Abstract
Due to the importance of preserving the genetic integrity of populations, strategies to restore damaged coral reefs should attempt to retain the allelic diversity of the disturbed population; however, genetic diversity estimates are not available for most coral populations. To provide a generalized estimate of genetic diversity (in terms of allelic richness) of scleractinian coral populations, the literature was surveyed for studies describing the genetic structure of coral populations using microsatellites. The mean number of alleles per locus across 72 surveyed scleractinian coral populations was 8.27 (±0.75 SE). In addition, population genetic datasets from four species (Acropora palmata, Montastraea cavernosa, Montastraea faveolata and Pocillopora damicornis) were analyzed to assess the minimum number of donor colonies required to retain specific proportions of the genetic diversity of the population. Rarefaction analysis of the population genetic datasets indicated that using 10 donor colonies randomly sampled from the original population would retain >50% of the allelic diversity, while 35 colonies would retain >90% of the original diversity. In general, scleractinian coral populations are genetically diverse and restoration methods utilizing few clonal genotypes to re-populate a reef will diminish the genetic integrity of the population. Coral restoration strategies using 10–35 randomly selected local donor colonies will retain at least 50–90% of the genetic diversity of the original population.
Keywords: Reef restoration, Conservation genetics, Genetic diversity, Scleractinian coral, Microsatellite, Rarefaction
Introduction
Increasing general degradation and acute physical disturbance (natural and anthropogenic) of coral reef habitats has led to increasing efforts to conserve and restore these ecosystems. Reef restoration approaches actively attempt to restore damaged reef areas to their “natural” or “original” state after a disturbance by increasing live coral cover and species diversity through larval seeding or coral transplantation of whole adult or juvenile colonies, colony fragments or nubbins from adjacent unaffected habitats or coral nurseries (Amar and Rinkevich 2007; Yeemin et al. 2006; reviewed in Rinkevich 2005 and Precht 2006). The success and value of reef restoration measures that aim to establish a viable adult coral community, however, is in debate (e.g., Precht et al. 2005; Precht 2006), and is dependent on the nature of the disturbance (acute versus chronic).
Although quantifying and protecting the genetic diversity (described as allelic richness throughout this paper, rather than heterozygosity) of a species is an important aspect of conservation biology and management (Haig 1998; Petit et al. 1998; Altizer et al. 2003; Reed and Frankham 2003; Spielman et al. 2004; Perez-Ruzafa et al. 2006; Jones et al. 2007; Baums 2008; DiBattista 2008), often stated as a goal when proposing and implementing restoration strategies, preserving the genetic integrity of a damaged coral population is generally not realized (but see Petersen and Tollrian 2001). This is primarily because levels of allelic diversity in natural coral populations have not been adequately described for this purpose, or have not been presented in a format amenable for incorporation into restoration strategies (Grober-Dunsmore et al. 2007). In situations where reef restoration via transplantation of corals is implemented (e.g., ship grounding and storm damage repair, coastal development mitigation), an idealized strategy would be to restore populations to pre-damage levels of genetic diversity. The primary concern is that restored populations will have less genetic diversity than the original population due to re-population with few clonal genotypes (i.e., using fragments from few donor colonies; Rinkevich 1995, 2000). Although natural coral populations often have a component of asexual reproduction contributing to the population (e.g., Stoddart 1983, 1984; Ayre and Resing 1986; LeGoff-Vitry et al. 2004; Baums et al. 2005, 2006; Foster et al. 2007), it is typically not a dominant reproductive strategy and naturally occurring monoclonal populations have not been common in literature. A potential consequence of genetically depauperate populations is increased vulnerability for local extinction due to localized environmental perturbations and variability in susceptibility to disease and bleaching (Nei et al. 1975; Petit et al. 1998; Eldridge et al. 1999; Altizer et al. 2003; Reed and Frankham 2003; Spielman et al. 2004; McKay et al. 2005). A proportion of individuals in genetically diverse populations may have a greater capacity to survive and acclimate to changing environmental conditions preventing local extinction events. Genotype-specific resistance to chronic perturbations in coral species is likely (Edmunds 1994), though not thoroughly studied, thus high levels of allelic diversity may increase resilience of coral populations (van Oppen and Gates 2006).
