Abstract
The protein-tyrosine phosphatase Shp1 negatively regulates insulin action on glucose homeostasis in liver and muscle, but its potential role in obesity-linked insulin resistance has not been examined. To investigate the role of Shp1 in hepatic insulin resistance, we generated hepatocyte-specific Shp1 knockout mice (Ptpn6H-KO), which were subjected to extensive metabolic monitoring throughout an 8-week standard chow diet (SD) or high-fat diet (HFD) feeding. We report for the first time that Shp1 expression is upregulated in metabolic tissues of HFD-fed obese mice. When compared with their Shp1-expressing Ptpn6f/f littermates, Ptpn6H-KO mice exhibited significantly lowered fasting glycemia and heightened hepatic insulin sensitivity. After HFD feeding, Ptpn6H-KO mice developed comparable levels of obesity as Ptpn6f/f mice, but they were remarkably protected from liver insulin resistance, as revealed by euglycemic clamps and hepatic insulin signaling determinations. Although Ptpn6H-KO mice still acquired diet-induced peripheral insulin resistance, they were less hyperinsulinemic during a glucose tolerance test because of reduced insulin secretion. Ptpn6H-KO mice also exhibited increased insulin clearance in line with enhanced CC1 tyrosine phosphorylation in liver. These results show that hepatocyte Shp1 plays a critical role in the development of hepatic insulin resistance and represents a novel therapeutic target for obesity-linked diabetes.
The protein-tyrosine phosphatase (PTP) superfamily includes classical and dual-specificity PTPs. The nonreceptor PTPs containing two SH2 domains (Shp), Shp1 and Shp2, are involved in the regulation of multiple cellular activities (1). Shp2 is a ubiquitously expressed positive modulator of insulin signaling (2–7), whereas Shp1 generally is recognized to negatively regulate signal transduction (8–15). Shp1 is expressed in most epithelial cells and in cells from the hematopoietic lineage (8–11), where it promotes the inactivation of phosphatidylinositol 3-kinase (PI3K) (16–18). However, much less is known about the metabolic role of Shp1 in insulin target cells.
We previously showed that Shp1 is expressed in liver and skeletal muscle. Furthermore, Ptpn6me-v/me-v (viable motheaten or mev) mice, which express low levels of catalytically impaired Shp1 protein, are highly insulin sensitive and glucose tolerant as a result of enhanced insulin receptor (IR) signaling to PI3K/Akt in these tissues (19). Interfering with Shp1 activity in liver by adenoviral expression of a dominant-negative Shp1 mutant, or by small hairpin RNA–mediated Shp1 silencing, improved hepatic insulin signaling and glucose tolerance (19). We also found that hepatic Shp1 deficiency markedly increased the tyrosine phosphorylation of the transmembrane glycoprotein carcinoembryonic antigen–related cell adhesion molecule-1 (CC1 or CEACAM1) (19), an essential modulator of hepatic insulin clearance (20,21). Additional studies by our group and others have established that CC1 also is a key regulator of hepatic glucose and lipid metabolism (22–25). More recent studies have shown that transcriptional activation of the Shp1 gene (Ptpn6) by overexpression of the homeodomain transcription factor Prep1 also attenuates insulin signaling and glucose storage in hepatic cells (26). In addition, Prep1-deficient mice showed improved hepatic insulin action in association with reduced Shp1 expression, further supporting a key role for Shp1 in the control of liver glucose metabolism.
Although it has been established that Shp1 regulates insulin sensitivity and glucose homeostasis in normal mice, its role in the development of insulin resistance and altered glucose metabolism in obesity remains unknown. Moreover, Shp1 is expressed not only in hepatocytes but in Kupffer cells (27,28), which also are targeted by adenoviral gene delivery (19). The major goal of the current study was to determine the function of hepatic parenchymal Shp1 in the modulation of glucose metabolism and to explore its potential effects on the development of obesity-linked insulin resistance. We found that Shp1 expression is increased in liver and other metabolic tissues of high-fat diet (HFD)-induced obese mice. Generation of mice with hepatocyte Shp1 deficiency (Ptpn6H-KO) fully protected those animals from developing hepatic insulin resistance.
RESEARCH DESIGN AND METHODS
Animals and genotyping.
All mice were housed under a controlled temperature (23°C) and 12-h light/dark cycle with water and food ad libitum. Mice were kept on a sterilized standard low-fat rodent diet (5.8% fat by kcal, total 3.1 kcal/g of diet, LM-485, Eq. 7912; Harlan Teklad). For diet studies, mice at 2 months were kept either on the standard diet (SD) or transferred to an HFD (54.8% fat by kcal, total 4.8 kcal/g of diet, TD93075; Harlan Teklad) for ~8 weeks until 4 months. All studies were conducted with the approved protocols by the animal care and handling committee of Laval University according to the standards defined by the Canadian Council on Animal Care. Only male mice were used for studies presented in this report.
Hepatocyte-specific Shp1 knockout mice (Ptpn6H-KO) were generated on a pure C57/BL6 background by crossing mice homozygous for floxed Shp1 (Ptpn6f/f) (29) with Alb-Cre mice (B6.Cg-Tg[Alb-cre]21Mgn/J, stock 3574; The Jackson Laboratory) (see Supplementary Fig. 1 for crossbreeding scheme). Genomic DNA was extracted from tail or ear samples using the DNA REDExtract-N-Amp PCR kit (Sigma), and genotyping was performed for floxed Ptpn6 (29) and Alb-Cre separately. Floxed Ptpn6 was measured by conventional PCR and agarose gel electrophoresis and Alb-Cre by TaqMan gene expression. Primers used for floxed Ptpn6 were 5′-ACC CTC CAg CTC CTC TTC-3′ and 5′-TgA ggT CCC ggT gAA ACC-3′. Homozygous, hemizygous, and the absence of Alb-Cre were detected as indicated by The Jackson Laboratory. Cre primers were 5′-gCg gTC Tgg CAg TAA AAA CTA TC-3′ and 5′-gTg AAA CAg CAT TgC TgT CAC TT-3; Cre TaqMan probe (TP) were 5′-6FAM-AAA CAT gCT TCA TCg TCg gTC Cgg-TAMRA-3′; Apob control primers were 5′-CAC gTg ggC TCC AgC ATT-3′ (forward) and 5′-TCA CCA gTC ATT TCT gCC TTT g-3′ (reverse); and Apob TP were 5′-Joe -CCA ATg gTC ggg CAC TgC TCA A-BHQ1-3′. All reactions were run in Roto-Gene (Corbett Life Science, Sydney, New South Wales, Australia).
