Abstract
We formed the GEnetics of Nephropathy–an International Effort (GENIE) consortium to examine previously reported genetic associations with diabetic nephropathy (DN) in type 1 diabetes. GENIE consists of 6,366 similarly ascertained participants of European ancestry with type 1 diabetes, with and without DN, from the All Ireland-Warren 3-Genetics of Kidneys in Diabetes U.K. and Republic of Ireland (U.K.-R.O.I.) collection and the Finnish Diabetic Nephropathy Study (FinnDiane), combined with reanalyzed data from the Genetics of Kidneys in Diabetes U.S. Study (U.S. GoKinD). We found little evidence for the association of the EPO promoter polymorphism, rs161740, with the combined phenotype of proliferative retinopathy and end-stage renal disease in U.K.-R.O.I. (odds ratio [OR] 1.14, P = 0.19) or FinnDiane (OR 1.06, P = 0.60). However, a fixed-effects meta-analysis that included the previously reported cohorts retained a genome-wide significant association with that phenotype (OR 1.31, P = 2 × 10−9). An expanded investigation of the ELMO1 locus and genetic regions reported to be associated with DN in the U.S. GoKinD yielded only nominal statistical significance for these loci. Finally, top candidates identified in a recent meta-analysis failed to reach genome-wide significance. In conclusion, we were unable to replicate most of the previously reported genetic associations for DN, and significance for the EPO promoter association was attenuated.
Type 1 diabetes has continuously increased worldwide, and the highest incidence is found in Finland (1). Diabetic nephropathy (DN) is a complication that develops in approximately 25–40% of patients with type 1 and type 2 diabetes. DN is the leading cause of end-stage renal disease (ESRD) in the developed world. Currently, 44% of the new cases of ESRD in the U.S. annually are attributable to DN (2). A better understanding of the causal factors of DN and its pathogenesis may lead to new strategies to decrease its incidence, preemptively treat the disorder, attenuate morbidity and mortality, and would be a valuable contribution to global public health.
Several observations suggest that DN, one of the major complications of type 1 and type 2 diabetes, has an inherent genetic susceptibility. Familial clustering of DN is evident for both type 1 and type 2 diabetes (3–6), and genetic risk factors are being sought in multiple populations (7–9). Unfortunately, robust replication of many initial associations has not been forthcoming (10).
This study recruited a large collection of individuals with type 1 diabetes as part of the GEnetics of Nephropathy–an International Effort (GENIE) consortium and examined selected candidate loci associated with DN from genome-wide case-control studies or other association studies that reported high levels of statistical significance. The variants examined and the rationale for their inclusion are as follows:
A single nucleotide polymorphism (SNP) (rs1617640) within the promoter region of the EPO gene (encoding erythropoietin) was identified as having a genome-wide significant (P < 5 × 10−8) association with ESRD and proliferative diabetic retinopathy (PDR) (11). Interestingly, erythropoietin levels were elevated sevenfold in the human vitreous fluid of nondiabetic individuals with the risk genotype TT compared with those with the wild-type GG genotype. In addition, EPO expression levels were significantly elevated above control in the tissues and vitreous fluid of animal models of DN (DN in db/db mice) and in proliferative retinopathy (murine oxygen-induced retinopathy model), respectively (11).
The engulfment and cell motility 1 gene (ELMO1) has been reported to be associated with DN in Japanese patients with type 2 diabetes (12). Recently, Pezzolesi et al. (13), using the Genetics of Kidneys in Diabetes U.S. Study (U.S. GoKinD) cohorts, also examined ELMO1 for association with DN and presented evidence of association of variants within this gene for the development of DN. However, the risk alleles for ELMO1 identified in their study differed from those reported in the original Japanese investigation. In the context of a genome-wide association study (GWAS), 118 SNPs were assessed in 1,705 individuals of European ancestry with type 1 diabetes (885 control subjects and 820 DN case subjects). The strongest associations in ELMO1 in the U.S. study occurred at rs11769038 (odds ratio [OR] 1.24; P = 1.7 × 10−3) and rs1882080 (OR 1.23; P = 3.2 × 10−3), located in intron 16. Two additional SNPs, located in introns 18 and 20, were also nominally associated with DN. In total, eight ELMO1 SNPs were reported to confer risk for DN, although none reached genome-wide significance (13). Supportive evidence was also found in African Americans with type 2 diabetes and ESRD (14).
The U.S. GoKinD GWAS analyzed 359,193 SNPs in 820 case subjects (284 with proteinuria and 536 with ESRD) and 885 control subjects with type 1 diabetes but no evidence of DN. Although no risk variant achieved genome-wide significance, the primary association analysis identified 11 SNPs representing four distinct chromosomal regions (P < 1 × 10−5). The strongest association with DN reported in this study was on chromosome 9q with rs10868025 (OR 1.45, P = 5.0 × 10−7) (15).
Finally, in an effort to systematically explore and comprehensively capture common genetic variations that might be associated with DN, we reviewed the largest meta-analysis published to date studying genetic associations with the DN phenotype (7). In GENIE, we examined the top-reported SNP (or proxy) for each gene in that report for an association with DN.
In this study, we have assembled the largest reported case-control sample of DN in type 1 diabetes to evaluate the previously reported genetic associations in newly genotyped samples from the U.K., Republic of Ireland (R.O.I.), and Finland, plus pre-existing data from the U.S. GoKinD.
RESEARCH DESIGN AND METHODS
Cohorts
U.K.-R.O.I. collection.
