Abstract
The functional consequences of genetic variation in mammalian regulatory elements are poorly understood. We report the in vivo dissection of three mammalian liver enhancers at single nucleotide resolution via a massively parallelized reporter assay. For each enhancer, we synthesized a library of >100,000 mutant haplotypes with 2–3% divergence from wild-type. Each haplotype was linked to a unique sequence tag embedded within a transcriptional cassette. We introduced each enhancer library into mouse liver and measured the relative activities of individual haplotypes en masse by sequencing of the transcribed tags. Linear regression yielded highly reproducible estimates of the impact of every possible single nucleotide change on enhancer activity. The functional impact of most mutations was modest, with ~22% impacting activity by >1.2-fold, and only ~3% by >2-fold. These results suggest that mammalian enhancers are relatively robust to single nucleotide changes. Several, but not all positions with higher impact showed evidence for purifying selection, or co-localized with known liver-associated transcription factor binding sites, demonstrating the value of empirical high-resolution functional analysis.
INTRODUCTION
Massively parallel sequencing has accelerated the cataloguing of cis-regulatory elements in mammalian genomes1–3. However, we still lack a high-resolution understanding of the functional basis of activity for the overwhelming majority of cis-regulatory elements. Understanding the architecture and internal grammar of cis-regulatory elements is essential for advancing our comprehension of the mechanistic basis for regulatory activity, for enabling the de novo design of synthetic regulatory elements, and for predicting the functional and phenotypic consequences of genetic variation within non-coding DNA.
Although diverse methods exist to predict the functional consequences of protein-altering variants, we remain poorly equipped to estimate the functional impact of regulatory variants4. Recent studies have highlighted the importance of regulatory variants. For example, disease-associated variants frequently coincide with regulatory regions, and in particular with enhancers2, 5–8. As a step forward, we sought to assess the spectrum of effect sizes of all possible single nucleotide variants (SNVs) on the functional activity of three mammalian enhancers with demonstrated in vivo activity in the liver, designated here ALDOB (hg19:chr9:104195570-104195828)9–11, ECR11 (hg19:chr2:169939082-169939701)12, 13, and LTV1 (mm9:chr7:29161443-29161744).
We previously reported a method called “synthetic saturation mutagenesis”14 in which programmable microarrays were used to synthesize variants of several regulatory elements (core promoters), each in cis with a downstream tag sequence. The population of core promoter variants was subjected to a cell-free in vitro assay, after which RNA-Seq of the tags was used to quantify the relative activity of specific core promoter variants. Although successful, several aspects of this approach limit its broader application and scalability: 1) When each regulatory element variant is synthesized as a separate array feature, the overall cost of synthesis remains high. 2) The separate synthesis of individual variants also limits how many combinations of mutations can be simultaneously programmed. 3) The maximum length of array-synthesized oligonucleotides is currently 200–300bp, whereas mammalian enhancers can be 1 kilobase or longer. 4) Access to array-derived oligonucleotide libraries remains restricted to a few groups. 5) The cell-free, in vitro assay that we used poorly captures biological context.
To circumvent these limitations and enable the high-resolution dissection of mammalian enhancers, we developed a new method (Figure 1). In brief, each enhancer of interest is synthetically constructed by polymerase cycling assembly using overlapping oligonucleotides (~90bp) containing a programmed level of degeneracy. At each position, 97% of molecules are expected to be synthesized correctly with 1% doping of each possible single nucleotide substitution (Online Methods). Therefore, each synthetic enhancer molecule contains, on average, 3 mutations per 100bp, randomly distributed along its length. The population of molecules is inherently complex, both with respect to representation of all possible SNVs of the wild-type enhancer as well as myriad unique combinations. Because nearly all synthetic enhancers contain multiple substitutions, they are referred to here as “enhancer haplotypes”. The enhancer haplotypes are cloned into a plasmid (Promega pGL4.23), which contains a minimal promoter upstream of the luciferase gene. In order to uniquely tag each enhancer haplotype, an oligonucleotide containing a 20bp fully degenerate subsequence is cloned to a separate site, in the 3′ UTR of the luciferase gene. The sequences of specific 20bp tags cloned in cis with specific enhancer haplotypes are determined by massively parallel sequencing. As the enhancer haplotypes are highly related sequences with lengths that exceed the maximum read-length of the Illumina platform, we use tag-guided subassembly15 to enable full-length, high-accuracy sequencing of individual enhancer haplotypes in association with their downstream tags. Each resulting library includes >100,000 fully sequenced enhancer haplotypes, with nearly all containing multiple substitutions and each associated with one or more unique tags. The library is then subjected to what is effectively a highly multiplexed reporter assay, either in vitro or in vivo. For the experiments described here, we used the hydrodynamic tail vein assay12, 16 to assess in vivo enhancer activity in the mouse liver. Mice were sacrificed 24 hours post-injection, at which time total RNA was extracted from each liver, followed by RT-PCR and massively parallel sequencing of cDNA from transcribed tags.
Figure 1. Schematic illustration of the method.
We used doped oligonucleotide synthesis and polymerase cycling assembly (PCA) to generate a highly complex library of enhancer haplotypes for each enhancer studied. On average, each enhancer haplotype diverged from wild-type by ~2–3% (red circles represent mutations). These mutant enhancers, along with 20bp random tags, were cloned into an expression vector (pGL4.23) containing a minimal promoter driving transcription of luciferase (minP/Luc). We performed “subassembly” on each library to determine the full sequence of each enhancer haplotype and to identify the 20bp tag to which each haplotype was cloned in cis. Each library was then introduced into two mice via hydrodynamic tail vein injection, livers were harvested after 24 hours, and RNA-seq was performed to quantify abundance of transcribed 20bp tags. These data were used to estimate the impact of each possible mutation on transcriptional activation.