Isolation and purification of primary hepatocytes and Kupffer cells.
Livers were perfused in anesthetized mice, and primary hepatic cells were isolated as described (30). Hepatocytes and Kupffer cells were then separated and purified by serial centrifugation with density selection prior to culture (31).
Metabolic phenotyping.
Body weight and body composition (Minispec NMR spectrophotometer; Bruker Optics) were measured every month for all mice on the SD. For mice in the diet studies (8 weeks), body weight and composition were taken before (2 months) and after (4 months) dietary treatment, whereas body weight was monitored every other week. Prior to being killed, mice were fasted for 6 h and anesthetized either by ketamine-based rodent cocktail or by isoflurane inhalant andthen administered either saline or insulin (3.8 units/kg body wt) through the tail vein for 5 min. Whole blood retrieved by direct cardiac puncture was used to measure plasma insulin, C-peptide, and glucagon by radioimmunoassay (Linco Research). Blood glucose was measured by glucometer (Accu-Chek). Intraperitoneal glucose and insulin tolerance tests were performed as previously described (23). Hyperinsulinemic-euglycemic clamp studies were conducted in mice at 4 months after the 8-week diet studies, as previously described (23,32), but with a lower liver-targeting insulin infusion rate of 2.5 mU/kg/min.
Tissue preparation.
Deep anesthesia was induced by isoflurane in mice, and various tissues were excised, weighed, and cleaned with PBS and then snap frozen in liquid nitrogen to be stored at −80°C. For livers collected, ~10 mg was homogenized fresh in 1 mL TRIzol (Sigma) for RNA extraction, and the rest was powdered in liquid nitrogen and stored at −80°C for further analysis.
Real-time PCR.
Total RNA was extracted and purified using the RNeasy Microarray Tissue Mini Kit (Qiagen) and used for cDNA synthesis using a reverse transcription PCR kit (Applied Biosystems). Real-time PCR was performed using the SYBR Green Jump-Start Gene Expression Kit (Sigma) with 1:25 diluted cDNA product from the reverse transcription. Primers used were Actb (5′-CTCTAgACTTCgAgCAggAg-3′ and 5′-AgAgTACTTgCgCTCAggAg-3′) and Ptpn6 (5′-TggTTTCACCgggACCTCAgC-3′ and 5′-AgTAAggCTgCCgCAggTAgA-3′).
Rapid-sampling in vivo insulin clearance assay.
The rapid-sampling in vivo insulin clearance (RIVIC) method, as previously developed (23), was used for the determination of insulin clearance. Radioactivity in 8-μL blood samples was counted in a gamma counter (Perkin Elmer), and clearance rate was measured in slopes calculated from linear-fitted curves.
Immunoprecipitation and Western blot analyses.
Immunoprecipitation and Western blotting were performed as previously described (19,23). CC1 and insulin receptor (IR) were immunoprecipitated from 1 mg of liver lysates and detected using polyclonal anti-CC1 or anti–CC1 L antibody (Beauchemin Laboratory, McGill University, Québec, Canada) and anti-IR antibodies (Santa Cruz Biotechnology or BD biosciences), respectively. Detection of pY was achieved with monoclonal anti-pY antibodies (Abcam and Upstate/Millipore). The following antibodies were used for general Western blotting: anti-Shp1 from Santa Cruz Biotechnology or BD Biosciences; anti-actin and anti-Akt1/2/3 from Santa Cruz Biotechnology; and anti-ACC, anti–β-actin, anti-eEF2, anti-pS473Akt, and anti-pT308Akt from Cell Signaling. Horseradish peroxidase–conjugated anti-mouse and anti-rabbit secondary antibodies were from Jackson ImmunoResearch Laboratories. Horseradish peroxidase of immunoreactive bands was illuminated by chemiluminescence agents (Millipore, Ontario, Canada) and scanned using the EC3 Imaging System with VisionWorks LS software (Ultra-Violet Products Ltd.). Densitometry was quantified using ImageQuant TL software (GE Healthcare Bio-Sciences).
Statistical analysis.
Statistical analyses were performed by either a two-tailed Student t test or two-way ANOVA using the JMP 8 program (JMP, Cary, NC). P values were considered significant if they were <0.05. SEMs are represented in the graphs.
RESULTS
Shp1 expression is augmented in insulin target tissues in obesity.
We first determined the effect of diet-induced obesity on Shpl expression. Mice were fed an SD or HFD for 8 weeks, and total tissue lysates were immunoblotted using a specific Shp1 antibody. We found that HFD feeding increases Shp1 protein levels in skeletal muscle (gastrocnemius), visceral fat depots (epididymal and perirenal white adipose tissues), and the liver (Fig. 1A). Shp1 mRNA levels also were increased (by 4.7-fold) in liver (Fig. 1B), indicating that obesity increases the expression of the PTP at the transcript level. This upregulation of Shp1 expression by HFD feeding suggests that this PTP may play a significant role in the development of obesity-linked insulin resistance.
FIG. 1.
