Abstract
The protein quality control system, composed of molecular chaperones and proteases, is of vital importance for the maintenance and function of the proteome and the health of the cell. To achieve this, the cellular proteostasis network integrates the protein folding machinery across all compartments of the eukaryotic cell to enable efficient communication and coordinate a rapid response of folding capacity. Quality control in the mitochondria, however, differs from its cytosolic counterpart due to its prokaryotic origin, and is entirely encoded by the nuclear genome. The control and regulatory cross-talk of mitochondrial function in cellular proteostasis is essential for cellular metabolism, organismal development, and lifespan. Consequently, mitochondrial dysfunction has dramatic effects on the development and progression of a number of neurodegenerative diseases, such as Friedreich’s ataxia and Parkinson’s disease. Studies using Caenorhabditis elegans as a model system have greatly contributed to our current knowledge of inter-compartmental proteostasis on the cellular and organismal levels.
Keywords: mtUPR, proteostasis, C. elegans, neurodegenerative diseases, aging
INTRODUCTION
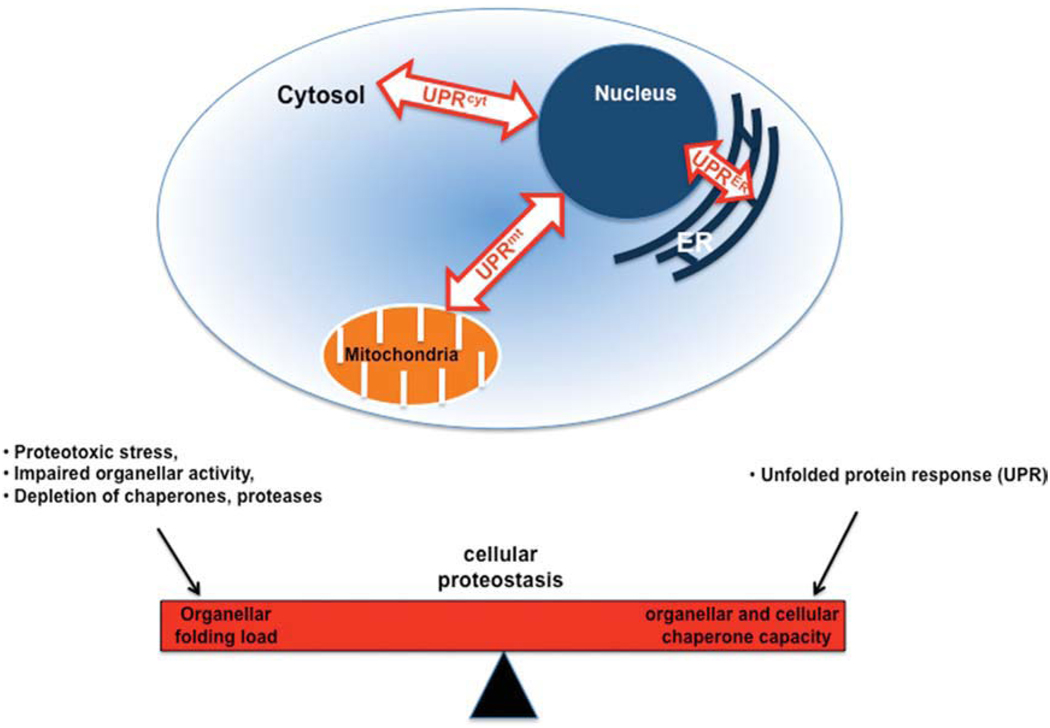
Subcellular compartmentalization in eukaryotes led to enhanced metabolic efficiency, which in metazoans resulted in an increase in cell size and cell number. This compartmentalization established a precise and rapid cross-talk between the cytoplasm and organelles to maintain cellular proteostasis and to ensure that the biosynthetic activity of the folding machinery is in balance with the folding load. Equally important was the response to diverse stress conditions that imbalance this cross-talk, and the adjustment achieved by reducing the folding load through attenuation of protein synthesis or by activating the protein quality control network. The cell encodes organellar-specific responses to these imbalances referred to as the unfolded protein response in the ER and mitochondria (UPRER/mt) and as heat shock response (HSR or UPRcyt) in the cytosol (Kaufman, 1999; Ron and Walter, 2007). The integration of the organellar UPR’s into the cellular proteostasis network is depicted in Figure 1.
