagnant waters, from brooks with salt water inflow of anthropogenic and natural origin and adjacent freshwater sites. Overall population structure was dominated by isolation by distance (IBD), which was significant across all populations, and analysis of molecular variance (AMOVA) revealed that 10.6% of the variation was explained by river catchment area. Populations in anthropogenic modified habitats deviated from the general IBD structure and in the AMOVA, grouping by habitat type running/stagnant water explained 4.9% of variation and 1.4% of the variation was explained by salt-/freshwater habitat. Sticklebacks in salt-polluted water systems seem to exhibit elevated migratory activity between fresh- and saltwater habitats, reducing IBD. In other situations, populations showed distinct signs of genetic isolation, which in some locations was attributed to mechanical migration barriers, but in others to potential anthropogenic induced bottleneck or founder effects. The present study shows that anthropogenic habitat alterations may have diverse effects on the population genetic structure of inhabiting species. Depending on the type of habitat change, increased genetic differentiation, diversification, or isolation are possible consequences.
Keywords: Anthropogenic impact, Gasterosteus aculeatus, genetic isolation, microsatellite, population structure
Introduction
In densely populated, industrialized, and agriculturally used areas of central Europe aquatic systems are altered by human activity in various ways. Water systems were engineered for industrial and agricultural purposes and are influenced by pollution with communal, industrial, and agricultural wastewaters. Since the 1970s, the far majority of sewage waters are subjected to mechanical and biological cleaning, and water quality was generally improved. However, stream-dwelling fish, in particular, seem to be susceptible to human habitat alterations (Hänfling and Weetman 2006; Raeymaekers et al. 2008; 2009; Koizumi 2011).
The three-spined stickleback (Gasterosteus aculeatus) is a well-suited model for studying population genetics in cultivated areas, since it is tolerant to a broad range of environmental conditions. Three-spined sticklebacks generally are euryhaline, but slight differences in salt tolerance between anadromous and resident freshwater sticklebacks were described (Wootton 1984). About 12,500 years ago, after the last deglaciation, freshwater habitats were recolonized by sticklebacks from marine ancestors (e.g., Walker and Bell 2000; Gow et al. 2006; Mäkinen et al. 2006; Caldera and Bolnick 2008; Wund et al. 2008). In the newly colonized water bodies, sticklebacks have evolved distinct ecotypes, such as lake and stream forms (e.g., Thompson et al. 1997; Reusch et al. 2001), within lakes limnetic and benthic forms (e.g., Taylor and McPhail 1999; Baker et al. 2005), or mud and lava forms (Kristjansson et al. 2002; Olafsdottir and Snorrason 2009; reviewed by Hendry et al. 2009).
Our present knowledge on stickleback radiation across different habitat types is mainly based on populations in (natural) habitats with minor postglacial anthropogenic influence and speciation ongoing since the last deglaciation, but biological species of G. aculeatus may also evolve within decades (Bell 2001). Anthropogenic changes of environments often are faster than natural changes and lead to a faster adaptive response of the inhabiting populations (Candolin 2009).
Three-spined sticklebacks were relatively fast in the establishment of stable populations in new human-created environments, for example when marine G. aculeatus were entrapped in freshwater lagoons created by the construction of dams (Klepaker 1993; Kristjansson et al. 2002; Olafsdottir et al. 2007) or when stream sticklebacks were transplanted to a lake (Vamosi 2006). Under certain conditions of anthropogenic induced isolation, stickleback populations seem to be vulnerable to bottleneck situations and inbreeding. Three-spined sticklebacks in northern Japan revealed signatures of genetic isolation due to weir construction and habitat deterioration (Takamura and Mori 2005). In the river Scheldt basin in Belgium, anthropogenic structures were the strongest determinant of population structure, when evaluated against a geographically baseline model accounting for natural effects (Raeymaekers et al. 2008). In the Zwalm subbasin (Scheldt basin, Belgium), effects of historical water mills (320–1000 years old), which prevent upstream migration but allow downstream drift, on genetic dispersal were lower in populations above than below mills (Raeymaekers et al. 2009). Water mills provoked an average loss of about 4% of the genetic variation, which accumulated to 40% across the entire system. The impact of individual mills increased with upstream distance and water mill height. One mill provoked significant genetic differentiation, despite the presence of a fish passage (Raeymaekers et al. 2009).
Population genetic structures of other sedentary river fish, such as the river sculpin (Cottus gobio) are also influenced by anthropogenic barriers, namely weirs (Hänfling and Weetman 2006). Here, a source–sink structure was evident since migration and genetic diversity in smaller upstream locations was emigration biased while it was immigration biased in larger downstream subpopulations. Asymmetry of population structure was partly attributable to the effects of flow direction, but was enhanced by weirs prohibiting compensatory upstream migration (Hänfling and Weetman 2006).
In sedentary river fish populations, the isolation by distance (IBD) model (Wright 1942) explains most of the genetic variation (Hänfling et al. 2002; Hänfling and Weetman 2006; Raeymaekers et al. 2008; 2009), but a migration–drift equilibrium is disturbed when artificial barriers are separating populations, resulting in upstream populations being more prone to genetic isolation and potentially more susceptible to bottleneck situations (Hänfling and Weetman 2006; Raeymaekers et al. 2008; 2009). However, a prominent counter example is given by stickleback populations in northern Germany (Schleswig Holstein), where stream and lake stickleback ecotypes are more closely related within ecotype across different drainage systems, than between ecotypes in the same drainage system, despite the absence of migratory barriers (Reusch et al. 2001). Here, IBD seems to be overruled by ecological speciation with different selection regimes and the formation of mating barriers being causative for the separation of stickleback populations, rather than distance.
Anthropogenic habitat modifications as well may influence selection in natural populations. For example, pulp mill effluence has acted as a selective agent on G. aculeatus populations, resulting in higher genetic differentiation between stickleback from polluted and not polluted sites, compared to not polluted reference sites (Lind and Grahn 2011).
Consequently, anthropogenic changes of aquatic habitats may alter population genetic dynamics of inhabiting sticklebacks in different ways. (1) Physical barriers (dams, weirs, sluices) may inhibit gene flow among stickleback populations along drainage systems and in extreme cases may result in genetic isolation. Habitat modifications, such as (2) construction of artificial “new” stagnant waters (e.g., ponds, ditches) may alter genetic differentiation, if founder populations separate (genetically) from their population of origin; and (3) pollution, depending on its quality and consistency, may subject populations to bottleneck situations or favor colonization of polluted areas by tolerant species. Finally (4), translocation of specimen by human activity as well might be imprinted in population genetic patterns. Overall, it is expected that genetic differentiation of populations (also in anthropogenic influenced landscapes) follows the IBD model, but populations with specific anthropogenic impact might deviate from general IBD (Koizumi et al. 2006). For example, isolated populations might show higher FST-values at lower geographical distance compared to not isolated populations. Also, populations that were subjected to bottleneck situations or founder events might deviate from a general IBD pattern. The latter may as well show reduced numbers of alleles and low M ratios. In these scenarios, effects of gene flow/drift might be stronger than selection. Alternatively, populations might be connected by effects of habitat type (e.g., running–stagnant, salt–freshwater), where habitat effects overrule IBD (or isolation by barrier) and effects of selection are stronger than effects of gene flow/drift.
In the present study, nine neutral microsatellite markers are used to investigate the population genetic structure of three-spined sticklebacks in a human-modified landscape and disentangling effects of selection and gene flow/drift is not possible. However, the present sampling strategy includes habitats with different anthropogenic alterations and comparison of microsatellite diversity in different habitat types will allow to detect situations that deviate from an overall IBD pattern. Three-spined sticklebacks were sampled in a densely human populated, industrialized, and agriculturally used area in North West Germany. Our main focus is (1) to investigate sticklebacks from human-created stagnant waters (ponds, rainwater retention basins, lakes) and adjacent ancient running waters, and (2) to investigate sticklebacks in brooks with consistent, long-term (decades) inflow of salty coal mine drainage water (and one natural salty spring) and adjacent freshwater habitats. We hypothesize that colonization of the new habitat type artificial pond and/or saltwater is detectable in the genetic differentiation of inhabiting sticklebacks.
Materials and Methods
Sampling sites
Three-spined sticklebacks (G. aculeatus) were collected exclusively from inland locations (36 in total), presumably recolonized by sticklebacks from marine ancestors after the last glaciation (Mäkinen et al. 2006). Sampling sites were chosen with the following criteria. (1) Pairs (groups) of sampling sites from artificial stagnant waters (ponds) with closely neighboring sampling sites in adjacent running waters in the area of Münster (brooks, ditches, small rivers) (Category 1 in Table 1) (2) plus additional sampling sites in running waters (brooks) to achieve a better network for testing the among-population differentiation (Category 2 in Table 1). (3) Pairs (groups) of sampling sites from running waters with anthropogenic and natural salt water influence with closely neighboring adjacent freshwater sampling sites (upstream and/or downstream from the saltwater discharge) (Category 3 in Table 1; see also maps in Figs. 1 and 2). Outgroups were a population from the “Grosse Plöner See” (GPS) in Schleswig Holstein (northern Germany) and a population from Northwest Spain (SPA) (Fig. 1A). Data of the outgroup populations were excluded from the majority of tests, but are left for comparison in Tables 1 and A1–A3 in the Appendix.
Table 1.
List of sampling sites sorted by catchment area.
| Sampling site | Code | n | MNA | HE | HO | M ratio | Latitude, Longitude | |
|---|---|---|---|---|---|---|---|---|
| Ems catchment area: | ||||||||
| Category 1. Running/stagnant waters: | ||||||||
| Meckelbach (ru) | MB | 20 | 6.0 | 0.62 | 0.61 | 0.61 ± 0.28 | 51°55′55.66″N,7°34′36.42″E | |
| Mecklenbeck, small pond (st) | SP° | 28 | 5.7 | 0.62 | 0.61* | 0.59 ± 0.29 | 51°56′00.15″N,7°34′33.39″E | |
| Mecklenbeck, large pond (st) | LP | 36 | 5.7 | 0.61 | 0.63* | 0.54 ± 0.25 | 51°55′59.81″N,7°34′30.28″E | |
| Münstersche Aa (ru) | MA | 19 | 6.2 | 0.69 | 0.66 | 0.53 ± 0.33 | 51°56′18.49″N,7°35′08.57″E | |
| Aasee (st) | AS | 21 | 5.7 | 0.66 | 0.63* | 0.57 ± 0.28 | 51°57′21.69″N,7°36′50.34″E | |
| Eschuss Bach (ru) | EB | 23 | 5.3 | 0.62 | 0.59 | 0.59 ± 0.29 | 52°03′20.98″N,7°30′31.40″E | |
| Rain storage reservoir (st) | RSR | 24 | 6.2 | 0.64 | 0.62* | 0.64 ± 0.24 | 52°02′58.76″N,7°29′23.51″E | |
| Hohnebach (ru) | HB | 18 | 4.8 | 0.62 | 0.63 | 0.57 ± 0.34 | 51°56′44.76″N,7°41′13.00″E | |