In addition, if restored populations are “founded” by fragments of a few donor colonies and these populations were primarily self-seeding, benefiting minimally from recruitment contributions from outside source populations, the future reproductive success of corals may be compromised due to effects of inbreeding depression, reproductive self-incompatibility and/or sex ratio bias. Although self-fertilization may be a viable reproductive strategy for some coral species (e.g., Heyward and Babcock 1986; Stoddart et al. 1988; Brazeau et al. 1998; Goffredo et al. 2004; Miller and Mundy 2005; Sherman 2007; Carlon and Lippe 2008), most corals, which include the major Caribbean reef builders of the Montastraea annularis complex and the threatened Acropora species, are partially or entirely reproductively self-incompatible (e.g., Heyward and Babcock 1986; Wallace and Willis 1994; Szmant et al. 1997; Willis et al. 1997; Fukami et al. 2004; Baums et al. 2005, 2006; reviewed in Carlon 1999). For gonochoric species, sampling from few random donor colonies may result in a restored population of a single gender or a skewed sex ratio that may limit the reproductive success of colonies. Consequently, a reef habitat resulting from previous restoration efforts may be ecologically functional and be aesthetically appealing, but may not be viable and unable to maintain or increase population sizes through local reproductive efforts over time. Thus, the motivation for understanding existing levels of genetic diversity and preserving the genetic integrity of a damaged coral population is multifaceted and will ultimately increase the effectiveness of the restoration efforts that may prove to be critical for the future of coral populations.
Ideally, levels of population genetic diversity for local species within a damaged reef would have been or could be directly estimated; however, this is not a practical expectation. This study surveys the literature for estimates of coral population genetic diversity using microsatellites and analyzes population genetic data sets from four scleractinian coral species to estimate conservative levels of genetic diversity, in terms of allelic richness, within contemporary coral populations. The goal is to provide estimates of genetic diversity of coral populations that is quantified in meaningful terms for restoration purposes and from that information provide guidelines related to number of donor colonies that should be sampled in attempts to approximate the genetic diversity of the original coral population when no prior knowledge of genetic diversity estimates are available.
Materials and methods
To describe the allelic richness (number of alleles per locus) of scleractinian coral populations, the literature was surveyed for studies that describe the development or use of microsatellite loci in corals (Table 1). Only studies that include allelic richness for individual loci and populations were included in this summary. For each population, number of loci, number of alleles per loci (range and mean) and the number of colonies sampled were recorded from each study. The number of alleles per species-specific locus of an “average” coral population was calculated as the mean number of alleles per locus across all populations and all species. Measures of allelic richness were recorded as actual observed numbers of alleles, not corrected for variation in samples size using rarefaction methods (e.g., Hurlbert 1971); therefore, the results represent a conservative estimate of allelic richness.
Table 1.
Allelic richness (number of alleles per locus) and sizes of scleractinian coral populations in surveyed literature (multiple records for a species indicate data for different populations)
| Species | No. of populations | No. of loci | No. of alleles per locus per population (mean) | No. of colonies sampled per population (total sampled) | Reference |
|---|---|---|---|---|---|
| Acropora millepora | 1 | 9 | 5–20 (8.7) | 23 (23) | vanOppen et al. (2007) |
| 1 | 1 | 11 (11.0) | 20 (20) | vanOppen et al. (2007) | |
| A. nasuta | 8 | 1 | 3–6 (3.9) | 10–39 (216) | Mackenzie et al. (2004) |
| A. palmata | 10 | 4 | 10–18 (14.4) | 39–127 (818) | Zubillaga (unpub. data) |
| Favia fragum | 2 | 15 | 2–16 (5.2) | 45–48 (93) | Carlon and Lippe (2008) |
| Goniastrea favulus | 1 | 5 | 2–10 (5.3) | 32–44 (32–44) | Miller and Howard (2004) |
| Montastraea annularis | 3 | 4 | 3–15 (9.8) | 45–48 (146) | Foster et al. (2007) |
| M. cavernosa | 1 | 5 | 9–17 (12.2) | 58 (58) | Shearer and Coffroth (2004) |
| 10 | 5 | 6–16 (10.1) | 21–56 (363) | Shearer (2004) | |
| M. faveolata | 4 | 6 | 4–29 (14.7) | 150–216 (692–780) | Porto (Table S1) |
| Platygyra daedalea | 1 | 5 | 4–11 (7.2) | 50–80 (50–80) | Miller and Howard (2004) |
| Pocillopora damicornis | 1 | 10 | 3–10 (5.6) | 21 (21) | Starger et al. (2007) |
| 2 | 7 | 6–10 (7.9) | 55–64 (119) | Shearer (Table S1) | |
| P. meandrina | 7 | 4 | 5–18 (10.2) | 22–49 (257) | Magalon et al. (2005) |
