Abstract
Amplification of the gene encoding the epidermal growth factor receptor (EGFR) occurs commonly in glioblastoma (GBM), leading to activation of downstream kinases, including phosphatidylinositol 3′-kinase (PI3K), Akt, and mammalian target of rapamycin (mTOR). A serine-threonine kinase, mTOR controls cell growth by regulating mRNA translation, metabolism, and autophagy; acting as both a downstream effector and upstream regulator of PI3K. These signaling functions are distributed between at least two distinct complexes, mTORC1 and mTORC2 with respect to pathway specificity. We have investigated mTOR signaling in glioma cells with the allosteric mTORC1 inhibitor rapamycin, the mTORC1/2 inhibitor Ku-0063794, a dual PI3K/mTORC1/2 kinase inhibitor PI-103, and siRNA against raptor, rictor, or mTOR, and evaluated the value of mTOR inhibitors for the treatment of glioblastoma.
Keywords: Glioblastoma, PI3-kinase, Akt:mTORC1, mTORC2, mTOR, EGFR
1. Introduction
Gliomas represent the most common primary brain tumor and are among the most lethal of all cancers. EGFR is commonly mutated in GBM, leading to overexpression and activation of downstream signaling pathways. EGFR signals through a complex network of intermediates, including PI3K/AKT/mTOR, MAPK, and PLCγ. Inactivation of PTEN and activating mutations in PI3K itself collectively occur in a majority of GBM tumors, effectively uncoupling PI3K from upstream control by EGFR (1).
PI3Ks are lipid kinases activated by a wide range of RTKs to generate the second messenger phosphatidylinositol-3,4,5-trispho- couples PI3K to downstream effectors, such as sphate (PIP3). PIP3 Akt, a serine-threonine kinase that suppresses apoptosis, promotes growth, and drives proliferation. PIP3 also indirectly activates the mammalian target of rapamycin (mTOR), a protein kinase critical for cell growth (1). The mTOR kinase contains a PI3K homology domain (making mTOR a PIK-related kinase – PIKK), although mTOR itself has no lipid kinase activity (2).
Signaling functions of mTOR are distributed between at least two distinct mTOR protein complexes: mTORC1 and mTORC2. In mTORC1, mTOR is associated with a number of proteins, including PRAS40 and the rapamycin-sensitive adapter protein of mTOR (Raptor), whereas in mTORC2, mTOR is associated with a separate protein complex, including the rapamycin-insensitive companion of mTOR (Rictor). Stimulation of PI3K in response to growth factors leads to phosphorylation and activation of Akt. In addition, phospho-Akt phosphorylates and separately inhibits PRAS40 and the Tsc1/2 (hamartin-tuberin) complex. PRAS40 is inhibitory to mTORC1, while tuberin is inhibitory to GTPase RHEB, which in turn is inhibitory to mTORC1. The detailed signaling leading to activation of mTORC2 is less clearly understood (3).
The activated mTORC1 complex phosphorylates substrates, including Thr-389 S6K, Ser-209 eIF4E, and 4EBP1. The mTORC2 complex phosphorylates Akt on Ser-473, and also phosphorylates additional substrates, including serum glucocorticoid-induced protein kinase (SGK) and PKCα Inhibition of mTORC1/S6K1 by allosteric inhibitors, including rapamycin (Sirolimus) (Fig. 1), CCI-779 (Tensirolimus), RAD001 (4), or other similar agents triggers a negative feedback loop through an IRS-I-dependent mechanism, resulting in increase phosphorylation of Akt. This negative feedback loop is prominent in glioma, however the robustness with which inhibition of mTORC1 activates Akt varies across multiple cancer types (5–7). Unlike rapamycin, ATP-competitive inhibitors of mTORC1/mTORC2 by mTOR inhibitors, including Torin, Ku-0063794 (8, 9), and pp242 (10) blocks the phosphorylation of Akt at Ser473. As an integrator of cell growth and proliferation, mTOR also regulates autophagy, a program of cellular self-digestion activated during periods of nutrient and growth factor deprivation (11). The signaling linking activation of mTOR signaling to blockade of autophagy in metazoan cells is poorly understood.