The enhancers studied here were identified by diverse methods (Supplementary Fig. 1). ALDOB (259bp) is a human intronic enhancer of the aldose B gene9–11. ECR11 (620bp) is a human enhancer located in an intron of dehydrogenase/reductase SDR family member 9 (DHRS9)12. LTV1 (302bp) is a candidate mouse enhancer located 3′ of zinc finger protein 36 (Zfp36) (Supplementary Fig. 2a,b). The functional activity of each wild-type enhancer was confirmed using a conventional hydrodynamic tail vein injection assay, in which luciferase activity in liver tissue was measured 24 hours post-injection (Supplementary Fig. 2c).
RESULTS
Synthetic saturation mutagenesis and massively parallel readout
To systematically dissect the functional consequences of all possible SNVs in these three enhancers, we applied to each the workflow described above (Figure 1). Sequencing with subassembly confirmed that the resulting libraries were complex, with a total of 641,135 distinct haplotypes associated with 1,186,696 tag sequences (Table 1). The observed number of mutations per haplotype approximated expectations, with ~2–3 substitutions per 100bp (Supplementary Fig. 3) and were well-distributed (Supplementary Fig. 4). All possible substitution variants of each enhancer were represented in ≥42 uniquely tagged haplotypes, with each position disrupted on an average of ~4,000 distinct enhancer haplotypes. Furthermore, all possible pairs of positions were disrupted in ≥1 haplotype with the exception of a single pair of positions in LTV1.
Table 1. Enhancer haplotype library characteristics.
For each library of enhancer haplotypes, we list the number of distinct haplotypes, the number of tags with which those distinct haplotypes are associated in cis, the percentage of possible single nucleotide substitutions that are present in at least one haplotype, the percentage of possible pairs of positions where both positions contain mutations together in at least one haplotype, and the per-base mutation rate in each library.
| Library | Number of haplotypes | Number of tags | % of possible substitutions in at least one haplotype | % of possible pairs of sites in at least one haplotype | Per-base mutation rate per haplotype (mean ± s.d.) |
|---|---|---|---|---|---|
| ALDOB | 378,450 | 406,071 | 100% (777 of 777) | 100% (33,411 of 33,411) | 0.021 ± 0.010 |
| ECR11 | 105,795 | 105,832 | 100% (1860 of 1860) | 100% (191,890 of 191,890) | 0.023 ± 0.006 |
| LTV1 rep. 1 | 119,950 | 403,869 | 100% (906 of 906) | 99.99% (45,449 of 45,451) | 0.031 ± 0.010 |
| LTV1 rep. 2 | 105,188 | 270,924 | 100% (906 of 906) | 99.99% (45,449 of 45,451) | 0.031 ± 0.010 |
We introduced each library (one each for ALDOB and ECR11, and two independently constructed libraries for LTV1) into two mice via hydrodynamic tail vein injection (Supplementary Fig. 2d). Total RNA from each mouse was split into several pools, with each aliquot separately subjected to RT-PCR with primers flanking the 20bp tag located in the 3′ UTR of the luciferase transcriptional cassette, and then to massively parallel sequencing on an Illumina GA2x. Because target RNA was very scarce relative to cellular RNA, a modest number of target RNA molecules contributed to each RT-PCR, leading to a complexity bottleneck. In other words, within each sequencing library, all reads corresponding to any single tag appeared to have been derived from amplification of a single RNA molecule. We therefore used the number of RNA pools in which a particular tag was observed, and not the total number of reads associated with a tag, as a measure of the relative transcriptional activity of its associated enhancer haplotype.
For each position in each enhancer, we constructed a linear model to assess the extent to which the presence of a mutation at that position is predictive of a change in the number of RNA pools in which an enhancer haplotype was observed, which is effectively a proxy for its impact on transcriptional activation, i.e. “effect size” (Online Methods). Specifically, we use the term “effect size” to describe the log2 fold change in the predicted transcriptional activity, as measured by the number of RNA pools in which a tag-associated haplotype appeared, relative to the wild-type. We first sought to assess reproducibility, so we calculated effect sizes separately for the two independently constructed LTV1 libraries (pooling data from the two mice subjected to each of these libraries). For ALDOB and ECR11, we calculated effect sizes separately on the data from each mouse. For these two types of biological replicates, the effect sizes were highly correlated (r=0.96 for LTV1, r=0.93 for ALDOB, r=0.96 for ECR11). Based on this high reproducibility and to increase resolving power, we performed all subsequent analyses after combining data across mice for each enhancer haplotype library (data for one of the two LTV1 replicate libraries is shown in Supplementary Fig. 5).
We next recalculated effect sizes in two ways (Online Methods). First, as for the reproducibility analysis, we constructed separate linear models for each position where mutational status was encoded as a single binary variable representing whether an enhancer haplotype was wild-type or mutant at that position (Fig. 2a,c,e, Supplementary Table 1a). Second, we constructed separate multiple linear regression models for each position with three variables, each corresponding to a particular nucleotide substitution at that position (Fig. 2b,d,f, Supplementary Table 1b). For each enhancer, we also constructed a multiple linear regression model incorporating all positions. These models were also significantly predictive (Supplementary Note 1; Supplementary Table 2), and yielded effect size profiles similar to models constructed independently for each position (Supplementary Fig. 6). As the coefficients from models constructed independently for each position are more naturally interpreted as position-specific effects, we used these models for subsequent analyses.