| Porites astreoides | 1 | 3 | 3–9 (6.0) | 50 (50) | Shearer and Coffroth (2004) |
| 7 | 2 | 2–6 (3.9) | 12–52 (192) | Shearer (unpub. data) | |
| Seriatopora hystrix | 1 | 10 | 3–14 (6.7) | 51 (51) | Underwood et al. (2006) |
| 1 | 9 | 2–15 (5.7) | 49 (49) | Underwood et al. (2006) | |
| 10 | 3 | 5–16 (8.6) | 11–32 (207) | Maier et al. (2005) |
Although expected heterozygosity (HO) is generally the more common estimate of the genetic diversity of a population, natural coral populations often deviate significantly from expected levels of heterozygosity due to deficits of heterozygotes (e.g., LeGoff-Vitry et al. 2004; Miller and Howard 2004; Shearer and Coffroth 2004; Magalon et al. 2005; Maier et al. 2005; Underwood et al. 2006; Carlon and Lippe 2008). In addition, a population with high levels of heterozygosity can have low allelic diversity since allelic richness measurements are more sensitive to population reductions than heterozygosity estimates (Cornuet and Luikart 1996; Kalinowski 2004). Therefore, allelic richness estimates are more appropriate for describing coral populations whose population sizes have decreased dramatically over the past several decades. Measurements of allelic richness are important for conservation issues since adaptation responses to selective forces of a changing environment are dependent on allelic variation. Although genetic markers used for population genetic studies (e.g., microsatellites) are putatively selectively neutral, it has been argued that allelic richness of these loci is reliably indicative of allelic richness of adaptively important loci (Bataillon et al. 1996). For efforts to maintain the genetic integrity of a population in an attempt to re-create a self-sustaining, resilient population, allelic richness, rather than heterozygosity, is a more appropriate descriptor of genetic variation in coral populations for this application and should be included as criteria for developing appropriate restoration strategies.
Population genetic data from four scleractinian species (Acropora palmata, Montastraea cavernosa, Montastraea faveolata and Pocillopora damicornis; See supplementry Table S1), with various reproductive strategies (Table 2) and local and widespread larval dispersal capabilities, were used to measure the genetic diversity (allelic richness) of the sampled coral population and calculate the number of randomly sampled colonies necessary to obtain specific proportions (25, 50 and 90%) of the genetic diversity of the sampled population. A. palmata, M. cavernosa and M. faveolata are among the most important reef-building corals in the Caribbean, western Atlantic Ocean and Gulf of Mexico. P. damicornis is a common scleractinian coral with highly variable life history characteristics throughout the Indo-Pacific.
Table 2.
Scleractinian coral species used in rarefaction analysis including modes of reproduction, number and locations of populations and loci sampled and the number of randomly sampled colonies required to retain 25, 50 and 90% of the allelic richness of the entire population and to retain the mean allelic richness (~8 alleles/locus) of an “average” coral population
| Species | Sexual reproduction | Asexual propagation (impact on population structure) | No. of populations | No. of loci | No. of colonies
|
|||
|---|---|---|---|---|---|---|---|---|
| 25% | 50% | 90% | 8 alleles/locus | |||||
| Acropora palmata | Hermaphroditic broadcast spawner | Fragmentation (occasionally significant) | 9a | 4 | 3 | 7–10 | 30–35 | 7 |
| Montastraea cavernosa | Gonochoric broadcast spawner | Fission (not significant) | 5b | 5 | 2 | 5–10 | 30–35 | 10–15 |
| Montastraea faveolata | Hermaphroditic broadcast spawner | Fission (not significant) | 4c | 5 | 2–3 | 8–9 | 35–40 | 10–15 |
| Pocillopora damicornis | Hermaphroditic brooder (self-fertilization occasionally significant) | Fragmentation, larval production (occasionally significant) | 2d | 7 | 1 | 3–4 | 30 | >50 |
Belize (4), Mexico (2), Panama (2), Puerto Rico (1)
Bahamas (1), Florida (2), Bermuda (2)
Belize (4)
Fiji (2)
The program HP-rare (Kalinowski 2005) was used to calculate the number of alleles observed at each locus within a population (minimum sample size = 50) and within each population sample (in terms of genes, the equivalent of 2 times the number of colonies) per locus. Rarefaction methods can be used to statistically analyze population genetic data from populations of different sizes to predict the number of alleles expected in a random subsample of the population. Rarefaction was used to calculate allelic richness (expected number of alleles in a sample of a number of genes taken from a population; Kalinowski 2004) for each locus and population for random subsamples of 2 to 100 genes (the equivalent of 1 to 50 colonies) based on population genetic data from each of the four species. From rarefaction analyses, the number of donor colonies necessary to approximate target goals of genetic diversity preservation of a restored population was determined.
