Abstract
Introduced species frequently show geographic differentiation, and when differentiation mirrors the ancestral range, it is often taken as evidence of adaptive evolution. The mouse-ear cress (Arabidopsis thaliana) was introduced to North America from Eurasia 150–200 years ago, providing an opportunity to study parallel adaptation in a genetic model organism. Here, we test for clinal variation in flowering time using 199 North American (NA) accessions of A. thaliana, and evaluate the contributions of major flowering time genes FRI, FLC, and PHYC as well as potential ecological mechanisms underlying differentiation. We find evidence for substantial within population genetic variation in quantitative traits and flowering time, and putatively adaptive longitudinal differentiation, despite low levels of variation at FRI, FLC, and PHYC and genome-wide reductions in population structure relative to Eurasian (EA) samples. The observed longitudinal cline in flowering time in North America is parallel to an EA cline, robust to the effects of population structure, and associated with geographic variation in winter precipitation and temperature. We detected major effects of FRI on quantitative traits associated with reproductive fitness, although the haplotype associated with higher fitness remains rare in North America. Collectively, our results suggest the evolution of parallel flowering time clines through novel genetic mechanisms.
Keywords: Arabidopsis, clines, ecological genomics, FRI, FLC, invasive species, parallel adaptation, PHYC
Introduction
The relative role of contingent versus deterministic forces continues to be an unresolved issue in evolutionary biology (Gould 1985, Travisano et al. 1995; Losos et al. 1998; Fenster and Galloway 2000; Simões et al. 2008). Echoes of this debate can be found in diverse research areas, including (but not limited to) the role of drift and selection shaping patterns of nucleotide polymorphism (Kimura 1983; Gillespie 1991), adaptive versus nonadaptive explanations for organismal features (Gould and Lewontin 1979; Mayr 1983), and mechanisms producing geographic clines within species (Endler 1977; Vasemägi 2006). Evaluating prevalence of convergent evolution (sensu Arendt and Reznick 2008) provides a clear approach for distinguishing these possibilities: it is unlikely that the same, seemingly adaptive features of organisms will evolve more than once by stochastic forces alone. Moreover, the expanding set of tools for investigating the molecular genetic basis to traits in model and nonmodel organisms offers the possibility of examining whether the same or independent genetic, developmental, and physiological mechanisms were used to produce similar phenotypes (Arendt and Reznick 2008). Here, we examine whether introduced populations of Arabidopsis thaliana have evolved geographic clines in flowering time and other life-history traits that are similar to those exhibited in the native range (Caicedo et al. 2004; Stinchcombe et al. 2004; Samis et al. 2008) and whether the same genes are associated with natural variation in flowering time.
Testing for parallel1 clines in phenotypic traits provides a straightforward way to examine the relative role of adaptive versus nonadaptive mechanisms in producing clinal variation. While systematic associations between phenotypes and environmental variables or geographic proxies (e.g., latitude, longitude, altitude) are traditionally interpreted as evidence for adaptive evolution, they can also be generated by nonadaptive processes (Endler 1977)—especially for traits with a relatively simple genetic basis (Endler 1977; Vasemägi 2006). Consequently, the occurrence of independent clines is strong evidence of adaptive mechanisms, particularly in traits with a complex, quantitative genetic basis. For example, if putatively adaptive phenotypes change in the same direction with increasing latitude and altitude, this suggests adaptation to climatic gradients that are either similar or favor similar phenotypes (see, e.g., Stinchcombe et al. 2004; Montesinos-Navaro et al. 2011).
Nonnative species commonly show evidence of parallel latitudinal or geographic clines (Johnston and Selander 1964; 1971; Lacey 1988; Weber and Schmid 1998; Huey et al. 2000; Gilchrist et al. 2001; Kollman and Banuelos 2004; Maron et al. 2004; 2007; Leger and Rice 2007; Etterson et al. 2008; Montague et al. 2008; Colautti et al. 2009; Keller et al. 2009; Gonzalez et al. 2010), which strongly suggests adaptive mechanisms in producing geographic differentiation in both ranges. However, founder events and unequal sampling from the ancestral range are also capable of producing the appearance of clines after introduction (Keller and Taylor 2008). Accordingly, determining whether geographic trends in the nonnative range represent adaptation requires integrating multilocus genetic data to infer the role of population structure, founder events, and other nonadaptive processes likely to be common in introduced and invasive species (Maron et al. 2004; Kliber and Eckert 2005; Kolbe et al. 2007; Keller and Taylor 2008; Keller et al. 2009).
Two approaches have been used to evaluate the genetic basis of clinal evolution. In the first, observing clear latitudinal or geographic patterns in allozyme, allele, or gene frequencies suggests natural selection on those or tightly linked loci. Numerous examples of clines of this type exist from a wide variety of species and genetic markers (Allard et al. 1972; Mitton et al. 1977; Lonn et al. 1996; Van Rossum et al. 1997; Schmidt and Rand 1999; 2001; Schmidt et al. 2000; Schmuths et al. 2004; Barker et al. 2011), with perhaps the most famous being the Adh in Drosophila (Oakeshott et al. 1982; Berry and Kreitman 1993). However, in most cases the ecological mechanism behind these patterns is unclear—that is how do different genotypes affect organismal phenotypes in a manner that differentially affects fitness in alternative environments? Evidence addressing this question is still relatively limited, but is accumulating from ecological experiments (Schmidt and Rand 2001), observed shifts in genetic clines in response to global climate change (Anderson et al. 2005; Umina et al. 2005; Balanya et al. 2006), and manipulative artificial selection experiments (Rand et al. 2010).
The second major approach to dissecting the genetic basis of clinal evolution has been to use quantitative trait locus (QTL), association, or candidate gene mapping to localize genetic regions, genes, or even nucleotides that contribute to the observed differentiation in traits. A benefit to this approach is that a focal phenotype, and presumably its connection to fitness, is well defined. Genetic mapping has been successful in classic genetic model systems (Arabidopsis: Caicedo et al. 2004; Stinchcombe et al. 2004; 2005; Lempe et al. 2005; Baxter et al. 2010; Li et al. 2010; Drosophila: McKechnie et al. 2010; Rand et al. 2010) as well as emerging models (Peromyscus: Storz and Dubach 2004; Storz et al. 2007; Mullen et al. 2009; Picea: Holliday et al. 2010; Populus: Ma et al. 2010). Current evidence is mixed between a common genetic basis to parallel clines (Calboli et al. 2003; Rus et al. 2006; Paaby et al. 2010) and unique genetic mechanisms producing adaptation to similar habitats (Hoekstra and Nachman 2003; Jeffery et al. 2003; Protas et al. 2006; Borowsky 2008; Steiner et al. 2009). Outside of clinal evolution, there are numerous examples of cases where diverse genetic mechanisms are associated with similar phenotypes within a species (see Table 1 of Arendt and Reznick 2008). Likewise, there is also substantial evidence for the same genes or developmental mechanisms being used to achieve similar phenotypes in unrelated species (e.g., pigmentation and Mc1r: Hoekstra et al. 2006; Rosenblum et al. 2010) or in several species of a clade (e.g., flower color in Ipomoea; Des Marais and Rausher 2010). Determining how frequently adaptive phenotypes evolve using the same genetic mechanisms directly informs debates about evolutionary constraints to adaptation (e.g., Wake 1991; Hoekstra 2006; Des Marais and Rausher 2008; 2010).
Table 1.
Number and frequency of haplotypes at three flowering time loci genotyped in North American (NA) and Eurasian (EA) lines of Arabidopsis thaliana. The frequency of different haplotypes was compared with a χ2 test, first using the entire sample, and second, by randomly sampling one accession per latitude and longitude in each continent to equalize sampling regimes. For the latter, the 2.5th and 97.5th percentile of the χ2 distribution are shown from 1000 random samples, with the P-value for the 97.5th percentile of the χ2 distribution.
| Locus | Allele | EA1 | NA | χ2 (P) | χ2, (P) (1 accession sampled per locality) |
|---|---|---|---|---|---|
| FRI | Wild type | 97 | 193 | 139.03 (<0.0001) | 21.85 < χ2 < 35.40 (<0.0001) |
| Null | 164 | 21 | |||
| Total | 261 | 215 | |||
| % NA rare | 62.8% | 9.8% | |||
| FLC | A | 109 | 194 | 76.30 (<0.0001) | 9.33 < χ2 < 19.75 (<0.002) |
| B | 103 | 22 | |||
| Total | 212 | 217 | |||
| % NA rare | 48.6% | 10.2% | |||
| PHYC | Col | 86 | 16 | 45.24 (<0.0001) | 3.56 < χ2 < 11.4 (<0.0591) |
| Ler | 174 | 197 | |||
| Total | 260 | 214 | |||
| % NA rare | 33.1% | 7.5% | |||
EA data from Caicedo et al. 2004, Stinchcombe et al. 2004, Samis et al. 2008. We excluded lab strains Col and Ler-1 (Landsberg erecta) from these calculations.
The mouse-ear cress (A. thaliana; Fig. 1, inset) represents a promising system to test for parallel clinal evolution. Numerous phenotypic clines have been detected within the native range in a host of phenotypes, including flowering time (Caicedo et al. 2004; Stinchcombe et al. 2004; Lempe et al. 2005; Samis et al. 2008; Li et al. 2010; Montesinos-Navaro et al. 2011), growth rate (Li et al. 1998), hypocotyl length (Maloof et al. 2001; Stenoien et al. 2002), circadian rhythm (Michael et al. 2003), salt tolerance (Baxter et al. 2010), and freezing tolerance (Zhen and Ungerer 2008). Of these phenotypic clines, flowering time appears robust to the effects of population structure (as inferred from patterns of SNP variation; Korves et al. 2007; Samis et al. 2008), although the effects of population structure on the other phenotypes has not been examined. Recent genome-wide association studies also show geographic differentiation of individual SNPs associated with fitness in common gardens (Fournier-Level et al. 2011) and of SNPs associated with environmental gradients (Hancock et al. 2011). Flowering time has also been intensely investigated on the genetic level, with numerous candidate genes identified, and evidence of three major-effect loci that are polymorphic in the ancestral range and contribute to clinal variation (FRI: Johanson et al. 2000; Stinchcombe et al. 2004; FLC: Michaels et al. 2003; PHYC: Balasubramanian et al. 2006; also see Werner et al. 2005). There is abundant evidence of population structure within European populations of A. thaliana (Bomblies et al. 2010; Platt et al. 2010). Finally, A. thaliana colonized North America approximately 150–200 years ago (Jorgensen and Mauricio 2004), providing the opportunity for the evolution of clines after introduction.
Figure 1.