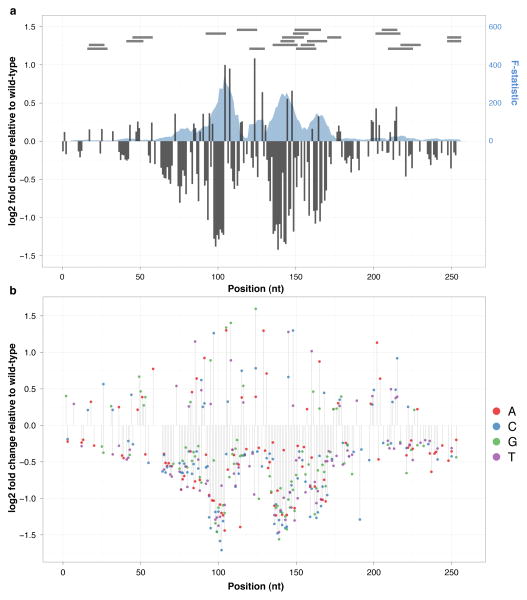
Figure 2. Effect size on transcriptional activity of all possible substitution mutations in three mammalian enhancers.
Estimated effect size of mutation at each position based on coefficients from univariate (grey columns, left axis) and trivariate (A:red, C:blue, G:green, T:purple) models are shown for ALDOB ((a) and (b) respectively), ECR11 ((c) and (d) respectively), and LTV1 ((e) and (f) respectively). Effect sizes were estimated by taking the log2 of the ratio of the number of pools predicted by the model with a mutation to the number of pools predicted for the wild-type nucleotide (total number of pools sequenced per library: ALDOB: 39; ECR11: 69; LTV1 Set 1: 10; LTV1 Set 2: 10). Effect sizes are only shown for positions where model coefficients had associated p-values ≤ 0.01. We also used multiple linear regression with sets of 10 adjacent positions as predictors. The F-statistic of these models, representing the extent to which the model is predictive of the outcome, is plotted (blue shadow, right axis) for ALDOB (a), ECR11 (c), and LTV1 (e). The locations of TFBS predictions using the MATCH web server (with restriction to TFs present in liver) are shown as horizontal grey bars at the top of the plot in (a), (c), and (e). The location of a partial LINE element in ECR11 is shown as an orange bar at the bottom of (c).
To provide further validation, we also performed site-directed mutagenesis to individually introduce the six mutations in ALDOB that were predicted to have amongst the largest effect sizes (three increasing activity, and three decreasing activity), and tested these individually via the hydrodynamic tail vein luciferase assay (Supplementary Fig. 7). Observed luciferase fold-changes were highly correlated with effect size predictions from the models (R=0.985).
Spatial clustering of high impact sites and concordance with known TFBSs
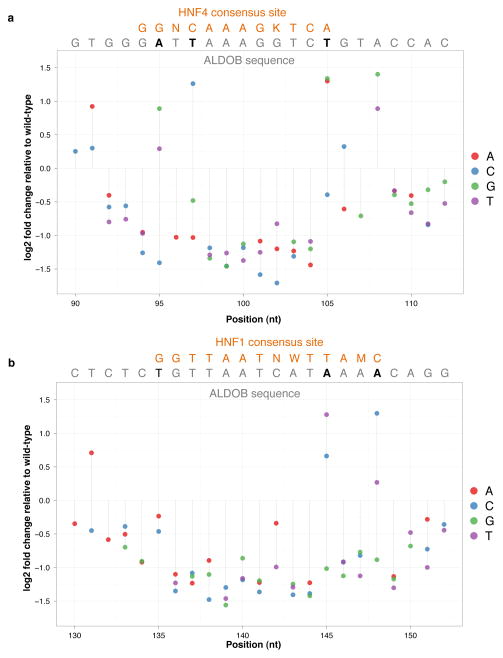
Across each enhancer, the effect size profiles exhibited spatial structure - i.e., a clustering of positions with larger effect sizes. Positions separated by less than ~6 nucleotides had significantly correlated effect sizes (Supplementary Fig. 8). To further explore this, we performed multiple linear regression using mutational status at 10 adjacent positions (i.e., a binary variable for wild-type or mutant) at a time (Online Methods). These models remained significantly predictive of transcriptional activity in a spatially resolved pattern (Fig. 2a,c,e). We suspected that these clusters of correlated positions might represent TFBSs. Indeed, when we predict TFBSs17 (Fig. 2a,c,e; Supplementary Table 3), we observe striking overlap between predicted binding sites and clusters of highly predictive positions (Fig. 2a,c,e). For example, a predicted binding site for HNF4 in the ALDOB enhancer (bases 94–105) coincides with a highly predictive localized model (Fig. 2a). Furthermore, all mutations in this region had negative effects on activity, with the notable exception of mutations that increased identity with the consensus HNF4 binding site, which were activating (e.g., 95A→G and 105T→A) (Fig. 3a). The same pattern was observed for other predicted sites as well, e.g. a predicted HNF1 binding site at bases 135–148 in ALDOB (Fig. 3b). Notably, independent experiments have established that these two transcription factors drive this element in vivo11. The spatial patterns may also reveal or refine broader features of activity – e.g. the boundaries of functional elements. For example, in ECR11, computational prediction yielded a large number of predicted liver-specific TFBSs in the proximal 300 bases12, but we observed that the highest impact SNVs were largely confined to the distal 160 bases (Fig. 2d; Supplementary Fig. 9).
Figure 3. Mutation effect size profiles in transcription factor binding sites.
For a predicted HNF4 site (positions 94–105) (a) and a predicted HNF1 site (positions 135–148) (b) in ALDOB, plotted is the effect size for each possible substitution, with the consensus TF binding sequence (orange) and the enhancer sequence (grey for consensus, black for non-consensus). Non-consensus positions where rescue is observed after mutating to consensus are shown in boldface. HNF4 binding to the ALDOB enhancer region in human liver has been previously demonstrated22, whereas in vivo occupancy data for HNF1 at this region is not yet available.