Results
A survey of the literature yielded microsatellite allelic richness data from 15 studies describing 72 populations from 13 scleractinian species (Table 1). Coral species included Caribbean and Indo-Pacific scleractinians with various reproductive strategies (broadcast spawners and brooders, with varying contributions of asexual reproduction and self-fertilization). Allelic diversity per population ranged from 2 to 29 alleles per species-specific locus with a mean of 8.27 (±0.75 SE) alleles per locus. A disadvantage of this allelic richness data is that this measure of genetic diversity is influenced by sample size (Kalinowski 2004) with higher numbers of alleles per locus with increasing sample size. Sample sizes of the surveyed populations varied widely (Table 1) and an R2 of 0.4274 was calculated for the relationship between allelic richness and sample size of all surveyed populations (data not shown). Studies with small sample sizes may underestimate the total genetic diversity of the population, and the overall mean of ~8 alleles per locus is actually a conservative estimate of allelic richness of an “average” coral population.
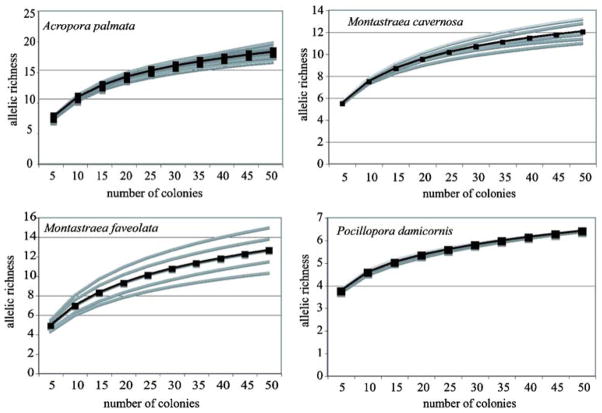
Rarefaction analysis calculated the allelic diversity of a randomly selected subset of colonies sampled from each population for each of the four species (Fig. 1). Randomly sampling 5–10 colonies from a population was sufficient to capture at least 50% of the genetic diversity of the original sampled population (Table 2). More than 30 sampled colonies retained 90% of the genetic diversity of the sampled populations. Small sample sizes (1–3 colonies) often utilized in restoration efforts captured less than 25% of the genetic diversity of the original population. These sample sizes are consistent across species. However, to obtain the mean allelic diversity of an “average” coral population (8 alleles per locus), sample sizes vary from 7 to >50 colonies across species (Table 2), which is a function of the total number of alleles per species-specific locus, as well as observed heterozygosity within the populations.
Fig. 1.
Rarefaction analysis of four scleractinian coral species. Gray lines represent individual populations and the black line represents the mean of the populations
For most populations, sample sizes of 50 colonies were insufficient to capture all of the genetic diversity within the population, as indicated by the increasing slope of the curve rather than leveling at the maximum allelic richness. Thus, 100% of the total population diversity has not been attained for these sampled populations, indicating these genetic diversity estimates as well as predictions from the rarefaction analysis underestimate the total genetic variation in coral populations.
Discussion
During periods of high population mortality, genetic bottlenecks are often observed, whereby rare neutral alleles are lost from the population and the genetic diversity, in terms of allelic richness (heterozygosity is less affected by bottlenecks; Cornuet and Luikart 1996; Kalinowski 2004), is diminished. Despite general degradation of coral reefs and recent reductions in coral population sizes due to natural and anthropogenic perturbations (e.g., bleaching events, disease outbreaks, storm damage, overfishing, increased coastal development and ship groundings), coral populations are genetically diverse with a conservative overall mean of ~8 alleles per species-specific microsatellite locus. Allozyme data also support the fact that coral populations, in general, are genetically diverse (Ayre and Hughes 2004) rather than monoclonal.
Of special interest is A. palmata, a Caribbean scleractinian listed as a threatened species under the US Endangered Species Act (Anonymous 2006) that has experienced high levels of mortality over last three decades (Aronson and Precht 1997, 2001; Jackson et al. 2001), and demonstrates strong clonal signatures in some areas of its distribution (Baums et al. 2005, 2006). Despite significant population reductions, this species demonstrated higher levels of allelic diversity per population (14.4 alleles/locus) than many other species (Table 1, Fig. 1), although lower allelic diversity in other species is partially related to small sample sizes. Most natural coral populations are not monoclonal and restoration efforts should avoid producing monoclonal populations. Clearly, fragments or asexual recruits from a single donor colony or even a few donor colonies are insufficient to replicate a significant proportion of the genetic diversity of any coral populations.