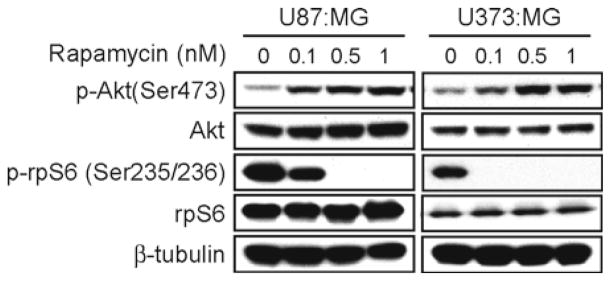
Fig. 1.
The allosteric mTORC1 inhibitor rapamycin induced p-Akt in glioma cells. PTEN mutant-type U87:MG and U373:MG cells were treated with rapamycin at doses shown and were assayed for total and phospho Akt and rpS6 by immunoblot. Rapamycin blocked p-rpS6 in both cell lines at a dose of 0.5 nM. Activation of Akt was dose-dependent, as indicated by increased levels of the p-Akt at Ser-473. β-tubulin is shown as loading control.
Preclinical evaluation of dual PI3K/mTOR inhibitors, such as PI-103 and NVP-BEZ235 have demonstrated efficacy for these agents in blocking the growth of glioblastoma (GBM) cells in vitro and in vivo (5, 12). NVP-BEZ235 and other dual inhibitors are therefore being evaluated in early clinical trials. Thus, inhibitors of mTOR and of PI3K/mTOR provide a new class of agents and therapeutic for glioma. Pure ATP-competitive inhibitors of mTORC1/2 inhibit both mTOR complexes, and have been evaluated in less detail in glioma. We have directly compared rapamycin, Ku-0063794, PI-103, or siRNA against mTOR in glioma cells. In contrast to rapamycin, both Ku-0063794 and PI-103 blocked the phosphorylation of Akt and prevented its activation. Ku-0063794 and PI-103 also decreased the phosphorylation of the mTORC1 target 4EBP1 and induced autophagy more effectively in comparison with the allosteric mTORC1 inhibitor rapamycin (Figs. 2 and 3).
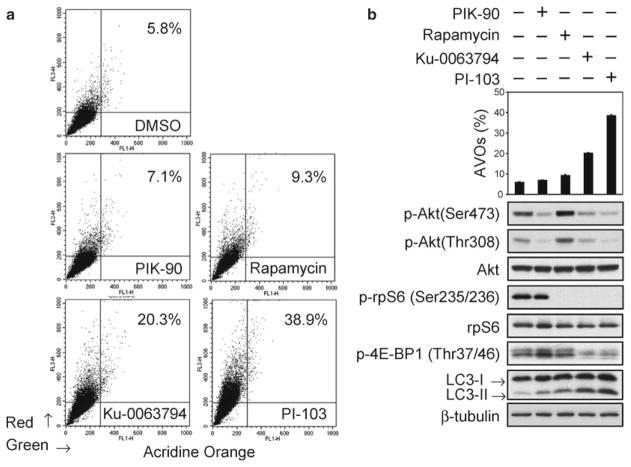
Fig. 2.