Increased Shp1 protein and gene expression in diet-induced obesity and generation of hepatocyte-specific Shp1 knockout mice (Ptpn6H-KO). A: Shp1 protein detection by Western blotting in gastrocnemius muscle (Gastroc), epididymal white adipose tissue (EWAT), perirenal white adipose tissue (PWAT), and liver of Ptpn6f/f mice fed an SD or HFD for 8 weeks (n = 12–13 per diet group), with eEF2 and β-actin as loading controls. B: Quantitative PCR analysis of hepatic Shp1 gene expression with Actb as the sample control (n = 11–12 per diet group). Dotted lines on blots separate noncontiguous sections of the same gel (^P < 0.05 and ^^^P < 0.005 diet effect). Western blot detection of Shp1 in liver (C), purified hepatocytes and Kupffer cells (D) isolated from Ptpn6H-KO mice with either hemizygous (+/−) or homozygous (+/+) expression of Alb-Cre (Alb-Cre), and from their Alb-Cre (−/−) Ptpn6f/f littermates. Shp1 level in J774 macrophages (M) acted as a blotting control. α-Tubulin (C) and β-actin (D) were loading controls.
Generation and phenotypic characterization of Ptpn6H-KO mice.
Because Shp1 was particularly increased in the liver of obese mice, and as we have shown previously that this PTP negatively modulates IR signaling and CC1 function in normal liver, we explored the role of parenchymal Shp1 in the regulation of hepatic glucose and lipid metabolism by generating hepatocyte-specific Shp1 knockout mice (Ptpn6H-KO) from homozygous Ptpn6f/f mice (29) and transgenic albumin-Cre (Alb-Cre) mice (Supplementary Fig. 1). Liver Shp1 protein expression was monitored in Ptpn6f/f and in Alb-Cre hemizygous and homozygous Ptpn6H-KO mice. A significant reduction in Shp1 protein was achieved in the Ptpn6H-KO animals, with residual detectable expression attributed to nonparenchymal Shp1 expression (Fig. 1C). A similar reduction was confirmed at the transcript level (data not shown). To verify the completeness of hepatocyte Shp1 knockout, we analyzed Shp1 protein levels in primary hepatocytes and Kupffer cells. These experiments confirmed the absence of Shp1 in the Ptpn6H-KO hepatocytes; hemizygous Alb-Cre expression deleted Ptpn6 using the Cre-LoxP system without affecting Kupffer cell Shp1 protein expression (Fig. 1D). Body and liver weight and gross anatomy in Ptpn6H-KO mice on the SD were compared with littermate Ptpn6f/f controls during the course of 1 year, and no significant changes were observed (data not shown).
Both Ptpn6f/f and Ptpn6H-KO mice were randomly assigned to chronic high-fat-feeding studies. Compared with mice fed a low-fat SD, Ptpn6f/f and Ptpn6H-KO mice subjected to an HFD for 8 weeks showed comparable energy intake increase and body weight gain (Fig. 2A and B). Body composition analysis of Ptpn6f/f and Ptpn6H-KO mice on an HFD revealed a similar gain of fat mass (approximately threefold increase over SD-fed mice) with comparable lean mass (Fig. 2C and D). The weights of different fat depot and gastrocnemius muscle also were similar in the two genotypes on both diets, although a trend toward higher liver weight, triglyceride, and cholesterol content was seen in HFD-fed Ptpn6H-KO mice (Supplementary Table 1). Plasma levels of adiponectin (9.0 ± 1.1 μg/mL wild type vs. 9.6 ± 0.9 μg/mL knockout), tumor necrosis factor-α (120.16 ± 6.58 pg/mL wild type vs. 133.94 ± 6.29 pg/mL knockout), and interleukin-6 (202.52 ± 18.75 pg/mL wild type vs. 191.68 ± 25.87 pg/mL knockout) also were similar in Ptpn6H-KO mice and their Ptpn6f/f controls.
FIG. 2.
Basic metabolic phenotyping of Ptpn6H-KO mice. A: Energy intake of Ptpn6f/f (white bar) and Ptpn6H-KO (black bar) mice fed either an SD or HFD for 8 weeks, expressed in kcal per day (n = 22–23 per genotype and diet group; ^P < 0.05 diet effect). B: Body weight (BW) gain in the percentage of basal body weight at 2 months before dietary treatments (n = 31–32 per genotype and diet group; ^^^P < 0.005 diet effect). Body composition (C and D) was measured at the end of the diet studies using a Minispec NMR rodent spectrometer (n = 36–39 per genotype and diet group; ^^^P < 0.005 diet effect). Fasting plasma glucose (E), insulin (G), and C-peptide (H) (n = 25–27 per genotype and diet group; ^^^P < 0.005 diet effect, ***P < 0.005, and *P < 0.05 genotype difference). F: Hepatic glycogen was detected by crude extraction followed by colorimetric measurement. Livers were collected from Ptpn6f/f and Ptpn6H-KO mice at the end of the 8-week diet studies after a 6-h fast (n = 12–13 per genotype and diet group; ***P < 0.005 genotype difference).
SD-fed Ptpn6H-KO mice exhibited lower fasting glycemia compared with Ptpn6f/f littermates (Fig. 2E). There also was a significant depletion in hepatic glycogen content in fasted SD-fed Ptpn6H-KO mice (Fig. 2F), possibly related to hypoglycemic compensation attributed to improved basal hepatic insulin sensitivity in the absence of hepatocyte Shp1 (33). After 8 weeks of HFD feeding, the fasting glycemia of Ptpn6H-KO mice remained significantly lower than that of Ptpn6f/f animals, although both groups developed the typical diet-induced increase in fasting glycemia, plasma insulin, and C-peptide levels (Fig. 2E, G, and H). Postprandial glycemia in HFD-fed Ptpn6H-KO mice also was significantly lower than in their Ptpn6f/f counterparts (data not shown). Fasting hepatic glycogen content was not altered in HFD-fed Ptpn6H-KO mice, possibly because they remained hyperglycemic despite the improved glycemia relative to their Ptpn6f/f littermates.
Ptpn6H-KO mice are protected from obesity-linked hepatic insulin resistance.