Restoration efforts should strive to preserve 100% of the genetic diversity of a population; however, this goal is impractical and virtually unattainable. An achievable genetic diversity standard based on knowledge from other populations of various species should be determined as priority for restoration efforts. Once this level of diversity is determined, results from this analysis can be used as a guideline to determine the number of donor colonies sampled from the original or adjacent population necessary to achieve this goal. For example, restoration of important reef-building Caribbean coral species (A. palmata, M. cavernosa and M. faveolata) would require sampling only 10 donor colonies to preserve the genetic diversity of an “average” coral population (8 alleles per locus), and specifically will preserve>50% of the estimated number of alleles in populations of these species. This number of donor colonies would retain ~70% of the alleles in the less genetically diverse P. damicornis populations. Because population genetic data are not available for most populations or species, sampling a minimum of 10 donor colonies is a reasonable target for any coral species. Sampling 35 colonies for use in restoration will retain >90% of the genetic diversity within an “average” population. Additional effort required to sample 10 colonies is minimal, while sampling 35 colonies may be less cost and/or time efficient, but the benefits of restoring a greater portion of the genetic diversity of the coral population are numerous (Baums 2008).
An assumption of the use of this data for restoration purposes is that a portion of the original population survived and these colonies are utilized as donor colonies. This may or may not be true depending on the nature of the disturbance. It is recommended that the donor colonies be selected from the existing remnant population, if possible, rather than from a distant, potentially genetically differentiated population. Even though alleles may be shared, it is important to maintain genotypes of local origin, especially in threatened (i.e., Caribbean Acropora spp.) or endangered populations (Fant et al. 2008). Correlations between genetic structure and habitat type have been documented in some corals (e.g., Benzie et al. 1995; Souter and Grahn 2008), indicating selective forces related to habitat may result in locally adapted genotypes. In some plant studies, the inclusion of non-local genotypes was detrimental to the restoration (Keller et al. 2000; Edmands and Timmerman 2003; Hufford and Mazer 2003; Baums 2008), and may change the ecological and evolutionary trajectories of the population or species.
The results of this study demonstrate that coral populations throughout the world are typically genetically diverse rather than monoclonal, and that when there is no prior knowledge of the genetic diversity of a damaged population, the “average” diversity calculated across species and populations can be used as an approximation of previous genetic diversity estimates. The observation that coral populations are genetically diverse indicates that restoration efforts may not utilize the minimum number of donor colonies necessary to retain a significant proportion of the genetic diversity within a coral population. Despite our limited knowledge of genetic diversity estimates for most coral species and our limited understanding of how genetic diversity directly influences sustainability of coral populations (Grober-Dunsmore et al. 2007), every practical effort should be made to maximize the success of restoration efforts, which includes assurance of sufficient genetic variation.
Supplementary Material
Acknowledgments
We thank the Florida Keys National Marine Sanctuary and the Flower Garden Banks National Marine Sanctuary, and the Governments of Bermuda, the Bahamas, Fiji, Mexico, Panama and Puerto Rico, for permission to collect scleractinian coral samples. Special thanks to S. Arnold, J. Azueta, C. Bastidas, M.A. Coffroth, J. Craig, A. Cróquer, M. del Carmen García, E. García, J. Gibson, C. Gutiérrez-Rodríguez, H. Guzmán, R.M. Loretto, M.I. Millet, D.G. Muñoz, C. Salazar, P. Sale, T. Snell, R. Steneck, A. Szmant, E. Weil and the people of Ba, Macuata, Nadroga, and Rewa provinces in Fiji for various contributions to this work. This research was supported by National Oceanic and Atmospheric Administration’s National Undersea Research Program (2000–15 and 2002–12), the National Science Foundation (OCE-95-30057 and OCE-99-07319), the National Coral Reef Institute, International Cooperative Biodiversity Group grant R21 TW006662-01 from the Fogarty International Center at the National Institutes of Health and the World Bank-Global Environment Fund Coral Reef Targeted Research program.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00338-009-0520-x) contains supplementary material, which is available to authorized users.
