Abstract
The major intention of the present study was to investigate whether an approach combining the use of niche-based palaeodistribution modeling and phylo-geography would support or modify hypotheses about the Quaternary distributional history derived from phylogeographic methods alone. Our study system comprised two closely related species of Alpine Primula. We used species distribution models based on the extant distribution of the species and last glacial maximum (LGM) climate models to predict the distribution of the two species during the LGM. Phylogeographic data were generated using amplified fragment length polymorphisms (AFLPs). In Primula hirsuta, models of past distribution and phylogeographic data are partly congruent and support the hypothesis of widespread nunatak survival in the Central Alps. Species distribution models (SDMs) allowed us to differentiate between alpine regions that harbor potential nunatak areas and regions that have been colonized from other areas. SDMs revealed that diversity is a good indicator for nunataks, while rarity is a good indicator for peripheral relict populations that were not source for the recolonization of the inner Alps. In P. daonensis, palaeo-distribution models and phylogeographic data are incongruent. Besides the uncertainty inherent to this type of modeling approach (e.g., relatively coarse 1-km grain size), disagreement of models and data may partly be caused by shifts of ecological niche in both species. Nevertheless, we demonstrate that the combination of palaeo-distribution modeling with phylogeographical approaches provides a more differentiated picture of the distributional history of species and partly supports (P. hirsuta) and partly modifies (P. daonensis and P. hirsuta) hypotheses of Quaternary distributional history. Some of the refugial area indicated by palaeodistribution models could not have been identified with phylogeographic data.
Keywords: AFLP, niche modeling, palaeodistribution, Quaternary
Introduction
Fossil evidence suggests that the climatic oscillations of the Quaternary strongly influenced the geographical distribution of plants in Europe (Lang 1994). The retreat of species to peripheral unglaciated refuges during glacial advances finds support from the fossil record, floristic and biogeographic evidence (Stehlik 2000), patterns of endemism (Tribsch and Schönswetter 2003; Tribsch 2004), palaeoenvironmental data on glacial cover during the last glacial maximum (LGM), and the location of nunataks (Schönswetter et al. 2005). Analyses of DNA sequence and fingerprint data have modified ideas about the location of these refugia. For example, “cryptic” northern refugia have been identified for various plant and animal taxa in the northern hemisphere. These northern refugia likely complemented southern refugia in allowing species survival (Stewart and Lister 2001; Bhagwat and Willis 2008; Birks and Willis 2008; Provan and Bennett 2008), and the persistence of several species in northern refugia likely reduced the importance of Mediterranean refugia for the recolonization of northern parts of Europe (beech: Magri et al. 2006; birch: Maliouchenko et al. 2007).
Two candidate hypotheses explain how alpine plant species survived glacial periods. The “tabula rasa” hypothesis postulates that plants survived outside the Alpine ice shield and recolonized vacant areas subsequent to glacial retreat (Brockmann-Jerosch and Brockmann-Jerosch 1926). Alternatively, the nunatak hypothesis holds that alpine plants survived glacial periods in ice-free localities above the glaciers (Stehlik 2000). Neither of these hypotheses find support from fossil evidence owing to the paucity of fossil pollen for most high-altitude plants of the European Alps. However, locations of glacial refugia have been proposed based on molecular phylogeographic analyses and other evidence. Phylogeographic studies support one or both scenarios for survival during glacial periods (nunatak or nunatak and peripheral survival: Ravazzi 1997; Fürchter et al. 2001; Stehlik et al. 2001; 2002; Stehlik 2002; Schönswetter et al. 2003b; Parisod and Besnard 2007; survival in peripheral refugia: Schönswetter et al. 2002, 2003a; 2004; Tribsch et al. 2002; Naciri and Gaudeul 2007; Paun et al. 2008). These studies demonstrate that phylogeographic methods have substantially improved our understanding of Quaternary plant biogeography.
Phylogeographic methods have, however, one inherent weakness: candidate refugia outside the present species range can rarely be identified. This requires subfossil DNA or other material to provide direct evidence of the paleodistribution of a species (Willerslev et al. 2007). While the availability of such DNA is unusual in the Alpine environment, the limitations of the phylogeographic approach could potentially be overcome through the use of species distribution models (SDMs; Guisan and Zimmermann 2000). SDMs quantify the relationship between species occurrences and geographic variation in the environment to describe the realized environmental niche of a species (sensu Hutchinson 1957; see Araujo and Guisan 2006). The quantified niche can then be projected across a geographic area to map suitable environmental conditions for a species and predict its potential distribution. Assuming that the species niche has remained unchanged over the time period of interest (Peterson et al. 1999), the palaeodistribution of species can be reconstructed by projecting SDMs to earlier time periods using palaeo-climate data. Diverse studies have combined SDMs and genetic data (e.g., Carstens and Richards 2007; Knowles et al. 2007; Richards et al. 2007; Jakob et al. 2009; Moussalli et al. 2009), for example, to assess past refugia for a snail in tropical Australia (Hugall et al. 2002), infer genetic diversity hotspots in the Atlantic forest of Brazil (Carnaval et al. 2009), and determine the historical distribution of dwarf willow in Europe (Alsos et al. 2009). To our knowledge, no study of the Quaternary history of high-mountain plants restricted to the European Alps has used SDMs to infer the glacial distribution of a species. In this paper, we assess the usefulness of this approach to infer the potential paleodistributions of species in an alpine environment.
Our study system comprises two species, Primula hirsuta and P. daonensis. These parapatrically distributed species of section Auricula are suitable for investigating glacial survival and comparing recolonization patterns. The species are closely related, predominantly silicicolous and grow on rocks, in rock crevices or in stony pastures at altitudes between 1500 and 3000 m for P. daonensis and 1400 (rarely 500 m) to 2800 m for P. hirsuta (Zhang and Kadereit 2004). As concluded from molecular clock dating, they presumably originated in isolated glacial refugia of the Quaternary (Kadereit et al. 2004; Zhang et al. 2004; Crema 2009). Recent studies indicated that the two species are not sister species as originally thought (Zhang and Kadereit 2004) but probably originated simultaneously along with several other species during an early cold phase of the Quaternary (Crema et al. unpubl. data).
The two species differ markedly in their current ranges. Primula hirsuta is widespread in the Alps (and also occurs in the Pyrenees), which may imply survival on nunataks. In contrast to this, P. daonensis is restricted to a few mountain ranges close to the periphery of the southern Central Alps. This suggests that the species survived in refuges that were peripheral to the alpine ice sheet. Here, we combine SDMs with phylogeographic methods to infer glacial refugia and to reconstruct patterns of postglacial recolonization of these two species. Our aim is twofold. First, we want to investigate whether these two approaches lead to congruent results or not. Second, we want to explore how the addition of SDM analyses supports and/or modifies hypotheses derived from phylogeographic data. We use species distribution models based on the extant distribution of the two species and LGM climate models to predict their distribution during the LGM 21,000 years ago. We compare findings from SDM models to patterns in population genetic data that were derived from the analysis of amplified fragment length polymorphisms (AFLPs) data. AFLP markers were chosen because sequence variation and phylogenetic resolution is generally low in Primula sect. Auricula, and few or no nucleotide substitutions are found between the study species (Zhang and Kadereit 2004 and unpubl. data from ETS and several cpDNA markers). We find that SDMs provide additional information that helps to clarify the Quaternary distribution history and survival of these alpine plants.
Material and Methods
Species distribution modeling
Climate data
We used current climate data with a spatial resolution of 30″ (ca. 1 km) from the WorldClim database (Hijmans et al. 2005). These climate data include monthly temperature and precipitation variables describing annual trends, seasonal variability and extreme, and represent potentially limiting environmental factors found to be meaningful to describe plant and animal species niches in several studies (Broennimann et al. 2007; Pearson et al. 2007; Waltari et al. 2007; Pearman et al. 2008b). The same variables for LGM climate data were drawn from general circulation model (GCM) simulations from two climate models: the Community Climate System Model (CCSM, version 3; Collins et al. 2006) and the Model for Interdisciplinary Research on Climate (MIROC, version 3.2; Hasumi and Emori 2004). The downscaled climate surfaces at 2.5′ (ca. 5 km) spatial resolution were provided by Robert Hijmans and derived from the original climate simulations as described in Waltari et al. (2007). These climate models differ in temperature and precipitation patterns. LGM climate as simulated by CCSM3 is colder and dryer than that of MIROC over Europe (study area [3.5–17°E, 43–49°N]: mean annual temperature: –16.5°C to 12.3°C for CCSM3 and –10.8°C to 14.7°C for MIROC; annual precipitation: 522–2720 mm/year for CCSM3 and 473–3016 mm/year for MIROC). Use of two different climate models enabled us to assess and account for modeling uncertainty due to LGM climate data.
In modeling species distributions, we used eight bio-climatic variables that are considered physiologically important for plants (Körner 1999) and for which all pairwise correlations were smaller than 0.8. These variables were: annual mean temperature (tam), mean diurnal temperature range (tmdr), temperature seasonality (ts), mean temperature of wettest quarter (tmwq), mean temperature of driest quarter (tmdq), annual precipitation (pa), precipitation seasonality (ps), and precipitation of warmest quarter (pwq).
Species distribution models and projection to the LGM
Several recent studies advise the combination of results from multiple modeling techniques (“ensemble forecasting”; Thuiller 2004; Pearson et al. 2006; Araujo and New 2007; Pearson et al. 2007) to account for between-techniques uncertainty in predictions. This seems particularly appealing when models are used to project the distribution of species to novel situations (Thuiller 2004), for example, where species distributions are projected into the LGM as is done here. An ensemble approach is beneficial because SDM techniques can differ in their ability to predict species distributions under different climates and, thus, yield different projected distributions. Such behavior makes the choice of one single modeling technique difficult if not arbitrary. Consensus models highlight areas that are predicted by several modeling techniques, making prediction maps more reliable.
We applied four very common SDM techniques, already used in previous alpine studies (e.g., Dubuis et al. 2011; Engler et al. 2011), using the latest release of the BIOMOD tool (Thuiller et al. 2009) implemented in R (R Development Core Team 2005): generalized linear models (GLM), generalized additive models (GAM), generalized boosting models (GBM), and random forests (RF). These techniques need presence–absence data for the species. Occurrences were gathered by combining several data sources (Ruffray et al. 2000–2009, unpubl. data; “ZDSF” [Zentrum des Datenverbundnetzes der Schweizer Flora, Altenbergrain 21, CH-3013 Bern, Switzerland. http://www.crsf.ch/], unpubl. data; A. Selvaggi, unpubl. data; F. Prosser, unpubl. data, and herbarium specimens from M, MSB, Z, ZT, RUEB, MJG), resulting in 257 localities for P. hirsuta and 168 for P. daonensis. As no absences could be inferred from these data, we created 5000 pseudoabsences that were randomly distributed over the entire study range as recommended in Elith et al. (2011) and weighted to equal the number of presences per species. Continuous model predictions ranging from 0 to 1 were transformed into binary predictions using a Receiver Operating Characteristics (ROC)-plot optimized cut-off threshold (Liu et al. 2005). Binary predictions of the four techniques were cumulated. As an ensemble forecast can also include different datasets (Araujo and New 2007) in addition to multiple SDM techniques, we created consensus maps from the predictions derived with the two LGM climate models. Our final distribution maps indicate those areas predicted by most modeling techniques under both estimates of LGM climate (i.e., five or more coincident models).