| Moat Haus Lütkenbeck (st) | MHL | 22 | 4.9 | 0.55 | 0.56 | 0.60 ± 0.34 | 51°56′34.98″N,7°39′35.69″E | |
| Werse (ru) | WE | 22 | 7.1 | 0.67 | 0.67 | 0.56 ± 0.22 | 51°57′47.35″N,7°42′15.30″E | |
| Rieselfelder (st) | RF | 22 | 4.0 | 0.56 | 0.60 | 0.47 ± 0.29 | 52°01′35.96″N,7°39′23.24″E | |
| Category 2. Running waters: | ||||||||
| Loddenbach (ru) | LB° | 20 | 3.3 | 0.37 | 0.36 | 0.49 ± 0.36 | 51°56′10.44″N,7°40′51.83″E | |
| Nienberger Bach (ru) | NB | 31 | 6.3 | 0.68 | 0.71 | 0.56 ± 0.24 | 52°00′15.56″N,7°35′40.66″E | |
| Kinderbach (ru) | KB° | 25 | 7.2 | 0.71 | 0.65 | 0.67 ± 0.24 | 52°00′08.17″N,7°36′48.87″E | |
| Category 3. Salt/freshwater: | ||||||||
| Ibb. Aa, upstream freshwater tributary (f) | IUF | 21 | 5.7 | 0.67 | 0.68* | 0.51 ± 0.24 | 52°15′38.28″N,7°44′24.72″E | |
| Ibb. Aa, freshwater tributary (f) | IF° | 15 | 4.7 | 0.57 | 0.51* | 0.56 ± 0.35 | 52°15′40.99″N,7°44′26.65″E | |
| Ibb. Aa, upstream Aasee (f) | IUA° | 22 | 5.8 | 0.59 | 0.59 | 0.52 ± 0.23 | 52°15′40.09″N,7°44′22.37″E | |
| Ibb. Aa, downstream freshwater tributary (f) | IDF | 20 | 5.2 | 0.62 | 0.62 | 0.53 ± 0.32 | 52°15′41.25″N,7°44′20.82″E | |
| Ibb. Aa, upstream saltwater discharge (f) | 2009 | IUS09° | 23 | 5.3 | 0.62 | 0.57* | 0,49 0,27 | 52°17′00.10″N,7°39′12.76″E |
| 05/2010 | IUS10a | 18 | 4.8 | 0.56 | 0.59 | 0,52 0,32 | ||
| 09/2010 | IUS10b | 18 | 4.9 | 0.58 | 0.60 | 0,50 0,31 | ||
| Saltwater discharge tributary (f) | SDT° | 19 | 3.1 | 0.48 | 0.48 | 0.46 ± 0.32 | 52°17′16.89″N,7°39′05.20″E | |
| Ibb. Aa, directly after saltwater discharge (s) | IDS | 20 | 5.1 | 0.60 | 0.63 | 0.46 ± 0.28 | 52°17′01.03″N,7°39′07.66″E | |
| Ibb. Aa, bridge at Püsselsbüren (s) | IP | 20 | 5.3 | 0.61 | 0.60 | 0.50 ± 0.27 | 52°17′22.16″N,7°38′01.81″E | |
| Ibb. Aa, upstream of Klosterbach (s) | 2009 | IUK09 | 23 | 5.1 | 0.60 | 0.61 | 0,51 0,30 | 52°17′32.47″N,7°36′52.90″E |
| 2010 | IUK10 | 19 | 5.3 | 0.61 | 0.63* | 0,58 0,34 | ||
| Klosterbach (s) | 2009 | KLB09° | 27 | 5.4 | 0.54 | 0.52 | 0,47 0,24 | 52°17′26.28″N,7°36′49.96″E |
| 2010 | KLB10° | 19 | 5.0 | 0.59 | 0.51* | 0,50 0,26 | ||
| Ibb. Aa, downstream of Klosterbach (s) | 05/2009 | IDK09a | 21 | 5.1 | 0.58 | 0.56 | 0,52 0,29 | 52°17′31.76″N,7°36′46.49″E |
| 09/2009 | IDK09b | 21 | 4.7 | 0.56 | 0.61 | 0,59 0,34 | ||
| 05/2010 | IDK10a | 20 | 4.8 | 0.59 | 0.68 | 0,56 0,34 | ||
| 09/2010 | IDK10b | 20 | 5.4 | 0.64 | 0.64 | 0,51 0,24 | ||
| Ibb. Aa, after Midland Channel culvert (s) | IDM | 17 | 5.4 | 0.62 | 0.63* | 0.44 ± 0.18 | 52°17′40.97″N,7°36′27.68″E | |
| Bever, upstream of Salzbach (f) | BUS | 15 | 5.8 | 0.63 | 0.63 | 0.52 ± 0.21 | 52°04′13.16″N,8°03′44.33″E | |
| Salzbach (s) | SB | 20 | 5.0 | 0.59 | 0.66 | 0.54 ± 0.28 | 52°04′12.55″N,8°03′20.45″E | |
| Bever, downstream Salzbach (s) | BDS | 22 | 5.1 | 0.59 | 0.60 | 0.52 ± 0.22 | 52°04′09.03″N,8°03′17.95″E | |
| Weser catchment area: | ||||||||
| Fuhse (f) | FU | 21 | 4.6 | 0.53 | 0.54 | 0.45 ± 0.25 | 52°07′21.91″N,10°22′21.93″E | |
| Mühlgraben (s) | MG° | 22 | 3.1 | 0.41 | 0.46 | 0.54 ± 0.29 | 52°07′05.05″N,10°22′05.72″E | |
| Rhine catchment area: | ||||||||
| Fossa Eugenia, upstream Große Goorley (f) | FUG | 20 | 7.4 | 0.72 | 0.74 | 0.48 ± 0.13 | 52°30′10.70″N,6°31′17.48″E | |
| Große Goorley (s) | GG | 20 | 7.4 | 0.74 | 0.77 | 0.48 ± 0.13 | 51°30′11.99″N,6°31′30.31″E | |
| Mühlenbach (f) | MUB | 21 | 7.8 | 0.73 | 0.77 | 0.52 ± 0.22 | 51°47′13.05″N,7°15′24.49″E | |
| Outgroups: | ||||||||
| Grosser Plöner See, Northern Germany (st) (f) | GPS | 23 | 5.4 | 0.69 | 0.68 | 54°8′38.36″N,10°24′53.23″E | ||
| Limia River, Spain (ru) (f) | SPA° | 20 | 10.0 | 0.67 | 0.63* | 42°8′1.27″N,7°39′47.801″W | ||
| Average | 21.3 | 5.5 | 0.61 | 0.61 | 0.53 | |||
| (standard deviation) | (3.8) | (1.2) | (0.07) | (0.08) | (0.27) | |||
st, stagnant; ru, running water; f, freshwater salinity (approximately 1 mS cm–1), (s) saltwater (> 4 mS cm–1); n, number of fish; MNA, mean number of alleles, HE, expected heterozygosity; HO, observed heterozygosity; Ibb. Aa, Ibbenbürener Aa.
°Population with null alleles (see also Appendix, Table A3).
Samples that deviate from Hardy–Weinberg equilibrium (HWE).
Figure 1.
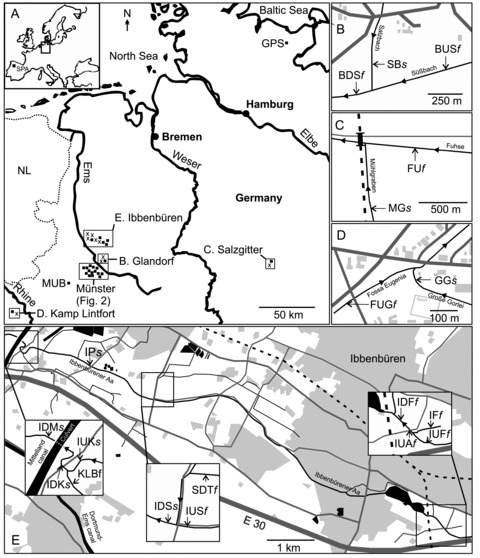
Sampling locations. (A) Sampling sites in North West Germany (▪, saltwater; ×, freshwater) and outgroups GPS and SPA (insert map). (B–E) Detailed maps of freshwater (f) and sampling locations with salt water influence (s). (B) Natural salt water inflow from a salty spring (SBs∼ 10,000 µS cm–1). (C) Salt pollution from potash mining (MGs∼ 4000 µS cm–1). (D, E) Salt pollution from coal mining (GGs∼ 6000 µS cm–1; IDMs, IUKs, IDKs, IPs, IDSs∼20,000 µS cm–1). For abbreviations of sampling sites see Table 1. Legend of detailed maps: housing scheme (light gray), streets (dark gray), railway (dashed line), surface water (black), sewage plant (black circles). Figures are modified maps from TIM-online.
Figure 2.
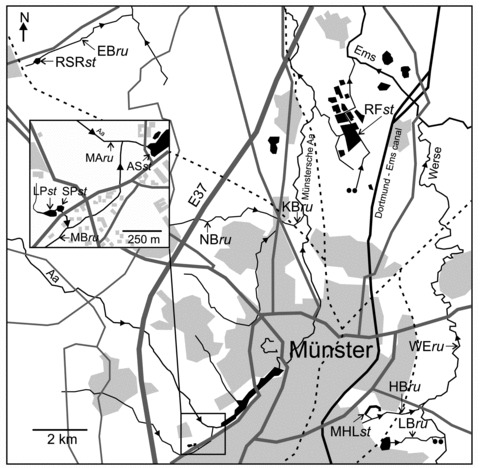
Sampling sites in the Münster area. For abbreviations of sampling sites see Table 1. (ru, running water; st, stagnant water; light gray, housing scheme; dark gray, streets, dashed line, railway; black, surface water; black circles, sewage plant). Figures are modified maps from TIM-online.
Most sampling sites were located in a densely human populated area in Northwest Germany around the city of Münster, a lowland area with sandy soil. Typical water bodies are lowland sandy brooks. Natural lakes and larger stagnant waters are absent in the area with the exception of the “Heilige Meer” and the “Erdfallsee,” where sticklebacks were not detected (Schmidt et al. 1985). Running waters in the area originate from the last deglaciation, but in their present shape show different levels of anthropogenic alterations such as river regulation and canalization, typical in landscapes with intensive agriculture and industrialization.
Water pollution is generally low in the area as all communal and industrial wastewaters are cleared before being discharged in the environment since the 1970s. Load with organic matter is low to intermediate in the investigated waters depending on natural and agricultural sources and organic remains in cleared wastewaters. Generally, sticklebacks were abundant in the investigated water bodies, indicating that ongoing drainage of heavily polluted wastewaters was absent. However, in some of the investigated waters, previous discharge of untreated wastewater might have affected the stickleback colonization history.
A prominent anthropogenic influence in some running waters of the area, in particular the brook “Ibbenbürener Aa” (Fig. 1E), is the discharge of salty coal mine drainage water. Here, the conductivity is increased from <1 mS cm–1 to > 20 mS cm–1 due to the inflow of the coal mine drainage water with a conductivity of about 45 mS cm–1 (compare seawater approximately 50 mS cm–1). This saltwater discharge exists since the 1970s. Additional salt water influenced sites were the brook “Grosse Gorley” (Fig. 1D), a brook with a conductivity of 8 mS cm–1, also due to inflow of coal mine drainage water since the 1970s, the brook “Mühlgraben” with approximately 4 mS cm–1 due to salty waste water from potash mining from 1910 to 1996, and as a naturally salt influenced brook, the “Salzbach” (Fig. 1B) with 10 mS cm–1 from a salty spring.
Sample collection
Three-spined sticklebacks (G. aculeatus) were collected at 36 locations, four of which were sampled repeatedly, resulting in 43 samplings with 21.1 ± 4.0 sticklebacks per sample (in total 908 sticklebacks; Table 1). Fish were collected with hand nets, transported to the laboratory, and killed with an overdose of MS 222 (tricaine, Sigma, Taufkirchen, Germany) 0.5 g L–1. From each stickleback, the caudal fin with a piece of the caudal peduncle was excised and stored at −20°C until extraction of genomic DNA.
DNA extraction and microsatellite genotyping
The genomic DNA was extracted with the DNA Tissue HTS 96 Kit/C (Invitek, Berlin, Germany) according to the manufacturer's instructions. Allelic variation was assessed at nine microsatellite loci organized in three multiplex and one single polymerase chain reaction (PCR). A first multiplex PCR contained primers for three microsatellites (GAC 1125, GAC 5196, GAC 7033), a second multiplex PCR primers for two microsatellites (GAC 1097, GAC 4170), all developed by Largiadèr et al. (1999). The microsatellite primers of the third multiplex PCR (Stn 18, Stn 32, Stn 84) as well as the single primer set PCR (Stn 75) were developed by Peichel et al. (2001). The PCR reaction mixes (10 μl) contained 1-μl template DNA, 0.075–0.6 µM primer with the forward primer fluorescently labeled with HEX, NED, or 6-FAM (Metabion, Martinsried, Germany), 0.25 U GoTaq Polymerase, 2 μl 5×Colorless GoTaq Reaction Buffer, 1.5 mM MgCl2 solution (all Promega, Mannheim, Germany), 1 μl dNTPs (Metabion), and RNAse-free distilled water. The PCR conditions were: 3 min at 94°C followed by 29 cycles of 1 min at 94°C, 1 min at 55°C, 1 min at 72°C, and a final elongation of 30 min at 72°C for multiplex PCR 1 and 2. The conditions of the third multiplex and the single PCR were 3 min at 94°C followed by 28 cycles (multiplex 3), respectively, 31 cycles (single) of 45 sec at 94°C, 45 sec at 56°C, 1 min at 72°C, and a final elongation of 40 min at 72°C. The PCR was performed in a Mastercycler ep gradient (Eppendorf, Wesseling-Berzdorf, Germany). For fragment length analysis, 350 ROX size standard and HiDi formamide (both Applied Biosystems, Darmstadt, Germany) were added to 0.5 μl of the PCR products according to the manufacturer's instruction. The analysis was performed in a 3130xl Genetic Analyzer (Applied Biosystems). The genotypes were scored automatically with Genemapper v. 4.0 (Applied Biosystems) and checked manually. Genotype data were controlled for null alleles with Microchecker v. 2.2.3 (Van Oosterhout et al. 2004) with 1000 randomizations and Bonferroni-adjusted confidence interval.
Genetic diversity
Genetic diversity of each sample site was measured as mean number of alleles, the observed and expected heterozygosity, and the percentage of polymorphic loci (0.99 criteria) with the software genetix v. 4.05 (Belkhirr et al. 2004). Probability of each sample site to be in Hardy–Weinberg equilibrium (HWE; Markov-Chain parameters 1000 dememorizations, 20 batches, 5000 iterations per batch) was calculated with genepop v. 4.0.11 (Rousset 2008). Sample sites were tested for pairwise linkage disequilibrium (LD) between the nine microsatellites with Arlequin v. 3.11 (Excoffier et al. 2005) under default settings and with Bonferroni-corrected significance levels. M ratios were calculated according to Garza and Williamson (2001) to detect possible reductions in population size. To test for recent migration in the Ems catchment area, BIMR v. 1.0 (Faubet and Gaggiotti 2008) was used. We used five replicates with a burn-in period of 50,000 iterations and additional 50,000 sampling iterations with default settings, a thinning interval of 50, and waterway distance as environmental factor.
Genetic structure
Pairwise fixation indices, FST with 1000 permutations and a significance level of P < 0.05, and absolute number of migrants were calculated with Arlequin (v. 3.11) adapted from Weir and Cockerham (1984). FSTAT (v. 2.9.3) was used to identify FST-values that were not significant with sequential Bonferroni correction (Goudet 2001). The hierarchical analysis of molecular variance (Amova) was done with Arlequin. Outgroups (SPA, GPS) were excluded from the Amova analysis. Hierarchical levels were catchment area, habitat type running/stagnant water, and habitat type salt-/freshwater across all habitats and within the Ems catchment area only (Table 2). Clustering of genotypes was tested with STRUCTURE v. 2.3.3 (Pritchard et al. 2000). Individual genotypes were assorted to a given number of clusters (K), which represent the populations of origin of individuals. All samples were 20 times tested with a burn-in period of 2.5 × 105 Monte Carlo Markov Chain iterations and a sampling of additional 106 iterations under default settings with K= 1 to K= 50. The post hoc ΔK function of Evanno et al. (2005) was used to estimate the likelihood of cluster formation.