Relationship between evolutionary conservation and functional constraint
Evolutionary constraint in non-coding, regulatory DNA has frequently served as a proxy for functional constraint18–20. However, recent studies have shown that many enhancers are evolving rapidly and that mammalian genomes contain large numbers of evolutionarily young, sometimes species-specific, enhancers21, 22. All three enhancers studied here are grossly conserved between human and mouse (Supplementary Fig. 1). We therefore investigated the relationship between functional constraint and evolutionary constraint at single nucleotide resolution. For two of three enhancers, linear models, constructed to assess whether evolutionary constraint (i.e. Genomic Evolutionary Rate Profiling (GERP)23) was predictive of functional constraint (i.e. the absolute value of univariate model coefficients that we obtained), were significantly predictive with modest explanatory power (ALDOB: R2=0.1232, p=6.31e-9; LTV1: R2=0.03911, p=5.47e-4). For both enhancers, positions with the highest functional effect sizes were significantly associated with elevated evolutionary constraint scores (Supplementary Fig. 10). However, not all positions with high GERP scores (≥4) had functional effect sizes in the top quartile for each enhancer (ALDOB: 33 of 61, 54%; ECR11: 5 of 25, 20%; LTV1: 0 positions with GERP≥4). These sites might have functions unrelated to the enhancer activity assayed here or might be of greater functional relevance in other contexts, e.g. other tissues or developmental time points. On the other hand, a small set of highly functional sites, e.g. most nucleotides within the distal-most C/EBP motif in ECR11, have low GERP scores, consistent with lineage or species-specific activity.
Effect size spectrum of of single nucleotide variants
A substantial proportion of polymorphisms and new mutations in mammalian genomes are single nucleotide substitutions24. However, the functional dissection of regulatory elements has historically relied on introducing nested or scanning deletions, limiting the extent to which they inform the interpretation of naturally occurring variation. Our results provided an opportunity to examine the distribution of effect sizes of SNVs in mammalian enhancers on the magnitude of transcriptional activation (Fig. 4). Surprisingly, we observed that the majority of SNVs result in only a modest change in transcription relative to the wild-type enhancer. Overall, fewer than 25% of the mutations alter transcriptional activity by >1.2-fold. Furthermore, only a few mutations, mostly in ALDOB, altered activity by a factor of >2-fold. These results suggest that these enhancers are highly robust to the vast majority of potential SNVs. Further application of this method will be needed to assess whether this is a general property of mammalian enhancers.
Figure 4. Distribution of effect sizes for all possible substitution mutations in three mammalian enhancers.
For the three enhancers studied (two replicate libraries for LTV1), plotted is the cumulative fraction of substitutions possessing a given effect size expressed as the absolute value of the effect size of a given substitution. For example, across the three enhancers, between ~80% and ~95% of substitutions influence transcriptional activity by less than a factor of 1.5.
Perhaps as expected, the majority of functionally significant mutations decreased activity (70% or 850/1211). In general, only one substitution at a given position was activating, e.g. substitutions that render a motif more consensus-like (Fig. 3). However, we observed some notable exceptions, including positions 83–93 and 272–278 in LTV1, where all or almost all substitutions are activating, consistent with binding of a repressive transcription factor. Positions 83–93 harbor a predicted binding site for NF-1, while there are no predicted sites in the immediate vicinity of positions 272–278, highlighting the value of experimental assessment of mutational impact.
Epistatic interactions
Finally, we sought to leverage the fact that our enhancer libraries contain multiple mutations on each haplotype to assess the degree of epistasis or interaction. To obtain adequate power, we restricted our analysis to pairs of positions that were both mutated in at least 20 haplotypes. For each pair of positions that passed this cutoff, we built a multiple linear regression model consisting of three binary variables where the first two variables encoded mutation status (wild-type or mutant) at each position independently and the third encoded whether both are mutant in a particular haplotype (Online Methods). With an FDR cutoff of 0.05, we observed few pairs with a significant interaction term (ALDOB: 82 of 33,389, 0.25%; ECR11: 199 of 184,206, 0.10%, LTV1: 45 of 43,975, 0.10%), suggesting that the effects of multiple SNVs on the same haplotype are generally additive, or that our study lacked power to identify subtle interactions. Interacting pairs were significantly enriched for proximity (i.e. pairs within 10bp versus pairs further apart) and we observed several different classes of interacting pairs with respect to the signs of the individual position effects and the sign of the interacting term (Supplementary Table 4).
DISCUSSION
In summary, we developed a strategy to construct complex libraries of mammalian enhancers that contain all possible single nucleotide substitutions, and hundreds of thousands of distinct haplotypes. This method surpasses its predecessor14 in terms of cost-effectiveness, tunability, applicability to full-length regulatory elements, and integration with an in vivo assay. We applied this method to empirically measure the distribution of effect sizes of all possible SNVs in three mammalian enhancers in an in vivo model. A key finding is that the vast majority of SNVs in these enhancers have highly reproducible yet remarkably modest effects on transcriptional activation. The distribution suggests that enhancers are highly robust to single nucleotide changes. We also find that the preponderance of combinations of single nucleotide changes have additive effects on function. As expected, there is a clear relationship between the magnitude of functional impact and the location of predicted TFBSs, although not all predicted TFBSs are functional, and not all functional motifs are associated with predicted TFBSs. Similarly, evolutionary constraint, although clearly correlated with the magnitude of functional impact, does not predict it well on a nucleotide-by-nucleotide basis.