Evaluating predictions
The predictive performance of the models was evaluated by repeating 100 times a split-sample cross-validation, each time using a random subset (70%) of the total dataset to calibrate the models while the remaining 30% were used to evaluate the model. Three different evaluation measures were calculated, that is, the area under the curve (AUC) of an ROC-plot, the true skill statistic (TSS), and the Kappa statistic.
Molecular methods
Sampling
Twenty-one populations of P. hirsuta and eight populations of P. daonensis were sampled (10–15 individuals per population) in 2005 and 2006, covering nearly the entire range of the two species (Table 1; Figs. 1A and 2B). Individuals sampled had a minimum distance of 3 m to avoid repeated collection of the same clone. Where this was not possible due to small population size, AFLP fingerprints were compared to exclude identical fingerprints. When possible, sampled individuals were evenly distributed across the entire population extent. In very large populations, sampling was limited to an extent comparable to most other populations. Voucher specimens were deposited in the herbarium of the Institut für Spezielle Botanik, Mainz (MJG).
Table 1.
Locations of the sampled populations of Primula daonensis and P. hirsuta. For localities, where several “populations” were sampled at a minimum distance of a few 100 m (locality code is underlined), the number of sampled individuals per subsample is given in brackets and differing altitudes of the subsamples are specified by small letters.
| Code | Locality, country | Altitude (m) | Lat/long | No. of individuals |
|---|---|---|---|---|
| P. hirsuta | ||||
| ARC | Arcegno, Switzerland | 450 | 8.744/46.158 | 11 |
| BER | Berninapass, Switzerland | 2250 | 10.005/46.415 | 12 |
| FLU | Flüelapass, Switzerland | ca. 2400 | 9.933/46.767 | 12 |
| GAL | Galzig, Austria | 2180 | 10.227/47.132 | 9 |
| GAU | Gauertal, Austria | ca. 2200 | 9.818/47.056 | 20 (11 + 9) |
| GRA | Grächen, Mattertal, Switzerland | 2100 | 7.868/46.208 | 9 |
| GRI | Grigna settentrionale, Italy | 1930 | 9.384/45.963 | 11 |
| GRU | Grütschalp, Lauterbrunnental, Switzerland | 2050 | 7.948/46.579 | 11 |
| IND | Indemini, Italy | 980 | 8.816/46.095 | 8 |
| LAU | Col du Lautaret, France | 2050–2100 | 6.415/45.031 | 12 |
| MAL | Malojapass, Switzerland | 1840 | 9.694/46.399 | 9 |
| OBE | Oberalbpass, Switzerland | ca. 1850 | 8.686/46.650 | 12 |
| OLA | Ref. d’Olan, Valgaudemar, France | ca. 1690 (a), 2320–2400 (b) | 6.204/44.842 | 9 (1 + 8) |
| SBE | San Bernadino, Switzerland | 1804 | 9.197/46.471 | 10 |
| SEL | Selva, Switzerland | ca. 1540 | 8.714/46.655 | 10 |
| SUL | Suldental, Italy | ca. 2360–2450 | 10.575/46.520 | 22 (9 + 10 + 3) |
| TIR1 | Neustift im Stubaital, Austria | 1870 | 11.185/47.092 | 9 |
| TIR2 | Vals im Valsertal, Austria | 1440 | 11.585/47.043 | 9 |
| TIR3 | Gries im Venntal, Austria | 1530 | 11.536/47.013 | 7 |
| VAL | Mte Nudo in Valcuvia, Italy | 1080 | 8.674/45.925 | 10 |
| ZEI | Zeinisjoch, Austria | ca. 1850 | 10.127/46.978 | 12 |
| P. daonensis | ||||
| VIV | Passo Vivione, Italy | ca. 1900 | 10.201/46.035 | 24 (12 + 12) |
| GAV | Passo Gavia, Italy | 2570 (a), ca. 2700 (b) | 10.485/46.339 | 13 (8 + 5) |
| STE | Passo Stelvio, Italy | 2560 | 10.439/46.535 | 13 |
| MUR | Alp Muraunza, Switzerland | ca. 2200 | 10.433/46.558 | 24 (5 + 4 + 6 + 9) |
| SCO | Monte Scorluzzo, Italy | 3050 | 10.442/46.521 | 8 |
| GOL | Goletto di Crocette, Italy | 2111 | 10.376/45.859 | 12 |
| LCA | Lago Casinei, Italy | 2090 | 10.513/45.947 | 9 |
| PED | Cascada Pedruc, Val Genova, Italy | 1530 | 10.623/46.196 | 10 |
Figure 1.
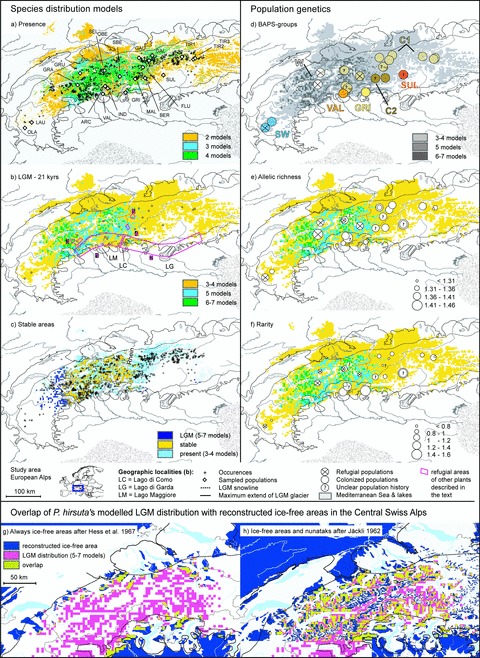
SDMs from consensus modeling (A, present; B, LGM [21 kyrs]; C, stable areas) for Primula hirsuta and population structure (D), allelic richness (E), and rarity (F) overlaid onto LGM distribution map (B); overlap of modeled LGM distribution of P. hirsuta in the Central Alps with always ice-free areas redrawn from Hess et al. (1967) (G) and ice-free areas and nunataks redrawn from Jäckli (1962) (H). For population assignment to BAPS groups and exact values of the population genetic indices, see Tables 1 and 3. Classification of populations as refugial or colonized is based on the comparison of present and past consensus models (Table 3). Species occurrences and sampled populations are shown in (A). Pink encircled areas in (B) are refugial areas of other plant species as discussed in the text; location of potential peripheral refugia between Valle d’Aoste and Lago di Como (1) and Lago di Como and the Dolomiti (2) as described by Schönswetter et al. (2005) and location of floristically rich nunatak areas in the high mountains of Monte Rosa and in the valleys of Visp (3), in Avers (4), and the Rothorn mountains near Arosa (5) as described by Stehlik (2000) are indicated by numbers.
Figure 2.
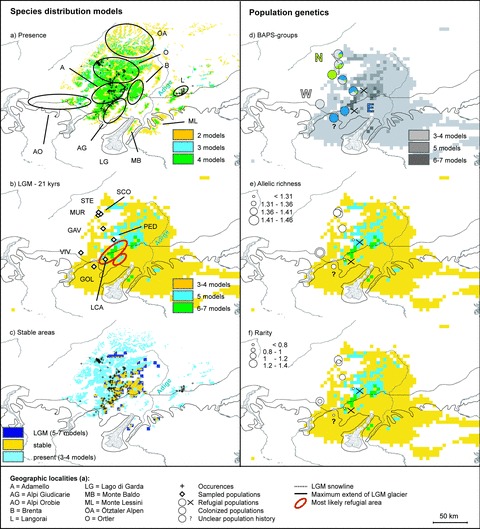
SDMs from consensus modeling (A, present; B, LGM [21 kyrs]; C, stable areas) for P. daonensis and population structure (D), allelic richness (E), and rarity (F) overlaid onto LGM distribution map (B). For population assignment to BAPS groups and exact values of the population genetic indices see Tables 1 and 3. Classification of populations as refugial or colonized is based on the comparison of present and past consensus models (Table 3). Species occurrences are shown in (A), sampled populations in (B). Red encircled areas in (B) are the most likely refugia (see Discussion).
DNA extraction
Total genomic DNA was extracted from desiccated leaf material (ca. 20 mg dry weight) using the QIAGEN DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the standard protocol with the modification that 10 mM sodium metadisulfide was added to the AP1-buffer. DNA was eluted two times using 40 μL TAE-buffer.
AFLP protocol
The AFLP protocol followed Vos et al. (1995) with the modification described in Kropf et al. (2003) except that incubation time was 2 h at 37°C and 8 h at 15°C, 1.56 μL 50 mM MgCl2, and 0.33 μL 20 mM dNTPs were used in the preselective polymerase chain reaction (PCR) where elongation time was 3 min at 72°C.
Primers used in the preselective PCR were E01 (5′-GACTGCGTACCAATTCA-3′) and M01 (5′-GATGAGTCCTGAGTAAC-3′), and in the selective PCRs E38-Hex (5′Hex-GACTGCGTACCAATTCACT-3′) combined with M52 (5′-GATGAGTCCTGAGTAACCC-3′), E38–6-Fam with M55 (5′-GATGAGTCCTGAGTAACGA-3′), and E39-NED (5′NED-GACTGCGTACCAATTCAGA-3′) with M55 (by Metabion International, Martinsried, Germany, except E39-NED by ABI, Life Technologies, Darmstadt, Germany). The three AFLP reactions were multiplexed and purified using Sephadex (Millipore, Schwalbach, Germany). AFLP fragments were separated using an ABI 3730 DNA Analyzer and the ROX500 size standard (ABI).