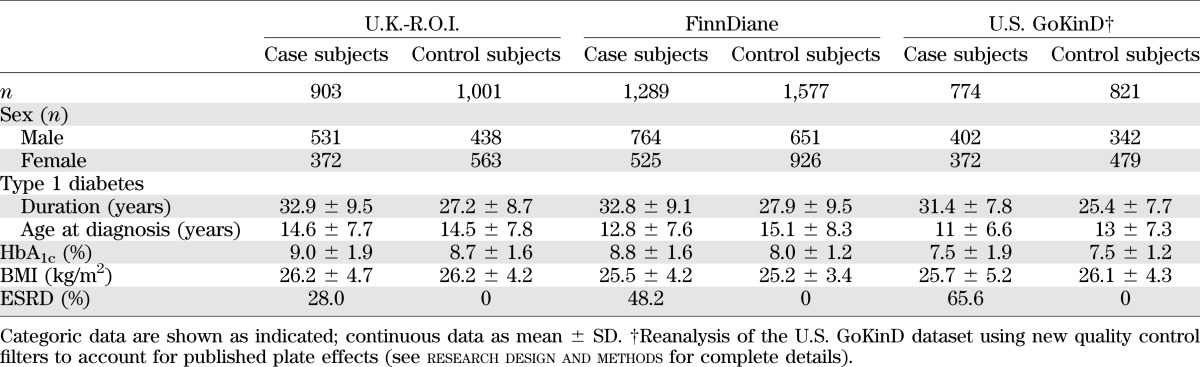
Recruited individuals were part of the All Ireland-Warren 3-Genetics of Kidneys in Diabetes U.K. collection (U.K.-R.O.I.). All were self-reported as white, with grandparents born in the U.K. or Ireland, and type 1 diabetes diagnosed before the age of 31 years requiring uninterrupted insulin treatment. Case subjects (n = 903) with DN had persistent proteinuria (>0.5 g/24 h), hypertension (>135/85 mmHg and/or treatment with antihypertensive medication), and diabetic retinopathy. ESRD (28%) was defined as requiring renal replacement therapy or having received a kidney transplant. Individuals in the control group (n = 1,001) had had type 1 diabetes for at least 15 years, had no evidence of microalbuminuria on repeated testing, and were not receiving antihypertensive medication (Table 1).
TABLE 1.
Phenotypic characteristics of the GENIE cohorts (U.K.-R.O.I., FinnDiane, and U.S. GoKinD)
Finnish Diabetic Nephropathy Study (FinnDiane).
The FinnDiane study is a nationwide multicenter study of >4,800 adult participants with type 1 diabetes (16). This study comprises genotype data for 2,914 patients with type 1 diabetes diagnosed before age 35 years and insulin treatment started within 1 year of diagnosis. The disease status was defined by urine albumin excretion rate (AER) or urine albumin-to-creatinine ratio (ACR) in at least two of three consecutive urine collections at local centers. Macroalbuminuria (n = 686) was defined as AER >200 μg/min or >300 mg/24 h or an ACR >25 mg/mmol for men and >35 mg/mmol for women in overnight, 24-h, or spot urine collections, respectively. Similarly, the limit for normal AER (n = 1,601) was <20 μg/min or <30 mg/24 h or ACR <2.5 mg/mmol for men and <3.5 mg/mmol for women. Control patients with normal AER were required to have type 1 diabetes duration of at least 15 years. ESRD (n = 627) was defined as ongoing dialysis treatment or receipt of a kidney transplant. From the total, 505 participants were included from an independent Finnish cohort collected by the National Institute for Health and Welfare (17). These participants met the FinnDiane diagnosis and selection criteria and were analyzed together with the FinnDiane cohort (Table 1).
U.S. GoKinD.
The U.S. GoKinD study consists of a DN case-control cohort of individuals diagnosed with type 1 diabetes before age 31 years, who were between 18 and 59 years of age at enrollment, and who began insulin treatment within 1 year after diagnosis (18). The 905 case subjects were defined as people aged 18–54, with type 1 diabetes for at least 10 years, and DN. The 898 control subjects were aged 18–59, had type 1 diabetes for at least 15 years, but did not have DN. The DN definition includes individuals with ESRD (on dialysis or having received a kidney transplant) or persistent macroalbuminuria (at least two of three tests positive for albuminuria by dipstick ≥1+ or ACR >300 μg albumin/mg of urine creatinine). The U.K. GoKinD inclusion criteria were used to recruit individuals to the control group. Individuals were recruited at two study centers, George Washington University (GWU) and the Joslin Diabetes Center (JDC) using differing methods of ascertainment and recruitment (see Pezzolesi et al. [15] for details). Analysis of the U.S. GoKinD cohort was limited to individuals whose reported primary ethnicity was white.
Phenotype definition: DN and EPO study outcomes.
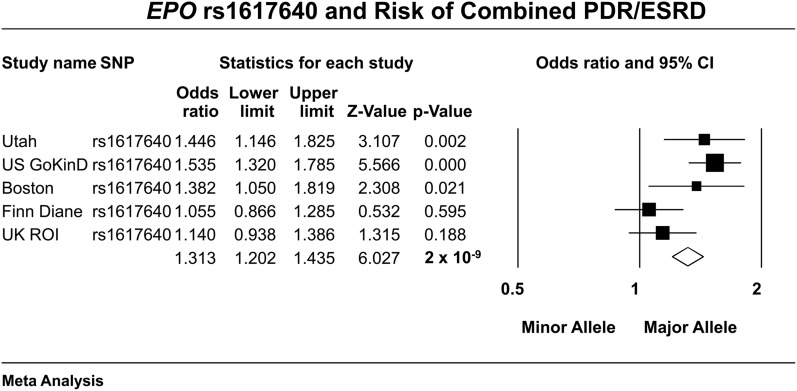
DN was the primary outcome for all association studies of the SNPs investigated. For the EPO study, we used the phenotypic definitions used in the original report (11); namely, ESRD as defined by case subjects who were on dialysis or who had received a renal transplant; concurrently, this ESRD population also had to have evidence of advanced PDR on physical examination and a history of laser treatment. Control subjects with evidence of PDR were excluded. We also examined PDR as an outcome independent of ESRD status. For this analysis, case subjects had clinical evidence of PDR, whereas control subjects had none (irrespective of DN status). Cohorts analyzed in the original EPO study (Fig. 1) were composed of European-American cases, and control subjects were collected from distinct geographic areas in the U.S. These included the GoKinD cohort (Boston, Pittsburgh, and Minnesota), the Utah cohort (Salt Lake City), and the Boston cohort (Boston Joslin Center for Diabetes) (11).
FIG. 1.
Previously published and new results from this study provide an estimate of the effect of the EPO promoter SNP (rs1617640) on the risk of the combined phenotype of PDR and ESRD in type 1 diabetes in five cohorts (3,162 case subjects and 3,845 control subjects) in a fixed-effects meta-analysis.
SNP selection.