There remain some limitations of method performance. First, while we exploited a mouse tail vein assay to assess function in vivo, the regulatory elements are episomal and therefore may not be subject to the same mechanisms governing elements residing on chromosomes. For example, because of the size of the synthetic construct, we are unable to assess the impact of mutations that may influence long-range interactions between regulatory elements. This might be addressed in part by transitioning to a lentiviral system, which would facilitate use in additional tissues and may also enable the application of other assays, e.g. ChIP-Seq, to enhancer variant libraries. Furthermore, our results must also be considered specific to the minimal promoter used here until other promoter classes are tested. Second, we have only assayed these enhancers in a single tissue and at a single time point. The activity profile of specific positions could well be different in other tissues, i.e. the long-standing context problem25. Third, because of the scarcity of the target transcript relative to total RNA, we observed complexity bottlenecking, limiting the precision of our effect size estimates. This can be addressed by optimization of RNA isolation, e.g. by hybridization-based enrichment. Fourth, we restricted our analysis to enhancer haplotypes containing only substitutions, as this was the dominant form of variation introduced during synthesis. To facilitate simultaneous dissection of the functional consequences of small insertions and deletions (indels), one could use reduced-fidelity oligonucleotide synthesis conditions, or polymerase cycling assembly with oligonucleotides containing scanning block indels. Current efforts are directed at implementing these improvements, scaling this method to more enhancers, and applying it to other classes of non-coding regulatory elements.
A fundamental goal of modern biology is to understand the human genome at single nucleotide resolution. Single nucleotide differences between genomes are causative for, or impact susceptibility to, a host of diseases, and single nucleotide mutations are a primary source of raw material for evolution. We anticipate that the high-throughput, empirical measurement of the functional impact of single nucleotide variants in enhancers will substantially facilitate the analysis of non-coding variants in GWAS hits, the study of the mechanistic basis for enhancer activity, and the engineering of enhancers with desired properties. Furthermore, with cost-effective, massively parallel methods for functional analysis, it may soon be realistic to empirically measure the functional impact of all possible single nucleotide changes in all non-coding regulatory elements in the human genome.
ONLINE METHODS
Construction of enhancer haplotypes from short doped oligonucleotides using PCA
Sets of overlapping oligonucleotides for each enhancer were designed either by manual inspection (LTV1) or using the program DNAWorks (ALDOB and ECR11). Common flanking sequences were included on either side to allow for amplification of the full length enhancer haplotypes during PCA. For LTV1, two versions of overlapping oligonucleotides were designed, such that the overlap region in each was different. Oligonucleotides were synthesized by Integrated DNA Technologies (IDT). All positions corresponding to the enhancer region were synthesized using a hand-mix doped at a ratio of 97:1:1:1 (i.e. designated base at a frequency of 97%, and every other base at a frequency of 1%). Sequences of all oligonucleotides are listed in Supplementary Methods.
For ALDOB as well as ECR11, the full length haplotypes were assembled in a single step. 50fmol of each oligonucleotide (ALDOB_PCA_OLIGO[1…6] or ECR11_PCA_OLIGO[1…12]) were used in a 25μl PCR reaction volume with 1X KapaHiFi Hot Start Ready Mix (Kapa BioSystems), and 0.5X SYBR Green II, with the following cycling conditions: 95°C for 3min; followed by 30 cycles of 98°C for 20s, 65°C for 15s, 72°C for 15s. Each sample was monitored and extracted from the PCR machine when fluorescence began to plateau. Four such reactions were carried out in parallel and then pooled together for each enhancer. The PCR product representing a complex pool of enhancer haplotypes was purified using QIAquick columns (Qiagen). The assembled enhancer haplotypes were then subjected to an additional around of PCR to add 15bp of vector homology on either side to render them competent for cloning using InFusion (Clontech). 20ng of template was used in a 25μl PCR reaction volume with 1X KapaHiFi Hot Start Ready Mix, 0.5x SYBR Green II, and each primer (VH_F and VH_R) at 0.3μM final concentration. Thermal cycling was performed with the following program: 95°C for 3min; followed by 30 cycles of 98°C for 20s, 65°C for 15s, 72°C for 15s. Each sample was monitored and extracted from the PCR machine when fluorescence began to plateau. Sixteen such reactions were carried out in parallel and then pooled together for each enhancer. The PCR product was purified using QIAquick columns (Qiagen).
The two LTV1 designs were assembled separately. For each design, pairs of oligonucleotides i.e. oligonucleotides 1 and 2, oligonucleotides 3 and 4, and oligonucleotides 5 and 6 were each assembled in parallel and the products of the three reactions were then assembled together into the final product in a single reaction The combinations of primers and oligonucleotides used in each reaction are listed in Supplemetary Methods. Each 50μl PCR reaction was prepared on ice with 1X iProof Ready Mix (Bio-Rad), 0.5x SYBR Green II, forward and reverse primers each at 0.5μM final concentration, and 50fmol of each template oligo. Thermal cycling was done in a MiniOpticon Real-time PCR system (Bio-Rad) with the following program: 98°C for 30s, followed by 30 cycles of 98°C for 10s, 62°C for 30s and 72°C for 15s. Each sample was monitored and extracted from the PCR machine when fluorescence began to plateau. PCR products were purified on a QIAquick column (Qiagen). The haplotypes obtained from each of the two LTV1 designs were pooled after the PCA step. Two aliquots were drawn from this pool, and then carried through subsequent steps as two independent samples and were associated with entirely different sets of tags.
Cloning of enhancer haplotypes and the degenerate tag into pGL4.23
For ALDOB and ECR11, we first cloned in the degenerate tag to create a complex library of tagged pGL4.23 plasmids. We then cloned in the enhancer haplotypes into these tagged pGL4.23 plasmids. For LTV1, we first cloned in the enhancer haplotypes and then cloned in the degenerate tag. Details of each cloning step remained the same, irrespective of the order in which they were carried out, and are described below.