Population genetic analyses
Scoring of AFLP fragments was done with GeneMarker Version 1.75 (GeneMarker, SoftGenetics, LLC, State College, PA, U.S.A.). Mismatch error rates were calculated to evaluate the quality of our analyses as described in Paun et al. (2008). Genetic variation was quantified using several diversity measures. Allelic richness Pb (ranging from 1 to 2) was calculated with AFLPDiv1.0 (Coart et al. 2005). For populations with population sizes that were smaller than the number of rarefactions used, the mean number of alleles per locus is given instead. Nei's gene diversity H (Nei 1973; also referred to as expected heterozygosity), Shannon's information index I (Shannon and Weaver 1949; Lewontin 1972), and the percentage of polymorphic loci PPL were calculated with Popgene 3.2 (Yeh and Boyle 1997) assuming Hardy–Weinberg equilibrium. Rare markers were previously found to correlate with refugia locations (Comps et al. 2001; Widmer and Lexer 2001; Paun et al. 2008). Therefore, we calculated a rarity index as described in Paun et al. (2008). Within-species genetic structure was analyzed using “Bayesian Analysis of Population Structure” (BAPS, version. 5.1; Corander et al. 2008). The procedure was run 10 times each for K= 2–10 as the assumed maximum number of populations present in the sample. Admixture analyses (Corander and Marttinen 2006) were run with 100 iterations to estimate admixture coefficients for individuals, 200 reference individuals from each population, and 20 iterations to estimate admixture co-efficients for reference individuals. To detect admixture between species, the analyses were run with the combined dataset with two predefined groups corresponding to the species. Analyses of molecular variance (AMOVA) were conducted using Arlequin version. 3.1 (Excoffier et al. 2005). Principal coordinate analyses (PCoA) using Dice (Sorensen) similarity were conducted with PAlaeontological STatistics (PAST, version.1.62; Ryan et al. 1995).
Evaluation of modeled LGM refugia using spatial patterns of genetic diversity, rarity, and the number of private fragments
Populations were classified into two types with respect to their population history as inferred from the SDMs: (1) refugial populations, located within LGM distribution areas and (2) colonized populations, in currently populated areas that are outside of refugia as modeled with SDMs. In order to identify regions that were colonized during the Holocene, we overlaid the present distribution maps with the estimated palaeo-distribution. This was done for each of the eight LGM models (four modeling techniques and two climates) for the two species separately. The resulting maps were then cumulated. For this, the resolution of the present distribution maps was transformed from a 1-km grid to a 5-km grid (the resolution of the palaeoclimatic maps) using the minimum criterion, that is, each new grid cell was considered occupied when at least one of the 25 component grid cells of a new grid cell was occupied by the species. This procedure probably resulted in an overestimation of postglacially colonized areas but ensured that no areas where the species was predicted as currently present were omitted due to change in resolution. This does not affect the number of models indicating refugial areas.
We classified extant populations as either refugial (occupying an LGM refuge) or colonized (founded subsequent to LGM) using the criterion established by the consensus map of SDM models for the LGM distribution of each species. We then compared refugial populations to colonized populations in terms of means of genetic diversity measures, allele rarity, and number of private alleles using randomization tests with 10,000 Monte Carlo runs (Poptools 3.0; Hood 2008). To ensure comparability between populations, only populations with similar numbers of sampled individuals were taken into account and sampling sites that were located a few 100 m apart from each other (e.g., GAUa, GAUb) were treated as separate populations. Populations with few individuals were omitted to reduce error.
Results
LGM distribution models
Under current climate conditions, AUC values indicated “good to excellent” model performance for both species across modeling techniques following Swets’ scale (Swets 1988; Table 2). For P. hirsuta, TSS indicated “good” to “high or excellent” performance, whereas Kappa indicated “fair” model performance, for P. daonensis TSS and Kappa indicated “high or excellent” model performance following Thuiller et al. (2008). The predictions for P. daonensis were generally better.
Table 2.
Evaluation of individual modeling techniques for the two species.
| AUC | Kappa | TSS | |
|---|---|---|---|
| P. hirsuta | |||
| GLM | 0.938 | 0.482 | 0.772 |
| GBM | 0.957 | 0.561 | 0.841 |
| GAM | 0.930 | 0.402 | 0.750 |
| RF | 0.963 | 0.582 | 0.855 |
| P. daonensis | |||
| GLM | 0.996 | 0.854 | 0.982 |
| GBM | 0.996 | 0.866 | 0.975 |
| GAM | 0.996 | 0.872 | 0.983 |
| RF | 0.997 | 0.877 | 0.980 |
Uncertainty in LGM distribution models due to the use of different climate models and modeling techniques can be recognized from the results from different individual models (one technique, one climate). In both species, LGM distributions using MIROC climate identified regions similar to present models, but often reduced in range; LGM distributions using the (colder) CCSM climate sometimes differed markedly from present models (data not shown). Despite this variation, the ensemble maps of both climate datasets identified similar regions. While LGM distribution using CCSM climate was highly variable between modeling techniques in both species, modeled LGM distribution of P. daonensis using MIROC climate data was very similar. LGM distribution of P. hirsuta using MIROC climate was more variable than of P. daonensis, but less than using CCSM climate and still overlapping.
Modeled glacial refugia of P. hirsuta and P. daonensis
The present distribution range of P. hirsuta was generally well recovered by the consensus models, except for the south-western part and some areas at the eastern distribution limit (Fig. 1A). Projection of the species niche to LGM climate predominantly indicated refugial areas within the major ice shield in the Central Swiss Alps (5–7 models) and in the neighboring southern Alps outside the snowline, that is, where snow melted during summer in climatically average years (5 models, Fig. 1B). In the Central Alps modeled LGM and present distributions overlap widely (stable areas, Fig. 1C), while the eastern part of the extant species range, east of a line from the river Rhein to Lago di Como, was postglacially colonized according to these models (Fig. 1C). Modeled LGM distribution in the Central Swiss Alps overlaps with “never glaciated areas” described by Hess et al. (1967) and ice-free areas and nunataks described by Jäckli (1962; Fig. 1G and H).
As in P. hirsuta, the present distribution range of P. daonensis was generally well recovered by the consensus models (Fig. 2A). The models were able to predict the region east of the Adige in Langorai (predominantly porphyry bedrock) even though no occurrences from this region were used to fit the models. The Brenta Massif (calcareous bedrock), which was indicated as an ecoclimatically suitable habitat, is, however, not populated by the species. Projecting the niche of the species to LGM climate predicted reductions in range size. Most of the models (6–7) predicted refugial areas in Brenta and the Alpi Giudicarie, both of which consist predominantly of calcareous rock, the eastern Adamello (siliceous rock), and small and discrete localities on Monte Baldo east of Lago di Garda (calcareous rock, Fig. 2B). Southwestern parts of the Ortler (siliceous rock) and southern parts of the Ötztaler Alpen (siliceous rock) as well as small discrete areas in Monte Lessini (calcareous rock) were predicted by at least half of the models (5–7). Combining present and past consensus models defines postglacially colonized areas north, west, and east of the refugial areas (Fig. 2C).
AFLP patterns
A total of 133 polymorphic fragments were scored un-ambiguously in P. hirsuta and P. daonensis. Nonpolymorphic markers and rare markers that were only found in less then four samples (a rather high threshold to exclude artifacts) were omitted from the analysis. One hundred thirty fragments were found in P. hirsuta (21 populations, 234 individuals) and 97 in P. daonensis (eight populations, 113 individuals). Thirty-six (27.1%) of these fragments were private to P. hirsuta and only three (2.3%) were private to P. daonensis. The mismatch error was 4.5% comparing 150 replicate pairs. These included replicates between the three runs, between different steps within one run (restriction/ligation, preselective, and selective PCR), and 19 DNA replicates from both species. DNA replicates (of which 16 were replicated between runs) had an error rate of 5.9%.
Population structure of P. hirsuta and P. daonensis
Primula hirsuta is divided into seven groups with a BAPS probability for seven clusters of 1. A group in the south-western Alps (SW), a group in the western Swiss Alps (W), two central alpine groups (C1, C2), and three populations from the southern margin of the species distribution range, which form three distinct groups, namely VAL east of Lago Maggiore, GRI east of Lago di Como, and SUL in the Ortler (Fig. 1D; Table 3; see Figs. 1 and 2 for localities). Only populations from C1, C2, and W were mixed with other genetic groups. Three populations of C1 were mixed with C2 and one with W, and one of the two populations of W was mixed with C1. Of all individuals of P. hirsuta, only one in population SEL contained genetic ancestry from different genetic groups (C1, C2, and SW, Bayesian P-value = 0.005, admixture analysis, BAPS).
Table 3.
Type of population, BAPS groups, and genetic indices for the populations of Primula hirsuta and P. daonensis. Sampling sites that were located a few 100 m from each other are underlined; populations not used to evaluate the palaeodistribution models (also not shown in Figs. 1 and 2) are given in brackets. Pop-type = specifying whether consensus models indicate that the population is situated in colonized area (C) or in refugial area (R), the number of models indicating one or the other are given in brackets, “?” populations with equal numbers of models indicating a refugial or a colonized area; BAPS group, affiliation to genetic groups inferred with BAPS, different BAPS groups in mixed populations are separated by a slash, brackets indicate that only one individual of a population belongs to the BAPS group named, mixture and admixture analyses with BAPS were based on all individuals sampled for a population including subsamples; Size, number of individuals per population; No. frag., number of AFLP-fragments; h, Nei's gene diversity; I, Shannon index; NPL, number of polymorphic loci; PPL, percentage of polymorphic loci; Pb, band richness with rarefaction sample size six for P. hirsuta and seven for P. daonensis; NPA, number of private alleles; Adm, number of individuals admixed between species.