SNP markers with evidence for association with DN susceptibility in reference studies (11) were selected for genotyping in GENIE. Where more than one SNP was associated at a particular locus with DN, the most strongly associated variant was selected for genotyping in the DN case-control cohorts. Where no genotyping assay could be developed for the index SNP, a proxy in strong linkage disequilibrium (LD) was genotyped using the CEU HapMap population. CEU is the official three letter code for the HapMap samples of Utah residents with ancestry from Northern and Western Europe (see http://hapmap.ncbi.nlm.nih.gov/citinghapmap.html). For the original U.S. GoKinD reported results and ELMO1, additional SNPs within 20-kb upstream and downstream of the locus (or index SNP) were selected using the SNP Annotation and Proxy Search (http://www.broadinstitute.org/mpg/snap/), specifying chromosome position, CEU samples, and 1000 Genome Pilot 1 data. The expanded SNP list was extracted from GWAS results for U.K.-R.O.I. and FinnDiane (N. Sandholm et al., submitted).
De novo genotyping.
For the U.K.-R.O.I. collection (n = 1,904 unique individuals), SNPs were genotyped using Sequenom iPLEX (Sequenom, Hamburg, Germany) or TaqMan (Applied Biosystems, Warrington, U.K.) technology. Duplicate and no-DNA-template samples were included on all plates as experimental controls.
In FinnDiane, the EPO locus was genotyped with TaqMan chemistry (Applied Biosystems, Foster City, CA) in 3,363 samples, of which 251 case subjects (ESRD + laser treatment) and 987 control subjects (no DN, no retinopathy) passed the phenotype criteria. All other SNPs were genotyped at the Institute of Molecular Medicine Finland (Helsinki, Finland) on the Illumina's BeadArray 610Quad array. Illumina’s BeadStudio clustering algorithm was used to call genotypes in FinnDiane. SNPs were filtered for those with call rates >95%, minor allele frequency (MAF) >1%, and test for Hardy Weinberg equilibrium (HWE; P > 1 × 10−3). Sample filters included individual call rates >95% and no first-degree relatives and resulted in 1,289 case subjects and 1,577 control subjects.
Existing genotype data for the U.S. GoKinD genotype data were downloaded from dbGAP (phs000018.v2.p1, retrieved June 2010), containing genotype data from the Affymetrix 500 K array (Affymetrix, Santa Clara, CA). The version 2 genotype data differed from the original U.S. GoKinD data, containing updated and recalled genotype calls for a previously reported problematic plate (19), and SNP call rate (>90%), MAF (>1%), and HWE performed by the National Heart, Lung, and Blood Institute. We applied additional quality control (QC) filters to the data, including removing samples with evidence of contamination (extreme heterozygosity, n = 16), known parents (n = 4), non–European-reported ancestry (n = 121), reported sex–genotype mismatch (n = 1), cryptic related subjects (n = 4), and principal component analysis admixture outliers (n = 5). Furthermore, SNP QC filters were applied to remove known problematic SNPs, HWE (P > 1 × 10−7), call rate (90%), missingness, MAF (1%), and SNPs plate effects.
GWAS genotyping.
Genotypes for U.K.-R.O.I. (n = 1,830) and FinnDiane (n = 3,651) were supplemented with SNPs retrieved from GWAS results for each study (Sandholm et al., submitted). The FinnDiane study samples were genotyped using the Illumina's BeadArray 610-Quad (Illumina, San Diego, CA) array at the Institute of Molecular Medicine Finland (FIMM, Helsinki, Finland). The U.K.-R.O.I. study samples were genotyped at the Broad Institute using the Illumina Omni1-Quad array. Samples with insufficient DNA quality, quantity, or poor genotype concordance with previous available genotypes at a fingerprinting stage were excluded. For all three discovery datasets (FinnDiane, U.K.-R.O.I., U.S. GoKinD), a standardized and detailed genotype QC procedures were applied using PLINK supplemented with perl and R scripts. These included selecting SNPs with a call rate >90%, MAF >1%, and concordance with HWE (P > 10−7). We discarded samples with a call rate <95%, extreme heterozygosity, cryptic relatedness, or ethnic outliers by principal components analysis, and tested for SNP missingness by haplotype (P < 10−8), by phenotype (P > 10−8), and by plate effects (P < 10−7). Probes for copy number variation as well as sex chromosome and mitochondrial SNPs were excluded from analyses. After applying the QC protocol, we had access to 549,530 SNPs in 3,370 FinnDiane samples, 791,687 SNPs in 1,726 U.K.-R.O.I. samples, and 360,899 SNPs in 1,595 U.S. GoKinD samples.
SNP imputation.
MACH 1.0 software (http://www.sph.umich.edu/csg/abecasis/MACH) with the HapMap phase II CEU reference panel was used to perform SNP imputation for GWAS results in each cohort. Estimates of the crossover and error rates were obtained via 50 iteration rounds in ∼300 randomly selected samples per cohort. A greedy algorithm was used for imputation, and the maximum likelihood method was specified to yield allele dosages. A filter was applied to exclude SNPs with low imputation quality (r2 < 0.6), resulting in ∼2.4 million SNPs per cohort.
Statistical analysis.
Association tests were conducted using PLINK v1.07 (20) (http://pngu.mgh.harvard.edu/purcell/plink), with logistic regression adjusted for sex and age. U.K.-R.O.I. was adjusted for recruitment center, but the two U.S. GoKinD centers, GWU and JDC, were analyzed separately as reported by Pezzolesi et al. (15). Data from the GWAS genotyping was adjusted additionally for duration of type 1 diabetes and principal components from Eigenstrat analysis. The EPO locus was analyzed with the Pearson χ2 test in the FinnDiane dataset without adjusting for any covariates. Fixed-effects meta-analyses were conducted with the software package Comprehensive Meta-Analysis (Version 2.2.040, Englewood, NJ) and the software package METAL (http://www.sph.umich.edu/csg/abecasis/Metal/) (21) under the additive genetic model. To determine the appropriate statistical cutoff for correction for multiple testing, we calculated the total number of effective tests (because a large portion of SNPs were in LD), using SNPSpD (http://gump.qimr.edu.au/general/daleN/SNPSpD/). SNPSpD uses correlation between analyzed SNPs to calculate the total number of independent tests (22). The total number of SNPs tested is 2,199, with 113.7 effect-independent tests. Thus the experiment-wide cutoff for statistical significance was set at 4.4 × 10−4 (0.05/113.7).
RESULTS
EPO promoter polymorphism.