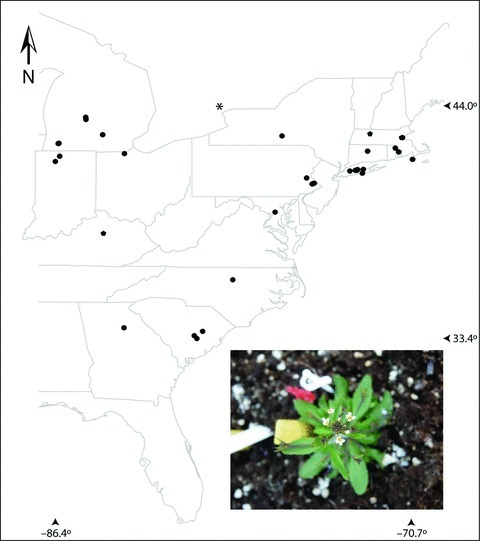
Distribution of North American (NA) Arabidopsis thaliana populations used for genotyping at functional loci associated with flowering time and the common garden experiment in Toronto, Canada (asterisk). N= 35 populations, 212 lines. Inset: Photo of A. thaliana flowering in the common garden.
Specifically, we use North American (NA) populations of A. thaliana to ask: (1) What is the pattern of population genetic structure in NA Arabidopsis populations? We use these data to establish a null model as a point of comparison for phenotypic traits. (2) What are the allele frequencies at the flowering time loci FRI, FLC, and PHYC, and are they different from the ancestral range? (3) Is there evidence for the evolution of parallel phenotypic clines in the introduced range, and if so, what are the likely ecological mechanisms? (4) What is the relative contribution of population structure, candidate genes (FRI, FLC, and PHYC), and climatic variation to the pattern of quantitative genetic differentiation in the introduced range?
Material and Methods
Study species
We used natural accessions of A. thaliana from the introduced NA range that we obtained from the Arabidopsis stock center (Arabidopsis Biological Resource Center [ABRC]; N= 92 lines, 16 populations) and recent collections from natural populations (N= 108 lines, 20 populations), with one population including lines from ABRC and recent collections (Fig. 1; Table S1). Geographic sampling was opportunistic, but spanned 10° latitude and 16° longitude. All new lines described here will be deposited with the ABRC, or available directly from the Stinchcombe lab. For comparison, we also obtained a sample of native range, Eurasian (EA) lines (N= 20 lines, 19 localities) from ABRC. We used these parental plants for DNA analysis, and to generate seeds for common garden experiments. After seven days wet stratification at 4°C to synchronize germination, we germinated and grew seeds from all collections in a growth chamber at the University of Toronto for one generation under the following standard conditions (Weigel and Glazebrook 2002): 23°C for 24 h with 19 h light at 200 µmol/m2/sec and 5 h dark, and without vernalization. All lines flowered and produced seed without vernalization, although a subset (∼6%) produced insufficient seeds in time to be included in the common garden.
Genotyping
We collected leaf material from parental lines and extracted DNA using the Qiagen DNeasy plant kit (Qiagen Inc., Toronto, Canada). We screened each line for variation at three functional loci, FRI, FLC, and PHYC. We designed primers to flank previously reported polymorphic regions in each locus (Table S2), and screened lines using PCR amplification (with Fermentas MBI reagents; Fermentas Canada Inc., Burlington, Canada). All loci were amplified using previously published, PCR-based genotyping protocols (FRI: Stinchcombe et al. 2004; FLC: Caicedo et al. 2004; PHYC: Samis et al. 2008). We sequenced a subset of amplification products from each locus (six to 15 lines per locus, 17 lines for more than 1 locus and 40 individuals overall) to verify sequence homology with published sequences from the native range. Because we focused on previously characterized variation at these loci, and no additional variants were detected in the sequenced subset, it should be noted with caution that other undetected variants may occur within these loci. All EA lines we used were FRI–wt (Table S1).
To evaluate genome-wide levels of polymorphism, we genotyped 188 NA lines (Table S1) using 149 SNP loci (Platt et al. 2010); these SNPs were developed for quantifying population diversification in A. thaliana (Platt et al. 2010). We cleaned the SNP data to remove poor quality loci (i.e., loci with ≥25% low-quality calls across 188 lines/locus) and lines (≥50% low-quality or missing calls of 149 loci/line). The final dataset included 136 SNP loci across 179 individuals. Additional SNP genotypes for EA lines were obtained from the dataset of Platt et al. (2010).
Common garden
We planted F1 seed from 35 NA populations (median = 2 lines per population), and 19 EA populations (1 line per population for all except 1) into plug trays (1.9 × 4.4 cm cells) with germination mix. We stratified seeds at 5°C for seven days in the dark, and then allowed them to germinate in the University of Toronto glasshouse (at approximately 22°C). Prior to transplanting, we exposed seedlings to two days of glasshouse growth at 15°C, and a day of natural conditions beside the gardens, to initiate hardening. We next transplanted 1095 seedlings with their soil plugs at 10 cm intervals into raised beds (2 m wide × 3 m long × 30 cm deep, and filled with Premier Promix PGX; Premier Tech Horticulture, Rivière-du-Loup, Canada) on the roof of the EEB Department (Earth Sciences Centre) at the University of Toronto on 2 October and 3 October 2008, when seeds naturally germinate in many NA populations. We used outdoor plantings to more realistically simulate natural cues and variability in light, water, snow, temperature, and other abiotic and biotic forces. We weeded soil plugs to one seedling (range 1–12 days old) per position prior to planting, and again as necessary, keeping the oldest seedling. Our experiment had five spatial blocks spread across two raised beds, with each block containing one replicate per line (219 plants = 199 NA, 20 EA). We covered the raised beds with shade cloth for the first seven nights, and watered as needed for one month to promote establishment.
We recorded survival three times before snow cover in late November, and opportunistically with snow melts throughout winter until the following April. Starting in spring, we tracked plants on a daily basis to determine bolting date (i.e., visible differentiation and elongation of the apical inflorescence), and first flower opening from April until August. At bolting, we measured rosette diameter and counted rosette leaves. At senescence, we harvested plants individually, dried them at 55°C, and then counted the total number of fruits per plant to estimate of plant fitness (Westerman and Lawrence 1970). The sample size of NA lines used in the common garden experiment (N= 199) is slightly reduced compared to the genotyping analysis (212), because not all genotyped plants produced sufficient seed in time for the start of the experiment.
Data analysis
Distribution and population structure
We used logistic regression to examine the geographic distribution of haplotypes at FRI, FLC, and PHYC, with latitude and longitude as predictors, where FRI was categorized as functional/null, FLC as A or B haplotype, and PHYC as Ler or Col haplotype. We used χ2 tests to compare the frequency of flowering time genotypes in North America to a similar-sized sample in the native range. To compare the allele frequency differences at flowering time loci to the “genome-wide” expectation, we also compared allele frequency changes at 136 SNPs. Because sampling regimes differed (with more within-population sampling in NA than EA), we randomly selected a single accession per geographic locality and ran the χ2 analyses again, equalizing within-population sampling per locality for both Europe and North America. We repeated random sampling and χ2 analysis 1000 times and present the range of χ2 values, and P-values from the 97.5th percentile of the χ2 statistic as an upper bound. EA flowering time genotypes were obtained from previous studies (Samis et al. 2008).
We evaluated the degree of admixture or differentiation among lines and regions for genome-wide polymorphisms using STRUCTURE (V2.2), a model-based clustering program (Pritchard et al. 2000). Although STRUCTURE is based on an evolutionary model, our goal was not to model the actual population history of the A. thaliana in North America based on its specifications, but instead to estimate aspects of population genetic structure that could lead to the spurious appearance of clines or gene–trait associations. It is important to note that alternative approaches, such as EIGENSTRAT, which is not based on an evolutionary model, typically recover similar levels population genetic structure, also with clear geographic interpretations (see, e.g., Patterson et al. 2006; Price et al. 2006; Zhao et al. 2007). We performed three separate analyses: (1) all NA lines, (2) NA and EA lines without any geographic information, and (3) with EA lines as a reference sample for NA lines. The first and second analyses grouped samples based on genotype information alone, while the third analysis forced introduced NA genotypes to cluster with inferred ancestral EA clusters. We interpreted each observed cluster (K) to represent ancestrally related genotypes where each individual is proportionally assigned to clusters with a predicted probability (ancestry coefficient). Assignment to more than one cluster suggests an admixed genome. Each algorithm ran with a burn-in of 10,000 Markov Chain Monte Carlo (MCMC) iterations, and parameters were estimated over 300,000 iterations for five replicates at K= 1–8 using correlated allele frequencies. For the third analysis, we used the PFROMPOPFLAGONLY option of STRUCTURE to ensure that ancestry coefficients for NA lines were estimated from the EA distribution (Murgia et al. 2006). We estimated the number of clusters, K, following Evanno et al. (2005). For clustering analyses, we included 179 NA lines from this study, and 228 EA lines from 139 populations genotyped at the same 136 SNP loci (from Platt et al. 2010). For all analyses, we defined populations by latitude and longitude. We analyzed our data as if A. thaliana were haploid, which is consistent with the largely selfing mating system of A. thaliana and its high homozygosity.
Morphological and phenological traits
We assessed survival as the proportion of five replicates per line surviving across two intervals: from two weeks after planting until the following April (winter survival), and until reproduction (survive to reproduce) using mixed-model analysis of variance (ANOVA) including continent (fixed) and population (random). Variation in phenotypes (days to bolting and flowering, rosette leaf number and rosette diameter at bolting, and lifetime fruit number) was assessed using nested, mixed effects ANOVAs. For initial models, we included region as a fixed effect (NA, EA), and the random effects of block, population nested within continent, and line nested within population within continent.
We subsequently performed separate analyses, by region, for traits that showed significant differences between EA and NA. For Europe, we tested for variation among lines and among blocks, omitting the population effect (only 1 locality in EA was represented by >1 line), while for NA samples we tested for variation among populations, lines within populations, and blocks. Significance of random effects was determined by calculating the difference between reduced and full model –2log-likelihood values, using a one-tailed χ2 test with 1 degrees of freedom (df). We evaluated covariation among phenotypes and fruit production using pairwise Pearson's correlations on inbred line means within each region separately; we compared correlation coefficients between regions with Z-tests.
Because none of the five replicates from one population (KYF) survived to reproduce, analyses of reproductive traits included 34 NA populations. We log10-transformed fruit number to reduce heterogeneity of variances. We analyzed data using SAS (V9.2) and JMP (V8.0.1, SAS Institute, Cary, NC, USA).