Hyperinsulinemic-euglycemic clamp studies were performed in conscious unrestrained Ptpn6H-KO mice and Ptpn6f/f controls to evaluate the impact of hepatocyte Shp1 deletion on whole-body insulin sensitivity, as well as hepatic and peripheral insulin action. A low dose of insulin infusion was used to target the liver in particular. Consistent with the fasting glycemia data, the clamp glucose infusion rate in Ptpn6H-KO mice was much higher than in Ptpn6f/f animals on both the SD and HFD (Fig. 3A and B). Nevertheless, whole-body insulin sensitivity was compromised in HFD-fed Ptpn6H-KO mice compared with their SD-fed controls, although it was similar to that of SD-fed Ptpn6f/f mice. Glucose turnover rates were determined (see Supplementary Fig. 2) for calculations of hepatic glucose production (HGP) and insulin action at both hepatic and peripheral levels during the clamp. HGP was markedly increased in HFD-fed Ptpn6f/f mice compared with that in SD-fed littermates. Conversely, HGP was fully inhibited during the clamp in both SD- and HFD-fed Ptpn6H-KO animals, indicating a full protection against obesity-linked liver insulin resistance (Fig. 3C). As a result, the percentage of insulin-suppressed HGP was dramatically higher in Ptpn6H-KO mice than in controls on both diets, whereas HFD-fed Ptpn6f/f mice displayed reduced insulin sensitivity (Fig. 3D). Although HFD promoted insulin resistance for glucose uptake in peripheral tissues (mostly reflecting that of skeletal muscle), no significant differences were observed between Ptpn6f/f and Ptpn6H-KO animals on either diet (Fig. 3E). These results from clamp studies demonstrate that the lack of hepatocyte Shp1 in the liver increases hepatic insulin sensitivity in SD-fed nonobese animals and also protects the liver against obesity-induced insulin resistance in HFD-fed animals.
FIG. 3.
Ptpn6H-KO mice display heightened hepatic insulin sensitivity. Hyperinsulinemic-euglycemic clamp studies were performed in both Ptpn6f/f (white bar) and Ptpn6H-KO (black bar) mice on the SD or HFD (n = 6–10 per genotype and diet group; ^^^P < 0.005 diet effect, ***P < 0.005 genotype difference). A and B: Clamp glucose infusion rate (GIR). C: Clamp HGP. D: Percentage of glucose production (GP) suppression by insulin. E: Percentage of clamp glucose uptake (GU).
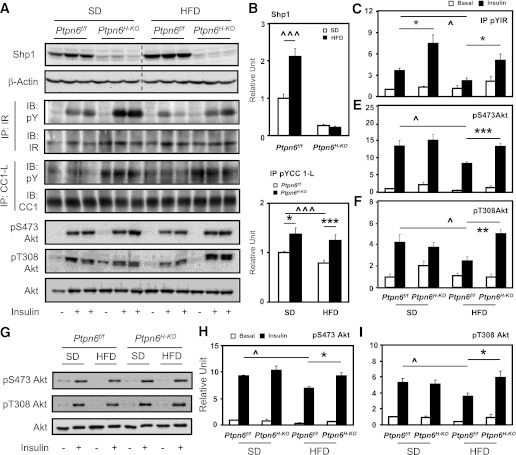
To explore the mechanisms of the enhanced hepatic insulin sensitivity in Ptpn6H-KO mice, liver lysates were subjected to Western blot analyses. As shown in Fig. 4A, hepatic Shp1 protein expression was upregulated in HFD-fed Ptpn6f/f animals compared with their littermates on the SD. As expected, Shp1 expression was reduced markedly in the Ptpn6H-KO liver, whereas the residual Shp1 protein contributed by other liver cells was not altered by HFD feeding (Fig. 4A and B). These data indicate that obesity-induced upregulation of hepatic Shp1 expression occurs mostly in the hepatocytes and not in nonparenchymal cells. Moreover, Shp1 protein levels were increased in both the gastrocnemius muscle and white adipose tissues of HFD-fed Ptpn6H-KO mice, as observed in their Ptpn6f/f littermate controls (data not shown).
FIG. 4.
Protected insulin signaling in Ptpn6H-KO mice. A–F: Western blot analysis of liver lysates from Ptpn6f/f and Ptpn6H-KO mice fed the SD or HFD for 8 week, fasted 6 h, and followed by tail vein administration of either saline or insulin (n = 12–13 per genotype and diet group; ^P < 0.05 and ^^^P < 0.005 diet effect, *P < 0.05, **P < 0.01, and ***P < 0.005 genotype difference). Dotted line on blot borders shows noncontiguous sections of the same gel. Shp1, pS473, pT308, and total Akt were detected directly using their respective antibodies, with β-actin as the loading control. IR and CC1-L were immunoprecipitated before being immunoblotted for pY, total IR, and CC1. G–I: Phosphorylation of Akt (pS473 and pT308) and total Akt in purified hepatocytes isolated from Ptpn6f/f and Ptpn6H-KO mice (8 weeks SD or HFD) were detected by immunoblotting with respective antibodies. Purified hepatocytes were cultured for 16 h, serum-deprived for 3 h, and treated with either PBS or insulin for 15 min before cell lysis (n = 3 per genotype and diet group; ^P < 0.05 diet effect; *P < 0.05 genotype difference).
Tyrosine phosphorylation of the IR and CC1 was increased in Ptpn6H-KO livers compared with the Ptpn6f/f controls on both the SD and HFD (Fig. 4A, C, and D). Insulin receptor substrate (IRS)-1 and IRS-2 tyrosine phosphorylation and their association with the p85 subunit of PI3K also were increased in livers of HFD-fed Ptpn6H-KO mice (Supplementary Fig. 3). In agreement with enhanced phosphorylation of IR, IRS-1, and IRS-2, and their association with p85 PI3K, insulin-stimulated Akt activation, as revealed by phosphorylation on residues S473 and T308, was increased in Ptpn6H-KO livers compared with that of Ptpn6f/f counterparts. This resulted in normalization of Akt activation in the livers of HFD-induced obese Ptpn6H-KO mice (Fig. 4A, E, and F). In SD-fed mice, however, only IRS-2 tyrosine phosphorylation and p85 PI3K association were increased in Ptpn6H-KO mice, which may explain the lack of significant improvements of Akt phosphorylation in these SD-fed animals. Improved hepatic Akt phosphorylation also was seen in isolated Ptpn6H-KO hepatocytes from HFD-fed mice (Fig. 4G–I), confirming the normalization of insulin signaling in hepatic parenchymal cells of these animals.