Fig. 1.
Balancing the cellular proteostasis by compartment-specific unfolded protein response (UPR). This cartoon depicts the integration of the organellar UPR’s into the cellular proteostasis network. The cellular proteostasis is a balance of the folding load of all organelles and their respective response mechanisms (UPR). Perturbance of the client to chaperone ratio, e.g., due to proteotoxic stress, could result in an imbalance of the cellular proteostasis. To rebalance the proteostasis, the cell can activate organelle-specific UPR’s to cope with the excess of client proteins.
In this review, we will address the role of mitochondria in the inter-compartmental proteostasis network through the use of C. elegans as a tool to provide a cellular and molecular systems approach. Imbalance of cellular protein homeostasis, or proteostasis, affects mitochondrial function with dramatic consequences for the health of the cell and the organism (Wallace, 2005). Mitochondria have been associated with aging, oxidative stress, and neurodegenerative diseases. The mitochondrial protein quality control system monitors and safeguards the mitochondrial proteome and, thereby, maintains the function of the mitochondria (Voos, 2009). This protein quality control system, however, is entirely encoded by the nuclear DNA, thus an up-regulation of the mitochondrial proteostasis system upon proteotoxic challenge requires retrograde signaling to the nucleus and a subsequent translocation of the chaperones into the mitochondria. An intriguing question, therefore, is how the mitochondrial protein quality control system senses perturbations of the folding environment and rebalances proteostasis via a signaling pathway that extends beyond its boundaries.
Mitochondria are highly dynamic organelles that constantly alter their shape and structure in response to different stimuli and metabolic demands of the cell. Mitochondria are the site of many metabolic pathways including the citric acid cycle, fatty acid oxidation, ATP generation via oxidative phosphorylation, ion homeostasis, and Fe-S cluster formation. Mitochondria originated in ancestral eukaryotic cells through endosymbiosis of proteobacteria. Thus, it is not surprising that mitochondria have a unique protein quality control system retaining many of the ancestral characteristics of prokaryotes. The outcome for the eukaryotic cell was to evolve a system in which the mitochondrial chaperone and proteolytic machinery was integrated into a coherent cellular proteostasis network. Because the majority of the mitochondrial proteins are encoded by the nucleus, proteins destined to the mitochondria undergo multiple rounds of unfolding and folding events to translocate through the mitochondrial membranes (Hartl and Neupert, 1990). These folding steps require the cooperation of the translocation machinery and multiple chaperones of the Hsp70 and Hsp90 families to facilitate the unfolding, the translocation, and the subsequent refolding and assembly on both the cytosolic interface as well as in the mitochondrial matrix (Voos and Rottgers, 2002). Molecular chaperones have multiple functions in the maintenance of cellular proteostasis, by assisting the folding of de novo synthesized proteins, during translocation into the ER or the mitochondria, or refolding of misfolded or aggregated proteins. Achieving this balance is critical, because the expression and accumulation of aggregation prone proteins or exposures to a plethora of stress conditions can lead to an enhanced progression of aging and the development of neurodegenerative diseases such as Huntington’s and Parkinson’s disease. The cellular proteostasis network affects all compartments and thus it is not surprising that neurodegenerative diseases are associated with organellar dysfunction.