| Code | Pop-type (C/R) | BAPS group | Size | No. frag. | h | I | NPL | PPL | Pb | Rarity | NPA | Adm. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. hirsuta | ||||||||||||
| GAL | C (5/3) | C1 | 9 | 77 | 0.15 | 0.22 | 57 | 43.85 | 1.37 | 0.9 | 0 | |
| GAUa | C (5/3) | C1 | 11 | 89 | 0.15 | 0.23 | 66 | 50.77 | 1.4 | 0.95 | 0 | |
| GAUb | C (5/3) | C1 | 9 | 80 | 0.12 | 0.19 | 52 | 40 | 1.34 | 0.84 | 0 | |
| (GAUa+b) | C (5/3) | C1 | 20 | 96 | 0.00 | 0.24 | 75 | 57.69 | 1.39 | 0.80 | 0 | |
| OBE | ? (4/4) | C1 | 12 | 81 | 0.13 | 0.2 | 55 | 42.31 | 1.33 | 0.83 | 0 | |
| SBE | R (3/5) | C1 | 10 | 83 | 0.15 | 0.23 | 61 | 46.92 | 1.39 | 0.88 | 0 | |
| SEL | R (3/5) | C1 | 10 | 85 | 0.14 | 0.21 | 60 | 46.15 | 1.39 | 1.09 | 0 | |
| TIR2 | C (5/3) | C1 | 9 | 75 | 0.15 | 0.22 | 54 | 41.54 | 1.36 | 0.72 | 0 | |
| TIR3 | C (5/3) | C1 | 7 | 69 | 0.11 | 0.17 | 40 | 30.77 | 1.29 | 1.04 | 1 | |
| ZEI | C (5/3) | C1 | 12 | 81 | 0.15 | 0.23 | 65 | 50 | 1.38 | 0.72 | 0 | |
| FLU | ? (4/4) | C1/(C2) | 12 | 84 | 0.14 | 0.22 | 64 | 49.23 | 1.38 | 0.78 | 0 | |
| IND | ? (4/4) | C1/(C2) | 8 | 86 | 0.15 | 0.23 | 62 | 47.69 | 1.42 | 1.29 | 1 | |
| TIR1 | C (5/3) | C1/(W) | 9 | 81 | 0.14 | 0.22 | 57 | 43.85 | 1.38 | 0.89 | 0 | |
| ARC | C (5/3) | C1/C2 | 11 | 84 | 0.14 | 0.22 | 61 | 46.92 | 1.39 | 0.84 | 0 | |
| BER | C (5/3) | C2 | 12 | 85 | 0.14 | 0.21 | 62 | 47.69 | 1.38 | 0.87 | 0 | |
| MAL | ? (4/4) | C2 | 9 | 75 | 0.16 | 0.23 | 56 | 43.08 | 1.38 | 1.07 | 0 | |
| GRI | C (5/3) | GRI | 11 | 80 | 0.15 | 0.22 | 59 | 45.38 | 1.35 | 1.43 | 1 | 3 |
| SULa | ? (3/3) | SUL | 9 | 78 | 0.14 | 0.22 | 58 | 44.62 | 1.39 | 1.39 | 3 | |
| SULb | ? (3/3) | SUL | 10 | 89 | 0.17 | 0.26 | 71 | 54.62 | 1.46 | 1.53 | 3 | |
| (SULc) | ? (3/3) | SUL | 3 | 71 | 0.11 | 0.17 | 39 | 30.00 | 1.31 | 1.49 | 2 | |
| (SULa+b+c) | ? (3/3) | SUL | 22 | 97 | 0.19 | 0.29 | 84 | 64.62 | 1.46 | 1.46 | 4 | 18 |
| LAU | C (4/3) | SW | 12 | 73 | 0.15 | 0.22 | 57 | 43.85 | 1.34 | 0.78 | 0 | |
| (OLAa) | R (0/3) | SW | 1 | 46 | - | - | - | - | - | 1.13 | 0 | |
| OLAb | R (0/3) | SW | 8 | 72 | 0.15 | 0.23 | 55 | 42.31 | 1.38 | 0.95 | 0 | |
| (OLAa+b) | R (0/3) | SW | 9 | 72 | 0.15 | 0.23 | 55 | 42.31 | 1.37 | 0.97 | 0 | |
| VAL | C (5/3) | VAL | 10 | 76 | 0.13 | 0.2 | 50 | 38.46 | 1.34 | 1.38 | 2 | |
| GRA | R (2/6) | W | 9 | 80 | 0.17 | 0.25 | 64 | 49.23 | 1.44 | 1.14 | 0 | |
| GRU | R (3/5) | W/(C1) | 11 | 80 | 0.16 | 0.24 | 64 | 49.23 | 1.39 | 0.89 | 0 | |
| P. daonensis | ||||||||||||
| GOL | ? (4/4) | E | 12 | 60 | 0.12 | 0.19 | 35 | 36.08 | 1.32 | 0.79 | 0 | |
| LCA | R (2/6) | E | 9 | 58 | 0.11 | 0.17 | 31 | 31.96 | 1.3 | 0.79 | 1 | |
| PED | R (3/5) | E/W/(N) | 10 | 62 | 0.12 | 0.17 | 34 | 35.05 | 1.31 | 0.71 | 0 | |
| SCO | C (6/2) | N | 8 | 63 | 0.15 | 0.22 | 40 | 41.24 | 1.39 | 1.06 | 0 | |
| VIVa | C (6/2) | W | 12 | 75 | 0.18 | 0.27 | 54 | 55.67 | 1.43 | 1.28 | 2 | |
| VIVb | C (6/2) | W | 12 | 70 | 0.16 | 0.24 | 47 | 48.45 | 1.38 | 0.99 | 0 | |
| (VIVa+b) | C (6/2) | W | 24 | 81 | 0.19 | 0.29 | 65 | 67.01 | 1.42 | 1.13 | 2 | |
| GAVa | C (6/2) | W/E/(N) | 8 | 64 | 0.13 | 0.2 | 38 | 39.18 | 1.37 | 1.06 | 1 | |
| (GAVb) | C (6/2) | W/E/(N) | 5 | 61 | 0.12 | 0.18 | 30 | 30.93 | 1.311 | 1.07 | 0 | |
| (GAVa+b) | C (6/2) | W/E/(N) | 13 | 70 | 0.16 | 0.24 | 48 | 49.48 | 1.4 | 1.07 | 1 | |
| STE | C (6/2) | W/N/(E) | 13 | 70 | 0.17 | 0.26 | 48 | 49.48 | 1.41 | 0.96 | 0 | |
| (MURa) | C (6/2) | W/N/E | 5 | 65 | 0.11 | 0.17 | 32 | 32.99 | 1.331 | 1.06 | 1 | |
| (MURba) | C (6/2) | W/N/E | 4 | 66 | 0.13 | 0.20 | 36 | 37.11 | 1.371 | 1.14 | 0 | |
| (MURbb) | C (6/2) | W/N/E | 6 | 71 | 0.15 | 0.23 | 45 | 46.39 | 1.461 | 1.04 | 1 | |
| MURc | C (6/2) | W/N/E | 9 | 71 | 0.13 | 0.21 | 43 | 44.33 | 1.4 | 1.23 | 4 | |
| (MURa+ba+bb+c) | C (6/2) | W/N/E | 24 | 84 | 0.18 | 0.28 | 66 | 48.04 | 1.46 | 1.13 | 4 |
Calculated without rarefaction, instead the mean number of allels is given.
In P. daonensis, the BAPS analysis detected three genetic groups with a probability for number of clusters of 1. These three groups, a northern (N), a western (W), and an eastern group (E), are distributed over wide areas of the species’ distribution range and overlap geographically (Fig. 2D; Table 3, see Schorr [2009] for more detailed geographic descriptions). While the three southernmost populations (VIV, GOL, and LCAS) and SCO contain individuals of only one genetic group, the northern populations contain individuals of all three genetic groups (see Fig. 2B for locations of sampled populations). No individuals have heterogeneous genetic ancestry (data of admixture analyses not shown).
In P. hirsuta, 16.17% of the overall variation is attributed to variation among BAPS groups, 11.88% to variation among populations within BAPS groups, and 71.94% to variation within populations (Table 4). In the PCoA (Fig. 3), the BAPS groups GRI, VAL, SUL, and SW can be distinguished from C1, C2, and W that cluster together. In P. daonensis, genetic variation among regional groups is lower than in P. hirsuta. Hierarchical AMOVAs attributed 7.72% of the overall variation to variation among the three major mountain ranges where the species is distributed, 9.16% to variation among populations within the mountain ranges, and 83.12% to variation within populations (Table 4). This is also reflected by lack of structure in the PCoA (Fig. 3). As expected for outcrossing species, most variation is found within populations, 75.66% in P. hirsuta and 84.94% in P. daonensis, whereas 24.34% and 15.06%, respectively, were found among populations (Table 4).
Table 4.
Analysis of molecular variance (AMOVA) for AFLP phenotypes of 21 populations of Primula hirsuta and eight populations of P. daonensis using 10,000 permutations.
| Source of variation | df | Sum of squares | Variance components* | Percent of variation | FST* |
|---|---|---|---|---|---|
| P. hirsuta | |||||
| Among populations | 20 | 1018.47 | 3.59 | 24.34 | 0.24 |
| Within populations | 213 | 2375.17 | 11.15 | 75.66 | |
| Among BAPS groups (SW, W, C1, C2, VAL, GRI, SUL) | 6 | 590.17 | 2.51 | 16.17 | 0.28 |
| Among populations within BAPS groups | 14 | 28.30 | 1.84 | 11.88 | |
| Within populations | 213 | 2375.17 | 11.15 | 71.94 | |
| P. daonensis | |||||
| Among populations | 7 | 192.57 | 1.42 | 15.06 | 0.15 |
| Within populations | 105 | 839.36 | 7.99 | 84.94 | |
| Among mountain ranges (VIV-GOL, LCA, PED-GAV, SCO, STE, MUR) | 2 | 98.27 | 0.74 | 7.72 | 0.17 |
| Among populations within mountain ranges | 5 | 94.30 | 0.88 | 9.16 | |
| Within populations | 105 | 839.36 | 7.99 | 83.12 |
All P-values < 0.001.
Figure 3.
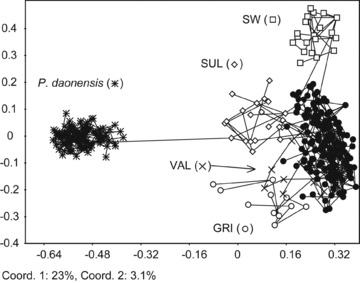
Principle coordinate analysis of Primula hirsuta and P. daonensis of 234 AFLP multilocus phenotypes from 21 populations and 113 AFLP multilocus phenotypes of eight populations, respectively. Symbols indicate the genetic groups obtained with BAPS for P. hirsuta (black circles = W, C1, C2), all P. daonensis individuals are marked identically. All individuals from P. hirsuta populations are assigned to the BAPS group to which most of the populations’ individuals belong. Lines indicate the minimum spanning tree. Axes are scaled using the square root of the eigenvalues.
Admixture between species
An admixture analysis using two predefined groups corresponding to the two species revealed significant admixture in two P. hirsuta populations, SUL (18 of 22 individuals were significantly admixed, P≤ 0.05) and GRI (three of 11 were significantly admixed, Table 3). In a PcoA, the first coordinate that explains 23% of the overall variation separated the two species (Fig. 3). Populations SUL and GRI of P. hirsuta have a somewhat intermediate position but are much closer to this species. A minimum spanning tree connects P. daonensis to SUL.
Evaluation of modeled refugia with population genetic patterns
In P. hirsuta refugial areas show significantly higher genetic diversities (expected heterozygosity, Shannon's information index, and allelic richness) than colonized areas (Figs. 1 and 4). In contrast, rarity is not significantly different between populations in refugial and colonized areas. Only five populations harbor private fragments and these did not originate from modeled refugial areas (Table 3). However, for two of them (SUL and IND, see Fig. 1A for locations) the consensus models did not allow us to discriminate between refugial and colonized populations.
Figure 4.