Geographic differentiation in flowering phenotypes and fitness
Based on previous observations that geographic clines in flowering time and correlated traits in the native range were associated with geographic variation in climate (Caicedo et al. 2004; Stinchcombe et al. 2004; Samis et al. 2008), we tested for signatures of geographic differentiation in our common garden experiment using three nested analysis of covariance (ANCOVA) models, analyzing NA and EA separately. In all analyses of geographic differentiation, we use inbred line mean phenotypes. (Using line means is akin to considering line a fixed effect to calculate an average, which avoids the attendant problems of analyzing BLUPs for random effects; see Hadfield et al. 2010.) First, we conducted a simple test by regressing traits on latitude and longitude (Model A; N= 196). Second, we tested for geographic clines after controlling for spurious associations due to population structure by including ancestry coefficients as a covariate in the first model (Model B; N= 176). We interpret geographic clines that remain significant with the inclusion of ancestry coefficients as robust to the effects of population structure—that is, they are less likely to be caused by spurious associations. Stochastic and demographic effects should be captured by the patterns inferred from a large number of presumably neutral SNPs (Keller and Taylor 2008; Keller et al. 2009). Ancestry coefficients, representing genome-wide patterns of differentiation inferred from the 136 SNPs, were derived from cluster analysis using only NA lines from this study (K= 2), and represent the proportional membership of each line to each inferred ancestral cluster. We transformed ancestry coefficients using log contrasts (log[p1/p2]; Samis et al. 2008). Third, we tested if geographic variation in plant traits above and beyond population structure was explained by the average climate at the site of origin (Model C, equation 1; N= 176):
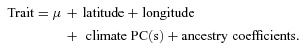 |
1 |
Principal components (PCs) were used to summarize the climate at the site of origin, generated from independent analyses of three climatic variables previously used in studies of Arabidopsis climatic adaptation (Stinchcombe et al. 2004; Rutter and Fenster 2007; Baird et al. 2011): mean monthly temperature, mean total precipitation, and mean diurnal temperature range (year range 1961–1990 climate normals, 5′ intervals; Mitchell and Jones 2005) from October to April, that is, over the critical phases of germination, vernalization, and the initiation of flowering in A. thaliana. Climate data for each NA A. thaliana population were extracted from the CRU dataset in ArcGIS 9.3 (ESRI). PCs were generated from correlation matrices, and those with eigenvalues greater than one (explaining >15% of the variation in climate variables) were saved from each principal component analysis (PCA; Quinn and Keough 2002). The loss of a significant latitudinal or longitudinal effect in Model C suggests that variation in the focal climate variable (or a highly correlated factor) may drive geographic variation in the trait; retention of a significant cline suggests that the focal climate variable does not explain the cline. Supplemental models using either mean monthly climate variables, or absolute differences between monthly climate at the site of origin and the common garden (the calibrated common garden approach of Rutter and Fenster 2007) showed similar results, and as such we only present results based on PCs.
We tested for geographic clines among EA lines in the experiment using the reduced geographic model (Model A), as well as after accounting for population structure (Model B). A supplementary cluster analysis with EA lines using published SNP data (Platt et al. 2010) was conducted to generate EA ancestry coefficients (K= 5, data not shown).
Relative effects of geographic origin, climate, and flowering time loci for phenotypes
We used a modified association mapping approach to examine the relative influence of site of origin, climate, and flowering time genes on phenotypes. We analyzed inbred line means for flowering time phenotypes, fitness (N= 172), and survival (N= 173) using the following ANCOVA model (Model D, Equation 2):
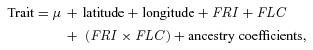 |
2 |
where µ is the model intercept (fit by default), latitude and longitude are continuous covariates representing the site of origin, and FRI and FLC represent genotype classes (functional or null, and A or B, respectively) where the interaction term accounts for epistatic effects among alleles at each locus (Caicedo et al. 2004). Haplotypes at the PHYC locus did not contribute significantly to the model, possibly because of the exceptionally low number of lines with the rare allele, and was subsequently excluded. Ancestry coefficients were the same as those for geographic cline ANCOVAs. We did not assess genetic associations in EA lines because of the limited sample size, and because all EA lines were selected for being FRI–wt. Because of the imbalanced distribution of flowering time genotypes for FRI and FLC, we verified the statistical significance of the FRI, FLC, and FRI×FLC terms using permutation testing (see, e.g., Cassell 2002; Heath 2010); permutation-based P-values had identical patterns of significance and marginal significance, and as such we present traditional hypothesis tests based on F-tests.
We elected not to use mixed model approaches that take advantage of both population structure and relative kinship (e.g., TASSEL: Bradbury et al. 2007; EMMA: Kang et al. 2008), because analyses including geographic covariates (latitude and longitude) either would not converge (TASSEL) or because it is unclear how to include such covariates (EMMA). While our modified structured association mapping approach might have greater false positives (at the level of locus detection) due to population structure, it is the most straightforward method for testing our hypotheses about the relative influence of site of origin, climate, and flowering time genes on phenotypes. Past studies (Nordborg et al. 2005) have documented extensive haplotype sharing between pairs of NA A. thaliana accessions, suggesting that the resolution of our candidate gene mapping is likely to be reduced compared to EA samples.
Results
Distribution and frequency of haplotypes
As suggested by previous investigations (see, e.g., Nordborg et al. 2005), our surveys of previously characterized allelic variation in NA lines revealed minimal variation in functional genes related to flowering time, with 85% of lines (180/212) exhibiting the common genotype, FRI–wt/FLC–A/PHYC–Ler. The frequencies of alternate genotypes were relatively low (<3.5% each), and no individuals exhibiting one genotype: FRI–wt/FLC–B/PHYC–Col (Table S3). Two alleles previously detected in EA samples of A. thaliana (Caicedo et al. 2004; Balasubramanian et al. 2006) were not detected in this NA sample: the 16-bp FRI deletion C and the Col-type FLC insertion. The Ler-type FLC insertion was detected in only three lines, and was not considered further. In contrast to the frequency of alleles in the native range, where FRI, FLC, and PHYC haplotypes occurred at frequencies close to 50%, one allele at each locus was significantly more rare in NA, occurring at frequencies of <10% (all P < 0.0001; Table 1). When we randomly sampled a single accession per locality, we still detected significant differences in haplotype frequencies between NA and EA, suggesting that differences in frequencies detected in the global sample are not due to sampling multiple accessions per locality in NA compared to Europe (Table 1). However, when compared to allele frequency changes at the 136 SNPs, the flowering time genes do not show greater differentiation than expected based on the “genome-wide” patterns of allele frequency changes (Fig. S3). Finally, while it is worth considering that our genotyping approach may have missed new variants either within or linked to the characterized loci, it is clear that variation characterized in EA samples differs in frequency from that occurring in North America.
The NA lines we sampled encompass a range of approximately 10° latitude and 16° longitude (Table S1). Within this range, PHYC haplotypes did not exhibit significant geographic variation in their distribution (Col 39.7 ± 0.65°N, –77.7 ± 1.49°W; Ler 39.6 ± 0.23°N, 78.5 ± 0.35°W; logistic regression, model P= 0.79). In contrast, both FRI and FLC genotype classes were differentiated geographically (FRI model P= 0.004, likelihood ratio χ2 tests, latitude χ2= 4.2, P= 0.041, longitude χ2= 3.2, P= 0.072; FLC model P < 0.0001, latitude χ2= 4.1, P= 0.043, longitude χ2= 12.0, P= 0.0005). Rarer genotype classes (FRI–null and FLC–B) exhibited significantly more northern and eastern distributions than the more common haplotypes at each locus, and in comparison to the mean of all lines (Fig. 2).
Figure 2.
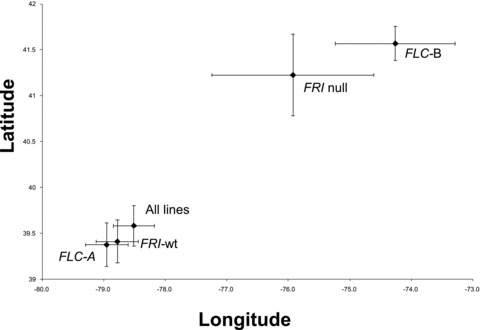
Mean (± SE) geographic location for all NA lines (N= 212), and for lines with each FRI (FRI–wt 192, FRI–null 20) and FLC (FLC–A 192, FLC–B 20) haplotype. Note that 187 lines were FRI–wt/FLC–A. There was a significant latitudinal differentiation in FRI haplotypes, and significant longitudinal and latitudinal differentiation in FLC haplotypes.
Population structure
Analysis of NA SNP genotypes alone revealed that lines cluster into two ancestral groups with no clear geographic structure (Fig. 3). Some populations exhibited little admixture (e.g., South Carolina, Maryland, and New York), while others exhibited variable levels of admixture (e.g., Pennsylvania, Long Island, and mid-western states). Inclusion of presumably ancestral lines from EA increased the degree of clustering (K= 3 without geographic information supplied), and also suggested higher admixture in North America than within Eurasia, and relatively less population structure within the native range (Fig. S2). The low level of clusters detected is unlikely to be due to a general lack of variation at the SNPs, as they were not generally lacking variation (Fig. S3). One striking finding of the structure analysis is that EA samples showed little to no structure and belonged almost entirely to a single cluster (K2), while membership in the remaining clusters (K1, K3) was restricted to North America. Both the number of clusters (K= 5) and the degree of admixture increased when NA lines were forced to cluster with EA lines as a learning set, but differences between regions remained (Fig. S1). When forced to cluster with EA lines, the dominant cluster in North America (K5) is also the dominant cluster in admixed accessions from France, Sweden, and Switzerland. These results are qualitatively similar to those of Nordborg et al. (2005) and Ostrowski et al. (2006), who found that NA lines cluster with those from Western Europe.
Figure 3.
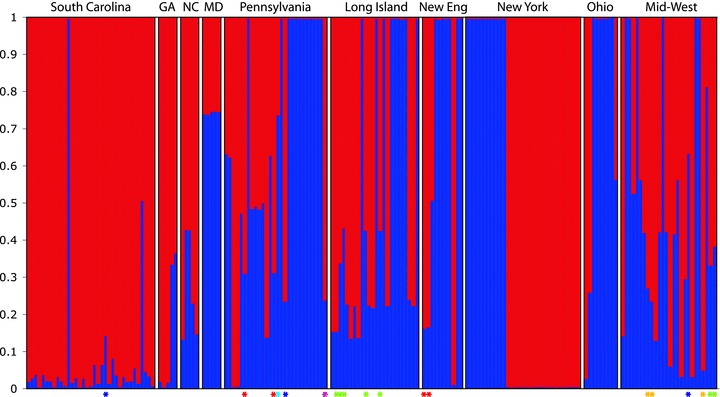
Population structure of 179 lines of Arabidopsis thaliana in eastern North America based on 136 SNP loci and the assumption that the number of ancestral clusters is two. Lines are organized randomly within U.S. states/regions of origin from south to north (details in Table S1). Lines are represented by vertical columns, where the proportion of assignment to each cluster (K1, K2) is indicated by color. Asterisks indicate lines with the noncommon genotype for three flowering time genes, where lines with the same color had the same genotype, and lines without asterisks had the common genotype (FRI–wt/FLC–A/PHYC–Ler).