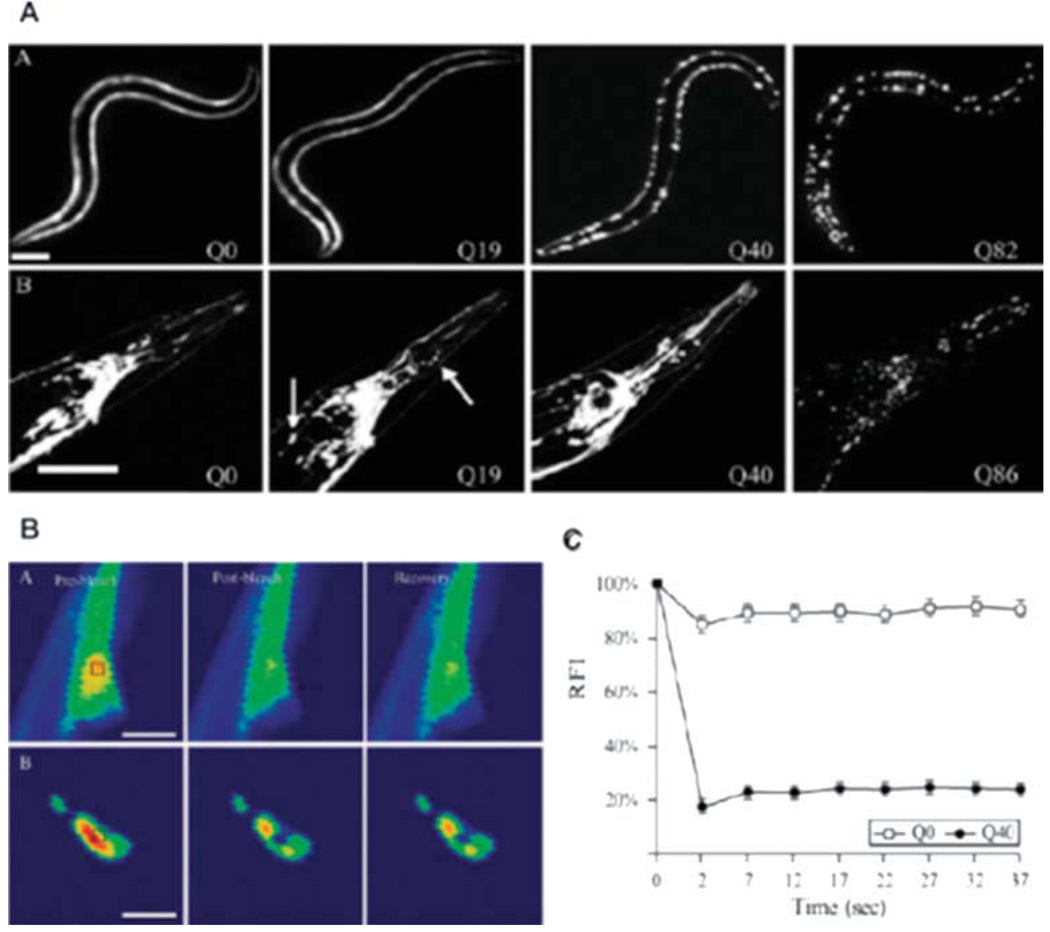
C. elegans is an excellent model for the study of proteostasis systems as it represents among the best-characterized metazoan model organisms with numerous advantages including a completed genome sequence, multiple genetic tools to assess function, and a defined cell lineage with a simple body plan, differentiated tissues, and diverse behavioral and physiological phenotypes available to study various biological and pathophysiological processes. For example, multiple C. elegans models of neurodegenerative diseases such as tissue-specific polyQ and mutant SOD1, have been generated by expression of fluorescently tagged protein fusions of the aggregation prone and disease causing proteins in C. elegans (Satyal et al., 2000; Morley et al., 2002; Brignull et al., 2006a; Gidalevitz et al., 2006, 2009). The transparent body of C. elegans allows visualization of aggregation-prone protein species and enables biophysical inspection using, e.g., FRAP (fluorescence recovery after photobleaching), FRET (Foerster resonance energy transfer), and FLIP (fluorescence loss in photobleaching) to understand the properties of different aggregate states. The neuronal and muscle-expression models for polyQ exhibit an age- and polyQ length-dependent aggregation and the biophysical characterization of the polyQ aggregates is depicted in Figure 2 (Morley et al., 2002; Brignull et al., 2006b). Additionally, C. elegans is amenable to genetic analysis such as gene knock-down by RNA interference (RNAi), which has been employed in genome-wide screens to identify genetic modifiers and pathways involved in the development and progression of aggregation and toxicity phenotypes (Nollen et al., 2004). C. elegans has a short lifespan and is, therefore, an attractive model to study aging. Early in adulthood, there is an accumulation of damaged proteins that occurs in multiple tissues revealing that proteostasis is already compromised well before other physiological and morphological markers of aging have been observed (Ben-Zvi et al., 2009).