Cloning of degenerate tag into pGL4.23 plasmid
The tag oligonucleotide (TAG_OLIGO) was made double-stranded using primer extension in a 50μl reaction volume with 1X iProof Master Mix, 0.5μg single stranded tag oligo, 0.5μg reverse primer (TAG_EXTEND). The reaction was incubated at 95°C for 3min, 61°C for 10min, and then 72°C for 5min. The product was purified using a QIAquick column (Qiagen) and eluted in 50μL EB. It was further subjected to ExoI treatment in 40μL reaction volume for 1h at 37°C to degrade any remaining single stranded DNA, and purified again using Qia-Quick columns. The resulting double stranded tag oligo was then cloned into pGL4.23 at the XbaI site (at 1799bp) using standard InFusion (Clontech) protocol. The InFusion reaction was diluted to 100μl using TE8. 1.5μl of this diluted cloning reaction was used to transform 50μl of chemically competent FusionBlue cells (Clontech) using the standard protocol. When the tag was being cloned in first, sixteen such transformation reactions were pooled and grown overnight in four 50ml liquid cultures at 37°C in a shaking incubator. DNA was extracted using the Invitrogen Charge Switch Mini Prep Kit for ALDOB and ECR11, and the Invitrogen Charge Switch Midi Prep Kit for LTV1.
Cloning enhancer haplotypes into pGL4.23 vector
The enhancer haplotypes were cloned into the EcoRV site (at 42bp) of the pGL4.23 plasmid, using standard InFusion protocol. 1.5μl of the cloning reaction was used to transform 50μl of chemically competent FusionBlue cells (Clontech) using standard protocol. Five transformations reactions were pooled and grown overnight in 50ml liquid cultures at 37°C in a shaking incubator. DNA was extracted using the Invitrogen Charge Switch Mini Prep Kit for ALDOB and ECR11, and the Invitrogen Charge Switch Midi Prep Kit for LTV1.
Tail vein injections
Enhancers were injected using methods as previously described12. Briefly, each library was injected in at least 3 mice using the TransIT EE Hydrodynamic Gene Delivery System (Mirus Bio) following the manufacturer’s protocol. 10ug of each library, alongside 2μg of pGL4.74[hRluc/TK] vector to correct for injection efficiency, was injected into the tail vein of CD1 mice (Charles River). Following 24h, mice were euthanized and livers were harvested.
Measurement of luciferase activity
Firefly and renilla luciferase activity were measured on a Synergy 2 Microplate Reader (BioTek Instruments, Inc.) for each liver using the Dual Luciferase Reporter Assay System (Promega). The firefly luciferase to renilla luciferase ratios were determined and expressed as relative luciferase activity. All mouse work was approved by the UCSF Institutional Animal Care and Use Committee.
Isolation of RNA from mouse livers
Fresh liver tissue was immediately stabilized in RNAlater solution (Ambion). Samples were homogenized in TRIzol reagent (Invitrogen) and RNA was isolated from the samples according to the manufacturer’s instructions.
DNase treatment of RNA
In order to remove any DNA contamination in the RNA extracted from mouse livers, it was subjected to DNaseI treatment using DNA-free (Ambion). Each reaction was prepared with 1x DNA-free Buffer, 1μl of rDNaseI enzyme, 10μg of RNA and RNase-free water to 50μL. The reactions were incubated at 37°C for 1h, with an additional 1μl of enzyme added mid-way through the incubation. The reaction was stopped by adding 7μl of the Inactivation reagent and incubating for 2min at room temperature with frequent shaking. The reaction was centrifuged in a microcentrifuge at >13,000rpm for 1.5min, and the supernatant containing RNA was carefully transferred to a fresh tube.
RT-PCR
Aliquots of RNA obtained after DNase treatment were reverse transcribed to cDNA and amplified by PCR using the Qiagen One-Step Kit. The PCR sought to amplify the 20bp degenerate tag encoded at the 3′ end of the luciferase transcript. The reactions were assembled on ice in a 25μL total volume with the following reagents: 1X Qiagen One-Step RT-PCR buffer, 400μM of each dNTP, 0.6μM of forward primer (BARCODE_PE_F), 0.6μM of relevant reverse primer (BARCODE_PE_R_ILMN_INDEX[1-8]), 0.5x SYBR Green II and 5μl (~1μg) of RNA template. Thermal cycling was performed on a Bio-Rad MiniOpticon Real-Time PCR system with the following program: 50°C for 30min (reverse transcription), 95°C for 15min (inactivation of reverse transcriptase and heat-activation of the DNA polymerase), then 30 cycles of 94°C for 30s, 65°C for 30s and 72°C for 30s. Each reaction was monitored and extracted from the PCR machine when the fluorescence began to plateau. The cDNA products were purified using the QIAquick PCR Purification Kit (Qiagen) and eluted in 35μl EB. The primers used for the RT-PCR contained the necessary sequences for compatibility with the Illumina flow-cell. Thus, the cDNA library obtained at the end of this step was sequencing-ready, eliminating the need for a separate sequencing library construction step. The reverse primer additionally included 6bp barcodes allowing for several RT-PCR reactions to be pooled into a single lane for sequencing.
Sequencing of tags
The pooled RT-PCR reaction products were sequenced on an Illumina GAIIx using a sequencing primer (BARCODE_SEQ_F) designed to read into the tag sequence. Each run was 36 cycles with an additional 6 cycles to read the indexing barcode (index sequencing primer: GTCTCCCCGTTTAATCCCATCTAGAGTCGA ).
For each pool, reads were filtered based on the quality scores for the first 20 bases which correspond to the degenerate tag. The numbers of occurrences of each tag were counted and tags that were supported by at least 10 reads were classified as being “present” in that pool.