The mTOR kinase inhibitor KU-0063794 and the dual PI3K/mTOR inhibitor PI-103 both block phosphorylation of Akt and induce autophagy. In comparison, the mTOCR1 inhibitor rapamycin-induced phosphorylation of Akt and was a less potent inducer of autophagy. (a) PTEN mutant U373:MG cells were treated with DMSO, PI3Kα inhibitor PIK-90 (1 μM), mTORC1 inhibitor rapamycin (100 nM), mTOR inhibitor Ku-0063794 (5 μM), or a dual PI3K/mTOR inhibitor PI-103 (1 μM) for 48 h and stained with acridine orange (1 μg/ml) for 15 min. Cells were analyzed by flow cytometry. Autophagy was quantified by the accumulation of acidic vescular organelles. Percentage of AVOs is indicated. (b) Percentages of cells positive for AVOs; mean ± S.E. for triplicate samples (top panel). Cells were treated as in (a) for 24 h and lysates examined by immunoblot using antibodies shown (bottom panel). In contrast to rapamycin, Ku-0063794 and PI-103 block both p-Akt and p-rpS6. Ku-0063794 and PI-103 also decreased the phosphorylation of 4EBP1 and induced autophagy more potent than rapamycin as indicated by increased levels of the autophagy marker LC3-II. The PI3Kα inhibitor PIK-90 blocked phosphorylation of Akt, but did not affect phosphorylation of 4EBP1 or of rpS6, demonstrating that inhibition of PI3K and Akt activation alone is not sufficient to block the phosphorylation of mTORC1 targets 4EBP1 and of rpS6. (Reproduced from ref. 13 with permission from AAAS).
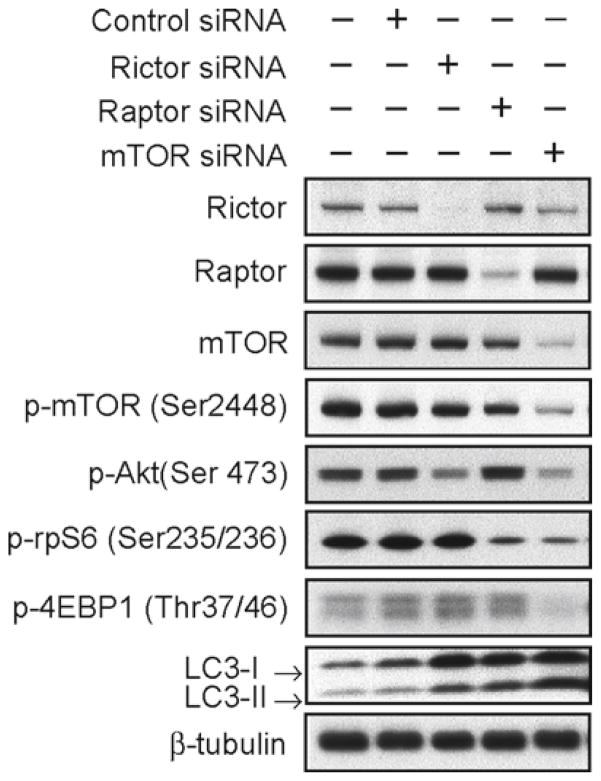
Fig. 3.
Knockdown of raptor, but not rictor or mTOR, activates Akt on Ser-473. We transfected U373:MG cells with control siRNA, siRNA against the mTORC2-associated mRNA rictor, the mTORC1-associated mRNA raptor, or to mTOR itself. Lysates were collected at 72 h and examined by immunoblot using antibodies shown. Knockdown of raptor blocked phosphorylation of rpS6 and increased the phosphorylation of Akt. In contrast, knockdown of rictor or mTOR both decreased the phosphorylation of Akt. Knockdown of rictor or raptor also induced autophagy, albeit to a lesser extent than that observed in response to knockdown of mTOR (as indicated by accumulation of LC3-II). (Reproduced from ref. 13 with permission from AAAS).
2. Materials
2.1. Cell Culture and Lysis
Dulbecco’s modified Eagle’s medium (DMEM H-21) supplemented with 10% fetal bovine serum (FBS, Gibco/BRL) and 1% penicillin–streptomycin and stored at +4°C.
Solution of trypsin (0.25%) stored at +4°C.
Rapamycin (Cell Signaling) is dissolved at 100 μM in methanol and stored in single use aliquots at −20°C.
Ku-0063794 is dissolved at 20 mM in DMSO and stored in single use aliquots at −20°C.
PIK-90 is dissolved at 20 mM in DMSO and stored in single use aliquots at −20°C.
PI-103 is dissolved at 20 mM in DMSO and stored in single use aliquots at −20°C.