Table 2.
Analysis of molecular variance (AMOVA) with ARLEQUIN.
| Hierarchical level | df | Percentage of variation | F-statistic | P |
|---|---|---|---|---|
| amova 1 Catchment areas | ||||
| Among catchment areas | 2 | 10.6 | 0.10635 | < 0.001 |
| Among populations within catchment areas | 38 | 11.2 | 0.1249 | < 0.001 |
| Within population | 1683 | 78.2 | 0.21797 | < 0.001 |
| amova 2 Habitat type: Running/stagnant water (all catchment areas) | ||||
| Among habitat types | 1 | 4.9 | 0.04936 | < 0.001 |
| Among population within habitat types | y39 | 12.8 | 0.12339 | < 0.001 |
| Within population | 1683 | 82.3 | 0.17712 | < 0.001 |
| amova 3 Habitat type: Salt-/freshwater (all catchment areas) | ||||
| Among habitat type | 1 | 1.4 | 0.01389 | 0.018 |
| Among populations within habitat type | 39 | 14.1 | 0.14247 | < 0.001 |
| Within population | 1683 | 84.6 | 0.15438 | < 0.001 |
| amova 4 Habitat type: Running/stagnant water (Ems catchment area only) | ||||
| Among habitat type | 1 | 5.4 | 0.05427 | 0.003 |
| Among populations within habitat type | 34 | 10.4 | 0.10997 | < 0.001 |
| Within population | 1480 | 84.2 | 0.15827 | < 0.001 |
| amova 5 Habitat type: Salt-/freshwater (Ems catchment area only) | ||||
| Among habitat type | 1 | 2.1 | 0.02026 | 0.015 |
| Among populations within habitat type | 34 | 11.7 | 0.11896 | < 0.001 |
| Within population | 1480 | 86.3 | 0.12681 | < 0.001 |
Phylogenetic relationship
The phylogenetic relationship of samples was analyzed with the program package Phylip v. 3.69 (Felsenstein 2005). The program Seqboot was used to create 105 datasets by bootstrapping, which were used to estimate the genetic chord distances DC (Cavalli-Sforza and Edwards 1967) with Gendist. The DC distance has the highest likelihood to find the right tree topology if the differentiation in recently diverged populations is dominated by genetic drift and gene flow (Reusch et al. 2001). Neighbor-joining trees were created with the program Neighbor and a consensus tree was constructed with Consense, all programs of the Phylip package. The tree was visualized with Treeview v. 1.6.6 (Page 1996).
Isolation by geographical distances
Genetic differentiation was measured as pairwise FST-values calculated with Arlequin and pairwise geographical distances were measured as waterway distances. To analyze potential isolation by geographical distance (IBD) patterns, correlations of waterway distance and genetic differentiation (FST-values) were analyzed by multiple Mantel tests (Mantel 1967; Manly 1986) using the IBD web service with 30,000 randomizations (Jensen et al. 2005) and Bonferroni correction for multiple testing. Mantel correlations were calculated with all sampling sites and within the Ems catchment area and, for further analyses, grouped into populations with “normal” migration rates (other), potentially high migration rates (mixing), and genetically isolated populations (isolated) (Table 3). Nonmetric multidimensional scaling (NMDS) plots based on genetic (pairwise FST) and geographic (waterline km) distance were generated for populations from the Ems catchment area using PASW v. 18 software. IBD blots were created with pairwise FST-values and waterway distance using SigmaPlot 11.
Table 3.
Multiple Mantel tests of correlations of genetic (FST) and geographic (waterway) distance with 30,000 randomizations (without outgroups). Populations of the Ems catchment area were also analyzed separately and grouped in mixing (migratory active), isolated (genetically and geographically), and other (normal).
| Populations | r | P | Slope | R2 | n |
|---|---|---|---|---|---|
| All | 0.549* | < 0.0001 | 0.036 ± 0.001 | 0.302 | 820 |
| Ems only | |||||
| All pairs | 0.667* | < 0.0001 | 0.133 ± 0.004 | 0.444 | 630 |
| Other | 0.452* | 0.0005 | 0.109 ± 0.007 | 0.205 | 190 |
| Other, mixing | 0.773* | < 0.0001 | 0.106 ± 0.003 | 0.597 | 528 |
| Other, isolated | 0.423* | < 0.0001 | 0.170 ± 0.010 | 0.179 | 253 |
| Mixing | −0.055 | 0.5204 | −0.401 ± 0.046 | 0.003 | 78 |
| Isolated | 0.353 | 0.4977 | 0.012 ± 0.011 | 0.125 | 3 |
| Isolated, mixing | 0.949* | 0.0025 | 0.212 ± 0.006 | 0.005 | 120 |
Slope of linear model (FST/100 km) ± SE.
Significant with Bonferroni correction for multiple testing P-value set to 0.006.
Results
Microsatellite amplification success
Across all samples and microsatellites investigated in the present study, 97.80% were amplified successfully (min.: 90.94% for the locus GAC 7033; max.: 99.67% for the locus GAC 4170). Due to a high percentage of missing data in GAC 7033, up to 66.67% in NB, this locus was excluded from the analyses with Arlequin, except for the analysis of the LD. All populations had an average amplification success above 95% for all microsatellites except for IDK09a (93%), IUF (91%), NB (87%), and IUA (86%). Null alleles were present in sticklebacks from 11 populations in the majority of catchment areas, except for the Rhine catchment area and the population GPS (Schlei/Trave catchment area), in single microsatellite loci except GAC 1125. In sticklebacks from the sampling sites LB and SPA, null alleles were present in two microsatellites.
Microsatellite diversity
The proportion of polymorphic loci (0.99 criteria, P0.99) was 1 except for LB, MB, RF, and MG with 0.89. Mean number of alleles (MNA) was highest (10.0) in the Spanish population (SPA) and lowest (3.1) in a brook (SDT), averaging 5.5 ± 1.2 across all populations (Table 1). Four populations (LB, RF, SDT, MG) had MNA <4.3 (average MNA minus standard deviation of MNA), suggesting bottlenecks or genetic isolation. Expected heterozygosity (HE) ranged from 0.37 to 0.74 and observed heterozygosity (HO) from 0.36 to 0.77 (Table 1).
HWE and LD
LD was significant after Bonferroni correction in 13 populations (Table A2), but LD was not associated with certain drainage systems or habitat types. Physical linkage is unlikely because the 13 cases are distributed across different locus pairs (Table A2) and seven of the nine microsatellites used here are located in different linkage groups (Peichel et al. 2001). From the 43 samplings, 11 showed a significant deviation from HWE (Table 1).
Phylogenetic relationship
The phylogenetic relationship of sampled populations (without outgroups; Fig. 3) mainly resembles the genetic differentiation according to water catchment areas. This was the case for main river catchment areas (Ems, Rhine, Weser), but also within the Ems catchment area for the investigated brook catchment areas.
Figure 3.
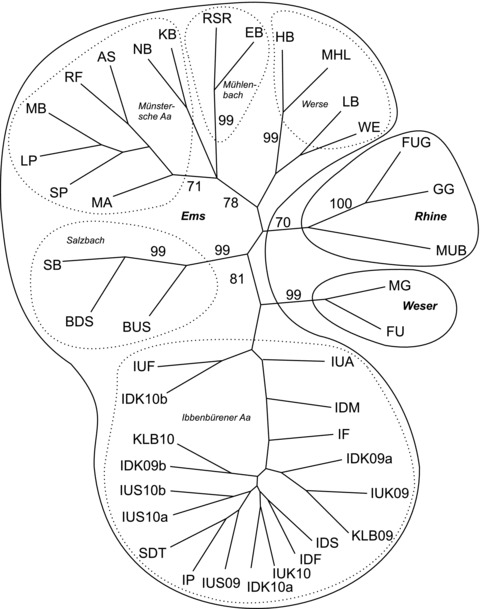
Unrooted neighbor-joining tree without outgroups (GPS, SPA). Consensus support based on 10,000 bootstraps higher than 70 is reported at relevant nodes. Populations are encircled according to catchment area (continuous lines) and within catchment area Ems according to brook catchment area (dotted lines). For abbreviations of sampling sites see Table 1.
Population structure
Genetic variation among populations was significant (P < 0.001) on all levels of the AMOVA. Grouping according to catchment area revealed that 10.6% of the overall genetic variation was distributed among the catchment areas. Grouping by habitat type explained 4.9% of variation between running verus stagnant water habitats and 1.4% between fresh verus saltwater habitats with all sampling sites (exclusive outgroups) included (Table 2). Within the Ems catchment area, 5.4% of variation was explained by grouping in habitat types running versus stagnant water and 2.1% between fresh versus saltwater habitats (Table 2). However, analysis of cluster formation of genotype data with STRUCTURE supported two clusters (K= 2) with the highest likelihood and lowest variation from the 20 repetitions (Fig. 7). Cluster 1 was formed with populations from the Ems (without Ibbenbürener Aa) and the Rhine catchment area, while Cluster 2 was formed with populations from the Ibbenbürener Aa and the Weser catchment area (Fig. 7). Details of STRUCTURE clustering analysis are shown in the Appendix (Fig. A1).
Overall, genetic differentiation among populations showed high variation, ranging from nonsignificant FST-values around 0.001 up to 0.45 in a pair with the Spanish outgroup (SPA-MB; Table A1). Of all 903 pairs, FST-values from 79 pairs (8.8%) were not significant (P < 0.05, Arlequin) and from 160 pairs (17.7%) with Bonferroni correction (P < 0.00061, FSTAT). Among these, groups with microsatellite amplification success below 95% (IDK09a, IUF, NB, IUA) had an elevated rate (26.2%) of not significant FST-values. Other not significant FST-values were present in geographically closely neighboring population pairs of (1) artificial stagnant (st)/running (ru) waters (e.g., RSRst–EBru, MHLst–HBru; Fig. 2; Table A1) and (2) salt (s) influenced/freshwater (f) habitats (e.g., SBs–BDSf, SBs–BUSf, GGs–FUGf; Fig. 1B and D, Table A1). Majority of nonsignificant FST-values were obtained with population pairs of the Ibbenbürener Aa, a brook strongly influenced by anthropogenic saltwater discharge. This is partly due to repeated sampling of the same sites (e.g., IUS09s–IUS10as, IUS09s–IUS10bs), but might also be explained by migration between geographically distant locations (e.g., IUS09s–IDMs, IUS09s–KLB09f) and in case of IDFf (e.g., IDFf–IDMs, IDFf–IPs, IDFf–IDSs) by potential anthropogenic translocation. In contrast, different (P < 0.05) FST-values were present in other population pairs on a relatively small geographical scale (<1 km), in pairs of stagnant/running waters (e.g., SPst–MBru, LPst-MBru, ASst-MAru), but also in pairs of connected running waters of similar type (e.g., IUAf–IUFf, NBru-KBru approximately 3 km). However, these FST-values were not significant with Bonferroni correction (P < 0.00061).
The M ratios (mean ± standard deviation) of the populations investigated here ranged from 0.44 ± 0.18 (IDM) to 0.67 ± 0.24 (KB) averaging 0.53 ± 0.27 across all populations (Table 1) and were below commonly used bottleneck threshold 0.68 proposed by Garza and Williamson (2001).
Isolation by distance
Correlation of genetic (FST) and waterway distance of all pairs (accept outgroups) of the populations investigated here (r= 0.549, P < 0.0001; Table 3) confirmed IBD. In the IBD blot (Fig. 4), separation of samples in pairs without populations from salt and stagnant waters (circle) (only running, ancient waters), pairs with stagnant and no salt waters (square), and pairs with salt but no stagnant waters (triangle) indicates that habitat type influences the overall IBD pattern. Population pairs with stagnant (no salt) waters result in a regression line lying parallel (short dash) above the regression of solely (ancient) running water pairs (continuous line), while regression line (long dash) of population pairs with salt water (no stagnant waters) has a higher slope compared with the other two (Fig. 4). Correlation of genetic and geographic distance was slightly stronger when only the samples from the Ems catchment area were tested (Table 3). Within the Ems catchment area, the populations SDTf, RFst, and LBru were categorized genetically “isolated” since they were outliers in NMDS plots of genetic differentiation (Fig. 5A), their MNA (<4.1; Table 1) was relatively low, and their FST-values were significant also with the geographically closely neighboring populations (Table A1). Populations IDKs, IDSs, IUKs, IUSf, KLBf, and IDFf were categorized genetically “mixing” since FST-values of more than 50% of sampling sites of the same brook catchment area (Ibbenbürener Aa only) were not significant or repeated sampling at the same site revealed significant (P < 0.05) FST-values (Table A1). Populations that were not “isolated” or “mixing” were categorized as “other” (Ems catchment area only). Multiple Mantel tests with categorized samplings revealed that the correlation of genetic and geographic distance was not significant when tested only among mixing and among isolated populations (Table 3). In the significantly correlated categorized pairs, the slope of the regression was always higher, when isolated populations were included, thus isolated populations showed higher genetic differentiation than would be expected by IBD only. In IBD blots of populations from the Ems catchment area (Fig. 6), pairs of isolated populations are present above (higher FST-values/distance) the category “other,” while “mixing” populations are present below (lower FST-values/distance) the latter category (Fig. 6B). Pairs of categories “other × isolated” are located in the upper range of the IBD blot, while pairs of categories “other × mixed” are present in the lower range (Fig. 6C). Pairs of the extreme categories, “isolated × mixed” are present in the lower left and the upper right area of the IBD blot (Fig. 6C).
Figure 4.