Fig. 2.
C. elegans polyglutamine model. A: Expression of polyQ in the body wall muscle (Punc-54Q(n)::YFP; top) and throughout the nervous system (PF25B3.3Q(n)::CFP; bottom). The expression of both models shows a polyQ length-dependent aggregation pattern. B: FRAP analysis of poly- Q::YFP in body wall muscle cells to analyze solubility of polyQ proteins. Images are taken before bleaching (pre-bleach, left), immediately after bleaching (post-bleach, middle), and after a recovery period (recovery, right). Q0::YFP recovers rapidly and is soluble, whereas Q40::YFP foci do not recover and are immobile. C: Quantification of FRAP results. The graphs show the relative fluorescence intensity (RFI) for each time point (Brignull et al., 2006b).
MITOCHONDRIAL DYSFUNCTION IS LINKED TO AN INCREASE OF ROS PRODUCTION IN NEURODEGENERATIVE DISEASES AND APOPTOSIS
Mitochondrial functionality is essential for the function of the cell, not only as the source of energy, but also as the major source of reactive oxygen species (ROS), which are thought to be causative agents of aging and diseases (Raha and Robinson, 2000). Although mitochondrial dysfunction has been associated with accelerated aging and the development of neurodegenerative diseases, little is known about the underlying molecular mechanisms. In the following sections, we will summarize our current knowledge of mitochondrial function and its cellular integrity in the light of its connection to ROS production and defense mechanisms, longevity, apoptosis, and the development of the neurodegenerative diseases taking advantage of the tools afforded by C. elegans.
ROS
Aging and age-associated damage to macromolecules correlates with increased production of free radical species and or decreased antioxidant defenses (Finkel and Holbrook, 2000). ROS are generated as a byproduct during oxidative phosphorylation with mitochondria as the major source of ROS and the major target of ROS damage. The pathogenic aggregation of proteins is associated with excessive production of ROS, which can cause oxidation of amino acid side chains, formation of hydroperoxides, carbonylation of proteins, and fragmentation of the protein backbone (Levine and Stadtman, 2001). This damage results in a loss of function associated with misfolding and aggregation, and thus is directly connected to the susceptibility to pathology and disease (see below). Mitochondrial DNA is also vulnerable to ROS-induced damage since it is located in immediate proximity to the source of ROS generation. In C. elegans, the mitochondrial genome expresses 37 genes, of which 13 are components of the respiratory chain, 22 encode tRNA’s, and 2 correspond to rRNA’s (Hoffmann et al., 2009). Mutation in any of these mitochondrial genes has dramatic consequences for oxidative phosphorylation and energy production. Therefore, not surprisingly, accumulation of mtDNA mutations and lesions is associated with many age-related diseases (Greaves and Turnbull, 2009). To protect themselves, cells have developed strategies to counteract ROS-induced damage employing antioxidant proteins and free radical scavengers including superoxide dismutases (SOD), catalases, peroxidases, thioredoxins, and ferredoxins (Apel and Hirt, 2004; Li et al., 2009). For example, superoxide is converted by SOD into the less reactive hydrogen peroxide. Hydrogen peroxide levels are then regulated by glutathione peroxidase and catalases. C. elegans encodes for five sod genes, each with a specific sub-cellular localization: sod-1 and sod-5 are cytoplasmic, sod-2 and sod-3 are localized in the mitochondria, and sod-4 is localized extracellularly. Mutations in the sod genes result in a higher sensitivity towards ROS generators such as paraquat or juglone, suggesting that the different SOD proteins might exhibit distinct substrate specificities for the detoxification of ROS species. However, mutations in sod genes do not decrease lifespan. An explanation for this could be that mutation in one sod gene is compensated by another sod gene (Van Raamsdonk and Hekimi, 2009). Interestingly, mutations of sod-2 encoding a mitochondrial SOD lead to an increase of lifespan. It is assumed that the sod-2 mutant extends lifespan by decreasing mitochondrial function, which supports the connection between mitochondrial function and longevity (Van Raamsdonk and Hekimi, 2009).