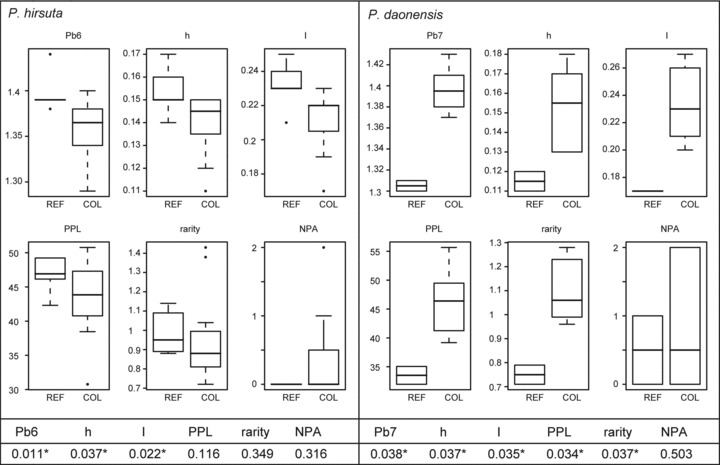
Boxplots of various genetic indices of Primula hirsuta and P.daonensis comparing populations situated in glacial refugia (“REF”: P. hirsuta: GRA, GRU, OLAb, SBE, SEL; P. daonensis: LCA, PED) and colonized areas (“COL”: P. hirsuta: ARC, BER, GAL, GAUa, GAUb, GRI, LAU, TIR1, TIR2, TIR3, ZEI, VAL; P. daonensis: GAVa, MURc, SCO, STE, VIVa, VIVb) as identified by distribution modeling. Pb, band richness with rarefaction sample size six for P. hirsuta and seven for P. daonensis; h, Nei's gene diversity; I, Shannon index; PPL, percentage of polymorphic loci; NPA, number of private alleles. Whiskers extend to the most extreme data point within the interquartile range from the box. P-values from one-tailed randomization tests using 10,000 Monte Carlo iterations comparing the two groups are given below the boxplots, significant differences are marked with an asterisk (*).
In P. daonensis, we found the opposite pattern for genetic diversity: populations modeled as colonized show significantly higher diversities (expected heterozygosity, Shannon's information index, allelic richness, and percentage of polymorphic markers) than populations in areas that were classified as refugial (Figs. 2 and 4). Also, rarity is significantly higher in colonized populations and they tend to harbor more private fragments.
Discussion
Uncertainties of hindcasting
Modeled refugial areas rely on how well climate models estimate the LGM climate in the Alps and on the accuracy of the SDM methodology. Several uncertainties are associated with the climate data. One major point is the reliability of global circulation models for the LGM climate in Europe. Earlier comparisons of model output to empirical climate data suggested that the LGM climate in Europe derived with global circulation models is generally warmer, especially in winter, than the climate reconstructed from pollen records, and it is believed that global circulation model reconstruction results are too wet. Differences were largest in western Europe and less pronounced in Central Europe and the Mediterranean basin (Kageyama et al. 2001; Pollard and Barron 2003; Jost et al. 2005). More recent studies, however, have shown that this discrepancy was mainly caused by an overestimation of LGM cooling by pollen-based climate reconstructions. This overestimation was caused by not accounting for the lower CO2 concentration during the LGM and its impact on vegetation—lower CO2 concentration decreased plant productivity and water-use efficiency probably resulting in a more open glacial vegetation than under similar climate today (Ramstein et al. 2007; Wu et al. 2007). Model simulated LGM temperatures are still warmer than the new pollen-based reconstructions, but the results from these two approaches are much closer to each other and within their error ranges.
In our data, uncertainty resulting from climate models and modeling techniques is evident from the different LGM species distributions obtained with single models. Despite this variation, the ensemble maps derived from the two climate datasets identified similar regions. This indicates that despite the differences between the two climate models we used, the ensemble models are robust with respect to the existing uncertainty in the climate data. Furthermore, our results show that using an ensemble approach rather than a single modeling technique to hindcaste species distributions may be particularly advantageous for species lacking a fossil record, since the comparison of different modeling techniques and climates allows partial assessment of hindcasting uncertainty.
Finally, high-resolution reconstructions of ice-free areas and nunataks for the entire study area that might be used to further refine LGM distributions were not available to us.
Primula hirsuta
Identification of glacial refugia with SDMs
Our palaeodistribution models suggested that refugia for P. hirsuta existed within the Central Alps and on peripheral nunataks at the southern Alpine margin. Several authors have found evidence for glacial survival of other plant species in areas that overlap with the refugia identified here (Fig. 1B, areas 1–5). The peripheral nunatak refuges predicted by our models are identical with “potential peripheral refugia” on siliceous bedrock identified by Schönswetter et al. (2005). These authors demonstrated that phylogeographic patterns of several alpine plant species confirmed the locations of their “potential peripheral refugia.” They concluded that the area between the Valle d’Aoste and Lago di Como (Fig. 1B, area 1) served as a refugium for Androsace alpina, and for some populations of A. brevis, Phyteuma globulariifolium, and Ranunculus glacialis. The area between Lago di Como and the Dolomit (Fig. 1B, area 2) was hypothesized as a refugium for A. alpina, and parts of A. brevis, A. wulfeniana, Eritrichium nanum, P. globulariifolium, R. glacialis, and Saponaria pumila. The central Alpine nunatak refugia that we identified for P. hirsuta partly overlap with three of five floristically rich nunatak areas proposed by Brockmann-Jerosch and Brockmann-Jerosch (1926) and reviewed and mapped by Stehlik (2000). Our species distribution models suggested that the glacial distribution of P. hirsuta included areas (1) around the high mountains of Monte Rosa and in the valleys of Visp (Fig. 1B, area 3), although more in the outer and lower parts of this area and not at the highest elevations, (2) in Avers (Fig. 1B, area 4), and (3) in the Rothorn mountains near Arosa (Fig. 1B, area 5). The models did not suggest glacial distribution of the species in (4) Simplon or in (5) very small and restricted areas of the high mountains of Engadin and Bernina, even though the species is frequent there today. The central Alpine refugium in the Monte Rosa mountains and the valleys of Simplon is also the location of a refugium for E. nanum, based on genetically distinct genotypes detected with AFLPs and cpDNA (Stehlik et al. 2001; 2002; Stehlik 2003). Based on results of the majority of SDMs, geological, palaeoenvironmental and floristic evidence, and other phylogeographic studies, P. hirsuta survived the LGM in central Alpine as well as in peripheral southern nunataks.
Congruence of approaches?
Are population genetic patterns found congruent with modeled scenarios of glacial survival of P. hirsuta? On the one hand, SDMs identified refugial areas within the inner Central Alps. The population genetic structure and diversity patterns we found in this area are congruent with what is expected for nunatak survival: restricted gene flow among refugial populations was found to result in well-defined refugial gene pools and high levels of regional diversity in refugial areas (Comes and Kadereit 1998; Petit et al. 2003; Hampe and Petit 2005). In case of nunatak survival, these gene pools should be distributed throughout the formerly glaciated inner Alps. In P. hirsuta, four genetic groups are situated within the formerly glaciated parts of the inner Alps, and variation among these groups is higher than variation among the populations within the groups. Additionally, all refugial populations in the inner Alps except SEL harbor high genetic diversity. On the other hand, rarity is low in these populations and hence seems to be a poor indicator for nunataks. Low rarity likely results from postglacial mixing of populations. Genetic mixture of nunatak populations with colonizing populations during recolonization of the inner Alps could have resulted in a loss of rare and private fragments. The elucidation of patterns in cpDNA data could strengthen evidence for nunatak survival and help to locate specific nunatak areas because rare cpDNA haplotypes that originated in nunatak areas are more resistant to being wiped out by gene flow from newly colonizing populations than are rare or private AFLP markers (Comes and Kadereit 1998; Stehlik 2003). In summary, the two approaches are largely congruent for the inner Alps and corroborate each other.
Consensus models support the argument that parts of the southern periphery of the Central Alps between Lago Maggiore and Lago di Como served as refugial areas. However, the four populations located there, GRI, VAL, ARC, and IND, are not situated within the predicted range but in close proximity to it. GRI and VAL are two genetically distinct populations. They harbor private alleles and have high rarity but low diversity (Fig. 1E and F; Table 3). High rarity and private alleles, as well as the location of these populations in refugial areas that are peripheral to the southern Alps (Schönswetter et al. 2005), and in one of the most important areas of endemism in the Eastern Alps (Tribsch 2004), imply that the regions where GRI and VAL are located were glacial refugia for these populations. Hence, our models did not predict the full extent of this refugial area. The refugial status of GRI and VAL is further supported by morphological divergence and geographic distance to the main distributional range of P. hirsuta; both populations have been described as endemic species or subspecies, that is, P. grignensis (Moser 1998) and P. hirsuta spp. valcuvianensis (Jeßen and Lehmann 2005). Re-colonization of the inner Alps by these populations is unlikely since they show no admixture with other groups. We regard them as relict populations and note that both populations grow on calcareous rather than siliceous rock, in contrast to other populations of the species. While high rarity and private alleles reflect the long-term isolation of these populations, low allelic diversity is at odds with their status as relict populations. This is probably a result of small population size and restricted gene flow within the small refugial areas.
Population genetic characteristics of the other two populations at the southern periphery of the Central Alps, ARC and IND, are different. High genetic diversity and high rarity, as well as one private allele in population IND, supports the refugial status of these populations (Fig. 1E and F; Table 3). Population ARC has an average genetic diversity but low rarity and no private fragments. All three P. hirsuta populations around Lago Maggiore (ARC, IND, VAL) occur at elevations lower than 1100 m (Table 1), which is low relative to most populations of the species. These patterns suggest that these populations are remnants of refugial populations. ARC and IND belong to genetic groups (C1, C2) that are mainly distributed in the inner Alps. This suggests that these populations, in contrast to VAL and GRI, were source populations for the recolonization of the inner Alps. Similar to the inner alpine refugial populations and in contrast to the relict populations VAL and GRI, these two peripheral refugial populations (ARC, IND) have rather high genetic diversity. Differential altitudinal distribution and partly different edaphic preferences indicate that a niche shift may have been associated with the glacial colonization of the refugial area at the southern periphery of the Central Alps between Lago Maggiore and Lago di Como. This could be a reason why the models fail to predict the full extent of the refugial area. If correct, this would illustrate that SDM output needs to be compared with additional data in order to produce robust inference of the locations of LGM refugia.
While large parts of the Central Alps were climatically stable (Fig. 1C), the SDM results suggest that P. hirsuta recolonized the eastern part of its current distribution range. However, many populations in the eastern Alps are also characterized by high genetic diversities. If the modeling results are correct, the high genetic diversity of populations in this area was caused by processes potentially unrelated to glacial survival. Extant factors that were not investigated here, such as large population sizes and/or high population densities and associated gene flow might explain the high genetic diversity we found in these eastern populations. A similar explanation has been suggested for the R. alpestris group (Paun et al. 2008). We can exclude admixture of divergent lineages from separate refugia (Petit et al. 2003; Heuertz et al. 2004; Schönswetter et al. 2004) as the cause of high diversity because most of the eastern populations belong to a single genetic group. The only exception is the distinct population (SUL) at the southeastern margin of the current distribution. It is highly diverse and contains high rarity (Fig. 1E and F; Table 3). Admixture analyses have revealed hybridization of SUL with P. daonensis (documented by herbarium specimens), which may explain the high divergence, diversity, and rarity in this population.