Associating tags with enhancer haplotypes
The enhancer haplotypes and tags were situated more than 1000bp away from each other on the pGL4.23 plasmid. To bring them adjacent and facilitate the subassembly method, we digested the pGL4.23 plasmids using HindIII, which had two cut sites, one just 3′ of the enhancer, and one just 5′ of the tag, thus resulting in excision of the intervening region. Cut site 1 was already a part of the pGL4.23 backbone. Cut site 2 was engineered in as a part of the tag oligo. The digest was carried out in a 50μl volume with 1X NEB Buffer 2, 1μg of plasmid and 1μl of HindIII Enzyme (New England BioLabs) and incubated at 37°C for 3h. The digested plasmid was purified using a QIAquick column (Qiagen).
The digested plasmids were then re-circularized using intra-molecular ligation, resulting in the tag becoming adjacent to the 3′ end of the enhancer. Ligation was performed using T4 DNA ligase (New England BioLabs) in a 20μl reaction with 15ng of template per reaction. The reaction was incubated for 15min at room temperature, followed 20min at 65°C to inactivate the ligase.
The enhancer and tag region were amplified from re-circularized plasmids using PCR with the forward primer targeting the region immediately 5′ of the enhancer (ENHANCER_F for ALDOB and ECR11, and LTV1_F for LTV1) and the reverse primer targeting the region immediately 3′ of the tag (BARCODE_PE_R). The reaction was carried out in a 25μl volume with 1X KapaHiFi Hot Start Ready Mix (Kapa BioSystems), 0.5x SYBR Green II, 5μl of the ligation reaction, and each primer at 0.3μM final concentration. Thermal cycling was carried out using Bio-Rad MiniOpticon Real-Time PCR system using the following program: 95°C for 3min; and then 30 cycles of 98°C for 20s, 65°C for 15s, 72°C for 15s. Each reaction was monitored and removed from the PCR machine when the fluorescence began to plateau. The reactions were then pooled and purified using QIAquick columns (Qiagen).
The amplicons were then subjected to the subassembly protocol as conceptually described in Hiatt et al15 with some modifications as follows. The random fragmentation step was carried out using the Nextera Tn5 transposase (EpiCentre) instead of mechanical shearing. The Nextera reaction was purified using MinElute column (Qiagen) and size-selected by PAGE (LTV1: 100+; ECR11:100-300,300+; ALDOB: no size-selection performed). The size-selected fragments were subjected to PCR in a 25μl reaction volume with 1X KapaHiFi Hot Start Ready Mix (Kapa BioSystems), 0.5x SYBR Green II, 5μl of the ligation reaction, Nextera Adapter 1 at 10nM final concentration, and primers Nextera BP1 and BARCODE_PE_R at 0.3μM final concentration each. Thermal cycling was carried out using BioRad Mini Opticon System using the following program: 95°C for 3min; and then 30 cycles of 98°C for 20s, 65°C for 15s, 72°C for 15s. Each reaction was monitored and removed from the PCR machine when the fluorescence began to plateau. The PCR products were purified using a QIAquick column and then sequenced on either an Illumina GAIIx or a Hi-Seq 2000. Read1 collected 76bp/101bp of the enhancer sequence staring at random breakpoints along the enhancer. Read 2 collected the 20bp tag sequence.
The reads were then grouped by tag. Reads belonging to each group were then aligned to the wild type enhancer sequence to identify the mutations on the haplotype associated with that tag using a custom analysis framework.
Estimation of effect size of mutation at each position along the enhancer (univariate model)
All linear regression analyses were performed using the lm() or lsfit() functions available in the R Statistical Package. To quantity the effect of mutation at any given position on the number of pools in which an enhancer haplotype was observed, we built a separate linear regression model at every position along the enhancer, with a single predictor representing whether the given position was wild-type or mutant. The predictor was thus a binary variable representing presence (1) or absence (0) of a mutation at that position.
where,
yi = number of pools in which the ith haplotype was observed (referred to as pool counts), and
Xij = 1 if position j was mutant and 0 if position j was wild-type in the ith haplotype.
To facilitate comparison between positions and between enhancers, we calculated the “effect size” of mutation at a position j as:
The p-value reported by the model for β1j was used to judge whether the effect size was significant.
For LTV1, since a single haplotype was typically associated with multiple tags, we normalized the pool counts for a given haplotype by dividing by the number of tags associated with that haplotype. In case of ALDOB and ECR11, since the enhancer haplotypes were cloned in second, almost all haplotypes were associated with single tags, and thus the pool counts for tags were used directly as the pool counts of their linked haplotypes.
Estimation of effect size of each specific nucleotide change at each position along the enhancer (trivariate model)
To explore whether the estimated effect sizes for each position were being driven by specific nucleotide substitutions, we modified the model just described to include three predictors, each representing one of the three possible nucleotide substitutions at that position. The factors were set up as binary variables representing the presence (1) or absence (0) of the particular change at that position.
Effect sizes were then calculated from the coefficients produced by the models as follows (for k=1,2,3):
The p-value reported by the model for βkj was used to judge whether the effect of a given nucleotide substitution at a given position was significant.
Spatial Structure
To quantify whether nearby sites tend to have similar effect sizes, we calculated the sum of the absolute values of the differences in effect sizes between positions located at a given distance (lag) from each other. In other words, we calculated , where k=1,2,…,20 denotes the lag, N denotes the length of the enhancer, and ri is the effect size of position i.
For each value of the lag k, we also calculated S1,*(k), …,S1000,*(k), each of which measures the sum of the absolute values of the differences in effect sizes between positions at a distance k from each other, after permuting the effect sizes (r1, …,rN). We then calculated a p-value associated with each value of the lag k as the fraction of the S1,*(k), …,S1000,*(k) that was as small or smaller than S(k).