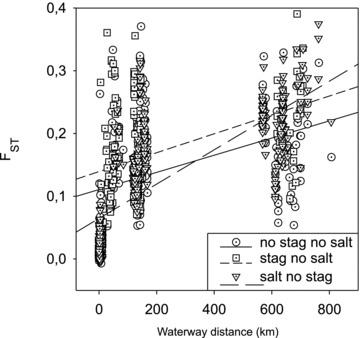
Isolation by distance (IBD) blot of all populations exclusive outgroups. Symbols represent population pairs from habitats without stagnant and salt water (circle) (running × running), pairs with at least one stagnant water but no salt water (square) (stagnant × running, stagnant × stagnant), and pairs with salt, but no stagnant waters (triangle) (salt × running, salt × salt).
Figure 5.
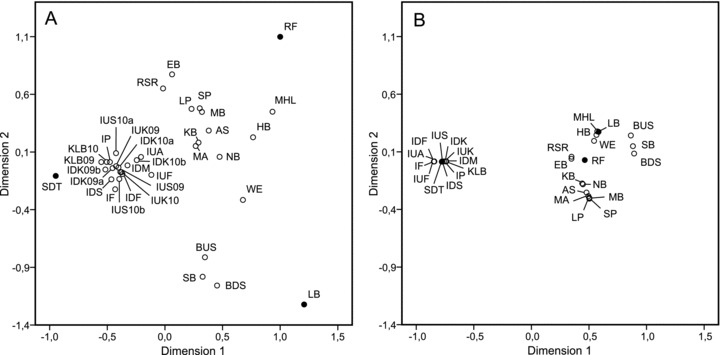
Nonmetric multidimensional scaling plots based on genetic (A) (pairwise FST) and geographic (B) (waterline km) distance of populations from the Ems catchment area. Note: (1) Populations RF, SDT, and LB (filled circles) are outliers in the genetic differentiation plot (A), but centred in the geographic distance plot (B). (2) Geographically closely neighboring populations (e.g., AS–MA, LP–MB, RSR–EB) are genetically distinct. (3) In the Ibbenbürener Aa cluster, with significant saltwater influence (Fig. 1E), consecutive samplings from same locations may have relatively high genetic differentiation (e.g., IUS10a–IUS10b, IDK10a–IDK10b; compare Table A1). For abbreviations of sampling sites see Table 1.
Figure 6.
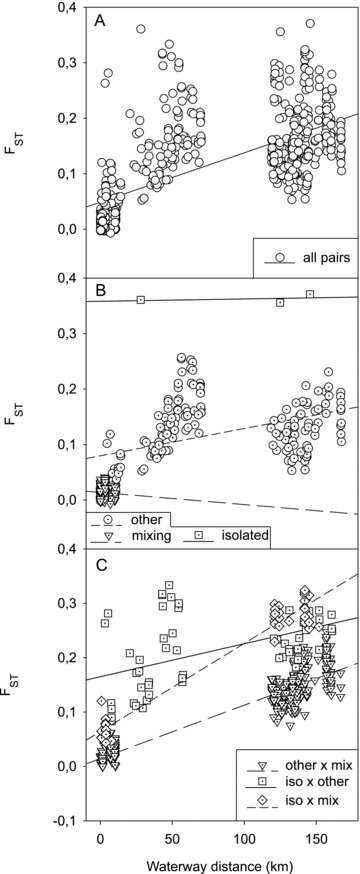
Isolation by distance (IBD) blots of populations from the Ems catchment area. (A) All population pairs, (B) pairs of populations within categories (other, mixing, isolated), and (C) pairs of populations across categories. For correlation analysis of categorized populations, see Table 3. Note that regression lines in (C) do not match with the regression analysis (Table 3), since pairs from single categories (B) are not included in (C).
In the NMDS plot of waterway distance (Fig. 5B) of sampling sites from the Ems catchment area, two main clusters were formed, one with sampling sites in the city of Münster and its closer surroundings and the other from the Ibbenbürener Aa brook sampling sites. This pattern is somewhat represented in the NMDS plot of genetic differentiation (Fig. 5A) with a distinct cluster of Ibbenbürener Aa sampling sites but a more dispersed pattern of the Münster sampling sites. Demographically isolated populations (SDTf, RFst, LBru) clearly were outliers in the NMDS plot of genetic differentiation (Fig. 5A).
However, no recent migration rate higher than 10–7 was detected with BIMR (Faubet and Gaggiotti 2008) between the populations of the Ems catchment area. Presumably because data of the present study do not fulfill all requirements (FST-values ≥0.01, 10 loci, and 50 samples per population) for reliable estimations of migration rates (Faubet and Gaggiotti 2008) and some FST-values, in particular in the Ibbenbürener Aa, were <0.01.
Discussion
In the present study, population genetics of sticklebacks (G. aculeatus) in a densely human populated, industrialized, and intensively agriculturally used area in North West Germany were analyzed with neutral genetic markers. Sampling sites were chosen in artificial ponds and neighboring brooks and in salt water polluted brooks with adjacent freshwater sites.
Isolation by distance
Over all populations, genetic differentiation of sticklebacks was significantly correlated to waterway distance and followed the IBD model. In the AMOVA analysis 10.6% (P < 0.001) of the variation among populations was explained by river catchment area and accordingly, in the phylogenetic analysis, populations were separated by river catchment areas. Thus, overall genetic differentiation of the stickleback populations investigated here is connected by an IBD model. However, considering the investigated anthropogenic habitat alterations, respective populations deviate from the general IBD pattern. Grouping of the data in the IBD blot by habitat type resulted in a steeper IBD regression for pairs with salt water habitats and a parallel but higher regression with stagnant water habitats compared to the IBD regression of solely running (ancient) water habitats (Fig. 4). Grouping of the data by habitat type running/artificial stagnant water in the AMOVA analysis explained 4.9% (P < 0.001) of genetic variation (Table 2). Changes of water salinity by coal mine drainage water as well seem to influence sticklebacks diversification, and 1.4% (P= 0.018) of variation (2.1%, P= 0.015, Ems only) was explained by grouping of populations by fresh-/saltwater habitat. Analysis of genetic clusters using STRUCTURE as well revealed potential disturbance of the “ancient” IBD pattern by human activity. Here populations are separated in two main clusters, which correspond only insufficiently with river catchment areas (Fig. 7). Both clusters are present in the Ems catchment area, one mainly in the Münster area (SP-WE + BUS-BDS; Fig. 7) and the other in the Ibbenbürener Aa (IUF-IDM; Fig. 7). Interestingly, populations from the Weser catchment area are in the “Ibbenbürener Aa” cluster and the Rhine catchment area populations are in the “Münster cluster.” If this is not an artifact, a possible explanation could be that sticklebacks were exchanged between the Ibbenbürener Aa and the Weser River via the Mitteland canal, which is crossed by the Ibbenbürener Aa with a culvert (Fig. 1E, insert map). Further West, the Ems is connected via the Mitteland canal to the Rhine, which may explain clustering of Rhine sticklebacks with the Ems stickleback from the Münster area. Generally, the IBD pattern of the investigated stickleback populations seems to be influenced by anthropogenic activity, but interpretation of the present data in more detail, as follows, reveals that in individual situations genetic diversification of sticklebacks might more specifically be influenced by anthropogenic habitat alterations.
Figure 7.
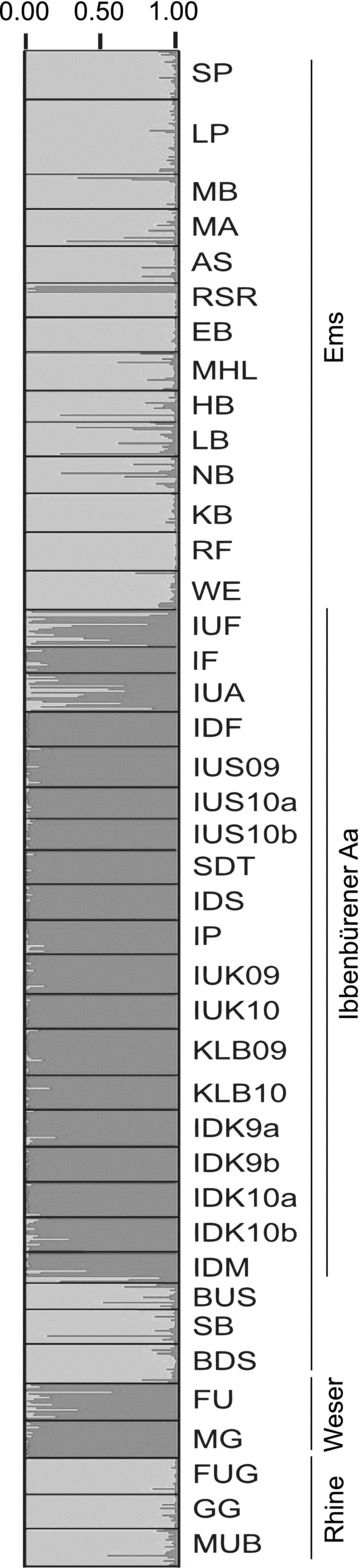
Bar plot of stickleback populations inferred by STRUCTURE (K= 2). Each individual per population is assigned proportionally to cluster 1 (dark gray) and 2 (light gray). River catchment areas are indicated by vertical lines.
Running/stagnant water
Two pairs of ponds and adjacent (<1 km) brooks (RSRst–EBru, MHLst–HBru) investigated here did not show significant FST-values, even though the moat MHLst already exists since 1720. In the RSRst–EBru and MHLst–HBru pairs, brooks are fed from the stagnant waters, the rain storage reservoir (RSRst) and the moat (MHLst) and migration of sticklebacks from the stagnant waters might have influenced the adjacent brook populations. Also in Lake Constance and a tributary brook, colonized with stickleback about 150 years ago, prominent population differentiation based on microsatellite analyses was not observed (Berner et al. 2010). The authors conclude that 150 years was too short time for ecological speciation (Berner et al. 2010).
In contrast, in the present study, three of the investigated artificial stagnant (st) waters (LPst, SPst, ASst) showed significant (P < 0.05) FST-values from closely neighboring (<1 km) running (ru) waters (brooks), suggesting that at least genetic exchange was reduced between these habitats. The ponds SP and LP were constructed in 1995 in a sports park and collect rain drainage water from the surrounding sports fields. During heavy rain, the ponds discharge into a brook (MBru). Regular migration of sticklebacks from the brook to the ponds is unlikely, due to a 1-m cataract in between. Presumably, ponds were colonized from the MBru during a flooding situation, with high water levels enabling the sticklebacks to pass the cascade from MBru. Mean numbers of alleles (MNA, 5.7) and heterozygosity (HO, 0.61 and 0.63; Table 1) were relatively high in SPst and LPst, suggesting absence of recent bottleneck situations. The genetic differentiation between SPst, respectively, LPst, and the brook MBru (FST 0.020 and 0.022, P < 0.05) could be explained by a founding event and an interrupted gene flow with the consequence of neutral genetic drift and/or habitat-specific selection.
The heterozygosity in SPst and LPst was equal to the neighboring brook (MBru), but the MNA was slightly lower in the ponds, which may support a recent founder event, where rare alleles got lost. Similar effects were also observed by Clegg et al. (2002) in populations of island-colonizing birds. Thus, diversification of the SPst and LPst populations from the MB on a relatively small time and geographical scale might be a consequence of adaptation to the pond versus brook environment and/or the absence of migratory activity between the habitats.
Significant FST-value between the pond ASst, construction started in 1660, completed in its present form in 1976, and its feeder brook MAru is more likely a result of differential adaptation since here migratory barriers are absent and the locations are only about 300 m apart from each other.
Habitat-specific selection and neutral genetic drift are both considered to be significant drivers in radiation and speciation of sticklebacks after the last deglaciation (Taylor and McPhail 2000; Hendry et al. 2002; McKinnon and Rundle 2002; Watanabe et al. 2003; Hendry and Taylor 2004; Cresko et al. 2007). Genetic diversification observed with the present study among distinct habitat types on a relatively small geographical scale might indicate that signatures of habitat-specific selection and/or neutral genetic drift are present already after decades.
Salinity
Changes in salinity due to natural and anthropogenic saltwater drainage in freshwater brooks also seem to influence genetic diversification of sticklebacks. AMOVA analysis grouped by habitat type salt-/freshwater explained 1.4% (P= 0.018) of variation (2.1%, P= 0.015, Ems only) among populations (Table 2). However, FST-values of neighboring freshwater (f)–saltwater (s) populations were not in all cases significant. Sticklebacks from the naturally saltwater influenced brook (SBs; Fig. 1B) did not show significant FST-values from the corresponding freshwater locations. This was also not the case for the brook GGs, which is subjected to drainage of salty coal mine drainage water (Fig. 1D).
In contrast, in the pair MGs–FUf, where MG is still salt polluted by remains of the potash mining until 1997, significant (also with Bonferroni correction, P < 0.00061) FST-values were observed. Here, both populations (MGs, FUf) showed relatively low MNA (3.1, 4.6) and HO (0.45, 0.53), (and in case of FUf, a low M ratio 0.45 ± 0.25, MGs= 0.54 ± 0.29), indicating that both populations were influenced by bottlenecks or might show founder effects. In this area, potash mining was extensive until 1997 and both MGs and FUf were heavily polluted. It is not known whether sticklebacks were completely eradicated by the pollution or when the sampling sites potentially were recolonized after the mining stopped, but low MNA and HO suggest that anthropogenic activity has influenced stickleback population genetics substantially in that region, which might also be causative for significant FST-values between MGs and FUf on a small geographical scale.
In the Ibbenbürener Aa, a brook with high saltwater burden due to drainage from a coal mine since the 1970s, a high number of population pairs had nonsignificant FST-values and high numbers of migrants (Table A1). This included pairs with relatively high geographic distance (approximately 5 km) such as IDMs–IUS09f and KLB09s–IUS09f. In contrast, significant (P < 0.05) FST-values were detected in two sites between repeated samplings within the same year (IUS10a–IUS10b, IDK10a–IDK10b; Table A1). Taken together, these observations suggest that the presence of fresh-/saltwater borders increases migratory activity of sticklebacks, rather than strictly separating populations in the two habitat types.