The association of the EPO promoter polymorphism, rs1617640, with DN was evaluated by de novo genotyping of the SNP in GENIE. For this analysis, case subjects were defined as having both ESRD and PDR because the initial report showed the polymorphism was robustly associated with DN when both of these “extreme” phenotypes were coexpressed. Significant association was not observed in the U.K.-R.O.I. (P = 0.19) or FinnDiane collections (P = 0.60), although the directions of effect were consistent with the original report. Fixed-effects meta-analysis of the association of rs1617640 with ESRD/PDR, including the previously reported cohorts (a total of 3,162 case and 3,845 control subjects across five separate cohorts of European and European-American ancestry) retained genome-wide statistical significance (OR 1.31 [95% CI 1.20–1.44], P = 2 × 10−9; Fig. 1).
As an additional experimental control, we examined the potential association of the EPO promoter polymorphism with the development of PDR in case subjects, irrespective of ESRD status. No association was observed between EPO and PDR for the individual cohorts or in the meta-analysis of the combined results for FinnDiane (OR 0.95 [95% CI 0.85–1.04], P = 0.25) or and U.K.-R.O.I. (0.96 [0.88–1.04], P = 0.29). Furthermore, no association was observed after inclusion of the restricted phenotype, PDR, in U.S. GoKinD case and control subjects, separately, or in the meta-analysis of all cohorts combined (results not shown).
ELMO1.
Neither single-center nor meta-analysis of de novo genotyping in U.K.-R.O.I., nor GWAS data for FinnDiane, revealed a significant association in subjects with type 1 diabetes between rs741301, the previously reported risk variant within ELMO1, and DN (OR 1.04 [95% CI 0.95–1.13], P = 0.46; Supplementary Fig. 1). Pezzolesi et al. (13) also tested rs741301 but did not replicate the reported association. They went on to test other SNPs in the region and reported nominal associations (P = 0.002–0.05) with eight other SNPs . We examined LD between rs741301 and the SNPs reported to be associated with DN by these investigators. The r2 statistic between rs741301 and the other SNPs revealed only low to moderate LD, ranging from 0.38 to 0.65 (Supplementary Table 1).
As an additional and more extensive test of variants in this region, we performed an expanded analysis capturing all available SNPs 20 kb upstream and downstream of the ELMO1 locus to account for LD differences presumably due to ancestry. Results of the expanded analysis did not reveal any significant SNPs for either cohort individually or for the two cohorts meta-analyzed after correcting for multiple testing (P < 4.3 × 10−4). Furthermore, no SNP achieved significance with inclusion of the U.S. GoKinD results in the meta-analysis (Supplementary Table 2).
Risk variants reported from U.S. GoKinD for DN in type 1 diabetes.
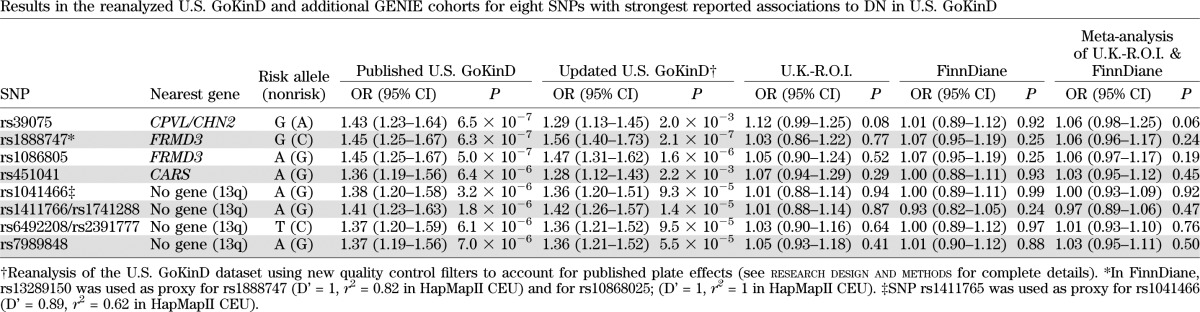
Eleven DN susceptibility SNPs that were first reported by the U.S. GoKinD investigators as highly associated with the risk of developing DN in type 1 diabetes were parsed into eight candidate loci. We selected one representative SNP for each region of strong LD in which multiple SNPs represented the same association signal. After performing additional QC checks (see research design and methods), we first tested the eight U.S. GoKinD potential DN susceptibility SNPs by reanalyzing the U.S. GoKinD dataset downloaded from dbGAP (19). As in the original report, none of the eight SNPs were associated with DN at genome-wide statistical significance (Table 2). The two SNPs in the FERM (F [Band 4.1], E [Ezrin], R [Radixin], M [Moesin]) domain 3 (FRMD3) region showed similar P values as in the original report (2.1 × 10−7 and 1.6 × 10−6, respectively; Table 2). In our analysis, the statistical significance of the P values for SNPs in the CPVL/CHN2 and CARS regions was reduced from 6.5 × 10−7 to 2 × 10−3 and 6.4 × 10−6 to 2.2 × 10−3, respectively. P values for the 6 SNPs in the 13q region were also changed from 1.8–7.0 × 10−6 to 1.4–9.5 × 10−5.
TABLE 2.
Results in the reanalyzed U.S. GoKinD and additional GENIE cohorts for eight SNPs with strongest reported associations to DN in U.S. GoKinD
We next examined these SNPs in our newly genotyped samples. Case-control association analysis for these loci in the U.K.-R.O.I. and FinnDiane samples revealed no significant associations in either cohort. The strongest signal was observed at rs39075 near CPVL/CHN2 in U.K.-R.O.I., (OR 1.12, P = 0.08) and in meta-analysis of the two replication cohorts (U.K.-R.O.I. and FinnDiane; OR 1.06, P = 0.06; Table 2). In expanded locus-region analyses (plus or minus 20 kb of the locus of interest), no SNP reached significance after adjustment for multiple testing (one-tailed P = 0.03, experiment-wide threshold P = 4.3 × 10−4) for the two cohorts separately or via meta-analysis. The combined meta-analysis including U.S. GoKinD revealed two SNPs downstream of FRMD3, rs1888747 (P = 1.5 × 10−4) and rs13288659 (P = 9.7 × 10−5), which showed significance after adjusting for experiment-wide multiple testing (P < 4.3 × 10−4). However, neither SNP achieved genome-wide significance (P < 5 × 10−8; Supplementary Table 3).
Pooled meta-analyses examining variants associated with DN in type 1 and type 2 diabetes.