Contributor Information
T. L. Shearer, Email: tonya.shearer@biology.gatech.edu, School of Biology, Georgia Institute of Technology, 310 Ferst Dr., Atlanta, GA 30332-0230, USA
I. Porto, Depto. Ciencias Biológicas, Universidad de los Andes, Carrera 1N° 18A 10, Bogotá, Colombia
A. L. Zubillaga, Depto. Biología de Organismos, Universidad Simón Bolívar, Apartado 1080-A, Caracas, Venezuela
References
- Altizer S, Harvell D, Friedle E. Rapid evolutionary dynamics and disease threats to biodiversity. Trends Ecol Evol. 2003;18:589–596. [Google Scholar]
- Amar KO, Rinkevich B. A floating mid-water coral nursery as larval dispersion hub: Testing an idea. Mar Biol. 2007;151:713–718. [Google Scholar]
- Anonymous. Endangered and threatened species: Final listing determinations for elkhorn coral and staghorn coral. Fed Regist. 2006;71:26852–26872. [Google Scholar]
- Aronson RB, Precht WF. Stasis, biological disturbance, and community structure of a Holocene coral reef. Paleobiology. 1997;23:326–346. [Google Scholar]
- Aronson RB, Precht WF. White-band disease and the changing face of Caribbean coral reefs. Hydrobiologia. 2001;460:25–38. [Google Scholar]
- Ayre DJ, Hughes TP. Climate change, genotypic diversity and gene flow in reef-building corals. Ecol Lett. 2004;7:273–278. [Google Scholar]
- Ayre DJ, Resing JM. Sexual and asexual production of planulae in reef corals. Mar Biol. 1986;90:187–190. [Google Scholar]
- Bataillon TM, David JL, Schoen DJ. Neutral genetic markers and conservation genetics: Simulated germplasm collections. Genetics. 1996;144:409–417. doi: 10.1093/genetics/144.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baums IB. A restoration genetics guide for coral reef conservation. Mol Ecol. 2008;17:2796–2811. doi: 10.1111/j.1365-294X.2008.03787.x. [DOI] [PubMed] [Google Scholar]
- Baums IB, Miller MW, Hellberg ME. Regionally isolated populations of an imperiled Caribbean coral, Acropora palmata. Mol Ecol. 2005;14:1377–1390. doi: 10.1111/j.1365-294X.2005.02489.x. [DOI] [PubMed] [Google Scholar]
- Baums IB, Miller MW, Hellberg ME. Geographic variation in clonal structure in a reef-building Caribbean coral, Acropora palmata. Ecol Monogr. 2006;76:503–519. [Google Scholar]
- Benzie JAH, Haskell A, Lehman H. Variation in the genetic composition of coral (Pocillopora damicornis and Acropora palifera) populations from different reef habitats. Mar Biol. 1995;121:731–739. [Google Scholar]
- Brazeau DA, Gleason DF, Morgan ME. Self-fertilization in brooding hermaphroditic Caribbean corals: Evidence from molecular markers. J Exp Mar Biol Ecol. 1998;231:225–238. [Google Scholar]
- Carlon DB. The evolution of mating systems in tropical reef corals. Trends Ecol Evol. 1999;14:491–495. doi: 10.1016/s0169-5347(99)01709-7. [DOI] [PubMed] [Google Scholar]
- Carlon DB, Lippe C. Fifteen new microsatellite markers for the reef coral Favia fragum and a new Symbiodinium microsatellite. Molecular Ecology Resources. 2008 doi: 10.1111/j.1471-8286.2008. 02095.x. [DOI] [PubMed] [Google Scholar]
- Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBattista JD. Patterns of genetic variation in anthropogenically impacted populations. Conserv Genet. 2008;9:141–156. [Google Scholar]
- Edmands S, Timmerman CC. Modeling factors affecting the severity of outbreeding depression. Conserv Biol. 2003;17:883–892. [Google Scholar]
- Edmunds PJ. Evidence that reef-wide patterns of coral bleaching may be the result of the distribution of bleaching susceptible clones. Mar Biol. 1994;121:137–142. [Google Scholar]
- Eldridge MDB, King JM, Loupis AK, Spencer PBS, Taylor AC, Pope LC, Hall GP. Unprecedented low levels of genetic variation and inbreeding depression in an island population of the black-footed rock-wallaby. Conserv Biol. 1999;13:531–541. [Google Scholar]
- Fant JB, Holmstrom RM, Sirkin EJRE, Masi S. Genetic structure of threatened native populations and propagules used for restoration in a clonal species, American Beachgrass (Ammophila breviligulata Fern.) Restor Ecol. 2008 doi: 10.1111/j.1526-100X.2007.00348.x. [DOI] [Google Scholar]
- Foster NL, Baums IB, Mumby PJ. Sexual vs. asexual reproduction in an ecosystem engineer: the massive coral Montastraea annularis. J Anim Ecol. 2007;76:384–391. doi: 10.1111/j.1365-2656.2006.01207.x. [DOI] [PubMed] [Google Scholar]