Primula daonensis
Identification of glacial refugia with SDMs
Palaeodistribution models indicated that refugial areas of P. daonensis were located east of the extant main distribution range and west of the Adige (see Fig. 2A for all localities). The three refugial areas predicted by most (6–7) of the SDM models—northern Alpi Giudicarie to southeastern Adamello, Brenta, and Monte Baldo—overlap with hypothetical refugial areas for silicicolous subnival, calcicolous subnival, and calcicolous upper alpine species, respectively, as reconstructed from geological and palaeoenvironmental data (Tribsch and Schönswetter 2003). The latter two predo-minantly calcareous mountain ranges (Brenta and Monte Baldo) seem unlikely to have served as refugia for P. daonensis as a predominantly silicicolous species. The species does grow in the predominantly calcareous Alpi Giudicarie, but always on granitic rock (Hegi 1966). Granitic and calcareous/dolomite rock alternate at a small scale in this area.
Areas from the northern Adamello to the Ötztaler Alpen and on Monte Lessini, which were predicted by 5 of 8 models, also are less likely refugial areas for the following reasons (Fig. 2B). Monte Lessini predominantly consists of calcareous rock, similar to Brenta and Monte Baldo. The elevation of the predicted area from the northern Adamello to the Ötztaler Alpen is more than 300 m above the LGM snowline (according to Tribsch and Schönswetter 2003). This elevation today represents the approximate upper limit of prospering populations of mountain plants in the central most part of the eastern Alps (Reisigl and Pitschmann 1958; Körner 1999; Tribsch and Schönswetter 2003). Based on model predictions as well as palaeoenvironmental and geological evidence, the area from the northern Alpi Giudicarie to the southeastern Adamello is the most likely refugium for P. daonensis during the LGM (encircled in red in Fig. 2B).
This area was climatically stable and consequently populations likely persisted, while the western and northern parts of the species extant distribution range were not suitable during the LGM and presumably were colonized in the Holocene. The area east of the Adige is a proposed glacial refugium for silicicolous upper alpine to subnival plants (southern Dolomiti, Alpi di Val di Fiemme; Tribsch and Schönswetter 2003), but SDMs suggest that P. daonensis colonized this area, located ca. 50 km east of the main distribution range (Fig. 2), in the Holocene.
Congruence of approaches?
The AFLP dataset of eight populations is small compared to the dataset for P. hirsuta, and the range of P. daonensis is also relatively small. Only two populations were located in modeled refugial areas and no populations were sampled from the isolated part of the range east of the Adige. In view of our small sample size from modeled refugial areas, we cannot exclude the possibility of having sampled genetically depleted populations from a refugial region with (undetected) high levels of genetic diversity. However, the fact that both populations show similarly low levels of genetic variation may indicate that our results do not represent sampling artifacts. Given the small range of the species, the phylogeographic dataset should be sufficient to infer glacial and postglacial history.
We identified a clear genetic pattern that is incongruent with the SDMs results. Based on theoretical and empirical studies, a decrease in genetic diversity from refugia toward newly colonized areas is expected, and such a pattern indicates the direction of migration (Hewitt 1996; Widmer and Lexer 2001; Stehlik et al. 2002; Tribsch et al. 2002). In contrast, populations of P. daonensis in the colonized area have high diversities and rarities and harbor private alleles, while populations in the refugial area as identified by SDM are genetically impoverished, have low numbers of rare alleles and only one of the populations has one private allele (Figs. 2E, F, and 4; Table 3).
Reasons for the incongruence in our results could be that refugial areas were incorrectly identified by SDMs because of a change of the ecological niche of P. daonensis (Pearman et al. 2008a) or due to some methodological limitations, such as the fact that the high habitat diversity of alpine environments may be insufficiently reflected in the 1 × 1 km resolution of the climate data used to build the models (see Trivedi et al. 2008; Randin et al. 2009) for example of scale mismatch of SDM projections in mountain areas) or additional nonclimatic variables being not taken into account (e.g., substrate). A change of ecological niche, caused by change in biotic interactions or dispersal limitations (see Pearman et al. 2008b), could be especially crucial for a species such as P. daonensis with a small distribution range and a narrow niche. Minor changes of niche breadth or position might have a strong impact on the potential distribution of the species. The reconstructed refugial areas are based on strongly consistent modeling results over all SDM techniques, and consensus maps of both climate models identified the same area. Thus the modeled refugia seem to be robust, and we cannot offer an explanation for the genetic depletion of populations in this area, except as an artifact of insufficient sampling.
If model-predicted colonized areas were correctly identified, an interpretation of the population genetic data different from what would be inferred from these data alone is required. Several processes associated with postglacial recolonization might explain the high genetic diversities found in colonized populations. One is secondary contact of different lineages from separate refugia having colonized previously uninhabited areas. This has been shown as a cause of high diversities in other studies (Petit et al. 2003; Heuertz et al. 2004; Schönswetter et al. 2004). All three genetic groups identified in P. daonensis are represented in these populations, and the northern area may therefore have been colonized by lineages from the Alpi Orobie as well as from the Adamello/Alpi Giudicarie. The western population VIV in the Alpi Orobie might represent a source population for the colonization, which seems plausible given its high diversity. However, like the northern part of the current distribution range, this area was not predicted as a refugial area by modeling. It is surprising that the refugial population PED also shows admixture from the three genetic groups but is genetically poor. Although the existence of three genetic groups indicates three refugia, overall low levels of genetic structuring in P. daonensis and the edaphic exclusion of two of three reconstructed refugia (see above) may imply survival in only one refugium. Improved sampling in all relevant mountain ranges including the Langorai could help to clarify this point. Another possible explanation for the high diversities found in colonized populations is associated with the mode of expansion. Following Hewitt (1996), slow postglacial expansion of P. daonensis retaining a broad advancing front (“phalanx”) would have allowed more alleles to survive with less genotypic divergence among populations. In contrast, rapid expansion (“pioneer”) could result in considerable homozygosity and derived genotypes spread over large areas of the colonized range. Salix herbacea likely underwent the first mode of expansion (Alsos et al. 2009). “Phalanx” expansion would explain the lack of genetic depletion associated with the colonization of western and northern parts of the current distribution of the species. Additionally, the rather small range shift that we suggest P. daonensis experienced would ensure gene flow between populations that colonized newly available habitats and the refugial populations, thus preventing loss of diversity and allele rarity with migration. The main distributional shift from the southernmost/easternmost limit of the glacial distribution to the northernmost/westernmost limit of the extant range is at the most approximately 60 km, and thus small compared to the greatest possible shift in P. hirsuta, ranging from the western to the eastern Alps. Still, the paucity of refugial populations remains surprising. Two further aspects should be considered: (1) extant factors such as population size and levels of gene flow strongly influence patterns of genetic variation and may confound the genetic imprints of glacial history, (2) complex patterns of consecutive range shifts during several glacial cycles and climatic instability during glacial periods (Ganopolski and Rahmstorf 2001; Paillard 2001) may have resulted in ambiguous genetic signals.
Conclusions
We demonstrated that hindcasting of past species distributions allowed constructing hypotheses for glacial and postglacial history that can be tested using molecular data. The lesson we learned from both investigated species is that the sampling design should be explicitly adjusted to the modeling output by sampling in all refugial and colonized areas. We have also shown that the incorporation of SDMs led to new interpretations of population genetic patterns and to new hypotheses of glacial survival and postglacial colonization.
The results for the two species are substantially different. In case of P. hirsuta, models of the past distribution of the species suggest widespread nunatak survival in the Central Alps, and this was generally congruent with patterns of genetic structure and diversity. Through the combination of the two approaches and the incorporation of morphological, floristic, and palaeoenvironmental information, shortcomings of the SDMs could be revealed. One of these was the inability to predict the full extent of the refugial area at the southern alpine periphery between Lago Maggiore and Lago di Como. This inability may have been caused by a niche shift associated with the glacial colonization of this area.
Comparison of colonized populations with refugial populations as inferred from SDMs revealed that high diversity seems to be a good indicator for refugial areas in the inner Alps. In contrast to this, the refugial history of southern alpine relict populations is reflected better by high levels of rare and private alleles rather than by high genetic diversity. On the other hand, high diversity was also found in the eastern populations that colonized this area according to modeling results. This indicates that LGM distribution models may be able to distinguish alpine regions that harbor potential nunatak areas and regions that were exclusively colonized from the alpine periphery or from other regions. This distinction could not easily have been made with phylogeographic data alone.
In P. daonensis, the location of refugia derived from SDMs and phylogeographic analysis clearly are incongruent. This opens the possibility that a shift of ecological niche might have been associated with Quaternary range shifts, but other explanations are conceivable. If the modeled refugia of P. daonensis should be correct, these areas could not have been located using phylogeographic data. SDMs revealed that patterns of genetic diversity and rarity for P. daonensis may be poor indicators of the location of refugia. Instead, high genetic diversity and rarity as found in recolonized areas as suggested by niche models are likely to be the result of processes other than refugial survival. Similarly, low genetic diversity and low levels of rarity found in modeled refugial areas imply that additional processes besides historical factors could have shaped the genetic patterns that we found.
With this study, we demonstrated that despite the uncertainties of niche-based palaeodistribution modeling, the integration of this approach with phylogeographic data provides a detailed picture of the history of the distribution of alpine species. Here, this combination of approaches helps to identify geographic areas in need of more detailed study, something that might be overlooked if one were to use a phylogeographic approach alone. Further studies may consider refining the spatial and temporal resolutions of the analyses, considering finer grained data and more time slices with climate reconstructions, and refining the classification of populations in more classes than refugia versus recolonized (e.g., along a stability metric as used in Hugall et al. 2002).
Acknowledgments
We thank A. Selvaggi of the “Istituto per le Piante da Legno e l’Ambiente s.p.a.c.so Casale,” Torino, Italia, and F. Prosser of the “Museo Civico de Rovereto,” Rovereto, Italia, who generously provided unpublished survey data on species occurrences. We thank the following herbaria for allowing the use of collection data: M, MSB, Z, ZT, RUEB, MJG, as well as H. Brisse and B. Bäumler for kindly providing species occurrence data from the data banks SOPYH and ZDSF, respectively.