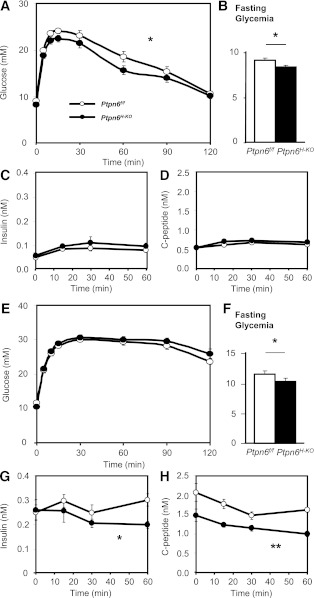
To further assess the impact of selective increase in Ptpn6H-KO hepatic insulin sensitivity on whole-body glucose and insulin homeostasis, Ptpn6f/f and Ptpn6H-KO animals on the SD and HFD were subjected to intraperitoneal glucose and insulin tolerance tests (IPGTTs and IPITTs, respectively). SD-fed Ptpn6H-KO mice showed moderate but significant improvement in glucose tolerance (Fig. 5A) with lower starting fasting glycemia (Fig. 5B) and no plasma insulin level alterations or glucose-stimulated insulin secretion, as assessed by C-peptide level (Fig. 5C and D). Although HFD-fed Ptpn6H-KO mice did not display improved glucose tolerance as compared with Ptpn6f/f controls (Fig. 5E), their fasting glycemia still was significantly improved (Fig. 5F). This may be partly linked to the fact that Shp1 still is increased in skeletal muscle and adipose tissues of the hepatocyte-specific Shp1 knockout mice. Of interest, to support similar glucose tolerance as their HFD-fed Ptpn6f/f controls, the HFD-fed Ptpn6H-KO mice were significantly less hyperinsulinemic during the IPGTT (Fig. 5G), which was at least partly explained by reduced insulin secretion as revealed by C-peptide concentrations (Fig. 5H). In the fasting state, plasma glucagon levels were comparable among experimental groups (data not shown), suggesting that overall islet functions are not altered in Ptpn6H-KO mice.
FIG. 5.
Glucose tolerance in Ptpn6H-KO and their Ptpn6f/f controls on the SD and HFD. IPGTT glucose (A and E), fasting glycemia (B and F), insulin (C and G), and C-peptide (D and H) levels of Ptpn6f/f and Ptpn6H-KO mice on the SD (n = 16–22 per genotype) or HFD (n = 10–17 per genotype), respectively. *P < 0.05 and **P < 0.01 genotype difference.
IPITT also revealed no significant differences in whole-body insulin sensitivity between Ptpn6f/f and Ptpn6H-KO mice on either diet (Fig. 6A and D), despite the consistent improvement of fasting glycemia (Fig. 6B and E). These data seemed to conflict with the improved insulin sensitivity seen in the euglycemic clamp studies. However, despite similar amounts of exogenous insulin administered, insulin disposal was faster in Ptpn6H-KO animals than in their Ptpn6f/f littermates on both the SD and HFD, resulting in lower insulin concentrations during most of the test (Fig. 6C and F). We suspected that increased CC1 tyrosine phosphorylation in the Ptpn6H-KO liver (Fig. 4A and D) might explain this augmented insulin clearance. This possibility was tested in a separate group of SD-fed animals using the previously established RIVIC assay (23). As shown in Fig. 6G–H, the insulin clearance rate was indeed significantly enhanced in Ptpn6H-KO mice.
FIG. 6.
Insulin tolerance and hepatic insulin clearance in Ptpn6H-KO and their Ptpn6f/f controls on the SD and HFD. Blood glucose (A and D), fasting glycemia (B and E), and plasma insulin levels (C and F) of Ptpn6f/f and Ptpn6H-KO mice fed the SD or HFD, respectively, during the IPITT (n = 28–29 per genotype and diet group; *P < 0.05, **P < 0.01, and ***P < 0.005 genotype difference). G: RIVIC was measured as the percentage of blood 125I counts per minute (CPM) to the reference counts per minute at 10 s after an intravenous bolus administration of human [125I-insulin] in Ptpn6f/f and Ptpn6H-KO mice fed the SD. Rate of RIVIC (H) was calculated as the slope of the linear curve fit in the logged graph of G (n = 6 per genotype; *P < 0.05 and ***P < 0.005 genotype difference).
DISCUSSION
In this report, we show for the first time that Shp1 protein expression is increased in metabolic tissues, such as skeletal muscle, white adipose tissue, and liver of diet-induced obese mice. We have previously shown that Shp1 is an important regulator of liver metabolism (19). We have now generated hepatocyte-specific Shp1-deficient mice to explore the role of this PTP in the regulation of hepatic glucose metabolism, as well as its contribution to the development of obesity-linked insulin resistance. This approach specifically eliminates hepatocyte Shp1 expression, leaving intact expression in other liver cells, especially in Kupffer cells, which also are implicated in obesity-related liver diseases (31,34–37). We demonstrate that the increased hepatic Shp1 expression in obese mice is likely confined to the hepatocytes, as the nonparenchymal Shp1 expression was not significantly altered between HFD-fed Ptpn6H-KO and Ptpn6f/f mice.