In natural coastal situations, typically resident salt- and freshwater stickleback populations are present, as well as anadromous sticklebacks that migrate to freshwater sites for mating (e.g. Raeymaekers et al. 2005; 2007; Takamura and Mori 2005; Mäkinen et al. 2008; Gelmond et al. 2009; Kume et al. 2010). In the Ibbenbürener Aa, mating sticklebacks were observed in both, salt- and freshwater parts, indicating that freshwater sticklebacks can adapt to mate in salty waters. At the moment, it is not clear what triggers the migratory activity of the Ibbenbürener Aa sticklebacks, supposedly spawning runs, but also environmental triggers such as availability of nutrients or intra- and interspecific competition could play a role.
In the salty parts of the Ibbenbürener Aa, three-spined sticklebacks are the almost exclusive fish species, with consequently low interspecific competition. After spawning and during growth of young of the year sticklebacks, intraspecific competition may increase, resulting in migration of saltwater born sticklebacks to freshwater habitats (Lugert, unpubl. data). It might also be possible that sticklebacks here are divided in resident fresh-/saltwater and migrating types, similar to coastal areas. Seasonal sampling, before, during, and after spawning of inhabiting sticklebacks may shed light on this hypothesis.
The significant AMOVA suggests that some genetic variation (1.4%, P= 0.018) can be explained by habitat type salt-/freshwater (Table 2). However, the AMOVA might be biased by the fact that saltwater populations show higher migratory activity and higher mixing rates, which may explain some deviation in genetic variation of saltwater sticklebacks from solely freshwater-inhabiting sticklebacks. Overall, inflow of salt water in inland freshwater brooks is a strong modifier of selective pressure, directly on osmoregulation and indirectly on the habitats ecology. A possible explanation of the present genetic data could be that sticklebacks cope relatively well with the salty environment, possibly due to rapid diversification and adaptation to the changed environment.
Gene flow
Two of the Ibbenbürener Aa populations were exceptional. This was (1) IDFf, also categorized as “mixing,” since FST-values of more than 50% of the other locations in the Ibbenbürener Aa brook catchment area were not significant (P < 0.05). IDFf is located >6 km upstream of the saltwater inflow, above an artificial lake with a cascade at its outlet, preventing upstream migration of sticklebacks (and other fish). Still, IDFf had insignificant FST-values with sticklebacks sampled at and below the saltwater inflow, suggesting close relatedness to the >6 km distant populations. Here, translocation of fish by human activity is the likely reason, since the section between the lake and the saltwater inflow is subjected to electric fishing 1 per year and collected fish (including sticklebacks) are released in the lake (modus operandi confirmed by local fisherman). Additionally, IDFf showed significant FST-values from closely neighboring (<500 m) IUAf (P < 0.00061) and IUFf (P < 0.05) locations (but not IF).
(2) The location SDTf had significant FST-values from all other Ibbenbürener Aa sampling sites (P < 0.00061). SDTf is a small (freshwater) brook that opens into the channel that transports the salty coal mine drainage water from the mines sewage plant to the Ibbenbürener Aa (insert map, Fig. 1E). In the channel, salinity is >40 mS cm–1, which is still tolerable for G. aculeatus, but its bed is flattened and the flow rate is high, presumably preventing upstream migration from the Ibbenbürener Aa to SDTf. Low MNA (3.1) and relatively low HE and HO (0.48) of SDTf sticklebacks could be a consequence of inbreeding of a small isolated population or of a bottleneck situation (M ratio 0.46 ± 0.32). A recent founder effect seems unlikely, due to relatively high genetic differentiation to the surrounding populations and the salt water channel as a migration barrier. Since SDTf is a relatively small brook (1 m in width), bottlenecks might have been caused by agricultural activity (inflow of dung and oxygen consumption) and/or low water levels during a dry period.
Low MNA (<4.1) were also detected in two other sites of the river Ems catchment area (RFst, LBru) and in MGru (Weser catchment area). Generally, genetic variation depends on population size (Frankham 1996), which in river-dwelling freshwater fish is reduced in upstream locations (Hänfling et al. 2002). In river systems, upstream–downstream asymmetry of genetic variation of populations is influenced by flow direction (Hänfling and Weetman 2006), which alters the theoretically expected migration–drift equilibrium among upstream–downstream populations (Hutchison and Templeton 1999). Low MNA in some populations of the present study could be a sign of upstream–downstream asymmetry, potentially influenced by migratory barriers.
Natural barriers, such as waterfalls (Crispo et al. 2006), but also anthropogenic barriers, such as weirs, may enhance genetic dilution (isolation or deviation from migration–drift equilibrium) of upstream populations if compensatory immigration from downstream populations is reduced (Hänfling and Weetman 2006). Three-spined stickleback populations inhabiting small upstream stretches and tributaries are vulnerable to genetic isolation by anthropogenic barriers and are more susceptible to bottleneck effects if compensatory immigration is inhibited (Raeymaekers et al. 2008).
The M ratios of the populations investigated here were generally below the commonly used bottleneck threshold 0.68 proposed by Garza and Williamson (2001), indicative for recent bottlenecks. Comparably low M ratios were observed in another stream-dwelling fish species, the European grayling (Thymallus thymallus) (Swatdipong et al. 2010). It is suggested that the low M ratio in European grayling is more consistent with the common historical genetic bottleneck scenario related to the last glacial period and subsequent postglacial recolonization, than in fact very recent bottleneck events (Swatdipong et al. 2010). Low M ratios observed with three-spined sticklebacks in the present study in their majority might as well be explained by the postglacial recolonization history. However, populations categorized “isolated” in the present study all had M ratios below the average M ratio of the investigated populations, confirming that isolated populations are more susceptible to bottlenecks as suggested by Raeymaekers et al. (2008).
In the present study, three populations in the river Ems catchment area, namely those with low MNA (SDTf, RFst, LBru), were distinct outliers in the NMDS plots of pairwise FST-values (Fig. 3A). The low MNA but high FST-values (see also Fig. 6B) of these populations suggest that they are genetically isolated. All three are located relatively close to the source of the respective water shed, which may support genetic isolation (Hänfling and Weetman 2006; Raeymaekers et al. 2008), but this was also the case for other locations (RSRst–EBru, MHLst–HBru) where distinct migratory barriers and signs of genetic isolation were absent.
In the case of RFst (Rieselfelder), immigration is inhibited. As a consequence of previous (1910–1975) use of the RFst wetland area for sewage cleaning and the since 1975 lasting drainage from a modern sewage plant, water flow across the RFst ponds is channeled and controlled rigorously. This includes weirs and several downfalls, before water reaches the downstream drainage systems. The present population was probably founded, either by survivors of the sewage swamp or by reintroduction of a (limited) number of sticklebacks (as a single event) after 1975. Given the relatively high FST-values, the low MNA of the RF sticklebacks, and a relatively low M ratio (0.47 ± 0.29), isolation of a relatively small RF stickleback population is likely, but their relatively high HE and HO indicate that the present population is outcrossing since a number of generations.
In case of the small brook Loddenbach (LBru), fed from a rainwater retention basin since 1978, mechanical migratory barriers to the adjacent (2 km) river Werse are absent, but 150 m before it opens up into the Werse River, a sewage purification plant discharges into the LB brook. In a single sampling trial with hand nets between the sewage plant and the river Werse, sticklebacks were not caught, but sunbleak (Leucaspius delineatus) and stone loach (Barbatula barbatula), indicating that netting was effective and that water conditions were not hostile to fish. We suspect that inflow of water from the sewage plant makes the habitat unattractive for immigration of sticklebacks from the Werse River and that upstream LB sticklebacks are enclosed.
Conclusions
The present study shows that three-spined stickleback population genetics are influenced by two principally different anthropogenic modifications of water systems in an industrialized and agriculturally used landscape. One process is the isolation of populations by mechanical barriers or creation of ponds that are completely isolated from the surrounding waterways. The introduction of salt into natural brooks is a second major change that allows us to test the rapid adaptation to the saline environment with documented history of salt influx. Habitat fragmentation and creation of new habitats seem to foster genetic differentiation of inhabiting sticklebacks. This might be attributed to the fact that three-spined sticklebacks are very successful in the colonization of and adaptation to new habitats. In general, migratory activity of freshwater sticklebacks in the area seems to be low, resulting in genetic differences among populations on relatively small geographical scales. Overall, the genetic population structure followed an IBD model, which was influenced by anthropogenic habitat alterations. Sticklebacks in salt-polluted water systems seem to exhibit elevated migratory activity between fresh- and saltwater habitats, reducing IBD. In other situations, populations showed distinct signs of genetic isolation, which in some locations was attributed to mechanical migration barriers, but in others to potential anthropogenic induced bottleneck or founder effects. The present study shows that anthropogenic habitat alterations may have diverse effects on the population genetic structure of inhabiting species. Depending on the type of habitat change, increased genetic differentiation, diversification, or isolation are possible consequences.
Acknowledgments
We are grateful to K. Keil, M. Zeidler, and in particular J. Raeymaekers for the fruitful discussions of the results of the present study. In addition, we thank three anonymous reviewers whose comments on an older version of the MS were extremely useful to shape it in its present form. We thank B. Hasert and I. Dankert for technical support in the microsatellite measurements and analysis and J. Lange for assistance in the fish collection. The sticklebacks for the outgroups were kindly provided by M. Kalbe (Max Planck Institute for Evolutionary Ecology, Plön, Germany) (GPS) and M. Hermida (University of Santiago de Compostela, Lugo, Spain) (SPA). We thank M. Schweyen for his support with the STRUCTURE analysis. The project was supported by the German Science Foundation (DFG, grant #SCHA 1257/2-1).
Figure A1.
Distribution of first- and second-order likelihood values of the cluster analysis with STRUCTURE. The K-values represent the numbers of clusters tested. (A) The Ln P (D) demonstrates the average value of 20 runs with 106 iterations each after a burn-in phase of 2.5 × 105 iterations. (B) The second-order likelihood values are given by ΔK, measured according to Evanno et al. (2005).
Table A1.
Pairwise genetic differentiation.