Variation and covariation in traits
The majority of plants survived at least two weeks after transplanting (84%), and the majority of those plants survived until spring (728 out of 925 plants, 79%), and became reproductive (78%). Because there was little difference between winter survival and survival to reproduction, we focus on survival to reproduction. EA plants displayed higher overall survival than NA plants (mean proportion of replicates per line surviving ± SE: EA 0.79 ± 0.054; NA 0.65 ± 0.023; P= 0.017), although we note that these differences were based on five replicates per line. ithin regions, significant variation in survival was detected among populations in North America (P= 0.03). All plants bolted over a relatively short timeframe in early spring, and the majority of plants (n= 634) completed flowering within 39 days of first flower (mean ± SD 26.7 ± 3.3 days).
Despite high overlap between regions, EA plants flowered significantly earlier (lsmeans ± SE: days to bolting EA 196.3 ± 0.72, NA 197.7 ± 0.58; P= 0.01, days to flowering EA 207.0 ± 0.64, NA 208.0 ± 0.54; P= 0.019), and were more productive (log10 (fruit) back-transformed lsmeans ± SE range: EA 428.5 + 92.55 – 76.11, NA 331.2 + 63.77 – 53.47; P= 0.012) than plants from North America. The sample of EA lines showed statistically significant variation among lines for days to bolting and days to flowering (Table 2), despite the limited power due to low sample size. Plants in both regions bolted at similar rosette leaf numbers (EA 29.2 ± 1.34, NA 29.2 ± 0.95; P= 0.99) and rosette diameters (EA 3.3 ± 0.18, NA 3.1 ± 0.15; P= 0.18). For rosette diameter and leaf number, there was significant genetic variation among lines within populations (P < 0.0001). Within North America, significant variation among lines within populations was detected for all traits (Table 2B). Significant among population variation was present for days to bolting and fruit number, and was marginally significant for days to flowering (P= 0.061; Table 2). These data indicate substantial quantitative genetic variation exists in North America, despite reduced variation at FRI, FLC, and PHYC. Although the block effects are appreciable, the presence of significant genetic variation, in the form of lines within populations, indicates that testing for geographic trends in inbred line mean phenotypes is possible.
Table 2.
Mixed model analysis of variance (ANOVA) for heterogeneity in individual plant traits among or within regions (native Eurasian or introduced North American range source), populations and lines for Arabidopsis thaliana planted in a common garden experiment in Toronto, Canada. The full model (A) includes the effect of NA and EA region, and subsequent models (B) partition variation within each region separately for those traits with a significant effect of region in the full model. For random effects, we report the variance component, as well as the likelihood ratio statistic and P-value associated with removing that term from the model.
| (A) Full model | Fixed effects F (P) | Random effects variance components (likelihood ratio statistic, P) | |||
|---|---|---|---|---|---|
| Trait | Region | Population (region) | Line (population) | Block | Residual |
| Rosette leaves | F < 0.001 (0.99) | 3.33 (χ2= 3, P= 0.042) | 10.81 (χ2= 30.3, P < 0.0001) | 3.06 (χ2= 26.5, P < 0.0001) | 33.4 |
| Rosette diameter | F= 1.82 (0.18) | 0.062 (χ2= 10.7, P < 0.0005) | 0.109 (χ2= 15.7, P < 0.0001) | 0.084 (χ2= 63, P < 0.0001) | 0.5442 |
| (B) Within regions | Population | Line (EA) or line (population) (NA) | Block | Residual | |
| Log10 (fruit) | EA | – | 0 (χ2= 0, P= 1) | 0.021 (χ2= 6.6, P= 0.005) | 0.098 |
| NA | 0.0042 (χ2= 6.5, P= 0.005) | 0.0091 (χ2= 4.4, P= 0.018) | 0.0277 (χ2= 128, P < 0.0001) | 0.0907 | |
| Days to flower | EA | – | 2.80 (χ2= 7.1, P= 0.004) | 1.24 (χ2= 4, P= 0.023) | 7.92 |
| NA | 0.262 (χ2= 2.4, P= 0.061) | 1.31, (χ2= 25.4, P < 0.0001) | 1.28 (χ2= 101.8, P < 0.0001) | 4.45 | |
| Days to bolting | EA | – | 5.71 (χ2= 8.7, P= 0.003) | 1.62 (χ2= 2.8, P= 0.047) | 12.84 |
| NA | 0.419 (χ2= 2.9, P= 0.044) | 2.33 (χ2= 49.9, P < 0.0001) | 1.34 (χ2= 94.3, P < 0.0001) | 5.11 | |
As expected for A. thaliana, within continents, early bolting was strongly correlated with early flowering, and larger rosette diameter was reflected in higher rosette leaf numbers at bolting (all P < 0.0001; Fig. 4). However, we detected qualitative differences in the patterns of trait covariation between plants in native and introduced regions. For instance, among EA lines, there was a positive correlation for bolting and flowering times with rosette diameter (both r > +0.62, P < 0.05), and with rosette leaf number (both r > +0.76, P < 0.0001) at bolting. In contrast, while NA flowering time was only moderately associated with rosette leaves (r=+0.15, P < 0.05) and rosette diameter (r=+0.20, P < 0.05); bolting time was not significantly associated with either trait (Fig. 4). Comparison of the correlation coefficients between EA and NA with Z-tests suggests that the coupling of flowering time with plant size and development is weaker in populations from the introduced range, but that correlations between rosette leaf number and size are stronger.
Figure 4.
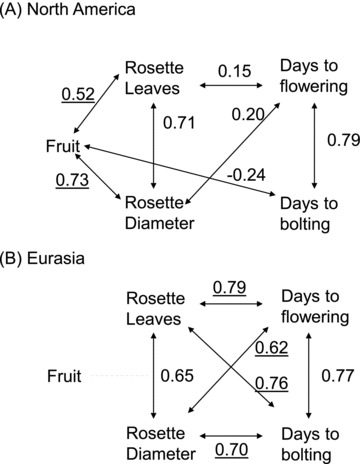
Pairwise Pearson's correlations among inbred line mean flowering time traits and fruit production for (A) North American and (B) Eurasian lines of Arabidopsis thaliana in a common garden experiment in Toronto, Canada. Correlation coefficients are shown for significant correlations only. In North America, three traits demonstrated significant correlations with fruit production, while no traits were associated with fruit production in Eurasia (dotted line). Underscored correlation coefficients are significantly larger than the same coefficient in the alternate geographic range, as determined by two-tailed Z-tests.
Geographic differentiation: flowering phenotypes and fitness
Multiple regression models revealed evidence for geographic differentiation. Furthermore, comparisons with EA lines in the experiment suggested a parallel cline in flowering time between regions. In the reduced model (Model A), mean inbred line days to bolting (estimate ± SE +0.094 ± 0.034, P= 0.0057) and days to flowering (+0.082 ± 0.030, P= 0.0061) exhibited significant regressions with longitude, and that remained significant above and beyond variation in neutral population structure (Model B; both P < 0.020, Table 3). A parallel, longitudinal cline was detected for days to flowering among EA lines (reduced model: –0.18 ± 0.061, P= 0.0091), although significance was reduced after controlling for population structure (P= 0.058). In both regions, inbred lines with more coastal origins (eastern in North America, western in Eurasia) flowered later than lines with more inland origins (Fig. 5). Reducing the NA sample to focus only on FRI functional genotypes (see, e.g., Stinchcombe et al. 2004) did not reveal additional geographic clines. In general, longitudinal clines were weak in overall magnitude, with considerable within-population variation in flowering time present, but statistically significant.
Table 3.
Multiple regression models testing for geographic clines in flowering time with and without control for neutral processes and climatic variation. All models were conducted on inbred line means for Arabidopsis thaliana plants grown in a common garden experiment in Toronto. Principal components (PC) describe mean monthly climate at home from October through to April, for the variable indicated in the column header. Gray shading indicates that the term was omitted from the model. Significant effects at α= 0.05 are bolded.
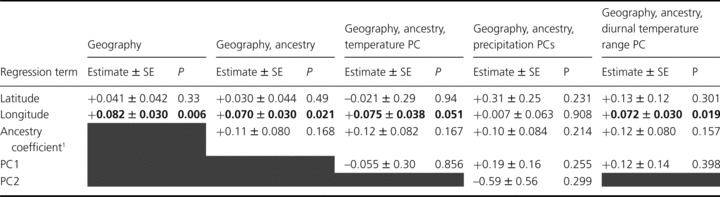 |
Figure 5.
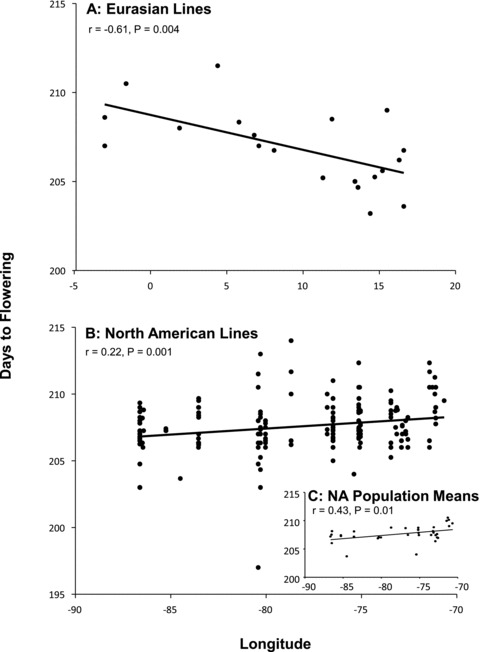
Parallel longitudinal clines in days to first flower displayed between populations from across the native (A) and introduced (B) ranges planted in a common garden experiment in Toronto, Canada. Population means from the introduced range are inset in panel (C). Statistics are presented for univariate, Pearson correlations; ANCOVA and multiple regression results are in Tables 3 and 4.
Significant positive latitudinal clines among NA samples were observed for mean inbred line rosette diameter (0.043 ± 0.014, P= 0.002), survival (0.016 ± 0.0052, P= 0.0023), and fruit production (0.019 ± 0.006, P= 0.0021) in the reduced model, although none approached significance after controlling for population structure (P > 0.12). Among samples from the native range (EA), we also observed a significant longitudinal cline for inbred line rosette leaf number (Full Model B: –0.42 ± 0.19, P= 0.044), suggesting that western lines that have functional FRI flower later and at a larger size than eastern populations.