Models to estimate combined predictive power of blocks of adjacent positions
To further characterize the nature of the spatial structure of the effect sizes and to explore whether certain regions along the enhancer were enriched for sites with larger effect sizes, we focused on blocks of adjacent sites in a 10bp sliding window along the length of the enhancer. For each window, we built a multiple linear regression model with one predictor for each position within the window. Each predictor was set up as a binary variable denoting the presence (1) or absence (0) of mutation at that site. The response variable y was the number of pools in which a given haplotype was seen.
The F-statistic from each model was used as a measure of the collective predictive power of positions within each window.
Multiple linear regression models based on the entire haplotype
The multiple linear regression model included one predictor for each position along the enhancer, encoded as a 1 or 0 to indicate presence or absence of a mutation at that site on a given haplotype, and the response variable y represented the number of pools in which the haplotype was observed. Here N is the number of sites within a given enhancer.
A p-value for the model was calculated by comparing the mean squared error (MSE) of the model to MSEs of 200 models built using randomly shuffled versions of the response variable. A p-value for the model was estimated by calculating the fraction of times that the MSE for models built using a shuffled response vector was at least as small as the MSE computed using real data.
We then expanded the model, such that each position was represented by three predictors to indicate which of the three possible nucleotide substitutions was observed at that position.
A p-value for the model was calculated by repeatedly permuting the outcome vector as described earlier in the context of the univariate regression model; however, only 100 permutations were used, due to the high computational burden of constructing this model.
Identification of epistatic interactions (i.e. non-additive effects) among pairs of mutations
For each pair of positions, we built a linear multiple regression model with three predictors: one predictor each to indicate the presence (1) or absence (0) of a mutation at each of the two positions and a third (referred to as the “interaction term”) whose value was set to 1 if both positions were mutant on the given haplotype and 0 otherwise. Only pairs of positions that were both mutant on at least twenty haplotypes were considered.
We used the p-values for the interaction terms for the resulting models to calculate a FDR for each interaction term (using the p.adjust() function in R, with method=”BH”). Interaction terms with FDR < 0.05 were considered significant and used for downstream analyses of epistatic interactions.
Supplementary Material
Acknowledgments
This work was supported in part by grants HG003988 from the National Human Genome Research Institute (LAP), NIH grant DP5OD009145 (DMW), NIGMS award number GM61390 (NA), NICHD grant number R01HD059862 (NA), the Pilot/Feasibility grant from the UCSF Liver Center (P30 DK026743) (NA), AG039173 from the National Institute on Aging (JBH) and a fellowship from the Achievement Rewards for College Scientists Foundation (JBH). MJK was supported in part by NIH Training Grant T32 GM007175 and the Amgen research excellence in bioengineering and therapeutic sciences fellowship. RPS is supported by a CIHR fellowship in the area of hepatology. Parts of the research were conducted at the E.O. Lawrence Berkeley National Laboratory and performed under Department of Energy Contract DE-AC02-05CH11231, University of California. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, NICHD, NHGRI or the NIGMS.
Footnotes
DATA RELEASE
A full list of mutations interrogated here, along with the associated effect sizes and p values are provided as Supplementary Data. Raw sequencing reads have been submitted to the NCBI Short Read Archive under accession number SRA049159.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Consortium EP, et al. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12:628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 5.Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noonan JP, McCallion AS. Genomics of long-range regulatory elements. Annu Rev Genomics Hum Genet. 2010;11:1–23. doi: 10.1146/annurev-genom-082509-141651. [DOI] [PubMed] [Google Scholar]
- 7.VanderMeer JE, Ahituv N. cis-regulatory mutations are a genetic cause of human limb malformations. Dev Dyn. 2011;240:920–930. doi: 10.1002/dvdy.22535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabourin JC, et al. An intronic enhancer essential for tissue-specific expression of the aldolase B transgenes. J Biol Chem. 1996;271:3469–3473. doi: 10.1074/jbc.271.7.3469. [DOI] [PubMed] [Google Scholar]
- 10.Gregori C, et al. Expression of the rat aldolase B gene: a liver-specific proximal promoter and an intronic activator. Biochem Biophys Res Commun. 1991;176:722–729. doi: 10.1016/s0006-291x(05)80244-x. [DOI] [PubMed] [Google Scholar]
- 11.Gregori C, Porteu A, Mitchell C, Kahn A, Pichard AL. In vivo functional characterization of the aldolase B gene enhancer. J Biol Chem. 2002;277:28618–28623. doi: 10.1074/jbc.M204047200. [DOI] [PubMed] [Google Scholar]
- 12.Kim MJ, et al. Functional characterization of liver enhancers that regulate drug-associated transporters. Clinical pharmacology and therapeutics. 2011;89:571–578. doi: 10.1038/clpt.2010.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang Q, et al. Structure of the hepatic control region of the human apolipoprotein E/C-I gene locus. J Biol Chem. 1995;270:22577–22585. doi: 10.1074/jbc.270.38.22577. [DOI] [PubMed] [Google Scholar]
- 14.Patwardhan RP, et al. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat Biotechnol. 2009;27:1173–1175. doi: 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiatt JB, Patwardhan RP, Turner EH, Lee C, Shendure J. Parallel, tag-directed assembly of locally derived short sequence reads. Nature methods. 2010;7:119–122. doi: 10.1038/nmeth.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang G, Budker V, Wolff JA. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther. 1999;10:1735–1737. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- 17.Kel AE, et al. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic acids research. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loots GG, et al. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 19.Margulies EH, et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–774. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Visel A, et al. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet. 2008;40:158–160. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blow MJ, et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet. 42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt D, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper GM, et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Botstein D, Shortle D. Strategies and applications of in vitro mutagenesis. Science. 1985;229:1193–1201. doi: 10.1126/science.2994214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.