| SP | LP | MB | MA | AS | RSR | EB | MHL | HB | LB | NB | KB | RF | WE | IUF | IF | IUA | IDF | IUS09 | IUS10a | IUS10b | SDT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SP | inf | 22.2 | 17.5 | 22.9 | 4.6 | 6.2 | 2.6 | 3.5 | 1.2 | 8.0 | 9.9 | 3.3 | 2.5 | 4.6 | 2.3 | 5.5 | 3.3 | 3.1 | 3.8 | 3.2 | 1.8 | |
| LP | 0.002 | 31.1 | 17.7 | 17.7 | 4.3 | 5.2 | 2.3 | 2.9 | 1.2 | 5.8 | 8.1 | 2.7 | 2.3 | 4.8 | 2.4 | 5.9 | 3.4 | 3.4 | 4.4 | 3.4 | 2.1 | |
| MB | 0.02 | 0.02 | 35.0 | 31.8 | 4.3 | 5.9 | 2.6 | 3.1 | 1.2 | 5.5 | 9.0 | 2.8 | 3.2 | 4.8 | 2.4 | 5.9 | 3.3 | 3.2 | 3.6 | 2.9 | 1.8 | |
| MA | 0.03 | 0.03 | 0.01 | 30.0 | 5.7 | 5.7 | 2.9 | 4.0 | 1.8 | 8.9 | 12.8 | 3.6 | 5.1 | 8.7 | 3.5 | 6.6 | 4.8 | 4.6 | 4.5 | 4.3 | 2.2 | |
| AS | 0.02 | 0.03 | 0.02 | 0.02 | 5.0 | 5.8 | 3.5 | 5.1 | 1.5 | 8.4 | 17.3 | 4.2 | 3.7 | 5.3 | 2.7 | 6.0 | 3.4 | 3.5 | 3.7 | 3.3 | 1.9 | |
| RSR | 0.01 | 0.10 | 0.10 | 0.08 | 0.09 | 35.6 | 3.1 | 3.7 | 1.1 | 4.3 | 8.4 | 2.9 | 3.3 | 4.8 | 3.4 | 6.5 | 3.9 | 3.6 | 3.5 | 3.2 | 2.0 | |
| EB | 0.08 | 0.09 | 0.08 | 0.08 | 0.08 | 0.01 | 2.3 | 2.9 | 1.0 | 4.2 | 9.1 | 2.4 | 2.8 | 3.6 | 2.6 | 5.1 | 3.2 | 2.9 | 3.1 | 2.6 | 1.6 | |
| MHL | 0.16 | 0.18 | 0.16 | 0.15 | 0.13 | 0.14 | 0.18 | 91.2 | 1.3 | 3.4 | 3.8 | 2.0 | 3.7 | 2.6 | 2.0 | 3.1 | 2.1 | 2.2 | 2.0 | 2.0 | 1.3 | |
| HB | 0.13 | 0.15 | 0.14 | 0.11 | 0.09 | 0.12 | 0.15 | 0.005 | 1.4 | 5.1 | 5.6 | 2.4 | 4.4 | 3.4 | 2.6 | 3.8 | 2.7 | 2.8 | 2.3 | 2.5 | 1.5 | |
| LB | 0.30 | 0.30 | 0.29 | 0.21 | 0.25 | 0.31 | 0.33 | 0.28 | 0.26 | 1.8 | 1.6 | 0.9 | 3.8 | 1.8 | 1.4 | 1.2 | 1.5 | 1.5 | 1.1 | 1.4 | 0.9 | |
| NB | 0.06 | 0.08 | 0.08 | 0.05 | 0.06 | 0.10 | 0.11 | 0.13 | 0.09 | 0.22 | 31.5 | 4.0 | 5.2 | 6.2 | 3.0 | 4.6 | 3.6 | 4.0 | 3.4 | 3.2 | 1.7 | |
| KB | 0.05 | 0.06 | 0.05 | 0.04 | 0.03 | 0.06 | 0.05 | 0.12 | 0.08 | 0.24 | 0.02 | 3.8 | 4.7 | 6.7 | 3.5 | 8.9 | 4.4 | 4.7 | 4.6 | 3.8 | 2.0 | |
| RF | 0.13 | 0.16 | 0.15 | 0.12 | 0.11 | 0.15 | 0.18 | 0.20 | 0.17 | 0.36 | 0.11 | 0.12 | 1.9 | 1.9 | 1.2 | 1.8 | 1.4 | 1.4 | 1.3 | 1.3 | 0.9 | |
| WE | 0.17 | 0.18 | 0.14 | 0.09 | 0.12 | 0.13 | 0.15 | 0.12 | 0.10 | 0.12 | 0.09 | 0.10 | 0.21 | 4.9 | 3.1 | 3.3 | 3.1 | 3.1 | 2.3 | 2.7 | 1.6 | |
| IUF | 0.10 | 0.09 | 0.09 | 0.05 | 0.09 | 0.09 | 0.12 | 0.16 | 0.13 | 0.21 | 0.08 | 0.07 | 0.21 | 0.09 | 16.1 | 21.7 | 8.8 | 18.1 | 7.5 | 13.6 | 5.1 | |
| IF | 0.18 | 0.18 | 0.17 | 0.13 | 0.16 | 0.13 | 0.16 | 0.20 | 0.16 | 0.27 | 0.14 | 0.13 | 0.29 | 0.14 | 0.03 | 14.9 | 71.3 | 18.1 | 7.8 | 31.9 | 5.5 | |
| IUA | 0.08 | 0.08 | 0.08 | 0.07 | 0.08 | 0.07 | 0.09 | 0.14 | 0.12 | 0.29 | 0.10 | 0.05 | 0.22 | 0.13 | 0.02 | 0.03 | 25.7 | 16.9 | 20.6 | 21.1 | 4.2 | |
| IDF | 0.13 | 0.13 | 0.13 | 0.10 | 0.13 | 0.12 | 0.14 | 0.20 | 0.16 | 0.26 | 0.12 | 0.10 | 0.27 | 0.14 | 0.02 | 0.007 | 0.02 | 181.8 | 18.8 | inf | 10.2 | |
| IUS09 | 0.14 | 0.13 | 0.13 | 0.10 | 0.12 | 0.12 | 0.15 | 0.18 | 0.15 | 0.25 | 0.11 | 0.10 | 0.26 | 0.14 | 0.03 | 0.03 | 0.03 | 0.003 | 33.2 | 266.6 | 9.0 | |
| IUS10a | 0.12 | 0.10 | 0.12 | 0.10 | 0.12 | 0.13 | 0.14 | 0.20 | 0.18 | 0.31 | 0.13 | 0.10 | 0.27 | 0.18 | 0.06 | 0.06 | 0.02 | 0.03 | 0.02 | 30.8 | 3.7 | |
| IUS10b | 0.14 | 0.13 | 0.15 | 0.11 | 0.13 | 0.14 | 0.16 | 0.20 | 0.16 | 0.27 | 0.14 | 0.12 | 0.28 | 0.16 | 0.04 | 0.02 | 0.02 | 0.001 | 0.002 | 0.02 | 7.2 | |
| SDT | 0.22 | 0.20 | 0.21 | 0.18 | 0.21 | 0.20 | 0.23 | 0.29 | 0.25 | 0.37 | 0.23 | 0.20 | 0.36 | 0.24 | 0.09 | 0.08 | 0.11 | 0.05 | 0.05 | 0.12 | 0.07 | |
| IDS | 0.15 | 0.14 | 0.15 | 0.12 | 0.15 | 0.13 | 0.16 | 0.21 | 0.18 | 0.28 | 0.15 | 0.12 | 0.30 | 0.17 | 0.04 | 0.03 | 0.03 | 0.001 | 0.006 | 0.04 | 0.007 | 0.05 |
| IP | 0.13 | 0.12 | 0.13 | 0.11 | 0.13 | 0.12 | 0.14 | 0.21 | 0.18 | 0.31 | 0.15 | 0.12 | 0.27 | 0.18 | 0.05 | 0.05 | 0.03 | 0.008 | 0.001 | 0.008 | 0.01 | 0.05 |
| IUK09 | 0.14 | 0.14 | 0.14 | 0.11 | 0.13 | 0.12 | 0.14 | 0.19 | 0.15 | 0.29 | 0.12 | 0.11 | 0.26 | 0.16 | 0.04 | 0.02 | 0.03 | 0.001 | 0.007 | 0.03 | 0.02 | 0.08 |
| IUK10 | 0.13 | 0.12 | 0.13 | 0.10 | 0.13 | 0.13 | 0.14 | 0.18 | 0.15 | 0.28 | 0.13 | 0.10 | 0.28 | 0.16 | 0.04 | 0.03 | 0.02 | 0.001 | 0.005 | 0.02 | 0.002 | 0.08 |
| KLB09 | 0.16 | 0.14 | 0.16 | 0.14 | 0.15 | 0.13 | 0.16 | 0.21 | 0.18 | 0.32 | 0.15 | 0.13 | 0.29 | 0.20 | 0.05 | 0.04 | 0.03 | 0.02 | 0.01 | 0.02 | 0.03 | 0.07 |
| KLB10 | 0.15 | 0.14 | 0.15 | 0.13 | 0.14 | 0.12 | 0.15 | 0.19 | 0.17 | 0.32 | 0.16 | 0.12 | 0.28 | 0.19 | 0.05 | 0.03 | 0.03 | 0.01 | 0.02 | 0.02 | 0.02 | 0.06 |
| IDK09a | 0.15 | 0.14 | 0.15 | 0.12 | 0.14 | 0.12 | 0.16 | 0.18 | 0.15 | 0.30 | 0.14 | 0.12 | 0.27 | 0.17 | 0.04 | 0.02 | 0.03 | 0.004 | 0.01 | 0.04 | 0.02 | 0.06 |
| IDK09b | 0.15 | 0.14 | 0.16 | 0.13 | 0.16 | 0.13 | 0.16 | 0.22 | 0.19 | 0.32 | 0.15 | 0.13 | 0.30 | 0.19 | 0.04 | 0.02 | 0.02 | 0.001 | 0.003 | 0.02 | 0.002 | 0.04 |
| IDK10a | 0.13 | 0.12 | 0.13 | 0.11 | 0.12 | 0.12 | 0.14 | 0.18 | 0.15 | 0.29 | 0.13 | 0.10 | 0.28 | 0.17 | 0.04 | 0.03 | 0.02 | 0.001 | 0.001 | 0.01 | 0.003 | 0.06 |
| IDK10b | 0.11 | 0.10 | 0.10 | 0.08 | 0.10 | 0.09 | 0.12 | 0.15 | 0.13 | 0.26 | 0.11 | 0.08 | 0.23 | 0.13 | 0.02 | 0.04 | 0.003 | 0.03 | 0.03 | 0.04 | 0.04 | 0.09 |
| IDM | 0.13 | 0.12 | 0.12 | 0.08 | 0.11 | 0.10 | 0.13 | 0.18 | 0.15 | 0.27 | 0.12 | 0.09 | 0.24 | 0.13 | 0.02 | 0.03 | 0.02 | 0.001 | 0.01 | 0.04 | 0.02 | 0.06 |
| BUS | 0.16 | 0.16 | 0.17 | 0.12 | 0.14 | 0.21 | 0.21 | 0.23 | 0.18 | 0.17 | 0.13 | 0.13 | 0.30 | 0.13 | 0.10 | 0.16 | 0.14 | 0.13 | 0.15 | 0.17 | 0.13 | 0.25 |
| SB | 0.19 | 0.19 | 0.20 | 0.15 | 0.16 | 0.23 | 0.24 | 0.26 | 0.21 | 0.16 | 0.15 | 0.15 | 0.32 | 0.15 | 0.12 | 0.18 | 0.17 | 0.15 | 0.16 | 0.19 | 0.14 | 0.25 |
| BDS | 0.21 | 0.21 | 0.21 | 0.16 | 0.16 | 0.25 | 0.25 | 0.25 | 0.20 | 0.16 | 0.16 | 0.17 | 0.32 | 0.14 | 0.14 | 0.19 | 0.19 | 0.17 | 0.19 | 0.22 | 0.17 | 0.28 |
| FU | 0.24 | 0.23 | 0.25 | 0.20 | 0.23 | 0.23 | 0.28 | 0.28 | 0.25 | 0.30 | 0.22 | 0.22 | 0.33 | 0.20 | 0.15 | 0.20 | 0.17 | 0.16 | 0.18 | 0.24 | 0.17 | 0.18 |
| MG | 0.28 | 0.27 | 0.29 | 0.24 | 0.29 | 0.30 | 0.34 | 0.34 | 0.31 | 0.34 | 0.28 | 0.28 | 0.39 | 0.25 | 0.21 | 0.26 | 0.24 | 0.21 | 0.23 | 0.29 | 0.21 | 0.25 |
| FUG | 0.22 | 0.23 | 0.21 | 0.18 | 0.19 | 0.23 | 0.24 | 0.27 | 0.20 | 0.33 | 0.19 | 0.16 | 0.29 | 0.22 | 0.19 | 0.26 | 0.21 | 0.23 | 0.23 | 0.25 | 0.24 | 0.32 |
| GG | 0.21 | 0.22 | 0.20 | 0.16 | 0.17 | 0.20 | 0.21 | 0.24 | 0.18 | 0.32 | 0.17 | 0.15 | 0.26 | 0.20 | 0.17 | 0.23 | 0.19 | 0.21 | 0.22 | 0.24 | 0.23 | 0.31 |
| MUB | 0.16 | 0.17 | 0.14 | 0.09 | 0.12 | 0.10 | 0.13 | 0.12 | 0.08 | 0.18 | 0.10 | 0.09 | 0.20 | 0.05 | 0.06 | 0.08 | 0.09 | 0.10 | 0.11 | 0.15 | 0.11 | 0.18 |
| GPS | 0.17 | 0.18 | 0.17 | 0.13 | 0.13 | 0.18 | 0.19 | 0.23 | 0.18 | 0.30 | 0.15 | 0.13 | 0.20 | 0.16 | 0.16 | 0.23 | 0.17 | 0.21 | 0.21 | 0.22 | 0.22 | 0.30 |
| SPA | 0.28 | 0.29 | 0.26 | 0.23 | 0.26 | 0.29 | 0.30 | 0.32 | 0.28 | 0.43 | 0.26 | 0.23 | 0.33 | 0.28 | 0.28 | 0.34 | 0.30 | 0.31 | 0.30 | 0.33 | 0.33 | 0.38 |
| SP | 2.8 | 3.3 | 3.1 | 3.4 | 2.7 | 3.0 | 2.9 | 2.8 | 3.4 | 4.2 | 3.5 | 2.7 | 2.1 | 1.9 | 1.6 | 1.3 | 1.8 | 1.9 | 2.7 | 2.4 | 1.3 | |
| LP | 3.0 | 3.7 | 3.1 | 3.5 | 3.0 | 3.2 | 3.1 | 3.0 | 3.8 | 4.5 | 3.7 | 2.6 | 2.1 | 1.9 | 1.7 | 1.4 | 1.7 | 1.8 | 2.4 | 2.3 | 1.2 | |