Congruent with the hypothesis that increased hepatocyte Shp1 expression is pathogenic in obesity, mice lacking hepatocyte Shp1 showed improved whole-body glucose homeostasis (fasting glycemia) and were fully protected from developing obesity-related hepatic insulin resistance. This was linked to restoration of normal hepatic insulin signaling as revealed by improved IR tyrosine phosphorylation and Akt Ser/Thr phosphorylation in obese Ptpn6H-KO mice. Akt activation is improved in isolated hepatocytes from these mice, confirming that hepatocyte Shp1 deletion is required for normalization of hepatic insulin sensitivity in Ptpn6H-KO animals.
As expected from hepatocyte-specific Shp1 deletion, Ptpn6H-KO mice displayed improved insulin-suppressed hepatic glucose output, although without any beneficial effects on peripheral insulin resistance in HFD-fed mice. Lack of improvement in peripheral insulin sensitivity may contribute to the finding of modest (SD fed) or lack of (HFD fed) ameliorations in glucose tolerance or insulin sensitivity when measured after a single insulin injection. However, this seemingly unaltered glucose and insulin tolerance in mice lacking hepatocyte Shp1 maybe a result of more rapid insulin clearance rate in the absence of hepatocyte Shp1, suggesting improved overall whole-body insulin sensitivity as less insulin was required to achieve similar systemic glycemia. This was confirmed in Ptpn6H-KO mice by enhanced liver CC1 tyrosine phosphorylation, representing the first direct genetic evidence that hepatocyte Shp1 plays a negative regulatory role in insulin clearance, most likely via CC1 dephosphorylation. More data will be needed to determine whether Shp1 and CC1 regulate hepatic insulin clearance through a Cdk2/β-catenin pathway as we recently reported in human embryonic kidney 293 cells (38).
It is noteworthy that insulin secretion was decreased in HFD-fed Ptpn6H-KO mice and contributed to the reduction in plasma insulin levels upon glucose challenge, thus revealing a novel regulation of β-cell insulin secretion by hepatocyte Shp1. Such a liver-pancreas cross-talk also has been reported in the liver-specific IR knockout mouse, where profound hepatic insulin resistance is associated with increased β-cell mass and insulin secretion (39–41). Using an inducible liver-specific IR knockout mouse model, Escribano et al. (42) reported that hepatic IGF-1 may be a primary factor driving this liver-pancreas endocrine axis. It also has been recently reported that liver-specific IR knockout mouse serum and liver explant- or hepatocyte-conditioned media enhanced β-cell proliferation in isolated wild-type mouse islets, providing further evidence that the liver is the source of circulating β-cell growth factor(s) (43). Our studies thus suggest that Shp1 may be involved in this liver-pancreas endocrine axis, regulating β-cell function through production of as yet unknown circulating factor(s).
It is of interest to compare the metabolic phenotypes of our hepatocyte-specific Shp1 knockout mice with that of mice lacking PTP-1B, also an immediate PTP for the IR. Whole-body deletion of PTP-1B was found to confer protection against HFD-mediated whole-body weight gain, insulin resistance, and glucose intolerance (44). Liver-specific PTP-1B knockout mice displayed improved glucose and lipid homeostasis with enhanced hepatic insulin action (45). However it is unlikely that hepatocyte PTP-1B modulates hepatic insulin clearance as we have seen in Ptpn6H-KO mice, because we have shown that CC1 is a substrate of Shp1 but not of PTP-1B in dephosphorylation assays in vitro (19).
In conclusion, the current study using Ptpn6H-KO mice provide genetic evidence that hepatocyte Shp1 is a novel mediator of hepatic insulin resistance through impairment of liver IR signaling for glucose metabolism. Moreover, Shp1 is overexpressed in other key tissues involved in glucose metabolism such as skeletal muscle and adipose tissue of obese mice. Taken together our data suggest that Shp1 represents a novel therapeutic target for obesity-linked diabetes.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by grants from the Canadian Institutes of Health Research (CIHR) (to K.A.S., N.B., and A.M.) and grant R37 CA49152 (to B.G.N.). E.X. received the Canadian Diabetes Association Doctoral Student Research Award and a studentship from the CIHR Training Program in Obesity at Laval University. K.A.S. is supported by a Canada Research Chair and the Sherman Family Chair in Genomic Medicine. B.G.N. is supported by a Canada Research Chair and also is partially supported by the Ontario Ministry of Health and Long-Term Care and the Princess Margaret Hospital Foundation. A.M. holds a CIHR/Pfizer Research Chair in the Pathogenesis of Insulin Resistance and Cardiovascular Diseases.
No potential conflicts of interest relevant to this article were reported.
E.X. researched data, contributed to the discussion, and wrote the manuscript. A.C. and Y.R. researched data and contributed to the discussion. K.B., K.A.S., and N.B. contributed to the discussion. L.P. generated the original Ptpn6f/f mice. B.G.N. provided the original Ptpn6f/f breeding couples and contributed to the discussion. A.M. contributed to the discussion and wrote the manuscript. All authors reviewed and edited the manuscript. A.M. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors acknowledge the gracious scientific support of Dr. Yves Deshaies from the Cardiology Axis of the Institut Universitaire de Cardiologie et de Pneumologie de Québec (CRIUCPQ), Hôpital Laval, Québec, Canada, and Dr. Robert M. O’Doherty from the Department of Medicine in the Division of Endocrinology and Metabolism, University of Pittsburgh, Pittsburgh, Pennsylvania. They also thank technicians Christine Dion, Jennifer Dumais, Patricia Pelletier, and Maryse Pitre; students Geneviève Chevrier, Phillip White, and Emmanuelle St-Amand; and postdoctoral fellow Dr. Philippe St-Pierre from the same group at CRIUCPQ, Hôpital Laval, for their technical assistance. The authors also thank the research associates in the same group at CRIUCPQ, Hôpital Laval, and Drs. Rita Kohen and Michael Schwab for their valuable opinions and advice.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-1502/-/DC1.