2.2. SDS-Polyacrylamide Gel Electrophoresis
Tris-buffered saline with Tween 20 (10× TBS-T).
NOVEX-NuPAGE LDS Sample Buffer (4×) (Invitrogen Corporation).
NOVEX-NuPAGE MOPS Running Buffer (20×) (Invitrogen Corporation).
NOVEX-NuPAGE Tris-Acetate Runing Buffer (25×) (Invitrogen Coropration).
NOVEX-NuPAGE Tris-Glycine Running Buffer (10×) (Invitrogen Corporation).
NOVEX-NuPAGE Transfer Buffer (20×) (Invitrogen Corporation) and NOVEX-NuPAGE Tris-Glycine Transfer Buffer (25×).
NOVEX-NuPAGE 4–12% BT SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gel, 1.0 mm 15 well, NOVEX-NuPAGE 3–8% Tris-Acetate gel, 1.0 mm, 15 well, and NOVEX-NuPAGE 16% Tris-cyclin gel, 1.0 mm, 15 well (Invitrogen Corporation).
Full-range rainbow molecular weight markers (GE Healthcare).
BCA™ Protein Assay Reagent (Thermo Scientific Corporation).
10× Cell lysis buffer (Cell Signaling Technology).
Complete protease inhibitor cocktail tables (Roche).
2.3. Western Blotting for PI3K-Akt-mTOR Pathway
NOVEX-NuPAGE Transfer Buffer (20×) (Invitrogen Corporation) and NOVEX-NuPAGE Tris-Glycine Transfer Buffer (25×).
Methanol (Sigma-Aldrich).
PVDF Transfer Membrane (GE Healthcare).
Blocking buffer: 5% (w/v) nonfat dry milk in TBS-T.
Membrane were blotted with p-Akt (Ser473), p-Akt (Thr308), Akt (pan), p-S6 ribosomal protein (Ser235/236), S6 ribosomal protein, rictor, raptor, p-mTOR (Ser 2448), mTOR, p-4E-BP1 (Thr37/46) (Cell Signaling Technology). LC3 (Novus) or β-tubulin (Millipore).
Primary antibody dilution buffer: TBS-T supplemented with 5% (w/v) fraction V bovine serum albumen (Sigma).
Secondary antibody: Goat anti-mouse IgG peroxidase conjugate and goat anti-rabbit IgG peroxidase conjugate (EMD Chemicals Inc).
Hybond™ ECL™ Western blotting detection Kit (GE Healthcare).
2.4. siRNA Transfection
Control small interfering RNA (siRNA) was purchased from Santa Cruz Biotechnology. Raptor siRNA (NM 020761 RAPTOR ON-TARGET plus SMARTpool), Rictor siRAN (NM 152756 RICTOR ON-TARGET plus SMARTpool), mTOR siRNA (NM 004958 FRAP1 ON-TARGET plus SMARTpool) were purchased from Thermo Scientific. 5× siRNA buffer (300 mM KCL, 30 mM HEPES-pH 7.5, 1.0 mM MgCL2); RNase-free water (molecular grade RNase-free water for dilution of 5× buffer). Resuspend siRNAs to 20 μM stock in 1× siRNA buffer and stored in single use aliquots at −20°C.
Lipofectamine 2000 (Invitrogen) stored at +4°C.
2.5. Detection and Quantification of AVOs
Acridine orange (Sigma Chemical Co.). Dissolve 1 g of acridine orange to 100 ml of distilled water. Store in a foil wrapped container at 4°C (see Note 1) and bring to room temperature prior to use. 5 ml polystyrene round-bottom tube with cell-strainer cap (BD Biosciences).
3. Methods
3.1. Preparation of Samples for Assay of PI3K Pathway by Western Blotting
U87:MG and U373:MG cells are grown in 10% FBS and are passaged every 3 days with trypsin/EDTA to provide new maintenance cultures on 100-mm tissue dishes and experimental culture on 6-well plates. Two 6-well plates are required for each data point.