We evaluated the contribution of climatic variation to the observed clines by incorporating climatic PCs into a multiple regression, with PCs for each climate variable estimated separately. PCs analyses generated one PC for mean October–April temperature (PC1: eigenvalue 6.9, 98.3% variation explained), one PC for mean diurnal temperature range (PC1: 6.5, 92.2%) and two PCs for mean total precipitation (PC1: 5.3, 76.4%; PC2: 1.2, 17%). Inclusion of PC1 for temperature or PCs for precipitation either reduced or eliminated the significance of longitude on days to flowering (Table 3), suggesting that either of these climatic factors or factors closely correlated with them account for the longitudinal cline in flowering time. Although inclusion of a temperature PC reduced the significance of the longitude effect (P= 0.051), the parameter estimate is largely unchanged (0.075 ± 0.038 vs. 0.082 ± 0.03 for the fully reduced model). In contrast, inclusion of precipitation PCs not only eliminated the longitude effect (P= 0.91), but also dramatically reduced the parameter estimate (0.007 ± 0.06). Inclusion of PC1 for diurnal temperature range had no effect (Table 3). Taken together, these results suggest that geographic variation in flowering time across the introduced range is driven by longitudinal variation in climate, with precipitation (or variables highly correlated with it) showing the strongest statistical evidence as the mechanism.
Relative effects of site of origin, climate, and flowering time genes on phenotypes
In the full, genetic model (Model D), all traits displayed significant associations with FRI or FLC genotypes, or their interaction (Table 4). Compared to lines with the common, FRI–wt genotype, FRI–null lines produced more fruit (back-transformed lsmeans ± SE: FRI–wt, 306.9 fruit/plant, SE range +35.74, –32.02; FRI–null, 512.9 fruit/plant, +96.56, –81.26), flowered later, and at a larger size (lsmeans ± SE; days to flowering: FRI–wt, 207.5 ± 0.34 days; FRI–null, 208.8 ± 0.53 days, rosette diameter: FRI–wt, 2.96 ± 0.11 cm; FRI–null, 3.5 ± 0.17 cm, rosette leaves: FRI–wt, 28.9 ±0.96 leaves; FRI–null, 32.8 ± 1.50 leaves), and had higher survival (lsmean ± SE; proportion of five replicates surviving to reproduction: FRI–wt, 2.83 ± 0.21; FRI–null, 3.86 ± 0.33). A significant association with FLC haplotype was apparent only for days to flowering, and revealed that the uncommon FLC–B haplotype flowered on average one to two days later than the common FLC–A haplotype (lsmeans ± SE: FLC–A, 207.4 ± 0.37 days, FLC–B, 208.9 ± 0.53 days).
Table 4.
Association mapping for genetic covariance with flowering time traits and fitness for North American inbred line means controlling for variation in geographic origin, genotype, and population structure.
| Trait | Latitude | Longitude | FRI | FLC | FRI×FLC | Ancestry coefficient | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F | P | F | P | F | P | F | P | F | P | F | P | ||||
| Fruit1 | 2.38 | 0.125 | 0.48 | 0.49 | 6.38 | 0.013 | 1.22 | 0.27 | 0.00 | 0.99 | 8.10 | 0.005 | |||
| Days to bolting | 0.20 | 0.65 | 2.84 | 0.094 | 0.74 | 0.39 | 2.85 | 0.093 | 5.31 | 0.022 | 4.61 | 0.033 | |||
| Days to flower | 0.19 | 0.66 | 3.47 | 0.064 | 4.25 | 0.041 | 5.14 | 0.025 | 3.96 | 0.048 | 2.80 | 0.096 | |||
| Rosette diameter | 0.70 | 0.40 | 1.16 | 0.28 | 6.99 | 0.009 | 0.04 | 0.84 | 0.35 | 0.55 | 23.05 | <0.0001 | |||
| Rosette leaves | 0.19 | 0.66 | 0.21 | 0.64 | 5.22 | 0.024 | 0.44 | 0.51 | 1.40 | 0.24 | 12.14 | 0.0006 | |||
| Survival | 2.92 | 0.089 | 0.00 | 0.99 | 7.10 | 0.0085 | 1.47 | 0.23 | 0.86 | 0.36 | 15.21 | <0.0001 | |||
Log10 (mean lifetime fruit production). Significance at α= 0.05 is bolded, at α= 0.10 in italics.
We also detected an epistatic association of FRI and FLC on flowering phenotypes. For both days to bolting and days to flowering, the interaction was associated with delayed phenology in lines with the FRI–null/FLC–B genotype compared to other genotypes (days to bolting, days to flowering: FRI–wt/FLC–A 197.2 ± 0.16, 207.4 ± 0.14; FRI–wt/FLC–B 196.7 ± 0.77, 207.6 ± 0.67; FRI–null/FLC–A 196.1 ± 0.84, 207.4 ± 0.73; FRI–null/FLC–B 199.1 ± 0.92, 210.2 ± 0.80).
Two aspects of the full, genetic model stand out. First, although the effects of genotype on flowering phenology range in strength from strong (e.g., effects of FRI on most traits, Table 4) to marginally significant (e.g., FLC on days to bolting P= 0.093; Table 4), the FRI×FLC interaction term should be interpreted with caution. Our common garden sample contained few lines that were not FRI–wt or FLC–A (6.4% and 6.9%, respectively), hence it contained few lines with genotypes other than FRI–wt/FLC–A (N for other genotype combinations, 6 FRI–null/FLC–A; 7 FRI–wt/FLC–B; 5 FRI–null/FLC–B). Second, in the full genetic model the effects of longitude on days to bolting and days to flowering were only marginally significant (P= 0.094, and P= 0.064, respectively; Table 4), although both were significant in the reduced, geographic models (Models A and B; all P < 0.020). However, parameter estimates for the longitude terms were similar between all three models (Model A, B vs. D parameter estimate ± SE for longitude: days to bolting A: +0.094 ± 0.034, B: +0.084 ± 0.034, vs. D: +0.061 ± 0.035; days to flowering A: +0.082 ± 0.029, B: +0.071 ± 0.030 vs. D: +0.058 ± 0.031). In addition, removing the FRI×FLC term from the full model restores the significance of the longitude effect on both days to bolting (P= 0.03) and days to flowering (P= 0.022). Finally, in an analysis restricted to lines with FRI–wt/FLC–A (N= 154) containing latitude, longitude, and ancestry coefficients, we still detect evidence of significant and marginally significant flowering time clines (longitude: days to bolting +0.095 ± 0.036, P= 0.009; days to flowering +0.061 ± 0.033, P= 0.069).
Discussion
Introduced model species offer the potential to study convergent evolution, distinguish between adaptive and nonadaptive explanations for geographic differentiation, and evaluate underlying genetic mechanisms. Three major results emerge from our study of the evolution and genetics of naturalized populations of A. thaliana: (1) introduction and geographic expansion has been accompanied by dramatic shifts in population genetic structure and allele frequencies at major flowering time genes, (2) abundant quantitative genetic variation (within populations) and longitudinal differentiation exists in the introduced range, despite reduced variation at three known, major-effect flowering time genes; and (3) strong effects of FRI are observed on several quantitative traits. We discuss these findings below.
Variation in population structure and allele frequencies between regions
Both our STRUCTURE analysis using SNPs and assessments of allele frequencies at functional loci suggest dramatic genetic differences between introduced and native populations of A. thaliana. For example, we detected fewer clusters in North America (K= 2; Fig. 3) than have previously been observed using EA samples (K= 5–8 for primarily EA samples: Nordborg et al. 2005; Schmid et al. 2006; Fig. S2). Given bottlenecks, founder effects, and past reports of reduced variation in North America (Jorgensen and Mauricio 2004; Nordborg et al. 2005; Platt et al. 2010), these results are unsurprising. The structure plots of both EA and NA when analyzed together (Figs. S1 and S2) indicate substantial differences in patterns of population structure.
It is likely that many sources of the colonization of North America were not included in our sample, therefore increasing the probability that a rare set of source populations were also not sampled (although other explanations still remain possible). Three lines of evidence support this view. First, when we analyzed NA and EA samples together without supplying geographic information, we obtained K= 3, with the predominant pattern that EA samples are almost exclusively cluster 2 (K2). In contrast, NA samples appear to be either cluster 1, cluster 3, or admixed combinations (i.e., K1–K2 or K2–K3; Fig S1). These data indicate qualitative and quantitative differences in clustering between North America and Eurasia. Second, any structure among EA samples disappears almost entirely when analyzed with NA samples (Fig. S1), suggesting that differences between regions are appreciably more pronounced than differences within EA samples alone. Finally, when NA samples are forced to cluster with EA samples, clusters that predominate in Eurasia are less frequent in North America, and vice-versa (Fig. S2).
Given differences in population genetic structure, how should the differences between regions in allele frequencies at FRI, FLC, and PHYC be interpreted? As these genes affect quantitative traits expected to be under selection, it is likely that natural selection has contributed, at least in part. However, while natural selection may drive changes in allele frequencies within ranges, it seems more likely that the dramatic differences between regions are due to stochastic effects. Functional loci such as FRI, FLC, and PHYC should be subject to the same evolutionary and demographic forces that produced the pervasive, genome-wide differences in population structure indicated by the structure plots. Moreover, comparisons of allele frequencies at 136 SNPs between the native and introduced range suggest that numerous loci throughout the genome exhibit large frequency differences (Fig. S3), again suggesting the prominent role of stochastic forces accompanying introduction.
Parallel clines and ecological mechanisms
Our common garden experiment detected statistically significant longitudinal clines in flowering time, both in NA and EA samples, although the clines for the NA samples are weaker with considerable variation present within populations. These data further strengthen past interpretations that adaptive clines in A. thaliana are associated with regional climatic forces. Samis et al. (2008) suggested that longitudinal clines among EA Arabidopsis lines were likely due to adaptation to seasonal winter conditions, leading to changes in critical photoperiods. Three additional findings from the present experiment support their interpretation: (1) we again detected longitudinal clines in EA samples, using a smaller subset of accessions and under seminatural garden conditions in Toronto (Fig. 5A), (2) we detected a weak, but statistically significant longitudinal cline in flowering time in NA samples that was mirrored in the EA samples (Fig. 5B), and (3) when we used multiple regression to explain flowering time in the introduced range, winter precipitation (or highly correlated variables) emerged as the strongest explanation of the NA cline (Tables 3 and 4), with weaker contributions from temperature. Winter temperature and precipitation (or highly correlated variables) also appeared to explain the EA cline (K. E. Samis and J. R. Stinchcombe, unpubl. data). In general, the stochastic forces and bottlenecks associated with the introduction history, along with the changes in allele frequency at SNPs and flowering time genes (see above), suggest that clines like the ones we have described here (with a great deal of scatter) should perhaps be the expectation for A. thaliana in North America.