- Fukami H, Budd AF, Levitan DR, Jara J, Kersanach R, Knowlton N. Geographic differences in species boundaries among members of the Montastraea annularis complex based on molecular and morphological markers. Evolution. 2004;58:324–337. [PubMed] [Google Scholar]
- Goffredo S, Mezzomonaco L, Zaccanti F. Genetic differentiation among populations of the Mediterranean hermaphroditic brooding coral Balanophyllia europaea (Scleractinia: Dendrophylliidae) Mar Biol. 2004;145:1075–1083. [Google Scholar]
- Grober-Dunsmore R, Bonito V, Frazer TK. Discernment of sexual recruits is not critical for assessing population recovery of Acropora palmata. Mar Ecol Prog Ser. 2007;335:233–236. [Google Scholar]
- Haig SM. Molecular contributions to conservation. Ecology. 1998;79:413–425. [Google Scholar]
- Heyward KJ, Babcock RC. Self- and cross-fertilization in scleractinian corals. Mar Biol. 1986;90:191–195. [Google Scholar]
- Hufford KM, Mazer SJ. Plant ecotypes: genetic differentiation in the age of ecological restoration. Trends Ecol Evol. 2003;18:147–155. [Google Scholar]
- Hurlbert SH. Nonconcept of species diversity: Critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- Jackson JBC, Kirby MX, Berger WH, Bjorndal KA, Botsford LW, Bourque BJ, Bradbury RH, Cooke R, Erlandson J, Estes JA, Hughes TP, Kidwell S, Lange CB, Lenihan HS, Pandolfi JM, Peterson CH, Steneck RS, Tegner MJ, Warner RR. Historical overfishing and the recent collapse of coastal ecosystems. Science. 2001;293:629–638. doi: 10.1126/science.1059199. [DOI] [PubMed] [Google Scholar]
- Jones ME, Jarman PJ, Lees CM, Hesterman H, Hamede RK, Mooney NJ, Mann D, Pukk CE, Bergfeld J, McCallum H. Conservation management of tasmanian devils in the context of an emerging, extinction-threatening disease: Devil facial tumor disease. EcoHealth. 2007;4:326–337. [Google Scholar]
- Kalinowski ST. Counting alleles with rarefaction: Private alleles and hierarchical sampling designs. Conserv Genet. 2004;5:539–543. [Google Scholar]
- Kalinowski ST. HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes. 2005;5:187–189. [Google Scholar]
- Keller M, Kollmann J, Edwards PJ. Genetic introgression from distant provenances reduces fitness in local weed populations. J Appl Ecol. 2000;37:647–659. [Google Scholar]
- LeGoff-Vitry MC, Pybus OG, Rogers AD. Genetic structure of the deep-sea coral Lophelia pertusa in the northeast Atlantic revealed by microsatellites and internal transcribed spacer sequences. Mol Ecol. 2004;13:537–549. doi: 10.1046/j.1365-294x.2004.2079.x. [DOI] [PubMed] [Google Scholar]
- Mackenzie JB, Munday PL, Willis BL, Miller DJ, van Oppen MJH. Unexpected patterns of genetic structuring among locations but not colour morphs in Acropora nasuta (Cnidaria; Scleractinia) Mol Ecol. 2004;13:9–20. doi: 10.1046/j.1365-294x.2003.02019.x. [DOI] [PubMed] [Google Scholar]
- Magalon H, Adjeroud M, Veuille M. Patterns of genetic variation do not correlate with geographical distance in the reef-building coral Pocillopora meandrina in the South Pacific. Mol Ecol. 2005;14:1861–1868. doi: 10.1111/j.1365-294X.2005.02430.x. [DOI] [PubMed] [Google Scholar]
- Maier E, Tollrian R, Rinkevich B, Nurnberger B. Isolation by distance in the scleractinian coral Seriatopora hystrix from the Red Sea. Mar Biol. 2005;147:1109–1120. [Google Scholar]
- McKay JK, Christian CE, Harrison S, Rice KJ. “How local is local?” - A review of practical and conceptual issues in the genetics of restoration. Restor Ecol. 2005;13:432–440. [Google Scholar]
- Miller KJ, Howard CG. Isolation of microsatellites from two species of scleractinian coral. Mol Ecol Notes. 2004;4:11–13. [Google Scholar]
- Miller KJ, Mundy CN. In situ fertilisation success in the scleractinian coral Goniastrea favulus. Coral Reefs. 2005;24:313–317. [Google Scholar]
- Nei M, Maruyama T, Chakraborty R. The bottleneck effect and genetic variability in populations. Evolution. 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- Perez-Ruzafa A, Gonzalez-Wanguemert M, Lenfant P, Marcos C, Garcia-Charton JA. Effects of fishing protection on the genetic structure of fish populations. Biol Conserv. 2006;129:244–255. [Google Scholar]