Longevity
The biogenesis and function of mitochondria are primary determinants of longevity. Energy production, via oxidative phosphorylation, generates a mitochondrial membrane potential ΔΨm across the inner membrane. Multiple longevity pathways converge on mitochondria and lead to a decreased ΔΨm, which is associated with increased lifespan in C. elegans (Lemire et al., 2009). Membrane potential and the generation of the membrane potential drive three important cellular processes: (1) ATP synthesis, (2) active transport of ions and metabolites, and (3) increased ROS production. Long-lived C. elegans mutants daf-2 (human ortholog: IGF1R), age-1 (PI3K), clk-1 (COQ7/CAT5), isp-1 (UQCRFS1), and eat-2 (CHRFAM7A) all exhibit lower ΔΨm. Lower membrane potential of daf-2 is daf-16 (FOXO3) dependent (Lemire et al., 2009), indicating that the insulin-like signaling pathway not only regulates lifespan, but can also affect mitochondrial energetics. Lifespan can also be extended by uncoupling the electron transport chain using CCCP (carbonyl cyanide m-chlorophenylhydrazone), which dissipates ΔΨmand diminishes ROS levels (“uncoupling to survive” theory). Mitochondrial uncoupling could also extend lifespan by changes in ion homeostasis and signaling molecules such as ATP, Ca2+, and succinate. Moreover, long-lived mutants could shift their metabolism towards alternative energy–generating pathways such as fermentation upon uncoupling. As a molecular basis for defense against ROS, the expression of molecular chaperones is up-regulated in long-lived mutants (Hsu et al., 2003; Murphy et al., 2003; Kuzmin et al., 2004).
Disruption of the electron transport chain (ETC) by knock-down of ETC components in C. elegans, which results in a lower ΔΨm, can result in life extension in C. elegans, whereas mutations that disrupt ETC function in humans shorten lifespan (Rea et al., 2007). These apparently contradictory observations are based on the extent of the dysfunction of the ETC. Using an RNAi dilution strategy, it was shown that knock-down of ETC components in C. elegans, such as atp-3 (ATP5O), nuo-2 (NDUFS3), isp-1 (UQCRFS1), cco-1 (COX5B), and frh-1 (FXN), have a gradual influence on lifespan. Moreover, partial inhibition of these genes extends lifespan, whereas complete inhibition shortens lifespan (Rea et al., 2007). Life extension in these RNAi mediated knock-down animals depends on a specific L3/L4 stage of development, characterized by the last somatic cell divisions and substantial mitochondrial DNA expansion. These observations suggest that cell-cycle checkpoints of somatic cell proliferations are critical to activate compensatory mitochondrial pathways that regulate longevity (Rea et al., 2007).