Cells are seeded in 6-well plates at 3 × 105/cm2 and will approach confluence after 72 h. Cells are treated by adding 2 ml fresh media containing inhibitors for desired time periods (typically 24 h in our experiments).
Media is aspirated, cells washed twice with ice-cold 1× PBS, and aspirated again. Cells are lysed by adding 1× ice-cold cell lysis buffer (50–100 μl per 6-well plate). Immediately scrape the cells off the plate and transfer the extract to a 1.5 ml microcentrifuge tube. Keep on ice for 30 min.
Sonicate for 5 s to shear DNA and reduce sample viscosity. Microcentrifuge at 12,700 rpm (15,000 × g) for 5 min at 4°C, and transfer supernatant to new tube.
Measure the protein concentration of samples by BCA protein Assay kit. Mix reagent A with Reagent B to generate a working reagent (WR). Load the 96-well microplate with WR (200 μL per well) and then add the protein samples (2 μL each) alone with BSA (bovine serum albumin) protein standard dilutions. Incubate 30 min at 37°C and read at 562 nm with a multimode microplate reader (BioTek Instruments Inc).
Heat sample by adding 25% 4× LDS sample buffer to 70°C on the heat block for 10 min. Cool on ice and microcentrifuge for 5 min at room temperature. Samples are now ready for separation by SDS-PAGE.
3.2. SDS-PAGE
These instructions assume the use of a Hoefer SE260B gel system.
Prepare the running buffer by diluting 20 ml of 20× running buffer (see Note 2) with 380 ml of Milli-Q water in a measuring cylinder. Cover with parafilm invert to mix.
Once the stacking gel has set, carefully remove the comb and use a 5 ml syringe fitted with a 22-gauge needle to wash wells with running buffer.
Add the running buffer to the upper and lower chambers of the gel unit and load equal amount of each sample in the well. Include one well for 10 μl of full-range rainbow molecular weight markers.
Complete the assembly of the gel unit and connect to a power supply. Two gels can be run at 110 mA (see Note 3) for 1 h. 126 V/two gels (start); 200 V/two gels (end).
3.3. Western Blotting for PI3K-Akt-mTOR Pathway
Samples that have been separated by SDS-PAGE are transferred electrophoretically to nitrocellulose membranes. These directions assume the use of a Bio-RAD Trans-Blot SD semi-dry transfer cell system. Prepare the transfer buffer (see Note 4) by diluting 15 ml of 20× transfer buffer with 285 ml of Milli-Q water in a measuring cylinder. Cover with parafilm. Invert to mix.
Cut a sheet of the PVDF-transfer membrane just larger than the size of the separating gel. Clip upper right corner to clarify orientation. Wet membrane in 100% methanol for 1 min. Rinse briefly in Milli-Q and then soak for 15 min in transfer buffer before use.
Cut filter paper to the dimensions of the gel. Two pieces of extra thick filter paper per gel are needed for each gel/membrane sandwich. You can substitute four pieces of thick or six pieces of thin filter paper. Completely saturate the filter paper by soaking in transfer buffer.
Place a presoaked sheet of extra thick filter paper onto the platinum anode. Roll a pipet over the surface of the filter paper to exclude all air bubbles.
Carefully place the equilibrated gel on top of the transfer membrane, aligning the gel on the center of the membrane.
Place the other sheet of presoaked filter paper on the top of the gel. Carefully roll a pipet over the filter paper to remove bubbles from between each layer.
Complete the assembly of the gel unit and connect to a power supply. Two gels can be run at 110 mA (see Note 5) for 1 h. 3–5 V/two gels (start); 10–12 V/two gels (end).
After transfer, wash the membrane with 25 ml TBS for 5 min at room temperature. The membrane is then incubated in 25 ml of TBS-0.5% Tween (TBS-T)/5% w/v nonfat dry milk for 1 h at room temperature. Wash three times with 25 ml of TBS-T, 10 min per wash.