In the most comprehensive literature search for DN associated genetic variants to date, Mooyaart et al. (7) identified 24 loci. In GENIE, we examined all the available top-reported SNPs (or their proxies) for each gene in that report. Three SNPs were nominally associated (P < 0.05) with DN: rs13293564 at UNC13B (P = 0.01) and rs179975 at the ACE (P = 0.03) in FinnDiane, and rs39075 at CPVL/CHN2 (P = 0.05) in the U.K.-R.O.I. samples. In a meta-analysis of the two cohorts, the ACE polymorphism remained nominally significant (P = 0.04). Including the U.S. GoKinD results, the FRMD3 signal at rs1888747 emerged as noted above; no other signals were significant after adjusting for multiple comparisons (see Supplementary Table 4 for full details).
DISCUSSION
Using a large, homogeneous population sample of European ancestry subjects with type 1 diabetes in the GENIE consortium, we were unable to replicate most of the previously reported genetic associations with DN that we examined.
Our findings do not support previously reported genetic associations with DN in type 1 diabetes in the largest GWAS published to date (15). None of those signals reached genome-wide statistical significance with the addition of larger, similarly ascertained datasets. Using ORs at the lower limit of the 95% CI from the original publication, our combined sample of U.K.-R.O.I. and FinnDiane cohorts in GENIE had 99.9% power (at α = 0.001) to detect the U.S. GoKinD reported effect sizes (Supplementary Table 5); thus, our negative results (even at α = 0.05) were unexpected. Further, after performing additional QC checks on the original U.S. GoKinD dataset and combining these samples with GENIE, we were unable to achieve genome-wide significant replication in the meta-analysis.
We investigated other previously reported genetic associations with DN in subjects with type 1 diabetes, namely the EPO promoter polymorphism rs1617640 and variants within the ELMO1 gene; these also failed to replicate in GENIE. The EPO promoter polymorphism retained genome-wide significance after meta-analysis of the prior data combined with ours, for the combined phenotype of ESRD and PDR. However, the overall P value attained was attenuated from 2.8 × 10−11 to 2 × 10−9.
We also did not observe evidence for replication of the association of rs741301 in the ELMO1 gene and DN in GENIE. Shimazaki et al. (12) first reported an association for this genetic variant with DN in Japanese subjects with type 2 diabetes. Subsequently, Pezzolesi et al. (13) failed to replicate the association of the same SNP in type 1 diabetes but reported that eight SNPs within the gene locus had nominal associations with DN in the European-derived subjects in U.S. GoKinD. Lack of replication may be due to different genomic patterns in populations of diverse ancestries or the distinct genetic architecture of DN in type 1 versus type 2 diabetes (14). To address the first concern, we expanded our analysis by interrogating all available SNPs plus or minus 20 kb of ELMO1. Because in the U.S. GoKinD analysis of ELMO1 the risk variants were only in weak to modest LD with the index SNP (r2 between rs741301 and the U.S. GoKinD SNPs ranged from 0.38 to 0.65), we reasoned that this expanded strategy would account for most of the differences in LD presumably due to ancestry. The results of this expanded analysis, however, did not reveal any significant association for the additional 670 SNPs interrogated in ELMO1 in the U.K.-R.O.I. or FinnDiane cohorts individually, for these two cohorts in meta-analysis, or when combined with previously reported risk variants reported from U.S. GoKinD.
As a final step to ensure a systematic and more comprehensive approach to candidate loci associated with DN, we reviewed the top SNPs from the 24 loci cited by the largest meta-analysis published to date that has examined candidate genetic variants for association with DN (7). From the results of the pooled meta-analyses, the strongest signal emerged at rs1888747 in FRMD3 (P = 1.5 × 10−4). Variants within this gene have been reported to be associated with DN in European cohorts (7,15,23), and Freedman et al. (24) recently reported evidence for a role for gene–gene interactions between myosin heavy-chain 9 (MYH9)–apolipoprotein L1 (APOL1) haplotypes and FRMD3 in African American subjects. It should be noted, however, that risk variants within this gene have achieved only nominal but not genome-wide statistical significance in all previous reports.
From the foregoing observations, we conclude that there is a high likelihood that many of the previously reported positive associations with DN represent potential false-positive findings (type I error). We emphasize that the combined sample of U.K.-R.O.I. and FinnDiane represents a substantially larger collection of case and control subjects than U.S. GoKinD and is well powered to detect the originally reported effect sizes, thereby decreasing the likelihood of a false-negative finding (type II error), even accounting for the likely overestimation of effect sizes due to the winner’s curse phenomenon (25). We also harmonized the ascertainment criteria for case-control definitions across all the study populations (including U.S. GoKinD), making it unlikely that phenotypic heterogeneity across study populations explains the lack of replication.
A crucial issue that bears on the interpretation of case-control studies of the genetics of DN concerns the adequacy of phenotype definition. In this and most studies cited to date, there is the presumption that long-duration diabetes exposure and the presence of frank protein in the urine—macroalbuminuria—defines DN and that phenotypic heterogeneity has been well controlled through this classification. These definitions are derived in large measure from the classic studies of Parving et al. (26), Viberti et al. (27), and Mogensen and Christensen (28), who documented 30 years ago a virtually inexorable progression to ESRD in patients who developed microalbuminuria after approximately 2 decades of exposure to the diabetic metabolic milieu. However, these longitudinal findings were based on small numbers of patients. The plasticity of DN phenotypes is reflected in more recent and much larger longitudinal studies showing that most patients with type 1 diabetes, categorized initially as having microalbuminuria, undergo regression to normoalbuminuria with preservation of renal function (29). It is not entirely clear that microalbuminuria versus macroalbuminuria, stage of chronic kidney disease and attendant renal function, the rate of renal decline, or the occurrence of extreme phenotypes, such as ESRD/PDR, represent one disease process along a continuum or many distinct disease states, each of which may be under distinct genetic control. As pointed out recently (24), genetic variants, such as those in MYH9 and APOL1 that are common in certain ethnic groups, may mask the effects at other loci unless methods such as multilocus modeling and interaction analyses are used to control for these effects.