Reduced ΔΨm also affects the vital process of mitochondrial translocation. The membrane potential with its negative charge in the mitochondrial matrix drives the initial transport of the positively charged pre-sequence before the matrix-localized Hsp70 facilitates the import by ATP-dependent cycles of transient binding of the incoming polypeptide (Martin et al., 1991; Voos et al., 1999). Consequently, a decrease of membrane potential will likely impair the mitochondrial import of those pre-proteins with a higher dependence on a membrane potential and thus contribute to a reduced mitochondrial function. Consistent with this, knock-down of components of the mitochondrial import machinery, tomm-7 (TOMM7), ddp-1 (TOMM8B), tin-9.1 (TIMM9), tin-9.2 (EXOSC4), and tin-10 (TIMM10) encoding for the Tim/Tom translocation complex, results in a defective formation of the gonad, reduced numbers of progeny, small body size, partial embryonic lethality, and thus precludes development (Curran et al., 2004). How does a reduced mitochondrial membrane potential result in an increase in lifespan when, at the same time, it impairs the translocation of mitochondrial pre-proteins? Perhaps the key here is to achieve a minimal level of mitochondrial function sufficient for basal translocation events without interfering with the overall flux of translocation.
An additional mechanism of mitochondrial regulation for C. elegans longevity has been demonstrated in a recent study that prohibitins, ubiquitious conserved proteins, that assemble at the inner mitochondrial membrane into ring-like high molecular weight complexes, enhance lifespan by modulating fat metabolism. The function of prohibitins is not well understood, but they are thought to act as assembly factors and protein scaffolds for the integrity of mitochondrial membranes (Osman et al., 2009). Depletion of prohibitin influences ATP levels, fat content, and mitochondrial proliferation in a genetic background and age-specific manner. Reduced prohibitin activity promotes survival of animals with a compromised mitochondrial function by triggering the fat metabolism (Artal-Sanz and Tavernarakis, 2009).
Apoptosis
Apoptosis is essential for the development and survival of most multicellular organisms. During C. elegans development, over 10% of somatic cells undergo apoptosis (Lettre and Hengartner, 2006). Mitochondria have a central role in programmed cell death in mammals, but in C. elegans this is less clear. Genetic studies have identified EGL-1, CED-9 (Bcl-2), CED-4, and CED-3 (CASP10) as key components of apoptosis (Metzstein et al., 1998). In living cells, CED-4 is associated with the Bcl-2 homologue, CED- 9. An apoptotic signal up-regulates the BH3-only protein, EGL-1, which binds to CED-9 and thus leads to a dissociation of CED-4. Subsequently, CED-4 oligomerizes into a tetramer and forms the CED-4 apoptosome, which facilitates the autoactivation of the caspase CED-3 (Yan et al., 2006; Shi, 2008).
The apoptotic mechanism in C. elegans differs from mammals in several aspects: although there is a functional conservation of CED-9/Bcl-2, C. elegans does not express IAP-like or SMAC/Diablo and Omi/HtrA2 homologues, and there is little evidence for an essential role of mitochondria in the induction of cell death in C. elegans. The Bcl-2 homologue CED-9 is localized to the mitochondrial outer membrane; however, the localization itself does not appear to be important for its function. Artificial tethering of CED-9 to the cytosolic face of the ER membrane rescues a ced-9 mutation (Tan et al., 2007), which suggests that mitochondrial localization of CED-9 may be required for alternate non-apoptotic functions.