Incubate the membrane and primary antibody at the appropriate dilution (see Note 6) in 10 ml primary antibody in TBS-T/5% BSA with gentle agitation on a rocking platform overnight at 4°C.
Wash three times for 10 min each with 25 ml of TBS-T. Incubate the membrane with HRP-conjugated secondary antibody (1:2,000) in 10 ml TBS-T/2%BSA with gentle agitation for 1 h at room temperature. Wash with TBS-T six times, 10 min per wash.
During the final wash, 5 ml aliquots of each portion of the ECL reagent are warmed separately to room temperature. Once the final wash is removed from the blot, the ECL reagents are mixed together and then immediately added to the blot, which is then rotated by hand for 1 min to ensure even coverage.
Remove blot from the ECL reagents. Drain membranes of excess developing solution (do not let dry), wrap in plastic warp and expose to X-ray film. An initial 10-s exposure should indicate the proper exposure time.
3.4. siRNA Transfection
Control siRNA was purchased from Santa Cruz Biotechnology. siRNAs against rictor, raptor, and mTOR were purchased from Dharmacon, and transfected using Lipofectamine 2000 (Invitrogen).
One day before transfection, cells were seeded in 6-well plates at 3 × 105/cm2 in 10% FBS growth medium without antibiotics (see Note 7).
Dilute 8 μl of 20 μM siRNA in 250 μl medium without serum (final concentration of RNA when added to cells is 80 nM). Mix gently. Incubate for 5 min at room temperature dilute 8 μl of lipofectamine in 250 μl medium without serum. Mix gently. Incubate for 5 min at room temperature.
After the 5 min incubation, combine the diluted siRNA with the diluted lipofectamine 2000. Mix gently and incubate for 20 min at room temperature to allow complex formation to occur.
Add 500 μl of the siRNA-lipofectamine 2000 complex to each well of a 6-well plate containing 1.5 ml 10% FBS growth medium without antibiotics. Mix gently by rocking the plate back and forth.
After 6 h (see Note 8), wash cells with PBS and adding 1.5 ml of 10% FBS growth media.
Incubate the cells at 37°C in a 5% CO2 gene knockdown. for 72 h, then assay for
3.5. Detection and Quantification of AVOs
In acridine orange-stained cells, the cytoplasm and nucleolus fluorescent bright green and dim red, respectively, whereas acidic compartments fluorescent bright red. The intensity of the red fluorescence is proportional to the degree of acidity. Therefore, the volume of the cellular acidic compartment can be quantified.
U373:MG cells were seeded in 6 well plates at 3 × 105/cm2 in 10% FBS growth medium. Cells were treated by adding 2 ml fresh media containing inhibitors for desired time periods (Typically 48 h in our experiments).
Cells were stained with 1 μg/ml of acridine orange for 15 min, wash cells once with PBS and incubate with 0.25% trypsin-EDTA for 3–5 min, add media with serum to stop trypsin-EDTA, removed from the plate with typsin-EDTA. Microcentrifuge at 500 rpm for 5 min at room temperature, the pellet was resuspended in 500 μl of phenol red-free growth medium, collected with a 5 ml in a foil wrapped polystyrene round-bottom tube with cell-strainer cap (see Note 1), and examined immediately by flow cytometry.
Analyze cells on a flow cytometry, green (510–530 nm) and red (650 nm) fluroscence emission from 1 × 104 cells illuminated with blue (488 nm) excitation light was measured with an FACSCalibur from (Becton Dickinson) using CellQuest software.
Acknowledgments
We thank Zachary Knight, Benjamin Houseman, Morri Feldman, and Kevan Shokat for providing PI-103, PIK-90, and Ku-0063794. We acknowledge support from NIH grants PCA133091, NS055750, CA102321, CA097257, CA128583, CA148699 P01 CA081403, Burroughs Wellcome Fund, American Brain Tumor Association, The Brain Tumor Society, Accelerate Brain Cancer Cure; Alex’s Lemonade Stand, Children’s National Brain Tumor, Katie Dougherty, Pediatric Brain Tumor, Samuel G. Waxman and V Foundations.