In addition, phenotypic variation may be a function of ethnicity and disease-specific gene expression. For example, Pima Indians with type 2 diabetes have very early-onset DN, characterized by an accelerated loss of renal function and progression to ESRD despite lower blood pressures and lipid levels, factors thought to be protective (30). This has been postulated to be due to structural differences in the nephron–podocyte number and density per glomerulus (“podocyte insufficiency”), a decrease in net nephron mass (glomerulopenia) resulting in glomerulomegaly, increased intraglomerular capillary pressure, and ultimately, hyperfiltration injury (30). Whether these structural and intrarenal hydraulic changes could be genetically regulated is ultimately a testable hypothesis; they warrant further investigation to continue the inquiry why certain populations have an apparent disproportional susceptibility to ESRD and, particularly, DN.
In summary, we have presented evidence that several previously reported genetic associations with DN in type 1 diabetes could not be replicated in a large, homogeneous sample of subjects with type 1 diabetes. Our failure to replicate these associations underscores the need to apply stringent statistical thresholds of significance, maximize power through meta-analysis of all available data, and seek replication in independent samples, as has been proposed by a number of different authors (31,32). Finally, the applicability and generalizability of DN risk loci from type 1 diabetes to type 2 diabetes, and the related question of shared genetic susceptibility for nephropathy between type 1 and type 2 diabetes, remain unresolved.
Supplementary Material
ACKNOWLEDGMENTS
Funding for this study was provided by National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases Grant R01-DK-081923 to J.N.H., P.-H.G., and J.C.F., a Science Foundation Ireland U.S.-Ireland R&D partnership, and The Health Research Board Ireland. R.M.S. was supported by a Juvenile Diabetes Research Foundation postdoctoral fellowship (#3-2011-70). The FinnDiane Study was supported by grants from the Folkhälsan Research Foundation, the Wilhelm and Else Stockmann Foundation, Liv och Hälsa Foundation, Helsinki University Central Hospital Research Funds (EVO), the Sigrid Juselius Foundation, the Signe and Arne Gyllenberg Foundation, Finska Läkaresällskapet, the European Commission, and the European Union’s Seventh Framework Program (FP7/2007-2013) for the Innovative Medicine Initiative under Grant Agreement No. IMI/115006 (the Surrogate Markers for Micro- and Macro-vascular Hard Endpoints for Innovative Diabetes Tools [SUMMIT] consortium).
J.C.F. has received consulting honoraria from Novartis, Eli Lilly, and Pfizer. No other potential conflicts of interest relevant to this article were reported.
W.W.W. contributed to study design and management, data acquisition, statistical and data analysis, and manuscript preparation and review. R.M.S. contributed to data acquisition, genotyping, statistical and data analysis, and manuscript review. A.J.M. contributed to data acquisition, statistical and data analysis, and manuscript preparation. N.S. contributed to data acquisition, statistical and data analysis, and manuscript preparation and review. C.F. contributed to data acquisition, statistical and data analysis, and manuscript review. A.T., C.Gu., and M.P. contributed to genotyping and manuscript review. J.B.M., E.F., V.H., R.L., D.G., K.H., J.K., M.R.-B., N.T., M.S., J.W., and B.H. contributed to data acquisition and manuscript review. G.J.M., T.I., E.P.B., D.M.S., C.P., S.B., F.M., C.Go., and A.P.M. contributed to manuscript review. J.S. contributed to genotyping and statistical and data analysis. L.T., J.P., C.S., J.T., and A.-M.Ö. contributed to data acquisition. A.S. contributed to data acquisition, genotyping, and manuscript review. K.T. contributed to statistical and data analysis and manuscript review. J.N.H. contributed to study design and manuscript review. P.-H.G. contributed to study design, statistical and data analysis, and to manuscript review. J.C.F. contributed to study design and management, statistical and data analysis, and manuscript preparation and review. J.C.F. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors acknowledge the physicians, nurses, and researchers at each center participating in the collection of patients: in the FinnDiane study group and at the Helsinki University Central Hospital, Department of Medicine, Division of Nephrology: C. Forsblom, A. Ahola, J. Fagerudd, M. Feodoroff, D. Gordin, V. Harjutsalo, O. Heikkilä, K. Hietala, J. Kytö, M. Lehto, S. Lindh, M. Parkkonen, K. Pettersson-Fernholm, M. Rosengård-Bärlund, M. Rönnback, A. Sandelin, A.-R. Salonen, L. Salovaara, M. Saraheimo, T. Soppela, A. Soro-Paavonen, P. Summanen, L. Thorn, N. Tolonen, J. Tuomikangas, T. Vesisenaho, and J. Wadén. Anjalankoski Health Centre: S. Koivula and T. Uggeldahl. Central Finland Central Hospital, Jyväskylä: T. Forslund, A. Halonen, A. Koistinen, P. Koskiaho, M. Laukkanen, J. Saltevo, and M. Tiihonen. Central Hospital of Åland Islands, Mariehamn: M. Forsen, H. Granlund, A.