L.P. is currently affiliated with Five Prime Therapeutics, South San Francisco, California.
REFERENCES
- 1.Neel BG, Gu H, Pao L. SH2-domain-containing protein-tyrosine phosphatases. In Handbook of Cell Signaling. Dennis EA, Ed. Burlington, Academic Press, 2003, p. 707–728 [Google Scholar]
- 2.Noguchi T, Matozaki T, Horita K, Fujioka Y, Kasuga M. Role of SH-PTP2, a protein-tyrosine phosphatase with Src homology 2 domains, in insulin-stimulated Ras activation. Mol Cell Biol 1994;14:6674–6682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milarski KL, Saltiel AR. Expression of catalytically inactive Syp phosphatase in 3T3 cells blocks stimulation of mitogen-activated protein kinase by insulin. J Biol Chem 1994;269:21239–21243 [PubMed] [Google Scholar]
- 4.Kharitonenkov A, Schnekenburger J, Chen Z, et al. Adapter function of protein-tyrosine phosphatase 1D in insulin receptor/insulin receptor substrate-1 interaction. J Biol Chem 1995;270:29189–29193 [DOI] [PubMed] [Google Scholar]
- 5.Ugi S, Maegawa H, Kashiwagi A, Adachi M, Olefsky JM, Kikkawa R. Expression of dominant negative mutant SHPTP2 attenuates phosphatidylinositol 3′-kinase activity via modulation of phosphorylation of insulin receptor substrate-1. J Biol Chem 1996;271:12595–12602 [DOI] [PubMed] [Google Scholar]
- 6.Yamauchi K, Milarski KL, Saltiel AR, Pessin JE. Protein-tyrosine-phosphatase SHPTP2 is a required positive effector for insulin downstream signaling. Proc Natl Acad Sci USA 1995;92:664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maegawa H, Hasegawa M, Sugai S, et al. Expression of a dominant negative SHP-2 in transgenic mice induces insulin resistance. J Biol Chem 1999;274:30236–30243 [DOI] [PubMed] [Google Scholar]
- 8.Shen SH, Bastien L, Posner BI, Chrétien P. A protein-tyrosine phosphatase with sequence similarity to the SH2 domain of the protein-tyrosine kinases. Nature 1991;352:736–739 [DOI] [PubMed] [Google Scholar]
- 9.Matthews RJ, Bowne DB, Flores E, Thomas ML. Characterization of hematopoietic intracellular protein tyrosine phosphatases: description of a phosphatase containing an SH2 domain and another enriched in proline-, glutamic acid-, serine-, and threonine-rich sequences. Mol Cell Biol 1992;12:2396–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plutzky J, Neel BG, Rosenberg RD. Isolation of a src homology 2-containing tyrosine phosphatase. Proc Natl Acad Sci USA 1992;89:1123–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yi TL, Cleveland JL, Ihle JN. Protein tyrosine phosphatase containing SH2 domains: characterization, preferential expression in hematopoietic cells, and localization to human chromosome 12p12-p13. Mol Cell Biol 1992;12:836–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen HE, Chang S, Trub T, Neel BG. Regulation of colony-stimulating factor 1 receptor signaling by the SH2 domain-containing tyrosine phosphatase SHPTP1. Mol Cell Biol 1996;16:3685–3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klingmüller U, Lorenz U, Cantley LC, Neel BG, Lodish HF. Specific recruitment of SH-PTP1 to the erythropoietin receptor causes inactivation of JAK2 and termination of proliferative signals. Cell 1995;80:729–738 [DOI] [PubMed] [Google Scholar]
- 14.Tomic S, Greiser U, Lammers R, et al. Association of SH2 domain protein tyrosine phosphatases with the epidermal growth factor receptor in human tumor cells. Phosphatidic acid activates receptor dephosphorylation by PTP1C. J Biol Chem 1995;270:21277–21284 [DOI] [PubMed] [Google Scholar]
- 15.Ren L, Chen X, Luechapanichkul R, et al. Substrate specificity of protein tyrosine phosphatases 1B, RPTPα, SHP-1, and SHP-2. Biochemistry 2011;50:2339–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imani F, Rager KJ, Catipovic B, Marsh DG. Interleukin-4 (IL-4) induces phosphatidylinositol 3-kinase (p85) dephosphorylation. Implications for the role of SHP-1 in the IL-4-induced signals in human B cells. J Biol Chem 1997;272:7927–7931 [DOI] [PubMed] [Google Scholar]
- 17.Yu Z, Su L, Hoglinger O, Jaramillo ML, Banville D, Shen SH. SHP-1 associates with both platelet-derived growth factor receptor and the p85 subunit of phosphatidylinositol 3-kinase. J Biol Chem 1998;273:3687–3694 [DOI] [PubMed] [Google Scholar]
- 18.Cuevas B, Lu Y, Watt S, et al. SHP-1 regulates Lck-induced phosphatidylinositol 3-kinase phosphorylation and activity. J Biol Chem 1999;274:27583–27589 [DOI] [PubMed] [Google Scholar]
- 19.Dubois MJ, Bergeron S, Kim HJ, et al. The SHP-1 protein tyrosine phosphatase negatively modulates glucose homeostasis. Nat Med 2006;12:549–556 [DOI] [PubMed] [Google Scholar]
- 20.Poy MN, Yang Y, Rezaei K, et al. CEACAM1 regulates insulin clearance in liver. Nat Genet 2002;30:270–276 [DOI] [PubMed] [Google Scholar]
- 21.Huang S, Kaw M, Harris MT, et al. Decreased osteoclastogenesis and high bone mass in mice with impaired insulin clearance due to liver-specific inactivation to CEACAM1. Bone 2010;46:1138–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Najjar SM, Yang Y, Fernström MA, et al. Insulin acutely decreases hepatic fatty acid synthase activity. Cell Metab 2005;2:43–53 [DOI] [PubMed] [Google Scholar]
- 23.Xu E, Dubois MJ, Leung N, et al. Targeted disruption of carcinoembryonic antigen-related cell adhesion molecule 1 promotes diet-induced hepatic steatosis and insulin resistance. Endocrinology 2009;150:3503–3512 [DOI] [PubMed] [Google Scholar]