Footnotes
Acridine orange is light sensitive.
MOPS SDS running buffer for NOVEX-NuPAGE 4–12% BT SDS-PAGE gel, Tris-acetate running buffer for NOVEX-NuPAGE 3–8% Tris-acetate gel (for high molecular weight protein detection, such as mTOR, p-mTOR (MW 289 kDa)), and Tris-glycine running buffer for NOVEX-NuPAGE 16% Tris-cyclin gel (for low molecular weight protein detection, such as LC3 (MW 18 kDa for LC3I and MW 16 kDa for LC3II)).
For NOVEX-NuPAGE 16% Tris-cyclin gel, two gels run at 125 V constant for 1.5 h. 30–40 mA (start); 8–12 mA (end).
1× NOVEX-NuPAGE transfer buffer with 10% methanol for NOVEX-NuPAGE 4–12% gel and 3–8% gel. 1× NOVEX-NuPAGE Tris-glycine transfer buffer with 20% methanol for NOVEX-NuPAGE 16% Tris-cyclin gel.
For NOVEX-NuPAGE 16% Tris-cyclin gel, run at 20 V constant for 1.5 h.
For the primary antibodies purchased from cell signaling was used at 1:1,000 dilution, for LC3 at 1:500 dilution, and for β-tubulin at 1:2,000 dilution.
Transfecting cells at lower density can minimize the loss of cell viability due to cell overgrowth after 3 days.
Removing the complexes after 6 h without loss of transfection activity, but can reduce the toxicity to cells.
References
- 1.Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: Targets for cancer therapy. Nat Rev Cancer. 2004;4:361–370. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- 2.Cheng CK, Fan QW, Weiss WA. PI3K signaling in glioma – animal models and therapeutic challenges. Brain Pathol. 2009;19:112–120. doi: 10.1111/j.1750-3639.2008.00233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 4.Faivre S, Kroemer G, Raymond E. Current development of mTOR inhibitors as anticancer agents. Nat Rev drug discov. 2006;5:671–688. doi: 10.1038/nrd2062. [DOI] [PubMed] [Google Scholar]
- 5.Fan QW, Knight ZA, Goldenberg DD, Yu W, Mostov KE, Stokoe D, Shokat KM, Weiss WA. A dual PI3 kinase/mTOR inhibitor reveals emergent efficacy in glioma. Cancer Cell. 2006;9:341–349. doi: 10.1016/j.ccr.2006.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun SY, Rosenberg LM, Wang X, Zhou Z, Yue P, Fu H, Khuri FR. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65:7052–7058. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- 7.Fan QW, Cheng CK, Nicolaides TP, Hackett CS, Knight ZA, Shokat KM, Weiss WA. A Dual Phosphoinositide-3-Kinase {alpha}/mTOR Inhibitor Cooperates with Blockade of Epidermal Growth Factor Receptor in PTEN-Mutant Glioma. Cancer Res. 2007;67:7960–7965. doi: 10.1158/0008-5472.CAN-07-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mTOR inhibitor reveals rapamycin-insensitive functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Martinez JM, Moran J, Clarke RG, Gray A, Cosulich SC, Chresta CM, Alessi DR. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR) Biochem J. 2009;421:29–42. doi: 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:0371–0383. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu TJ, Koul D, LaFortune T, Tiao N, Shen RJ, Maira SM, Garcia-Echevrria C, Yung WK. NVP-BEZ235, a novel dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor, elicits multifaceted antitumor activities in human gliomas. Mol Cancer Ther. 2009;8:2204–2210. doi: 10.1158/1535-7163.MCT-09-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan QW, Cheng CK, Hackett CS, Feldman ME, Houseman BT, Nicolaides TP, Haas-Kogan DA, James CD, Oakes SA, Debnath J, Shokat KM, Weiss WA. Akt and autophagy cooperate to promote survival of drug-resistant glioma. Sci Signal. 2010;3(147):ra81. doi: 10.1126/scisignal.2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]