References
- Alsos IG, Alm T, Normand S, Brochmann C. Past and future range shifts and loss of diversity in dwarf willow (Salix herbacea L.) inferred from genetics, fossils and modeling. Global Ecol. Biogeogr. 2009;18:223–239. [Google Scholar]
- Araujo MB, Guisan A. Five (or so) challenges for species distribution modeling. J. Biogeogr. 2006;33:1677–1688. [Google Scholar]
- Araujo MB, New M. Ensemble forecasting of species distributions. Trends Ecol. Evol. 2007;22:42–47. doi: 10.1016/j.tree.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Bhagwat SA, Willis KJ. Species persistence in northerly glacial refugia of Europe: a matter of chance or biogeographical traits? J. Biogeogr. 2008;35:464–482. [Google Scholar]
- Birks HJB, Willis KJ. Alpines, trees, and refugia in Europe. Plant Ecol. Divers. 2008;1:147–160. [Google Scholar]
- Brockmann-Jerosch H, Brockmann-Jerosch M. Die Geschichte der Schweizerischen Alpenflora. In: Schröter C, editor. Das Pflanzenleben der Alpen. Zürich: Raustein; 1926. pp. 1110–1215. [Google Scholar]
- Broennimann O, Treier UA, Müller-Schärer H, Thuille W, Peterson AT, Guisan A. Evidence of climatic niche shift during biological invasion. Ecol. Lett. 2007;10:701–709. doi: 10.1111/j.1461-0248.2007.01060.x. [DOI] [PubMed] [Google Scholar]
- Carnaval AC, Hickerson MJ, Haddad CB, Rodrigues MT, Moritz C. Stability predicts genetic diversity in the Brazilian Atlantic forest hotspot. Science. 2009;323:785–789. doi: 10.1126/science.1166955. [DOI] [PubMed] [Google Scholar]
- Carstens BC, Richards CL. Integrating coalescent and ecological niche modeling in comparative phylogeography. Evolution. 2007;61:1439–1454. doi: 10.1111/j.1558-5646.2007.00117.x. [DOI] [PubMed] [Google Scholar]
- Coart E, van Glabeke S, Petit RJ, van Bockstaele E, Roldan-Ruiz I. Range wide versus local patterns of genetic diversity in hornbeam (Carpinus betulus L.) Conserv. Genet. 2005;6:259–273. [Google Scholar]
- Collins WD, Bitz CM, Blackmon ML, Bonan GB, Bretherton CS, Carton JA, Chang P, Doney SC, Hack JJ, Henderson TB, et al. The Community Climate System Model version 3 (CCSM3) J. Climate. 2006;19:2122–2143. [Google Scholar]
- Comes HP, Kadereit JW. The effect of quaternary climatic changes on plant distribution and evolution. Trends Plant Sci. 1998;3:432–438. [Google Scholar]
- Comps B, Gomory D, Letouzey J, Thiebaut B, Petit RJ. Diverging trends between heterozygosity and allelic richness during postglacial colonization in the European beech. Genetics. 2001;157:389–397. doi: 10.1093/genetics/157.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corander J, Marttinen P. Bayesian identification of admixture events using multilocus molecular markers. Mol. Ecol. 2006;15:2833–2843. doi: 10.1111/j.1365-294X.2006.02994.x. [DOI] [PubMed] [Google Scholar]
- Corander J, Marttinen P, Siren J, Tang J. Enhanced Bayesian modeling in BAPS software for learning genetic structures of populations. BMC Bioinformatics. 2008;9:539. doi: 10.1186/1471-2105-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crema S. Ph.D. thesis. Bologna, Italy: Università degli Studi di Bologna; 2009. Molecular diversity of Primula apennina and phylogeny of Primula subsection Euauricula. [Google Scholar]
- Dubuis A, Pottier J, Rion V, Pellissier L, Theurillat J-P, Guisan A. Predicting spatial patterns of plant species richness: a comparison of direct macroecological and species stacking approaches. Divers. Distrib. 2011;17:1122–1131. [Google Scholar]
- Elith J, Graham CH, Anderson RP, Dudik M, Ferrier S, Guisan A, Hijmans RJ, Huettmann F, Leathwick JR, Lehmann A, et al. Climate change impacts on European mountain plant diversity. Global Change Biol. 2011;17:2330–2341. [Google Scholar]
- Engler R, Randin CF, Thuiller W, Dullinger S, Zimmermann NE, Araújo MB, Pearman PB, Le Lay G, Piedallu C, Albert CH, et al. 21st century climate change threatens moutnain flora unequally across Europe. Global Change Biol. 2011;17:2330–2341. [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fürchter R, Widmer A, Baltisberger M. Nunataks as glacial refugia? A molecular phylogeographical case study with Draba aizoides L. Bauhinia. 2001;15:78. [Google Scholar]
- Ganopolski A, Rahmstorf S. Rapid changes of glacial climate simulated in a coupled climate model. Nature. 2001;409:153–158. doi: 10.1038/35051500. [DOI] [PubMed] [Google Scholar]
- Guisan A, Zimmermann NE. Predictive habitat distribution models in ecology. Ecol. Model. 2000;135:147–186. [Google Scholar]
- Hampe A, Petit RJ. Conserving biodiversity under climate change: the rear edge matters. Ecol. Lett. 2005;8:461–467. doi: 10.1111/j.1461-0248.2005.00739.x. [DOI] [PubMed] [Google Scholar]
- Hasumi H, Emori S. K-1 coupled GCM (MIROC) description. . Tokyo: University of Tokyo; 2004. [Google Scholar]
- Hegi G. Illustrierte Flora von Mitteleuropa, Bd. 5/3. Berlin: P. Parey; 1966. [Google Scholar]
- Hess H, Landolt E, Hirzel R. Flora der Schweiz und angrenzender Gebiete, Bd. 1. Basel: Birkhäuser; 1967. [Google Scholar]
- Heuertz M, Hausman JF, Hardy OJ, Vendramin GG, Frascaria-Lacoste N, Vekemans X. Nuclear microsatellites reveal contrasting patterns of genetic structure between western and southeastern European populations of the common ash (Fraxinus excelsior L.) Evolution. 2004;58:976–988. doi: 10.1111/j.0014-3820.2004.tb00432.x. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 1996;58:247–276. [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005;25:1965–1978. [Google Scholar]
- Hood GM. PopTools version 3.0. 2008. Available at: http://www.poptools.org.
- Hugall A, Moritz C, Moussalli A, Stanisic J. Reconciling palaeodistribution models and comparative phylogeography in the Wet Tropics rainforest land snail Gnarosophia bellendenkerensis (Brazier 1875) Proc. Natl. Acad. Sci. U.S.A. 2002;99:6112–6117. doi: 10.1073/pnas.092538699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson GE. Concluding remarks. Cold Spring Harbor Symp. Quant. Biol. 1957;22:415–427. [Google Scholar]
- Jakob SS, Martinez-Meyer E, Blattner FR. Phylogeographic analyses and palaeodistribution modeling indicate Pleistocene in situ survival of Hordeum species (Poaceae) in southern Patagonia without genetic or spatial restriction. Mol. Biol. Evol. 2009;26:907–923. doi: 10.1093/molbev/msp012. [DOI] [PubMed] [Google Scholar]
- Jeßen S, Lehmann L. Primula hirsuta subsp valcuvianensis subsp. nov. – ein Lokalendemit der südlichen Voralpen, Provinz Varese (sect. Auricula, Primulaceae) 2005. Sammelblätter Gebirgspflanzen Blatt 3.22.01.2.
- Jost A, Lunt D, Kageyama M, Abe-Ouchi A, Peyron O, Valdes PJ, Ramstein G. High-resolution simulations of the last glacial maximum climate over Europe: a solution to discrepancies with continental palaeoclimatic reconstruction? Clim. Dynam. 2005;24:577–590. [Google Scholar]
- Jäckli H. Die Vergletscherung der Schweiz im Würmmaximum. Eclogae Geologicae Helvetiae. 1962;55:185–294. [Google Scholar]
- Kadereit JW, Griebeler EM, Comes HP. Quaternary diversification in European alpine plants: pattern and process. Philos. Trans. R. Soc. Lond. Ser. B. 2004;359:265–274. doi: 10.1098/rstb.2003.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama M, Peyron P, Pinot S, Tarasov P, Guiot J, Joussaume S, Ramstein G. The last glacial maximum climate over Europe and wetsern Siberia: the PMIP comparison between modes and data. Clim. Dynam. 2001;17:23–43. [Google Scholar]
- Knowles LL, Carstens BC, Keat ML. Coupling genetic and ecological-niche models to examine how past population distributions contribute to divergence. Curr. Biol. 2007;17:940–946. doi: 10.1016/j.cub.2007.04.033. [DOI] [PubMed] [Google Scholar]
- Kropf M, Kadereit JW, Comes HP. Differential cycles of range contraction and expansion in European high mountainh plants during the late Quaternary: insights form Pritzelago alpina (L.) O. Kunze (Brassicaceae) Mol. Ecol. 2003;12:931–949. doi: 10.1046/j.1365-294x.2003.01781.x. [DOI] [PubMed] [Google Scholar]
- Körner C. Alpine plant life. 1st ed. Berlin: Springer; 1999. [Google Scholar]
- Lang G. Quartäre Vegetationsgeschichte Europas. Stuttgart: Gustav Fischer; 1994. [Google Scholar]
- Lewontin RC. The apportionment of human diversity. Evol. Biol. 1972;6:381–398. [Google Scholar]
- Liu CR, Berry PM, Dawson TP, Pearson RG. Selecting thresholds of occurrence in the prediction of species distributions. Ecography. 2005;28:385–393. [Google Scholar]
- Magri D, Vendramin GG, Comps B, Dupanloup I, Geburek T, Gömöry D, Latalowa M, Litt T, Paule L, Roure JM, et al. A new scenario for the Quaternary history of European beech populations: palaeobotanical evidence and genetic consequences. New Phytol. 2006;171:199–221. doi: 10.1111/j.1469-8137.2006.01740.x. [DOI] [PubMed] [Google Scholar]
- Maliouchenko O, Palme AE, Buonamici A, Vendramin GG, Lascoux M. Comparative phylogeography and population structure of European Betula species, with particular focus on B. pendula and B. pubescens. J. Biogeogr. 2007;34:1601–1610. [Google Scholar]
- Moser DM. Ein neuer Reliktendemit der Grigna Meridionale, Provincia di Como, Italien: Primula grignensis D. M. Moser (sect. Auricula, subsect. Erythrodrosum Schott) Candollea. 1998;53:387–393. [Google Scholar]
- Moussalli A, Moritz C, Williams SE, Carnaval AC. Variable responses of skinks to a common history of rainforest fluctuation: concordance between phylogeography and palaeo-distribution models. Mol. Ecol. 2009;18:483–499. doi: 10.1111/j.1365-294X.2008.04035.x. [DOI] [PubMed] [Google Scholar]
- Naciri Y, Gaudeul M. Phylogeography of the endangered Eryngium alpinum L. (Apiaceae) in the European Alps. Mol. Ecol. 2007;16:2721–2733. doi: 10.1111/j.1365-294X.2007.03269.x. [DOI] [PubMed] [Google Scholar]
- Nei M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. U.S.A. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillard D. Climatology – glacial hiccups. Nature. 2001;409:147–148. doi: 10.1038/35051691. [DOI] [PubMed] [Google Scholar]