Friedreich’s Ataxia
Friedreich’s ataxia is the most common heritable ataxia and is caused by the defective expression of frataxin, e.g., due to a GAA triplet repeat expansion in intron 1. The function of this mitochondrial localized, nuclear encoded protein is not well understood and frataxin-deficient cells exhibit impaired biosynthesis and function of Fe-S cluster-containing proteins (Rotig et al., 1997). Since several proteins of the electron transport chain contain Fe-S clusters, it is therefore not surprising that frataxin deficiency results in impaired mitochondrial respiration and lower ATP levels, reduced mitochondrial membrane potential, ΔΨm, and an impaired Ca2+ buffering. The failure of Fe-S cluster biosynthesis leads to iron accumulation, which causes free radical damage. Although knockdown of frataxin (frh-1, FXN) in C. elegans using RNAi results in a reduced body and brood size, lifespan is enhanced by 25% (Pastore et al., 2003; Ventura et al., 2005; Condo et al., 2006). However, retaining some lower level of expression of frh-1 seems to be critical as a strong knockdown results in reduced lifespan (Rea et al., 2007; see above).
Parkinson’s Disease
Parkinson’s disease (PD) is characterized by the degeneration of dopaminergic neurons. C. elegans has been used as a model system for PD (Schmidt et al., 2007; van Ham et al., 2008) and the relationship between mitochondria and PD has been supported by (1) a proteomic study identifying 75 proteins possibly involved in PD, of which more than 90% are subunits of the five complexes of the respiratory chain (Li et al., 2009), (2) that PD causes a decrease of complex I activity (Schapira et al., 1989), (3) that inhibition of complex I by the neurotoxin MPTP (1-methyl-4-phenyl- 1,2,3,6-tetrahydropyridine) or rotenone or mutations in mt DNA causes PD (Betarbet et al., 2000; Fornai et al., 2005), (4) that α-synuclein immunostaining is observed in degenerating mitochondria (Martin et al., 2006), (5) that pan-neuronal overexpression of α-synuclein, knock-down using RNAi of DJ-1 orthologue (B0432.2, encoding for a redox-reactive signaling protein) or deletion of the parkin orthologue (pdr-1, encoding for an E3 ubiquitin–protein ligase), impairs mitochondrial function and enhances the toxicity of the complex I inhibitor, rotenone (Song et al., 2004; Ved et al., 2005), and (6) that several PD-associated proteins are localized in mitochondria and/or interact with mitochondrial proteins such as: Parkin, DJ-1, and PINK (PTEN-induced kinase; Dodson and Guo, 2007; Narendra et al., 2008). Mutations in the Leucine-rich repeat kinase 2 (LRRK-2) are the most common genetic cause of PD, but it was unknown for a long time how LRRK-2 contributes to the pathophysiology of PD. Using C. elegans, it has been established that the LRRK-2 orthologue, lrk-1, protects against the complex I inhibitor, rotenone, suggesting a role for regulating mitochondrial physiology (Wolozin et al., 2008). In addition, C. elegans PD models were used to identify modifiers of PD based on the folding of the natively unfolded α-synuclein. Misfolding of α-synuclein was enhanced upon knock-down of genes encoding for trafficking proteins (endocytic pathway), whereas suppressors of α-synuclein inclusions are aging-associated genes and genes encoding for the ubiquitin proteasome system and chaperones (Hamamichi et al., 2008; Kuwahara et al., 2008; van Ham et al., 2008).
An increasing amount of evidence suggests a correlation of the PD-associated proteins, Parkin and PINK, with the autophagy of mitochondria (mitophagy) (Whitworth and Pallanck, 2009). PINK rapidly accumulates on damaged mitochondria and specifically recruits Parkin from the cytosol to damaged mitochondria and thereby promotes mitophagy in D. melanogaster and mammalian cell culture (Narendra et al., 2010; Vives-Bauza et al., 2010). It remains to be shown if these observations extend to C. elegans.