| MB | 2.7 | 3.3 | 3.1 | 3.4 | 2.6 | 2.9 | 2.8 | 2.7 | 3.5 | 4.6 | 3.7 | 2.5 | 2.0 | 1.9 | 1.5 | 1.2 | 1.8 | 2.1 | 3.0 | 2.5 | 1.4 | |
| MA | 3.8 | 4.2 | 4.0 | 4.4 | 3.1 | 3.5 | 3.8 | 3.3 | 4.2 | 6.1 | 5.5 | 3.7 | 2.8 | 2.7 | 2.1 | 1.6 | 2.3 | 2.7 | 4.9 | 3.5 | 1.7 | |
| AS | 2.8 | 3.3 | 3.2 | 3.4 | 2.8 | 3.2 | 3.2 | 2.7 | 3.8 | 4.3 | 4.1 | 3.2 | 2.6 | 2.6 | 1.7 | 1.2 | 2.2 | 2.4 | 3.8 | 3.3 | 1.5 | |
| RSR | 3.3 | 3.7 | 3.7 | 3.5 | 3.3 | 3.6 | 3.7 | 3.3 | 3.6 | 5.0 | 4.6 | 1.9 | 1.6 | 1.5 | 1.7 | 1.2 | 1.7 | 2.0 | 4.7 | 2.3 | 1.2 | |
| EB | 2.7 | 3.0 | 3.1 | 3.1 | 2.6 | 2.9 | 2.6 | 2.6 | 3.0 | 3.6 | 3.4 | 1.9 | 1.6 | 1.5 | 1.3 | 1.0 | 1.6 | 1.8 | 3.3 | 2.1 | 1.2 | |
| MHL | 1.8 | 1.9 | 2.2 | 2.2 | 1.9 | 2.1 | 2.3 | 1.8 | 2.3 | 2.9 | 2.3 | 1.7 | 1.5 | 1.5 | 1.3 | 1.0 | 1.4 | 1.6 | 3.6 | 1.6 | 1.0 | |
| HB | 2.3 | 2.3 | 2.8 | 2.9 | 2.2 | 2.5 | 2.8 | 2.2 | 2.8 | 3.4 | 2.8 | 2.3 | 1.9 | 2.0 | 1.5 | 1.1 | 2.0 | 2.3 | 5.5 | 2.3 | 1.3 | |
| LB | 1.3 | 1.1 | 1.2 | 1.3 | 1.1 | 1.0 | 1.1 | 1.1 | 1.2 | 1.4 | 1.4 | 2.5 | 2.6 | 2.7 | 1.2 | 1.0 | 1.0 | 1.1 | 2.3 | 1.2 | 0.7 | |
| NB | 3.0 | 2.9 | 3.6 | 3.4 | 2.8 | 2.7 | 3.0 | 2.8 | 3.3 | 4.2 | 3.8 | 3.3 | 2.8 | 2.6 | 1.7 | 1.3 | 2.1 | 2.4 | 4.4 | 2.8 | 1.4 | |
| KB | 3.7 | 3.9 | 4.2 | 4.4 | 3.4 | 3.6 | 3.8 | 3.4 | 4.4 | 5.6 | 5.1 | 3.5 | 2.8 | 2.5 | 1.8 | 1.3 | 2.6 | 2.9 | 4.9 | 3.3 | 1.7 | |
| RF | 1.2 | 1.3 | 1.4 | 1.3 | 1.2 | 1.3 | 1.4 | 1.2 | 1.3 | 1.7 | 1.6 | 1.2 | 1.1 | 1.1 | 1.0 | 0.8 | 1.3 | 1.4 | 2.0 | 2.0 | 1.0 | |
| WE | 2.5 | 2.4 | 2.6 | 2.7 | 2.1 | 2.2 | 2.5 | 2.1 | 2.5 | 3.3 | 3.3 | 3.4 | 2.9 | 3.0 | 2.1 | 1.5 | 1.7 | 2.0 | 8.8 | 2.7 | 1.3 | |
| IUF | 12.1 | 9.9 | 13.1 | 11.1 | 9.1 | 9.7 | 13.8 | 10.8 | 13.3 | 26.5 | 26.2 | 4.3 | 3.7 | 3.2 | 2.9 | 1.9 | 2.2 | 2.5 | 8.2 | 2.7 | 1.3 | |
| IF | 15.9 | 10.2 | 24.1 | 14.5 | 12.1 | 16.2 | 21.4 | 27.7 | 16.8 | 12.1 | 18.2 | 2.6 | 2.3 | 2.1 | 2.0 | 1.4 | 1.5 | 1.6 | 6.1 | 1.7 | 1.0 | |
| IUA | 16.2 | 14.4 | 18.2 | 25.5 | 14.6 | 16.4 | 19.6 | 21.1 | 29.7 | 180.7 | 33.5 | 3.1 | 2.5 | 2.2 | 2.4 | 1.6 | 1.9 | 2.2 | 5.1 | 2.4 | 1.2 | |
| IDF | inf | 63.2 | 1323.1 | 900.3 | 21.2 | 40.2 | 125.1 | inf | 784.9 | 19.0 | 926.6 | 3.4 | 2.9 | 2.4 | 2.7 | 1.9 | 1.7 | 1.9 | 4.7 | 1.9 | 1.1 | |
| IUS09 | 80.9 | inf | 73.0 | 104.8 | 36.3 | 32.4 | 38.1 | 196.6 | inf | 15.6 | 34.8 | 2.8 | 2.6 | 2.1 | 2.3 | 1.6 | 1.7 | 1.8 | 3.9 | 1.9 | 1.2 | |
| IUS10a | 13.0 | 58.7 | 17.7 | 31.9 | 21.1 | 22.0 | 13.7 | 21.7 | 51.2 | 13.6 | 13.9 | 2.4 | 2.2 | 1.8 | 1.6 | 1.2 | 1.5 | 1.6 | 2.8 | 1.8 | 1.0 | |
| IUS10b | 73.5 | 42.9 | 26.9 | 265.3 | 14.9 | 22.9 | 31.8 | 239.7 | 150.9 | 12.1 | 22.7 | 3.3 | 3.0 | 2.4 | 2.5 | 1.9 | 1.6 | 1.7 | 3.9 | 1.7 | 1.0 | |
| SDT | 8.9 | 9.9 | 5.9 | 6.0 | 6.9 | 8.0 | 7.5 | 11.0 | 7.9 | 5.3 | 7.4 | 1.5 | 1.5 | 1.3 | 2.3 | 1.5 | 1.1 | 1.1 | 2.3 | 1.2 | 0.8 | |
| IDS | 42.6 | 21.2 | inf | 13.2 | 16.1 | 2.0 | inf | 28.8 | 11.7 | 3.9 | 2.9 | 2.5 | 2.0 | 2.3 | 1.7 | 1.6 | 1.8 | 3.7 | 1.7 | 1.1 | ||
| IP | 0.01 | 31.3 | 69.6 | 54.5 | 3063.6 | 24.0 | 218.9 | inf | 19.1 | 30.0 | 2.2 | 2.0 | 1.7 | 2.0 | 1.4 | 1.6 | 1.7 | 3.1 | 1.8 | 1.2 | ||
| IUK09 | 0.02 | 0.02 | 41.1 | 73.2 | 89.1 | 189.1 | 76.8 | 246.0 | 8.0 | 59.3 | 2.3 | 2.1 | 1.8 | 2.0 | 1.5 | 1.5 | 1.7 | 3.7 | 1.7 | 1.1 | ||
| IUK10 | 0.001 | 0.007 | 0.01 | 19.1 | 35.0 | 2.9 | 333.9 | 159.1 | 1.5 | 6.9 | 3.1 | 2.5 | 2.1 | 2.2 | 1.7 | 1.8 | 2.0 | 4.0 | 1.8 | 1.2 | ||
| KLB09 | 0.04 | 0.01 | 0.007 | 0.03 | inf | 79.9 | 617.2 | 75.9 | 17.3 | 68.3 | 1.9 | 1.8 | 1.5 | 2.0 | 1.4 | 1.3 | 1.4 | 2.8 | 1.5 | 1.0 | ||
| KLB10 | 0.03 | 0.001 | 0.006 | 0.01 | 0.001 | 91.1 | 251.6 | inf | 20.5 | 60.3 | 2.0 | 1.8 | 1.6 | 1.8 | 1.3 | 1.5 | 1.6 | 3.2 | 1.6 | 1.1 | ||
| IDK09a | 0.02 | 0.02 | 0.003 | 0.02 | 0.006 | 0.005 | 72.2 | 53.3 | 7.8 | 5.3 | 2.3 | 2.0 | 1.8 | 2.4 | 1.6 | 1.5 | 1.8 | 4.0 | 1.7 | 1.1 | ||
| IDK09b | 0.001 | 0.002 | 0.006 | 0.002 | 0.001 | 0.002 | 0.001 | 571.2 | 15.4 | 3.7 | 2.2 | 2.0 | 1.6 | 2.2 | 1.6 | 1.4 | 1.5 | 3.2 | 1.5 | 1.0 | ||
| IDK10a | 0.02 | 0.001 | 0.002 | 0.003 | 0.007 | 0.001 | 0.009 | 0.001 | 28.2 | 75.9 | 2.7 | 2.5 | 2.1 | 2.1 | 1.5 | 1.6 | 1.8 | 3.5 | 1.8 | 1.1 | ||
| IDK10b | 0.04 | 0.03 | 0.03 | 0.02 | 0.03 | 0.02 | 0.02 | 0.03 | 0.02 | 25.9 | 2.9 | 2.3 | 2.0 | 2.7 | 1.9 | 1.9 | 2.2 | 4.5 | 2.3 | 1.3 | ||
| IDM | 0.04 | 0.02 | 0.008 | 0.03 | 0.007 | 0.008 | 0.006 | 0.02 | 0.006 | 0.004 | 2.7 | 2.4 | 2.1 | 2.9 | 1.8 | 1.6 | 1.9 | 4.7 | 2.1 | 1.2 | ||
| BUS | 0.15 | 0.19 | 0.18 | 0.14 | 0.21 | 0.20 | 0.18 | 0.19 | 0.16 | 0.15 | 0.16 | inf | 80.1 | 1.8 | 1.5 | 2.5 | 2.6 | 3.5 | 2.1 | 1.1 | ||
| SB | 0.17 | 0.20 | 0.19 | 0.17 | 0.22 | 0.22 | 0.20 | 0.20 | 0.17 | 0.18 | 0.17 | 0.001 | inf | 1.7 | 1.4 | 2.1 | 2.1 | 2.9 | 1.9 | 1.0 | ||
| BDS | 0.20 | 0.23 | 0.22 | 0.20 | 0.25 | 0.24 | 0.22 | 0.23 | 0.20 | 0.20 | 0.20 | 0.006 | 0.001 | 1.5 | 1.2 | 2.3 | 2.3 | 3.0 | 2.0 | 1.0 | ||
| FU | 0.18 | 0.20 | 0.20 | 0.18 | 0.20 | 0.21 | 0.17 | 0.19 | 0.19 | 0.15 | 0.15 | 0.22 | 0.23 | 0.25 | 8.7 | 1.1 | 1.2 | 2.6 | 1.5 | 0.8 | ||
| MG | 0.23 | 0.26 | 0.25 | 0.23 | 0.27 | 0.28 | 0.24 | 0.24 | 0.25 | 0.21 | 0.22 | 0.25 | 0.27 | 0.29 | 0.06 | 0.8 | 0.9 | 1.8 | 1.0 | 0.6 | ||
| FUG | 0.24 | 0.24 | 0.25 | 0.21 | 0.28 | 0.26 | 0.25 | 0.27 | 0.24 | 0.21 | 0.24 | 0.17 | 0.19 | 0.18 | 0.31 | 0.38 | 68.8 | 2.4 | 2.8 | 1.7 | ||
| GG | 0.22 | 0.23 | 0.23 | 0.20 | 0.26 | 0.24 | 0.22 | 0.25 | 0.22 | 0.18 | 0.21 | 0.16 | 0.19 | 0.18 | 0.29 | 0.35 | 0.007 | 2.8 | 3.1 | 1.6 | ||
| MUB | 0.12 | 0.14 | 0.12 | 0.11 | 0.15 | 0.14 | 0.11 | 0.14 | 0.13 | 0.10 | 0.10 | 0.12 | 0.15 | 0.15 | 0.16 | 0.22 | 0.17 | 0.15 | 2.9 | 1.5 | ||
| GPS | 0.23 | 0.22 | 0.23 | 0.22 | 0.25 | 0.23 | 0.22 | 0.25 | 0.22 | 0.18 | 0.19 | 0.19 | 0.21 | 0.20 | 0.26 | 0.33 | 0.15 | 0.14 | 0.15 | 1.5 | ||
| SPA | 0.32 | 0.30 | 0.32 | 0.30 | 0.34 | 0.32 | 0.32 | 0.34 | 0.31 | 0.28 | 0.30 | 0.32 | 0.33 | 0.34 | 0.39 | 0.45 | 0.22 | 0.24 | 0.26 | 0.26 |
Absolute number of migrants (upper triangular matrix) and FST-values (lower triangular matrix) calculated with Arlequin. Nonsignificant FST-values (P > 0.05) are bolded. Nonsignificant FST-values with Bonferroni correction (P > 0.00061) according to FSTAT are given in italics. Coarse evaluation of FST-values: 0.001–0.05 = little, 0.05–0.15 = moderate, 0.15–0.25 = high, and >0.25 = very high genetic differentiation (inf, infinity).
Table A2.
Linkage disequilibrium (LD) analyzed with Arlequin. The upper triangular matrix shows the percentage of sampling sites that had significant LD after Bonferroni correction for 36 tests (P= 0.00142). The lower triangular matrix shows the sites with significant LD. For abbreviations of sampling sites see Table 1.
| Stn 18 | Stn 32 | Stn 75 | Stn 84 | GAC 1097 | GAC 1125 | GAC 4170 | GAC 5096 | GAC 7033 | |
|---|---|---|---|---|---|---|---|---|---|
| Stn 18 | - | 0 | 0 | 2.32 | 0 | 0 | 2.32 | 0 | 2.32 |
| Stn 32 | - | 4.65 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Stn 75 | SPA, GG | - | 0 | 0 | 0 | 0 | 0 | 2.32 | |
| Stn 84 | IUF | - | 0 | 2.32 | 2.32 | 0 | 0 | ||
| GAC 1097 | - | 0 | 4.65 | 2.32 | 0 | ||||
| GAC 1125 | KLB 10 | - | 0 | 2.32 | 0 | ||||
| GAC 4170 | IF | MA | RSR, MG | - | 0 | 0 | |||
| GAC 5096 | MUB | IDK 09a | - | 0 | |||||
| GAC 7033 | KLB 10 | IUF | EB | - |
Table A3.