- Parisod C, Besnard G. Glacial in situ survival in the Western Alps and polytopic autopolyploidy in Biscutella laevigata L. (Brassicaceae) Mol. Ecol. 2007;16:2755–2767. doi: 10.1111/j.1365-294X.2007.03315.x. [DOI] [PubMed] [Google Scholar]
- Paun O, Schönswetter P, Winkler M, Intrabiodiv Consortium. Tribsch A. Historical divergence vs. contemporary gene flow: evolutionary history of the calcicole Ranunculus alpestris group (Ranunculaceae) in the European Alps and the Carpathians. Mol. Ecol. 2008;17:4263–4275. doi: 10.1111/j.1365-294x.2008.03908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearman PB, Guisan A, Broennimann O, Randin CF. Niche dynamics in space and time. Trends Ecol. Evol. 2008a;23:149–158. doi: 10.1016/j.tree.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Pearman PB, Randin CF, Broennimann O, Vittoz P, van der Knaap WO, Engler R, Le Lay G, Zimmermann NE, Guisan A. Prediction of plant species distributions across six millennia. Ecol. Lett. 2008b;11:357–369. doi: 10.1111/j.1461-0248.2007.01150.x. [DOI] [PubMed] [Google Scholar]
- Pearson RG, Raxworthy CJ, Nakamura M, Peterson AT. Predicting species distributions from small numbers of occurrence records: a test case using cryptic geckos in Madagascar. J. Biogeogr. 2007;34:102–117. [Google Scholar]
- Pearson RG, Thuiller W, Araujo MB, Marttinen P, Brotons L, McClean C, Miles L, Seguardo P, Dawson TP, Lees DC. Model-based uncertainty in species range prediction. J. Biogeogr. 2006;33:1704–1711. [Google Scholar]
- Peterson AT, Soberon J, Sanchez-Cordero V. Conservatism of ecological niches in evolutionary time. Science. 1999;285:1265–1267. doi: 10.1126/science.285.5431.1265. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Aguinagalde I, de Beaulieu JL, Bittkau C, Brewer S, Cheddadi R, Ennos R, Fineschi S, Grivet D, Lascoux M, et al. Glacial refugia: hotspots but not melting pots of genetic diversity. Science. 2003;300:1563–1565. doi: 10.1126/science.1083264. [DOI] [PubMed] [Google Scholar]
- Pollard D, Barron EJ. Causes of model-data discrepancies in European climate during oxygen isotope stage 3 with insights from the last glacial maximum. Q. Res. 2003;59:108–113. [Google Scholar]
- Provan J, Bennett KD. Phylogeographic insights into cryptic glacial refugia. Trends Ecol. Evol. 2008;23:564–571. doi: 10.1016/j.tree.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Ramstein G, Kageyama M, Guiot J, Hély C, Krinner G, Brewer S. How cold was Europe at the last glacial maximum? A synthesis of the progress achieved since the first PMIP model-data comparison. Clim. Past. 2007;3:331–339. [Google Scholar]
- Randin CF, Engler R, Normand S, Zappa M, Zimmermann NE, Pearman PB, Vittoz P, Thuiller W, Guisan A. Climate change and plant distribution: local models predict high-elevation persistence. Global Change Biol. 2009;15:1557–1569. [Google Scholar]
- Ravazzi C. Distribuzione ed ecologia di due primule endemiche delle prealpi calcaree meridionali, Primula glaucescens e P. spectabilis, e considerazioni sulla loro corogenesi. Archivio Geobotanico. 1997;3:125–148. [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. . Vienna, Austria: R Foundation for Statistical Computing; 2005. [Google Scholar]
- Reisigl H, Pitschmann H. Obere Grenzen der Flora und Vegetation in der Nivalstufe der zentralen Ötztaler Alpen (Tirol) Vegetatio. 1958;8:93–129. [Google Scholar]
- Richards CL, Carstens BC, Knowles LL. Distribution modeling and statistical phylogeography: an integrative framework for generating and testing alternative biogeographical hypotheses. J. Biogeogr. 2007;34:1833–1845. [Google Scholar]
- Ruffray P, Brisse H, Grandjouan G. “SOPHY” une banque de données botaniques et écologiques. 2000–2009. Available at: http://sophy.u-3mrs.fr/sophy.htm.
- Ryan PD, Harper DAT, Whalley JS. PALSTAT, statistics for palaeontologists. . London: Chapman & Hall (now Kluwer Academic Publishers); 1995. [Google Scholar]
- Schorr G. Ph.D. thesis. Mainz, Germany: Institut für Spezielle Botanik, Johannes Gutenberg-Universität Mainz; 2009. Rekonstruktion der eiszeitlichen Verbreitung und Artbildung vier alpiner Primeln durch Artenverbreitungsmodelle und Phylogeographie. [Google Scholar]
- Schönswetter P, Tribsch A, Niklfeld H. Phylogeography of the high alpine cushion plant Androsace alpina (Primulaceae) in the European Alps. Plant Biol. 2003a;5:623–630. [Google Scholar]
- Schönswetter P, Tribsch A, Schneeweiss GM, Niklfeld H. Disjunctions in relict alpine plants: phylogeography of Androsace brevis and A. wulfeniana (Primulaceae) Bot. J. Linn. Soc. 2003b;141:437–446. [Google Scholar]
- Schönswetter P, Tribsch A, Stehlik I, Niklfeld H. Glacial history of high alpine Ranunculus glacialis (Ranunculaceae) in the European Alps in a comparative phylogeographical context. Biol. J. Linn. Soc. 2004;81:183–195. [Google Scholar]
- Schönswetter P, Tribsch A, Barfuss M, Niklfeld H. Several Pleistocene refugia detected in the high alpine plant Phyteuma globulariifolium Sternb. & Hoppe (Campanulaceae) in the European Alps. Mol. Ecol. 2002;11:2637–2647. doi: 10.1046/j.1365-294x.2002.01651.x. [DOI] [PubMed] [Google Scholar]
- Schönswetter P, Stehlik I, Holderegger R, Tribsch A. Molecular evidence for glacial refugia of mountain plants in the European Alps. Mol. Ecol. 2005;14:3547–3555. doi: 10.1111/j.1365-294X.2005.02683.x. [DOI] [PubMed] [Google Scholar]
- Shannon CE, Weaver W. The mathematical theory of communication. Urbana: Univ. of Illinois Press; 1949. [Google Scholar]
- Stehlik I, Blattner FR, Holderegger R, Bachmann K. Nunatak survival of the high Alpine plant Eritrichium nanum (L.) Gaudin in the Central Alps during the ice ages. Mol. Ecol. 2002;11:2027–2036. doi: 10.1046/j.1365-294x.2002.01595.x. [DOI] [PubMed] [Google Scholar]
- Stehlik I, Schneller JJ, Bachmann K. Resistance or emigration: response of the high-alpine plant Eritrichium nanum (L.) Gaudin to the ice age within the Central Alps. Mol. Ecol. 2001;10:357–370. doi: 10.1046/j.1365-294x.2001.01179.x. [DOI] [PubMed] [Google Scholar]
- Stehlik I. Nunataks and peripheral refugia for alpine plants during Quaternary glaciation in the middle part of the Alps. Bot. Helv. 2000;110:25–30. [Google Scholar]
- Stehlik I. Glacial history of the alpine herb Rumex nivalis (Polygonaceae): a comparison of common phylogeographic methods with nested clade analysis. Am. J. Bot. 2002;89:2007–2016. doi: 10.3732/ajb.89.12.2007. [DOI] [PubMed] [Google Scholar]
- Stehlik I. Resistance or emigration? Response of alpine plants to the ice ages. Taxon. 2003;52:499–510. [Google Scholar]
- Stewart JR, Lister AM. Cryptic northern refugia and the origins of the modern biota. Trends Ecol. Evol. 2001;16:608–613. [Google Scholar]
- Swets JA. Measuring the accuracy of diagnostic systems. Science. 1988;240:1285–1293. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- Thuiller W, Lafourcade B, Araujo MB. ModOperating Manual for BIOMOD. Grenoble, France: Laboratoire d'Écologie Alpine, Université Joseph Fourier; 2008. [Google Scholar]
- Thuiller W, Lafourcade B, Engler R, Araujo MB. BIOMOD – a platform for ensemble forecasting of species distributions. Ecography. 2009;32:369–373. [Google Scholar]
- Thuiller W. Patterns and uncertainties of species’ range shifts under climate change. Global Change Biol. 2004;10:2020–2027. doi: 10.1111/gcb.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribsch A, Schönswetter P. Patterns of endemism and comparative phylogeography confirm palaeoenvironmental evidence for Pleistocene refugia in the Eastern Alps. Taxon. 2003;52:477–497. [Google Scholar]
- Tribsch A, Schönswetter P, Stuessy TF. Saponaria pumila (Caryophyllaceae) and the Ice Age in the European Alps. Am. J. Bot. 2002;89:2024–2033. doi: 10.3732/ajb.89.12.2024. [DOI] [PubMed] [Google Scholar]
- Tribsch A. Areas of endemism of vascular plants in the Eastern Alps in relation to Pleistocene glaciation. J. Biogeogr. 2004;31:747–760. [Google Scholar]
- Trivedi MR, Berry PM, Morecroft MD, Dawson TP. Spatial scale affects bioclimate model projections of climate change impacts on mountain plants. Global Change Biol. 2008;14:1089–1103. [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, et al. AFLP – a new technique for DNA-Fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltari E, Hijmans RJ, Peterson AT, Nyari AS, Perkins SL, Guralnick RP. Locating Pleistocene refugia: comparing phylogeographic and ecological niche model predictions. PLoS One. 2007;2:e563. doi: 10.1371/journal.pone.0000563. doi: 10.1371/journal.pone.0000563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer A, Lexer C. Glacial refugia: sanctuaries for allelic richness, but not for gene diversity. Trends Ecol. Evol. 2001;16:267–269. doi: 10.1016/s0169-5347(01)02163-2. [DOI] [PubMed] [Google Scholar]
- Willerslev E, Cappellini E, Boomsma W, Nielsen R, Hebsgaard MB, Brand TB, Hofreiter M, Bunce M, Poinar HN, Dahl-Jensen D, et al. Ancient biomolecules from deep ice cores reveal a forested southern Greenland. Science. 2007;317:111–114. doi: 10.1126/science.1141758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Guiot J, Brewer S, Guo Z. Climatic changes in Eurasia and Africa at the last glacial maximum and mid-Holocene: reconstruction from pollen data using inverse vegetation modeling. Clim. Dynam. 2007;29:211–229. [Google Scholar]
- Yeh FC, Boyle TJB. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg. J. Bot. 1997;129:157. [Google Scholar]
- Zhang LB, Kadereit JW. Classification of Primula sect. Auricula (Primulaceae) based on two molecular data sets (ITS, AFLPs), morphology and geographical distribution. Bot. J. Linn. Soc. 2004;146:1–26. [Google Scholar]
- Zhang LB, Comes HP, Kadereit JW. The temporal course of quaternary diversification in the European high mountain endemic Primula sect. Auricula (Primulaceae) Intl. J. Plant Sci. 2004;165:191–207. [Google Scholar]