- 24.Lee SJ, Heinrich G, Fedorova L, et al. Development of nonalcoholic steatohepatitis in insulin-resistant liver-specific S503A carcinoembryonic antigen-related cell adhesion molecule 1 mutant mice. Gastroenterology 2008;135:2084–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeAngelis AM, Heinrich G, Dai T, et al. Carcinoembryonic antigen-related cell adhesion molecule 1: a link between insulin and lipid metabolism. Diabetes 2008;57:2296–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oriente F, Iovino S, Cabaro S, et al. Prep1 controls insulin glucoregulatory function in liver by transcriptional targeting of SHP1 tyrosine phosphatase. Diabetes 2011;60:138–147 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Angata T, Kerr SC, Greaves DR, Varki NM, Crocker PR, Varki A. Cloning and characterization of human Siglec-11: a recently evolved signaling molecule that can interact with SHP-1 and SHP-2 and is expressed by tissue macrophages, including brain microglia. J Biol Chem 2002;277:24466–24474 [DOI] [PubMed] [Google Scholar]
- 28.Ruff SJ, Chen K, Cohen S. Peroxovanadate induces tyrosine phosphorylation of multiple signaling proteins in mouse liver and kidney. J Biol Chem 1997;272:1263–1267 [DOI] [PubMed] [Google Scholar]
- 29.Pao LI, Lam KP, Henderson JM, et al. B cell-specific deletion of protein-tyrosine phosphatase Shp1 promotes B-1a cell development and causes systemic autoimmunity. Immunity 2007;27:35–48 [DOI] [PubMed] [Google Scholar]
- 30.Qiu W, Avramoglu RK, Dubé N, et al. Hepatic PTP-1B expression regulates the assembly and secretion of apolipoprotein B-containing lipoproteins: evidence from protein tyrosine phosphatase-1B overexpression, knockout, and RNAi studies. Diabetes 2004;53:3057–3066 [DOI] [PubMed] [Google Scholar]
- 31.Huang W, Metlakunta A, Dedousis N, et al. Depletion of liver Kupffer cells prevents the development of diet-induced hepatic steatosis and insulin resistance. Diabetes 2010;59:347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charbonneau A, Marette A. Inducible nitric oxide synthase induction underlies lipid-induced hepatic insulin resistance in mice: potential role of tyrosine nitration of insulin signaling proteins. Diabetes 2010;59:861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kishore P, Gabriely I, Cui MH, et al. Role of hepatic glycogen breakdown in defective counterregulation of hypoglycemia in intensively treated type 1 diabetes. Diabetes 2006;55:659–666 [DOI] [PubMed] [Google Scholar]
- 34.Clementi AH, Gaudy AM, van Rooijen N, Pierce RH, Mooney RA. Loss of Kupffer cells in diet-induced obesity is associated with increased hepatic steatosis, STAT3 signaling, and further decreases in insulin signaling. Biochim Biophys Acta 2009;1792:1062–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lanthier N, Molendi-Coste O, Horsmans Y, van Rooijen N, Cani PD, Leclercq IA. Kupffer cell activation is a causal factor for hepatic insulin resistance. Am J Physiol Gastrointest Liver Physiol 2010;298:G107–G116 [DOI] [PubMed] [Google Scholar]
- 36.Wunderlich FT, Ströhle P, Könner AC, et al. Interleukin-6 signaling in liver-parenchymal cells suppresses hepatic inflammation and improves systemic insulin action. Cell Metab 2010;12:237–249 [DOI] [PubMed] [Google Scholar]
- 37.Stienstra R, Saudale F, Duval C, et al. Kupffer cells promote hepatic steatosis via interleukin-1beta-dependent suppression of peroxisome proliferator-activated receptor alpha activity. Hepatology 2010;51:511–522 [DOI] [PubMed] [Google Scholar]
- 38.Fiset A, Xu E, Bergeron S, et al. Compartmentalized CDK2 is connected with SHP-1 and β-catenin and regulates insulin internalization. Cell Signal 2011;23:911–919 [DOI] [PubMed] [Google Scholar]
- 39.Michael MD, Kulkarni RN, Postic C, et al. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol Cell 2000;6:87–97 [PubMed] [Google Scholar]
- 40.Kulkarni RN, Jhala US, Winnay JN, Krajewski S, Montminy M, Kahn CR. PDX-1 haploinsufficiency limits the compensatory islet hyperplasia that occurs in response to insulin resistance. J Clin Invest 2004;114:828–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okada T, Liew CW, Hu J, et al. Insulin receptors in beta-cells are critical for islet compensatory growth response to insulin resistance. Proc Natl Acad Sci USA 2007;104:8977–8982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Escribano O, Guillén C, Nevado C, Gómez-Hernández A, Kahn CR, Benito M. β-Cell hyperplasia induced by hepatic insulin resistance: role of a liver-pancreas endocrine axis through insulin receptor A isoform. Diabetes 2009;58:820–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.El Ouaamari A, Kawamori K, Liew CW, et al. Liver-derived factor(s) drive β-cell hyperplasia in insulin resistant mice. Late-breaking abstract presented at the 71st Scientific Sessions of the American Diabetes Association 24–28 June 2011, at the San Diego Convention Center, San Diego, California [Google Scholar]
- 44.Elchebly M, Payette P, Michaliszyn E, et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 1999;283:1544–1548 [DOI] [PubMed] [Google Scholar]
- 45.Delibegovic M, Zimmer D, Kauffman C, et al. Liver-specific deletion of protein-tyrosine phosphatase 1B (PTP1B) improves metabolic syndrome and attenuates diet-induced endoplasmic reticulum stress. Diabetes 2009;58:590–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.