Significant null alleles with Bonferroni-adjusted confidence level performed with MICROCHECKER. Null alleles were distributed across the majority of drainage systems and microsatellites. Geographically neighboring populations did not have null alleles in the same microsatellites like IF and IUA.
| Population | Microsatellite |
|---|---|
| IF | Stn 84 |
| IUS09 | GAC 4170 |
| KLB09 | GAC 4170 |
| KLB10 | Stn 18 |
| SDT | Stn 75 |
| MG | GAC 1097 |
| KB | GAC 5196 |
| LB | Stn 32/GAC 7033 |
| SP | Stn 32 |
| IUA | GAC 7033 |
| SPA | Stn 18/GAC 1097 |
References
- Baker JA, Cresko WA, Foster SA, Heins DC. Life-history differentiation of benthic and limnetic ecotypes in a polytypic population of threespine stickleback (Gasterosteus aculeatus. Evol. Ecol. Res. 2005;7:121–131. [Google Scholar]
- Belkhirr K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions. Montpellier, France: Université de Montpellier II; 2004. CNRS UMR 5000. [Google Scholar]
- Bell MA. Lateral plate evolution in the threespine stickleback: getting nowhere fast. Genetica. 2001;113:445–461. [PubMed] [Google Scholar]
- Berner D, Roesti M, Hendry AP, Salzburger W. Constraints on speciation suggested by comparing lake-stream stickleback divergence across two continents. Mol. Ecol. 2010;19:4963–4978. doi: 10.1111/j.1365-294X.2010.04858.x. [DOI] [PubMed] [Google Scholar]
- Caldera EJ, Bolnick DI. Effects of colonization history and landscape structure on genetic variation within and among threespine stickleback (Gasterosteus aculeatus) populations in a single watershed. Evol. Ecol. Res. 2008;10:575–598. [Google Scholar]
- Candolin U. Population responses to anthropogenic disturbance: lessons from three-spined sticklebacks Gasterosteus aculeatus in eutrophic habitats. J. Fish Biol. 2009;75:2108–2121. doi: 10.1111/j.1095-8649.2009.02405.x. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Edwards AWF. Phylogenetic analysis models and estimation procedures. Am. J. Hum. Genet. 1967;19:233–257. [PMC free article] [PubMed] [Google Scholar]
- Clegg SM, Degnan SM, Kikkawa J, Moritz C, Estoup A, Owens IPF. Genetic consequences of sequential founder events by an island-colonizing bird. Proc. Natl. Acad. Sci. USA. 2002;99:8127–8132. doi: 10.1073/pnas.102583399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresko WA, McGuigan KL, Phillips PC, Postlethwait JH. Studies of threespine stickleback developmental evolution: progress and promise. Genetica. 2007;129:105–126. doi: 10.1007/s10709-006-0036-z. [DOI] [PubMed] [Google Scholar]
- Crispo E, Bentzen P, Reznick DN, Kinnison MT, Hendry AP. The relative influence of natural selection and geography on gene flow in guppies. Mol. Ecol. 2006;15:49–62. doi: 10.1111/j.1365-294X.2005.02764.x. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Faubet P, Gaggiotti OE. A new Bayesian method to identify the environmental factors that influence recent migartion. Genetics. 2008;178:1491–1504. doi: 10.1534/genetics.107.082560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Seattle, WA: Department of Genome Sciences, University of Washington; 2005. [Google Scholar]
- Frankham R. Relationship of genetic variation to population size in wildlife. Conserv. Biol. 1996;10:1500–1508. [Google Scholar]
- Garza JC, Williamson EG. Detection of reduction in population size using data from microsatellite loci. Mol. Ecol. 2001;10:305–318. doi: 10.1046/j.1365-294x.2001.01190.x. [DOI] [PubMed] [Google Scholar]
- Gelmond O, von Hippel FA, Christy MS. Rapid ecological speciation in three-spined stickleback Gasterosteus aculeatus from Middleton Island, Alaska: the roles of selection and geographic isolation. J. Fish Biol. 2009;75:2037–2051. doi: 10.1111/j.1095-8649.2009.02417.x. [DOI] [PubMed] [Google Scholar]
- Goudet J. 2001. FSTAT: a computer program to estimate and test gene diversities and fixation indices (version 2.9.3). Available at http://www2.unil.ch/izea/softwares/fstat.html.
- Gow JL, Peichel CL, Taylor EB. Contrasting hybridization rates between sympatric three-spined sticklebacks highlight the fragility of reproductive barriers between evolutionarily young species. Mol. Ecol. 2006;15:739–752. doi: 10.1111/j.1365-294X.2006.02825.x. [DOI] [PubMed] [Google Scholar]
- Hänfling B, Weetman D. Concordant genetic estimators of migration reveal anthropogenically enhanced source-sink population structure in the river Sculpin, Cottus gobio. Genetics. 2006;173:1487–1501. doi: 10.1534/genetics.105.054296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänfling B, Hellemans B, Volckaert FAM, Carvalho GR. Late glacial history of the cold-adapted freshwater fish Cottus gobio, revealed by microsatellites. Mol. Ecol. 2002;11:1717–1729. doi: 10.1046/j.1365-294x.2002.01563.x. [DOI] [PubMed] [Google Scholar]
- Hendry AP, Taylor EB. How much of the variation in adaptive divergence can be explained by gene flow: an evaluation using lake-stream stickleback pairs. Evolution. 2004;58:2319–2331. doi: 10.1111/j.0014-3820.2004.tb01606.x. [DOI] [PubMed] [Google Scholar]
- Hendry AP, Taylor EB, McPhail JD. Adaptive divergence and the balance between selection and gene flow: lake and stream stickleback in the Misty system. Evol. Int. J. Org. Evol. 2002;56:1199–1216. doi: 10.1111/j.0014-3820.2002.tb01432.x. [DOI] [PubMed] [Google Scholar]
- Hendry AP, Bolnick DI, Berner D, Peichel –. Along the speciation continuum in sticklebacks. J. Fish Biol. 2009;75:2000–2036. doi: 10.1111/j.1095-8649.2009.02419.x. [DOI] [PubMed] [Google Scholar]
- Hutchison DW, Templeton AR. Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution. 1999;53:1898–1914. doi: 10.1111/j.1558-5646.1999.tb04571.x. [DOI] [PubMed] [Google Scholar]
- Jensen JL, Bohonak AJ, Kelley ST. Isolation by distance, web service Ver. 3.16 BMC Genet. 2005;6:13. doi: 10.1186/1471-2156-6-13. Available at http://ibdws.sdsu.edu/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepaker T. Morphological-changes in a marine population of threespine stickleback, Gasterosteus aculeatus, recently isolated in fresh water. Can. J. Zool. 1993;71:1251–1258. [Google Scholar]
- Koizumi I. Integration of ecology, demography and genetics to reveal population structure and persistence: a mini review and case study of stream-dwelling Dolly Varden. Ecol. Freshwat. Fish. 2011 doi: 10.1111/j.1600-0633.2010.00480.x. [Google Scholar]
- Koizumi I, Shoichiro Y, Maekawa K. Decomposed pairwise regression analysis of genetic and geographic distances reveals a metapopulation structure of stream-dwelling Dolly Varden charr. Mol. Ecol. 2006;15:3175–3189. doi: 10.1111/j.1365-294X.2006.03019.x. [DOI] [PubMed] [Google Scholar]
- Kristjansson BK, Noakes DLG, Skulason S. Rapid divergence in a recently isolated population of threespine stickleback (Gasterosteus aculeatus L.) Evol. Ecol. Res. 2002;4:659–672. [Google Scholar]
- Kume M, Kitano J, Mori S, Shibuya T. Ecological divergence and habitat isolation between two migratory forms of Japanese threespine stickleback (Gasterosteus aculeatus. J. Evol. Biol. 2010;23:1436–1446. doi: 10.1111/j.1420-9101.2010.02009.x. [DOI] [PubMed] [Google Scholar]
- Largiadèr CR, Fries V, Kobler B, Bakker TCM. Isolation and characterization of microsatellite loci from the three-spined stickleback (Gasterosteus aculeatus L.) Mol. Ecol. 1999;8:342–344. [PubMed] [Google Scholar]
- Lind E, Grahn M. Directional genetic selection by pulp mill effluent on multiple natural populations of three-spined stickleback (Gasterosteus aculeatus. Ecotoxicology. 2011;20:503–512. doi: 10.1007/s10646-011-0639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkinen HS, Cano JM, Merilä J. Genetic relationships among marine and freshwater populations of the European three-spined stickleback (Gasterosteus aculeatus) revealed by microsatellites. Mol. Ecol. 2006;15:1519–1534. doi: 10.1111/j.1365-294X.2006.02871.x. [DOI] [PubMed] [Google Scholar]
- Mäkinen HS, Cano JM, Merilä J. Identifying footprints of directional and balancing selection in marine and freshwater three-spined stickleback (Gasterosteus aculeatus) populations. Mol. Ecol. 2008;17:3565–3582. doi: 10.1111/j.1365-294X.2008.03714.x. [DOI] [PubMed] [Google Scholar]
- Manly BFJ. Randomization and regression methods for testing for associations with geographical, environmental and biological distances between populations. Res. Popul. Ecol. 1986;28:201–218. [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- McKinnon JS, Rundle HD. Speciation in nature: the threespine stickleback model systems. Trends Ecol. Evol. 2002;17:480–488. [Google Scholar]
- Olafsdottir GA, Snorrason SS. Parallels, nonparallels, and plasticity in population differentiation of threespine stickleback within a lake. Biol. J. Linn. Soc. 2009;98:803–813. [Google Scholar]
- Olafsdottir GA, Snorrason SS, Ritchie MG. Parallel evolution? Microsatellite variation of recently isolated marine and freshwater three-spined stickleback. J. Fish Biol. 2007;70:125–131. [Google Scholar]
- Page RDM. TREEVIEW: an application display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Peichel CL, Nereng KS, Ohgi KA, Cole BL, Colosimo PF, Buerkle CA, Schluter D, Kingsley DM. The genetic architecture of divergence between threespine stickleback species. Nature. 2001;414:901–905. doi: 10.1038/414901a. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeymaekers JAM, Maes GE, Audenaert E, Volckaert FAM. Detecting Holocene divergence in the anadromous-freshwater three-spined stickleback (Gasterosteus aculeatus) system. Mol. Ecol. 2005;14:1001–1014. doi: 10.1111/j.1365-294X.2005.02456.x. [DOI] [PubMed] [Google Scholar]
- Raeymaekers JAM, Van Houdt JK, Larmuseau MH, Geldof S, Volckaert FA. Divergent selection as revealed by P(ST) and QTL-based F(ST) in three-spined stickleback (Gasterosteus aculeatus) populations along a coastal-inland gradient. Mol. Ecol. 2007;16:891–905. doi: 10.1111/j.1365-294X.2006.03190.x. [DOI] [PubMed] [Google Scholar]
- Raeymaekers JAM, Maes GE, Geldof S, Hontis I, Nackaerts K, Volckaert FAM. Modeling genetic connectivity in sticklebacks as a guideline for river restoration. Evol. Appl. 2008;1:475–488. doi: 10.1111/j.1752-4571.2008.00019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeymaekers JAM, Raeymaekers D, Koizumi I, Geldof S, Volckaert FAM. Guidelines for restoring connectivity around water mills: a population genetic approach to the management of riverine fish. J. Appl. Ecol. 2009;46:562–571. [Google Scholar]
- Reusch TB, Wegner KM, Kalbe M. Rapid genetic divergence in postglacial populations of threespine stickleback (Gasterosteus aculeatus): the role of habitat type, drainage and geographical proximity. Mol. Ecol. 2001;10:2435–2445. doi: 10.1046/j.0962-1083.2001.01366.x. [DOI] [PubMed] [Google Scholar]
- Rousset F. GENEPOP ‘ 007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Res. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Schmidt GW, Brenner T, Steinberg LU, Wolff U. Zur Fischfauna der Naturschutzseen Großes Heiliges Meer und Erdfallsee in Hopsten, Nordrhein Westfalen. Natur und Landschaft. 1985;60:87–89. [Google Scholar]
- Swatdipong A, Primmer CR, Vasemägi A. Historical and recent genetic bottlenecks in European grayling, Thymallus thymallus. Conserv. Genet. 2010;11:279–292. [Google Scholar]
- Takamura KA, Mori SB. Heterozygosity and phylogenetic relationship of Japanese threespine stickleback (Gasterosteus aculeatus) populations revealed by microsatellite analysis. Conserv. Genet. 2005;6:485–494. [Google Scholar]
- Taylor EB, McPhail JD. Evolutionary history of an adaptive radiation in species pairs of threespine sticklebacks (Gasterosteus): insights from mitochondrial DNA. Biol. J. Linn. Soc. 1999;66:271–291. [Google Scholar]
- Taylor EB, McPhail JD. Historical contingency and ecological determinism interact to prime speciation in sticklebacks, Gasterosteus. Proc. R. Soc. B Biol. Sci. 2000;267:2375–2384. doi: 10.1098/rspb.2000.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CE, Taylor EB, McPhail JD. Parallel evolution of lake-stream pairs of threespine sticklebacks (Gasterosteus) inferred from mitochondrial DNA variation. Evolution. 1997;51:1955–1965. doi: 10.1111/j.1558-5646.1997.tb05117.x. [DOI] [PubMed] [Google Scholar]
- TIM-online, ©. 2011. Geobasisdaten der Kommunen und des Landes NRW @Geobasis NRW.
- Vamosi SM. Contemporary evolution of armour and body size in a recently introduced population of threespine stickleback Gasterosteus aculeatus. Acta Zool. Sin. 2006;52:483–490. [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 2004;4:535–538. [Google Scholar]
- Walker JA, Bell MA. Net evolutionary trajectories of body shape evolution within a microgeographic radiation of threespine sticklebacks (Gasterosteus aculeatus. J. Zool. 2000;252:293–302. [Google Scholar]
- Watanabe K, Mori S, Nishida M. Genetic relationships and origin of two geographic groups of the freshwater threespine stickleback, “hariyo.”. Zool. Sci. 2003;20:265–274. doi: 10.2108/zsj.20.265. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wootton RJ. A functional biology of sticklebacks. Berkeley and Los Angeles, CA: Univ. of California Press; 1984. [Google Scholar]
- Wright S. Isolation by distance. Genetics. 1942;38:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wund MA, Baker JA, Clancy B, Golub –, Fosterk SA. A test of the “Flexible stem” model of evolution: ancestral plasticity, genetic accommodation, and morphological divergence in the threespine stickleback radiation. Am. Nat. 2008;172:449–462. doi: 10.1086/590966. [DOI] [PubMed] [Google Scholar]