-C. Jonsson, and B. Nyroos. Central Hospital of Kanta-Häme, Hämeenlinna: P. Kinnunen, A. Orvola, T. Salonen, and A. Vähänen. Central Hospital of Länsi-Pohja, Kemi: H. Laukkanen, P. Nyländen, and A. Sademies. Central Ostrabothnian Hospital District, Kokkola: S. Anderson, B. Asplund, U. Byskata, P. Liedes, M. Kuusela, and T. Virkkala. City of Espoo Health Centre Espoonlahti: A. Nikkola and E. Ritola; Tapiola: M. Niska and H. Saarinen; Samaria: E. Oukko-Ruponen and T. Virtanen; and Viherlaakso: A. Lyytinen. City of Helsinki Health Centre Puistola: H. Kari and T. Simonen; Suutarila: A. Kaprio, J. Kärkkäinen, and B. Rantaeskola; and Töölö: P. Kääriäinen, J. Haaga, and A.-L. Pietiläinen. City of Hyvinkää Health Centre: S. Klemetti, T. Nyandoto, E. Rontu, and S. Satuli-Autere. City of Vantaa Health Centre Korso: R. Toivonen and H. Virtanen; Länsimäki: R. Ahonen, M. Ivaska-Suomela, and A. Jauhiainen; Martinlaakso: M. Laine, T. Pellonpää, and R. Puranen; Myyrmäki: A. Airas, J. Laakso, and K. Rautavaara; Rekola: M. Erola and E. Jatkola; and Tikkurila: R. Lönnblad, A. Malm, J. Mäkelä, and E. Rautamo. Heinola Health Centre: P. Hentunen and J. Lagerstam. Herttoniemi Hospital, Helsinki: V. Sipilä. Hospital of Lounais-Häme, Forssa: T. Kalliomäki, J. Koskelainen, R. Nikkanen, N. Savolainen, H. Sulonen, and E. Valtonen. Iisalmi Hospital: E. Toivanen. Jokilaakso Hospital, Jämsä: A. Parta and I. Pirttiniemi. Jorvi Hospital, Helsinki University Central Hospital: S. Aranko, S. Ervasti, R. Kauppinen-Mäkelin, A. Kuusisto, T. Leppälä, K. Nikkilä, and L. Pekkonen. Jyväskylä Health Centre, Kyllö: K. Nuorva and M. Tiihonen. Kainuu Central Hospital, Kajaani: S. Jokelainen, P. Kemppainen, A.-M. Mankinen, and M. Sankari. Kerava Health Centre: H. Stuckey and P. Suominen. Kirkkonummi Health Centre: A. Lappalainen, M. Liimatainen, and J. Santaholma. Kivelä Hospital, Helsinki: A. Aimolahti and E. Huovinen. Koskela Hospital, Helsinki: V. Ilkka and M. Lehtimäki. Kotka Health Centre: E. Pälikkö-Kontinen and A. Vanhanen. Kouvola Health Centre: E. Koskinen and T. Siitonen. Kuopio University Hospital: E. Huttunen, R. Ikäheimo, P. Karhapää, P. Kekäläinen, M. Laakso, T. Lakka, E. Lampainen, L. Moilanen, L. Niskanen, U. Tuovinen, I. Vauhkonen, and E. Voutilainen. Kuusamo Health Centre: T. Kääriäinen and E. Isopoussu. Kuusankoski Hospital: E. Kilkki, I. Koskinen, and L. Riihelä. Laakso Hospital, Helsinki: T. Meriläinen, P. Poukka, R. Savolainen, and N. Uhlenius. Lahti City Hospital: A. Mäkelä and M. Tanner. Lapland Central Hospital, Rovaniemi: L. Hyvärinen, S. Severinkangas, and T. Tulokas. Lappeenranta Health Centre: P. Linkola and I. Pulli. Lohja Hospital: T. Granlund, M. Saari, and T. Salonen. Loimaa Health Centre: A. Mäkelä and P. Eloranta. Länsi-Uusimaa Hospital, Tammisaari: I.-M. Jousmaa and J. Rinne. Malmi Hospital, Helsinki: H. Lanki, S. Moilanen, and M. Tilly-Kiesi. Mikkeli Central Hospital: A. Gynther, R. Manninen, P. Nironen, M. Salminen, and T. Vänttinen. Mänttä Regional Hospital: I. Pirttiniemi and A.-M. Hänninen. North Karelian Hospital, Joensuu: U.-M. Henttula, P. Kekäläinen, M. Pietarinen, A. Rissanen, and M. Voutilainen. Nurmijärvi Health Centre: A. Burgos and K. Urtamo. Oulaskangas Hospital, Oulainen: E. Jokelainen, P.-L. Jylkkä, E. Kaarlela, and J. Vuolaspuro. Oulu Health Centre: L. Hiltunen, R. Häkkinen, and S. Keinänen-Kiukaanniemi. Oulu University Hospital: R. Ikäheimo. Päijät-Häme Central Hospital: H. Haapamäki, A. Helanterä, S. Hämäläinen, V. Ilvesmäki, and H. Miettinen. Palokka Health Centre: P. Sopanen and L. Welling. Pieksämäki Hospital: V. Javtsenko and M. Tamminen. Pietarsaari Hospital: M.-L. Holmbäck, B. Isomaa, and L. Sarelin. Pori City Hospital: P. Ahonen, P. Merensalo, and K. Sävelä. Porvoo Hospital: M. Kallio, B. Rask, and S. Rämö. Raahe Hospital: A. Holma, M. Honkala, A. Tuomivaara, and R. Vainionpää. Rauma Hospital: K. Laine, K. Saarinen, and T. Salminen. Riihimäki Hospital: P. Aalto, E. Immonen, and L. Juurinen. Salo Hospital: A. Alanko, J. Lapinleimu, P. Rautio, and M. Virtanen. Satakunta Central Hospital, Pori: M. Asola, M. Juhola, P. Kunelius, M.-L. Lahdenmäki, P. Pääkkönen, and M. Rautavirta. Savonlinna Central Hospital: E. Korpi-Hyövälti, T. Latvala, and E. Leijala. South Karelia Central Hospital, Lappeenranta: T. Ensala, E. Hussi, R. Härkönen, U. Nyholm, and J. Toivanen. Tampere Health Centre: A. Vaden, P. Alarotu, E. Kujansuu, H. Kirkkopelto-Jokinen, M. Helin, S. Gummerus, L. Calonius, T. Niskanen, T. Kaitala, and T. Vatanen. Tampere University Hospital: I. Ala-Houhala, T. Kuningas, P. Lampinen, M. Määttä, H. Oksala, T. Oksanen, K. Salonen, H. Tauriainen, and S. Tulokas. Tiirismaa Health Centre, Hollola: T. Kivelä, L. Petlin, and L. Savolainen. Turku Health Centre: I. Hämäläinen, H. Virtamo, and M. Vähätalo. Turku University Central Hospital: K. Breitholz, R. Eskola, K. Metsärinne, U. Pietilä, P. Saarinen, R. Tuominen, and S. Äyräpää. Vaajakoski Health Centre: K. Mäkinen and P. Sopanen. Valkeakoski Regional Hospital: S. Ojanen, E. Valtonen, H. Ylönen, M. Rautiainen, and T. Immonen. Vammala Regional Hospital: I. Isomäki, R. Kroneld, and M. Tapiolinna-Mäkelä. Vaasa Central Hospital: S. Bergkulla, U. Hautamäki, V.-A. Myllyniemi, and I. Rusk.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-0751/-/DC1
See accompanying commentary, p. 1923.