Identifying adaptive or parallel clines may be complicated by the potential problems of nonoverlapping latitudinal ranges (Colautti et al. 2009) and cryptic population structure associated with neutral processes (Keller and Taylor 2008). Neither factor appears to influence the flowering time clines we detected. First, the primary axis of differentiation we detected was longitudinal rather than latitudinal (and we included latitude as a covariate in all models). Second, our full models accounted for population genetic structure, and suggested that both neutral and adaptive processes contributed to geographic variation in flowering time. In contrast, any latitudinal clines we detected disappeared when ancestry coefficients were considered, suggesting largely neutral or demographic mechanisms (also see Keller et al. 2009).
One noteworthy aspect of our results is that we failed to detect latitudinal clines in flowering time, in contrast to past reports (Caicedo et al. 2004; Stinchcombe et al. 2004; Lempe et al. 2005; Brachi et al. 2010; Li et al. 2010). Our relatively sparse latitudinal sampling could have contributed, as past studies have shown that sample composition can affect power to detect clines and genetic associations (Samis et al. 2008). It is also likely that climatic differences among experiments also contributed. Although our study, Stinchcombe et al. (2004), and Caicedo et al. (2004) were each carried out under seminatural conditions, the climate differs markedly between locations (and growth chamber programs used in other studies). For example, prolonged, subzero winter temperatures and snow-thaw cycles were more common in Toronto during this experiment than past studies (Samis and Stinchcombe, unpub. data). The timing and duration of seasonal cues can be extremely important in determining genetic associations (Brachi et al. 2010) and life-history transitions (even in an isogenic background, Wilczek et al. 2009). As such, our failure to find latitudinal clines in flowering time under different conditions may be environment specific.
Phenotypic associations with major effect loci
Two important results emerge from our modified structured association analyses. First, the observed longitudinal cline for flowering time in North America is not accounted for by the effects of FRI, FLC, or their epistatic interaction (Table 4). Although we observed some evidence of differing geographic centers for different allele classes (Fig. 2), these differences were insufficient to account for the observed clines. These data suggest that the evolution of flowering time clines in North America has been achieved largely through other, unknown mutations. Although any linkage disequilibrium (LD) or haplotype sharing among NA samples (cf. Nordborg et al. 2005; Platt et al. 2010) may inhibit the resolution of association mapping, and make this conclusion conservative, we still detect geographic differentiation even after accounting for observed variation at FRI and FLC, and implicitly, other unknown loci in LD with them.
Second, we observed pervasive and dramatic effects of FRI (or genes in LD with FRI) on several quantitative traits (Table 4). Similar to previous studies (Caicedo et al. 2004; Korves et al. 2007; Scarcelli et al. 2007), we detected evidence of an epistatic interaction between FRI and FLC, although caution is warranted because of small sample sizes. The minor allele frequencies for many genotype combinations are quite low: the underrepresented combinations contain between 5 and 7 inbred lines, while the dominant genotype combination (FRI–wt/FLC-A) contained 181 inbred lines. Previous studies have shown that false-positive genetic associations increase in frequency when the minor allele frequency is < 10% (Bracchi et al. 2010). Second, the phenotypic effects of genetic associations (here FRI–null/FLC–B genotypes bolted 1.5–3 days later and flowered 2.6–2.8 days later than the others) are likely to be overestimated due to the Beavis effect.
Aside from the effects on phenology, the effects of FRI on other quantitative traits was more dramatic: plants with the FRI–null allele had larger rosette diameters, produced more rosette leaves at flowering, produced significantly more fruit, and had higher survival (Table 4). Despite these apparent performance and fitness advantages, FRI–null alleles are relatively rare in our sample. Three potential mechanisms could explain this discrepancy: (1) FRI–null alleles may indeed have a fitness advantage, but have not had sufficient time to increase in frequency since introduction; (2) while the seminatural conditions of our experiment may have lead to a fitness advantage of FRI–null alleles (also see Korves et al. 2007), FRIwt alleles may be favored elsewhere in the introduced range or seasonal environments; and (3) other unknown loci in LD with FRI led to the observed performance and fitness differences (which could also explain the later flowering of FRI–null alleles, which typically flower earlier). With the data in hand, it is not possible to distinguish these possibilities.
Conclusions
Our results point to three major conclusions: (1) Introduced NA samples of A. thaliana show qualitatively and quantitatively different patterns of genetic variation than EA samples—based on population structure (Fig. 3), allele frequencies at functional loci (Table 1), and covariation in quantitative traits (Fig. 4); (2) introduction to North America appears to have been accompanied by the evolution of weak but significant longitudinal clines in flowering time (Fig. 5), associated with geographic variation in winter precipitation, temperature, or closely correlated environmental variables (Table 3); and (3) variation at major flowering time genes FRI, FLC, and PHYC is insufficient to explain the cline, although variation in FRI (or factors in LD with it) appears to have dramatic effects on a host of quantitative traits. Several crucial next steps are necessary to confirm these results. First, reciprocal transplant experiments across the longitudinal range in North America are necessary, and manipulations of precipitation (either in the field or growth chambers) will be required to confirm or reject its role as a selective agent. Second, crosses and traditional QTL mapping approaches will still be necessary (see, e.g., Bracchi et al. 2010; Nemri et al. 2010) to confirm or reject the associations between FRI and associated quantitative traits. Careful selection of accessions should make it possible to evaluate the genetic basis of geographic differentiation in the introduced range.
Acknowledgments
We thank NSERC Canada and CFI for funding and B. Hall, D. Tam, and A. Petrie for logistical support. B. Harrett, A. J. Stock, H. van Tol, and K. Wang helped to collect data. We thank J. Borevitz and Y. Li for SNP genotyping assistance and manuscript comments. Comments by A. Buerkle, L. Delph, and four anonymous reviewers improved the manuscript.
Footnotes
We use “parallel” in the geometric sense, without implication about genetic mechanisms.
Support information
Figure S1. Haplotype cluster analysis (K= 3) of Arabidopsis thaliana lines from North America and Euarasia conducted without geographic origin information for lines.
Figure S2. Haplotype cluster analysis (K= 5) of Arabidopsis thaliana lines from North America clustered into ancestral populations based on Eurasian haplotypes.
Figure S3. Changes in allele frequency between the native and introduced range for 136 SNPs.
Table S1. Origins and genotypes of Arabidopsisthaliana lines used in the genotyping experiment.
Table S2. Primer descriptions used for genotyping NorthAmerican A. thaliana for variation in three loci previously demonstrated to be associated with flowering time.
Table S3. The frequency of genotypes across threefunctional loci (n = 213) for lines genotyped at all three loci.
References
- Allard RW, Babbel GR, Clegg MT, Kahler AL. Evidence for coadaptation in Avena barbata. Proc. Natl. Acad. Sci. U.S.A. 1972;69:3043–3048. doi: 10.1073/pnas.69.10.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson AR, Hoffmann AA, McKechnie SW, Umina PA, Weeks AR. The latitudinal cline in the In(3R)Payne inversion polymorphism has shifted in the last 20 years in Australian Drosophila melanogaster populations. Mol. Ecol. 2005;14:851–858. doi: 10.1111/j.1365-294X.2005.02445.x. [DOI] [PubMed] [Google Scholar]
- Arendt J, Reznick D. Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends Ecol. Evol. 2008;23:26–32. doi: 10.1016/j.tree.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Baird TD, Henson CA, Daily NM, Baccari GV, Murren CJ. Differential patterns of plasticity to water availability along native and naturalized latitudinal gradients. Evol. Ecol. Res. 2011;13:55–73. [Google Scholar]
- Balanya J, Oller JM, Huey RB, Gilchrist GW, Serra L. Global genetic change tracks global climate warming in Drosophila subobscura. Science. 2006;313:1773–1775. doi: 10.1126/science.1131002. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Agrawal M, Michael TP, Wessinger C, Maloof JN, Clark R, Warthmann N, Chory J, Weigel D. The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat. Genet. 2006;38:711–715. doi: 10.1038/ng1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker JSF, Frydenberg J, Sarup P, Loeschcke V. Altitudinal and seasonal variation in microsatellite allele frequencies of Drosophila buzzatii. J. Evol. Biol. 2011;24:430–439. doi: 10.1111/j.1420-9101.2010.02180.x. [DOI] [PubMed] [Google Scholar]
- Baxter I, Brazelton JN, Yu D, Huang YS, Lahner B, Yakubova E, Li Y, et al. A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genet. 2010;6:e1001193. doi: 10.1371/journal.pgen.1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry A, Kreitman M. Molecular analysis of an allozyme cline: alcohol dehydrogenase in Drosophila melanogaster on the east coast of North America. Genetics. 1993;134:869–893. doi: 10.1093/genetics/134.3.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Yant L, Laitinen RA, Kim S-T, Hollister JD, Warthmann N, Fitz J, et al. Local-scale patterns of genetic variability, outcrossing, and spatial structure in natural stands of Arabidopsis thaliana. PLoS Genet. 2010;6:e1000890. doi: 10.1371/journal.pgen.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowsky R. Restoring sight in blind cavefish. Curr. Biol. 2008;18:R23–R24. doi: 10.1016/j.cub.2007.11.023. [DOI] [PubMed] [Google Scholar]