- Petersen D, Tollrian R. Methods to enhance sexual recruitment for restoration of damaged reefs. Bull Mar Sci. 2001;69:989–1000. [Google Scholar]
- Petit RJ, El Mousadik A, Pons O. Identifying populations for conservation on the basis of genetic markers. Conserv Biol. 1998;12:844–855. [Google Scholar]
- Precht WF. Coral reef restoration handbook. CRC Press; Boca Raton, FL: 2006. [Google Scholar]
- Precht WF, Aronson RB, Miller SL, Keller BD, Causey B. The folly of coral restoration programs following natural disturbances in the Florida Keys National Marine Sanctuary. Ecol Restor. 2005;23:24–28. [Google Scholar]
- Reed DH, Frankham R. Correlation between fitness and genetic diversity. Conserv Biol. 2003;17:230–237. [Google Scholar]
- Rinkevich B. Restoration strategies for coral reefs damaged by recreational activities: The use of sexual and asexual recruits. Restor Ecol. 1995;3:241–251. [Google Scholar]
- Rinkevich B. Steps towards the evaluation of coral reef restoration by using small branch fragments. Mar Biol. 2000;136:807–812. [Google Scholar]
- Rinkevich B. Conservation of coral reefs through active restoration measures: Recent approaches and last decade progress. Environ Sci Technol. 2005;39:4333–4342. doi: 10.1021/es0482583. [DOI] [PubMed] [Google Scholar]
- Shearer TL. Reef connectivity: genetic analysis of recruitment and gene flow among Caribbean scleractinian corals. University; Buffalo: 2004. p. 202. [Google Scholar]
- Shearer TL, Coffroth MA. Isolation of microsatellite loci from the scleractinian corals, Montastraea cavemosa and Porites astreoides. Mol Ecol Notes. 2004;4:435–437. [Google Scholar]
- Sherman CDH. Mating system variation in the hermaphroditic brooding coral, Seriatopora hystrix. Heredity. 2007;100:296–303. doi: 10.1038/sj.hdy.6801076. [DOI] [PubMed] [Google Scholar]
- Souter P, Grahn M. Spatial genetic patterns in lagoonal, reef-slope and island populations of the coral Platygyra daedalea in Kenya and Tanzania. Coral Reefs. 2008;27:433–439. [Google Scholar]
- Spielman D, Brook BW, Briscoe DA, Frankham R. Does inbreeding and loss of genetic diversity decrease disease resistance? Conserv Genet. 2004;5:439–448. [Google Scholar]
- Starger CJ, Yeoh SSR, Dai CF, Bakero AC, Desalle R. Ten polymorphic STR loci in the cosmopolitan reef coral, Pocillopora damicornis. Mol Ecol Resour. 2007;8:619–621. doi: 10.1111/j.1471-8286.2007.02017.x. [DOI] [PubMed] [Google Scholar]
- Stoddart JA. Asexual production of planulae in the coral Pocillopora damicornis. Mar Biol. 1983;76:279–284. [Google Scholar]
- Stoddart JA. Genetical structure within populations of the coral Pocillopora damicornis. Mar Biol. 1984;81:19–30. [Google Scholar]
- Stoddart JA, Babcock RC, Heyward AJ. Self-fertilization and maternal enzymes in the planulae of the coral Goniastrea favulus. Mar Biol. 1988;99:489–494. [Google Scholar]
- Szmant AM, Weil E, Miller MW, Colon DE. Hybridization within the species complex of the scleractinan coral Montastraea annularis. Mar Biol. 1997;129:561–572. [Google Scholar]
- Underwood JN, Souter PB, Ballment ER, Lutz AH, van Oppen MJH. Development of 10 polymorphic microsatellite markers from herbicide-bleached tissues of the brooding pocilloporid coral Seriatopora hystrix. Mol Ecol Notes. 2006;6:176–178. [Google Scholar]
- van Oppen MJH, Gates RD. Conservation genetics and the resilience of reef-building corals. Mol Ecol. 2006;15:3863–3883. doi: 10.1111/j.1365-294X.2006.03026.x. [DOI] [PubMed] [Google Scholar]
- van Oppen MJH, Underwood JN, Muirhead AN, Peplow L. Ten microsatellite loci for the reef-building coral Acropora millepora (Cnidaria, Scleractinia) from the Great Barrier Reef, Australia. Mol Ecol Notes. 2007;7:436–438. [Google Scholar]
- Wallace CC, Willis BL. Systematics of the coral genus Acropora: Implications of new biological findings for species concepts. Annu Rev Ecol Syst. 1994;25:237–262. [Google Scholar]
- Willis BL, Babcock RC, Harrison PL, Wallace CC. Experimental hybridization and breeding incompatibilities within the mating systems of mass spawning reef corals. Coral Reefs. 1997;16:S53–S65. [Google Scholar]
- Yeemin T, Sutthacheep M, Pettongma R. Coral reef restoration projects in Thailand. Ocean Coast Manage. 2006;49:562–575. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.