REFERENCES
- 1.DIAMOND Project Group . Incidence and trends of childhood Type 1 diabetes worldwide 1990–1999. Diabet Med 2006;23:857–866 [DOI] [PubMed] [Google Scholar]
- 2.U.S. Renal Data System. USRDS 2010 Annual Data Report: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States. Bethesda, MD, National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, 2010 [Google Scholar]
- 3.Seaquist ER, Goetz FC, Rich S, Barbosa J. Familial clustering of diabetic kidney disease. Evidence for genetic susceptibility to diabetic nephropathy. N Engl J Med 1989;320:1161–1165 [DOI] [PubMed] [Google Scholar]
- 4.Quinn M, Angelico MC, Warram JH, Krolewski AS. Familial factors determine the development of diabetic nephropathy in patients with IDDM. Diabetologia 1996;39:940–945 [DOI] [PubMed] [Google Scholar]
- 5.Harjutsalo V, Katoh S, Sarti C, Tajima N, Tuomilehto J. Population-based assessment of familial clustering of diabetic nephropathy in type 1 diabetes. Diabetes 2004;53:2449–2454 [DOI] [PubMed] [Google Scholar]
- 6.McKnight AJ, Currie D, Maxwell AP. Unravelling the genetic basis of renal diseases; from single gene to multifactorial disorders. J Pathol 2010;220:198–216 [DOI] [PubMed] [Google Scholar]
- 7.Mooyaart AL, Valk EJ, van Es LA, et al. Genetic associations in diabetic nephropathy: a meta-analysis. Diabetologia 2011;54:544–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maeda S, Osawa N, Hayashi T, Tsukada S, Kobayashi M, Kikkawa R. Genetic variations associated with diabetic nephropathy and type II diabetes in a Japanese population. Kidney Int Suppl 2007:S43–S48 [DOI] [PubMed] [Google Scholar]
- 9.Nelson RG, Newman JM, Knowler WC, et al. Incidence of end-stage renal disease in type 2 (non-insulin-dependent) diabetes mellitus in Pima Indians. Diabetologia 1988;31:730–736 [DOI] [PubMed] [Google Scholar]
- 10.Conway BR, Maxwell AP. Genetics of diabetic nephropathy: are there clues to the understanding of common kidney diseases? Nephron Clin Pract 2009;112:c213–c221 [DOI] [PubMed] [Google Scholar]
- 11.Tong Z, Yang Z, Patel S, et al. Genetics of Diabetes and Diabetic Complication Study Group . Promoter polymorphism of the erythropoietin gene in severe diabetic eye and kidney complications. Proc Natl Acad Sci USA 2008;105:6998–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimazaki A, Kawamura Y, Kanazawa A, et al. Genetic variations in the gene encoding ELMO1 are associated with susceptibility to diabetic nephropathy. Diabetes 2005;54:1171–1178 [DOI] [PubMed] [Google Scholar]
- 13.Pezzolesi MG, Katavetin P, Kure M, et al. Confirmation of genetic associations at ELMO1 in the GoKinD collection supports its role as a susceptibility gene in diabetic nephropathy. Diabetes 2009;58:2698–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leak TS, Perlegas PS, Smith SG, et al. Variants in intron 13 of the ELMO1 gene are associated with diabetic nephropathy in African Americans. Ann Hum Genet 2009;73:152–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pezzolesi MG, Poznik GD, Mychaleckyj JC, et al. DCCT/EDIC Research Group . Genome-wide association scan for diabetic nephropathy susceptibility genes in type 1 diabetes. Diabetes 2009;58:1403–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thorn LM, Forsblom C, Fagerudd J, et al. FinnDiane Study Group . Metabolic syndrome in type 1 diabetes: association with diabetic nephropathy and glycemic control (the FinnDiane study). Diabetes Care 2005;28:2019–2024 [DOI] [PubMed] [Google Scholar]
- 17.Osterholm AM, He B, Pitkaniemi J, et al. Genome-wide scan for type 1 diabetic nephropathy in the Finnish population reveals suggestive linkage to a single locus on chromosome 3q. Kidney Int 2007;71:140–145 [DOI] [PubMed] [Google Scholar]
- 18.Mueller PW, Rogus JJ, Cleary PA, et al. Genetics of Kidneys in Diabetes (GoKinD) study: a genetics collection available for identifying genetic susceptibility factors for diabetic nephropathy in type 1 diabetes. J Am Soc Nephrol 2006;17:1782–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pluzhnikov A, Below JE, Konkashbaev A, et al. Spoiling the whole bunch: quality control aimed at preserving the integrity of high-throughput genotyping. Am J Hum Genet 2010;87:123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 2004;74:765–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeda S, Araki S, Babazono T, et al. Replication study for the association between four Loci identified by a genome-wide association study on European American subjects with type 1 diabetes and susceptibility to diabetic nephropathy in Japanese subjects with type 2 diabetes. Diabetes 2010;59:2075–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freedman BI, Langefeld CD, Lu L, et al. Differential effects of MYH9 and APOL1 risk variants on FRMD3 Association with Diabetic ESRD in African Americans. PLoS Genet 2011;7:e1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet 2003;33:177–182 [DOI] [PubMed] [Google Scholar]
- 26.Parving HH, Oxenbøll B, Svendsen PA, Christiansen JS, Andersen AR. Early detection of patients at risk of developing diabetic nephropathy. A longitudinal study of urinary albumin excretion. Acta Endocrinol (Copenh) 1982;100:550–555 [DOI] [PubMed] [Google Scholar]
- 27.Viberti GC, Hill RD, Jarrett RJ, Argyropoulos A, Mahmud U, Keen H. Microalbuminuria as a predictor of clinical nephropathy in insulin-dependent diabetes mellitus. Lancet 1982;1:1430–1432 [DOI] [PubMed] [Google Scholar]
- 28.Mogensen CE, Christensen CK. Predicting diabetic nephropathy in insulin-dependent patients. N Engl J Med 1984;311:89–93 [DOI] [PubMed] [Google Scholar]
- 29.Perkins BA, Ficociello LH, Silva KH, Finkelstein DM, Warram JH, Krolewski AS. Regression of microalbuminuria in type 1 diabetes. N Engl J Med 2003;348:2285–2293 [DOI] [PubMed] [Google Scholar]
- 30.Lemley KV. A basis for accelerated progression of diabetic nephropathy in Pima Indians. Kidney Int Suppl 2003;S38–S42 [DOI] [PubMed] [Google Scholar]
- 31.Chanock SJ, Manolio T, Boehnke M, et al. NCI-NHGRI Working Group on Replication in Association Studies . Replicating genotype-phenotype associations. Nature 2007;447:655–660 [DOI] [PubMed] [Google Scholar]
- 32.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 2008;9:356–369 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.