- Brachi B, Faure N, Horton M, Flahauw E, Vazquez A, Nordborg M, Bergelson J, Cuguen J, Roux F. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet. 2010;6:e1000940. doi: 10.1371/journal.pgen.1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Caicedo AL, Stinchcombe JR, Olsen KM, Schmitt J, Purugganan MD. Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15670–15675. doi: 10.1073/pnas.0406232101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calboli FCF, Kennington WJ, Partridge L. QTL mapping reveals a striking coincidence in the positions of genomic regions associated with adaptive variation in body size in parallel clines of Drosophila melanogaster on different continents. Evolution. 2003;57:2653–2658. doi: 10.1111/j.0014-3820.2003.tb01509.x. [DOI] [PubMed] [Google Scholar]
- Cassell DL. Proceedings of the twenty-seventh annual SAS users group international conference (SUGI 27) Cary, NC: SAS Institute; 2002. A randomized-test wrapper for SAS PROCs; pp. 1–4. [Google Scholar]
- Colautti RI, Maron JL, Barrett SCH. Common garden comparisons of native and introduced plant populations: latitudinal clines can obscure evolutionary inferences. Evol. Appl. 2009;2:187–199. doi: 10.1111/j.1752-4571.2008.00053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais DL, Rausher MD. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature. 2008;454:762–765. doi: 10.1038/nature07092. [DOI] [PubMed] [Google Scholar]
- Des Marais DL, Rausher MD. Parallel evolution at multiple levels in the origin of hummingbird pollinated flowers in Ipomoea. Evolution. 2010;64:2044–2054. doi: 10.1111/j.1558-5646.2010.00972.x. [DOI] [PubMed] [Google Scholar]
- Endler JA. Geographic variation, speciation, and clines. Princeton, NJ, USA: Princeton Univ. Press; 1977. [PubMed] [Google Scholar]
- Etterson JR, Delf DE, Craig TP, Ando Y, Ohgushi T. Parallel patterns of clinal variation in Solidago altissima in its native range in central USA and its invasive range in Japan. Botany-Botanique. 2008;86:91–97. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Fenster CB, Galloway LF. Population differentiation in an annual legume: genetic architecture. Evolution. 2000;54:1157–1172. doi: 10.1111/j.0014-3820.2000.tb00551.x. [DOI] [PubMed] [Google Scholar]
- Fournier-Level A, Korte A, Cooper MD, Nordborg M, Schmitt J, Wilczek AM. A map of local adaptation in Arabidopsis thaliana. Science. 2011;334:86–89. doi: 10.1126/science.1209271. [DOI] [PubMed] [Google Scholar]
- Gilchrist GW, Huey RB, Serra L. Rapid evolution of wing size clines in Drosophila subobscura. Genetica. 2001;112:273–286. [PubMed] [Google Scholar]
- Gillespie JH. The causes of molecular evolution. New York: Oxford Univ. Press; 1991. [Google Scholar]
- Gonzalez J, Karasov TL, Messer PW, Petrov DA. Genome-wide patterns of adaptation to temperate environments associated with transposable elements in Drosophila. PLoS Genet. 2010;6:e1000905. doi: 10.1371/journal.pgen.1000905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ. The paradox of the first tier: an agenda for paleobiology. Paleobiology. 1985;11:2–12. [Google Scholar]
- Gould SJ, Lewontin RC. The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist program. Proc. R. Soc. Lond. B. 1979;205:581–598. doi: 10.1098/rspb.1979.0086. [DOI] [PubMed] [Google Scholar]
- Hadfield JD, Wilson AJ, Garant D, Sheldon BC, Kruuk LEB. The misuse of BLUP in ecology and evolution. Am. Nat. 2010;175:116–125. doi: 10.1086/648604. [DOI] [PubMed] [Google Scholar]
- Hancock AM, Brachi B, Faure N, Horton MW, Jarymowycz LB, Sperone FG, Toomajian C, et al. Adaptation to climate across the Arabidopsis thaliana genome. Science. 2011;334:83–86. doi: 10.1126/science.1209244. [DOI] [PubMed] [Google Scholar]
- Heath KD. Intergenomic epistasis and coevolutionary constraints in plants and rhizobia. Evolution. 2010;64:1446–1458. doi: 10.1111/j.1558-5646.2009.00913.x. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity. 2006;97:222–234. doi: 10.1038/sj.hdy.6800861. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Nachman MW. Different genes underlie adaptive melanism in different populations of rock pocket mice. Mol. Ecol. 2003;12:1185–1194. doi: 10.1046/j.1365-294x.2003.01788.x. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science. 2006;313:101–104. doi: 10.1126/science.1126121. [DOI] [PubMed] [Google Scholar]
- Holliday JA, Ritland K, Aitken SN. Widespread, ecologically relevant genetic markers developed from association mapping of climate-related traits in Sitka spruce (Picea sitchensis. New Phytol. 2010;188:501–514. doi: 10.1111/j.1469-8137.2010.03380.x. [DOI] [PubMed] [Google Scholar]
- Huey RB, Gilchrist GW, Carlson ML, Berrigan D, Serra L. Rapid evolution of a geographic cline in size in an introduced fly. Science. 2000;287:308–309. doi: 10.1126/science.287.5451.308. [DOI] [PubMed] [Google Scholar]
- Jeffery WR, Strickler AG, Yamamoto Y. To see or not to see: evolution of eye degeneration in Mexican blind cavefish. Integr. Comp. Biol. 2003;43:531–541. doi: 10.1093/icb/43.4.531. [DOI] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science. 2000;290:344–347. doi: 10.1126/science.290.5490.344. [DOI] [PubMed] [Google Scholar]
- Johnston RF, Selander RK. House sparrows: rapid evolution of races in North America. Science. 1964;144:548–550. doi: 10.1126/science.144.3618.548. [DOI] [PubMed] [Google Scholar]
- Johnston RF, Selander RK. Evolution in the house sparrow. II. Adaptive differentiation in North American populations. Evolution. 1971;25:1–28. doi: 10.1111/j.1558-5646.1971.tb01855.x. [DOI] [PubMed] [Google Scholar]
- Jorgensen S, Mauricio R. Neutral genetic variation among wild North American populations of the weedy plant Arabidopsis thaliana is not geographically structured. Mol. Ecol. 2004;13:3403–3413. doi: 10.1111/j.1365-294X.2004.02329.x. [DOI] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller SR, Taylor DR. History, chance and adaptation during biological invasion: separating stochastic phenotypic evolution from response to selection. Ecol. Lett. 2008;11:852–866. doi: 10.1111/j.1461-0248.2008.01188.x. [DOI] [PubMed] [Google Scholar]
- Keller SR, Sowell DR, Neiman M, Wolfe LM, Taylor DR. Adaptation and colonization history affect the evolution of clines in two introduced species. New Phytol. 2009;183:678–690. doi: 10.1111/j.1469-8137.2009.02892.x. [DOI] [PubMed] [Google Scholar]
- Kliber A, Eckert CG. Interaction between founder effect and selection during biological invasion in an aquatic plant. Evolution. 2005;59:1900–1913. [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge, U.K: Cambridge Univ. Press; 1983. [Google Scholar]
- Kolbe JJ, Larson A, Losos JB. Differential admixture shapes morphological variation among invasive populations of the lizard Anolis sagrei. Mol. Ecol. 2007;16:1579–1591. doi: 10.1111/j.1365-294X.2006.03135.x. [DOI] [PubMed] [Google Scholar]
- Kollmann J, Banuelos MJ. Latitudinal trends in growth and phenology of the invasive alien plant Impatiens glandulifera (Balsaminaceae) Divers. Distrib. 2004;10:377–385. [Google Scholar]
- Korves TM, Schmid KJ, Caicedo AL, Mays C, Stinchcombe JR, Purugganan MD, Schmitt J. Fitness effects associated with the major flowering time gene FRIGIDA in Arabidopsis thaliana in the field. Am. Nat. 2007;169:E141–E157. doi: 10.1086/513111. [DOI] [PubMed] [Google Scholar]
- Lacey EP. Latitudinal variation in reproductive timing of a short-lived monocarp, Daucus carota (Apiaceae) Ecology. 1988;69:220–232. [Google Scholar]
- Leger EA, Rice KJ. Assessing the speed and predictability of local adaptation in invasive California poppies (Eschscholzia californica. J. Evol. Biol. 2007;20:1090–1103. doi: 10.1111/j.1420-9101.2006.01292.x. [DOI] [PubMed] [Google Scholar]
- Lempe J, Balasubramanian S, Sureshkumar S, Singh A, Schmid M, Weigel D, Doebley J. Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 2005;1:e6. doi: 10.1371/journal.pgen.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Suzuki JI, Hara T. Latitudinal variation in plant size and relative growth rate in Arabidopsis thaliana. Oecologia. 1998;115:293–301. doi: 10.1007/s004420050519. [DOI] [PubMed] [Google Scholar]
- Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO. Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 2010;107:21199–21204. doi: 10.1073/pnas.1007431107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonn M, Prentice HC, Bengtsson K. Genetic structure, allozyme habitat associations and reproductive fitness in Gypsophila fastigiata (Caryophyllaceae) Oecologia. 1996;106:308–316. doi: 10.1007/BF00334558. [DOI] [PubMed] [Google Scholar]
- Losos JB, Jackman TR, Larson A, de Queiroz K, Rodriguez-Schettino L. Contingency and determinism in replicated adaptive radiations of island lizards. Science. 1998;279:2115–2118. doi: 10.1126/science.279.5359.2115. [DOI] [PubMed] [Google Scholar]
- Ma X-F, Hall D, Onge KRS, Jansson S, Ingvarsson PK. Genetic differentiation, clinal variation and phenotypic associations with growth cessation across the Populus tremula photoperiodic pathway. Genetics. 2010;186:1033–1044. doi: 10.1534/genetics.110.120873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloof JN, Borevitz JO, Dabi T, Lutes J, Nehring RB, Redfern JL, Trainer GT, et al. Natural variation in light sensitivity of Arabidopsis. Nat. Genet. 2001;29:441–446. doi: 10.1038/ng777. [DOI] [PubMed] [Google Scholar]
- Maron JL, Vila M, Bommarco R, Elmendorf S, Beardsley P. Rapid evolution of an invasive plant. Ecol. Monogr. 2004;74:261–280. [Google Scholar]
- Maron JL, Elmendorf SC, Vila M. Contrasting plant physiological adaptation to climate in the native and introduced range of Hypericum perforatum. Evolution. 2007;61:1912–1924. doi: 10.1111/j.1558-5646.2007.00153.x. [DOI] [PubMed] [Google Scholar]
- Mayr E. How to carry out the adaptationist program? Am. Nat. 1983;121:324–334. [Google Scholar]
- McKechnie SW, Blacket MJ, Song SV, Rako L, Carroll X, Johnson TK, Jensen LT, Lee SF, Wee CW, Hoffmann AA. A clinally varying promoter polymorphism associated with adaptive variation in wing size in Drosophila. Mol. Ecol. 2010;19:775–784. doi: 10.1111/j.1365-294X.2009.04509.x. [DOI] [PubMed] [Google Scholar]
- Michael TP, Salome PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, Alonso JM, et al. Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science. 2003;302:1049–1053. doi: 10.1126/science.1082971. [DOI] [PubMed] [Google Scholar]
- Michaels SD, He Y, Scortecci KC, Amasino RM. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:10102–10107. doi: 10.1073/pnas.1531467100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell TD, Jones PD. An improved method of constructing a database of monthly climate observations and associated high-resolution grids. Int. J. Climatol. 2005;25:693–712. [Google Scholar]
- Mitton JB, Linhart YB, Hamrick JL, Beckman JS. Observations on the genetic structure and mating system of Ponderosa pine in the Colorado Front Range. Theor. Appl. Genet. 1977;51:5–13. doi: 10.1007/BF00306055. [DOI] [PubMed] [Google Scholar]
- Montague JL, Barrett SCH, Eckert CG. Re-establishment of clinal variation in flowering time among introduced populations of purple loosestrife (Lythrum salicaria, Lythraceae) J. Evol. Biol. 2007;21:234–245. doi: 10.1111/j.1420-9101.2007.01456.x. [DOI] [PubMed] [Google Scholar]
- Montesinos-Navarro A, Wig J, Xavier Pico F, Tonsor SJ. Arabidopsis thaliana populations show clinal variation in a climatic gradient associated with altitude. New Phytol. 2011;189:282–294. doi: 10.1111/j.1469-8137.2010.03479.x. [DOI] [PubMed] [Google Scholar]
- Mullen LM, Vignieri SN, Gore JA, Hoekstra HE. Adaptive basis of geographic variation: genetic, phenotypic and environmental differences among beach mouse populations. Proc. R. Soc. B. 2009;276:3809–3818. doi: 10.1098/rspb.2009.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgia C, Pritchard JK, Kim SY, Fassati A, Weiss RA. Clonal origin and evolution of a transmissible cancer. Cell. 2006;126:477–487. doi: 10.1016/j.cell.2006.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemri A, Atwell S, Tarone AM, Huang YS, Zhao K, Studholme DJ, Nordborg M, Jones JDG. Genome-wide survey of Arabidopsis natural variation in downy mildew resistance using combined association and linkage mapping. Proc. Natl. Acad. Sci. 2010;107:10302–10307. doi: 10.1073/pnas.0913160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:1289–1299. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakeshott JG, Gibson JB, Anderson PR, Knibb WR, Anderson DG, Chambers GK. Alcohol dehydrogenase and glycerol-3-phosphate dehydrogenase clines in Drosophila melanogaster on different continents. Evolution. 1982;36:86–96. doi: 10.1111/j.1558-5646.1982.tb05013.x. [DOI] [PubMed] [Google Scholar]
- Ostrowski M-F, David J, Santoni S, McKhann H, Reboud X, Le Corre V, Camilleri C, et al. Evidence for a large-scale population structure among accessions of Arabidopsis thaliana: possible causes and consequences for the distribution of linkage disequilibrium. Mol. Ecol. 2006;15:1507–1517. doi: 10.1111/j.1365-294X.2006.02865.x. [DOI] [PubMed] [Google Scholar]
- Paaby AB, Blacket MJ, Hoffmann AA, Schmidt PS. Identification of a candidate adaptive polymorphism for Drosophila life history by parallel independent clines on two continents. Mol. Ecol. 2010;19:760–774. doi: 10.1111/j.1365-294X.2009.04508.x. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigen analysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A, Horton M, Huang YS, Li Y, Anastasio AE, Mulyati NW, Agren J, Bossdorf O, Byers D, Donohue K, et al. The scale of population structure in Arabidopsis thaliana. PLoS Genet. 2010;6:e1000843. doi: 10.1371/journal.pgen.1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protas ME, Hersey C, Kochanek D, Zhou Y, Wilkens H, Jeffery WR, Zon LI, et al. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat. Genet. 2006;38:107–111. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- Quinn GP, Keough MJ. Experimental design and data analysis for biologists. New York: Cambridge Univ. Press; 2002. [Google Scholar]
- Rand DM, Weinreich DM, Lerman D, Folk D, Gilchrist GW. Three selections are better than one: clinal variation of thermal QTL from independent selection experiments in Drosophila. Evolution. 2010;64:2921–2934. doi: 10.1111/j.1558-5646.2010.01039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum EB, Rompler H, Schoneberg T, Hoekstra HE. Molecular and functional basis of phenotypic convergence in white lizards at White Sands. Proc. Natl. Acad. Sci. U.S.A. 2010;107:2113–2117. doi: 10.1073/pnas.0911042107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A, Baxter I, Muthukumar B, Gustin J, Lahner B, Yakubova E, Salt DE. Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet. 2006;2:1964–1973. doi: 10.1371/journal.pgen.0020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter MT, Fenster CB. Testing for adaptation to climate in Arabidopsis thaliana: a calibrated common garden approach. Ann. Bot. 2007;99:529–536. doi: 10.1093/aob/mcl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samis KE, Heath KD, Stinchcombe JR. Discordant longitudinal clines in flowering time and Phytochrome C in Arabidopsis thaliana. Evolution. 2008;62:2971–2983. doi: 10.1111/j.1558-5646.2008.00484.x. [DOI] [PubMed] [Google Scholar]
- Scarcelli N, Cheverud JM, Schaal BA, Kover PX. Antagonistic pleiotropic effects reduce the potential adaptive value of the FRIGIDA locus. Proc. Natl. Acad. Sci. U.S.A. 2007;104:16986–16991. doi: 10.1073/pnas.0708209104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid K, Törjék O, Meyer R, Schmuths H, Hoffmann M, Altmann T. Evidence for a large-scale population structure of Arabidopsis thaliana from genome-wide single nucleotide polymorphism markers. Theor. Appl. Genet. 2006;112:1104–1114. doi: 10.1007/s00122-006-0212-7. [DOI] [PubMed] [Google Scholar]
- Schmidt PS, Rand DM. Intertidal microhabitat and selection at Mpi: interlocus contrasts in the northern acorn barnacle, Semibalanus balanoides. Evolution. 1999;53:135–146. doi: 10.1111/j.1558-5646.1999.tb05339.x. [DOI] [PubMed] [Google Scholar]
- Schmidt PS, Rand DM. Adaptive maintenance of genetic polymorphism in an intertidal barnacle: habitat- and life-stage-specific survivorship of Mpi genotypes. Evolution. 2001;55:1336–1344. doi: 10.1111/j.0014-3820.2001.tb00656.x. [DOI] [PubMed] [Google Scholar]
- Schmidt PS, Bertness MD, Rand DM. Environmental heterogeneity and balancing selection in the acorn barnacle Semibalanus balanoides. Proc. R. Soc. Lond. Ser. B. 2000;267:379–384. doi: 10.1098/rspb.2000.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmuths H, Hoffmann MH, Bachmann K. Geographic distribution and recombination of genomic fragments on the short arm of chromosome 2 of Arabidopsis thaliana. Plant Biol. 2004;6:128–139. doi: 10.1055/s-2004-817837. [DOI] [PubMed] [Google Scholar]
- Simões P, Santos J, Fragata I, Mueller LD, Rose MR, Matos M. How repeatable is adaptive evolution? The role of geographical origin and founder effects in laboratory adaptation. Evolution. 2008;62:1817–1829. doi: 10.1111/j.1558-5646.2008.00423.x. [DOI] [PubMed] [Google Scholar]
- Steiner CC, Rompler H, Boettger LM, Schoneberg T, Hoekstra HE. The genetic basis of phenotypic convergence in beach mice: similar pigment patterns but different genes. Mol. Biol. Evol. 2009;26:35–45. doi: 10.1093/molbev/msn218. [DOI] [PubMed] [Google Scholar]
- Stenøien HK, Fenster CB, Kuittinen H, Savolainen O. Quantifying latitudinal clines to light responses in natural populations of Arabidopsis thaliana (Brassicaceae) Am. J. Bot. 2002;89:1604–1608. doi: 10.3732/ajb.89.10.1604. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JR, Weinig C, Ungerer M, Olsen KM, Mays C, Halldorsdottir SS, Purugganan MD, Schmitt J. A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4712–4717. doi: 10.1073/pnas.0306401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe JR, Caicedo AL, Hopkins R, Mays C, Boyd EW, Purugganan MD, Schmitt J. Vernalization sensitivity in Arabidopsis thaliana (Brassicaceae): the effects of latitude and FLC variation. Am. J. Bot. 2005;92:1701–1707. doi: 10.3732/ajb.92.10.1701. [DOI] [PubMed] [Google Scholar]
- Storz JF, Dubach JM. Natural selection drives altitudinal divergence at the albumin locus in deer mice, Peromyscus maniculatus. Evolution. 2004;58:1342–1352. doi: 10.1111/j.0014-3820.2004.tb01712.x. [DOI] [PubMed] [Google Scholar]
- Storz JF, Sabatino SJ, Hoffmann FG, Gering EJ, Moriyama H, Ferrand N, Monteiro B, et al. The molecular basis of high-altitude adaptation in deer mice. PLoS Genet. 2007;3:e45. doi: 10.1371/journal.pgen.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travisano M, Mongold JA, Bennett AF, Lenski RE. Experimental tests of the roles of adaptation, chance, and history in evolution. Science. 1995;267:87–90. doi: 10.1126/science.7809610. [DOI] [PubMed] [Google Scholar]
- Umina PA, Weeks AR, Kearney MR, McKechnie SW, Hoffmann AA. A rapid shift in a classic clinal pattern in Drosophila reflecting climate change. Science. 2005;308:691–693. doi: 10.1126/science.1109523. [DOI] [PubMed] [Google Scholar]
- Van Rossum F, Vekemans X, Meerts P, Gratia E, Lefebvre C. Allozyme variation in relation to ecotypic differentiation and population size in marginal populations of Silene nutans. Heredity. 1997;78:552–560. [Google Scholar]
- Vasemägi A. The adaptive hypothesis of clinal variation revisited: single-locus clines as a result of spatially restricted gene flow. Genetics. 2006;173:2411–2414. doi: 10.1534/genetics.106.059881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake DB. Homoplasy: the result of natural selection, or evidence of design limitations? Am. Nat. 1991;138:543–567. [Google Scholar]
- Weber E, Schmid B. Latitudinal population differentiation in two species of Solidago (Asteraceae) introduced into Europe. Am. J. Bot. 1998;85:1110–1121. [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. Arabidopsis: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- Werner JD, Borevitz JO, Uhlenhaut NH, Ecker JR, Chory J, Weigel D. FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics. 2005;170:1197–1207. doi: 10.1534/genetics.104.036533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerman JM, Lawrence MJ. Genotype-environment interaction and developmental regulation in Arabidopsis thaliana. I. Inbred lines; description. Heredity. 1970;25:609–627. [Google Scholar]
- Wilczek AM, Roe JL, Knapp MC, Cooper MD, Lopez-Gallego C, Martin LJ, Muir CD, Sim S, Walker A, Anderson J, et al. Effects of genetic perturbation on seasonal life history plasticity. Science. 2009;323:930–934. doi: 10.1126/science.1165826. [DOI] [PubMed] [Google Scholar]
- Zhao K, Aranzana MJ, Kim S, Lister C, Shindo C, Tang C, Toomajian C, Zheng H, Dean C, Marjoram P, et al. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 2007;3:e4. doi: 10.1371/journal.pgen.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y, Ungerer MC. Clinal variation in freezing tolerance among natural accessions of Arabidopsis thaliana. New Phytol. 2008;177:419–427. doi: 10.1111/j.1469-8137.2007.02262.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