Huntington’s Disease
A number of observations suggest a link between Huntington’s disease (HD) and mitochondrial function: (1) HD patients exhibit an increased lactate production, which indicates a compromised mitochondrial function (Jenkins et al., 1993), (2) mitochondria from HD patients have reduced levels of complex II and II–III activity (Mann et al., 1990), (3) mitochondria from HD patients display a polyQ-length-dependent perturbation of the mitochondrial membrane potential and in Ca2+ homeostasis (Panov et al., 2002), and (4) over-expression of Htt proteins with extended polyQ stretches increases oxidative stress-induced mitochondrial fragmentation in Hela cells, correlating with increased caspase-3 activation and cell death (Wang et al., 2009). It is assumed that the polyQ expansion that interacts with mitofusin (Mfn) proteins located on the outer mitochondrial membrane is required for mitochondrial fusion. This interaction is thought to compromise Mfn function, causing enhanced mitochondrial fragmentation and hence loss of function. C. elegans was used to test the hypothesis that polyQ expansion proteins interfere with the fusion and fission of mitochondria. RNAi-mediated knock-down of drp-1 (DNM1L), which is required for mitochondrial fission, indeed reduced the polyQ-induced motility defects in C. elegans (Wang et al., 2009).
MITOCHONDRIAL PROTEOSTASIS NETWORK
The importance of the mitochondrial proteostasis network in C. elegans has been established by genetic and proteomic studies that have identified members of the major chaperone families Hsp70/40, Hsp60/10, Hsp90, and Hsp100/AAA+, proteases, and assembly factors homologous to their bacterial or mammalian counterparts (Heschl and Baillie, 1989; Li et al., 2009; Wormbase: www.wormbase. org). These are listed in Table 1. The protein quality control system is essential to maintain the functionality of the mitochondrial proteome by facilitating the translocation of mitochondrial proteins, assisting in their refolding and assembly into functional multi-subunit complexes in the matrix, and to maintain the functional folded state or directing damaged proteins to the clearance machinery. Decreased expression of mitochondrial chaperones results in severe developmental defects, for example, knockdown of hsp-6 (mt Hsp70), which is essential for the import of all nuclear encoded mitochondrial proteins that leads to abnormal mitochondrial morphology, lower ATP levels, defects in embryogenesis, and shorter lifespan (Kimura et al., 2007). Similarly, knock-down of hsp-60 (mt chaperonin) causes severe growth defects and embryonic and early developmental arrest (Yoneda et al., 2004), and likewise down-regulation of clpp-1 (ClpP) has effects on embryonic viability, growth, and locomotion (Simmer et al., 2003). These findings underscore the importance of the mitochondrial proteostasis network for the development and health of the organism.
TABLE 1.
C. elegans Mitochondrial Chaperones, Proteases, and Assembly Factorsa
| Mitochondrial chaperones, proteases and assembly factors | |
|---|---|
| C. elegans gene/sequence name | Homologue |
| Hsp70/40 family | |
| hsp-6 | hsp-70 |
| dnj-10 | hsp-40 |
| dnj-15 | hsp-40 |
| dnj-21 | hsp-40 |
| C34C12.8 | grpE |
| Hsp60/10 family | |
| hsp-60 | hsp-60/mt chaperonin |
| hsp-10 | hsp-10 |
| Proteases | |
| clpP-1 | clpP |
| C34B2.6 | lon/pim1 |
| spg-7 | paraplegin/mtAAA |
| cbn-ymel-1 | AAA+ATPase/FtsH |
| ZK550.3 | mip1/metalloedopeptidase |
| Hsp100 and AAA+ATPases | |
| K07A3.3 | clpX (1) |
| D2030.2 | clpX (2) |
| atad-3 | ATAD3 |
| bcs-1 | AAA+ATPase |
| Hsp90 family | |
| R151.7α | hsp-75/TRAP1 |
| Specific assembly factors | |
| eat-3 | opa-1 |
| phb-1 | phb |
| phb-2 | phb |
The components of the mitochondrial protein quality control were identified using genetic and proteomic studies and are comprised of chaperones of the Hsp70/40, Hsp60/10, Hsp100, and other AAA+ and Hsp90 family, proteases, and assembly factors. The respective bacterial, yeast or mammalian homologues are depicted in the right column.
The expression of a single mt Hsp70 and multiple Hsp40 proteins (encoded by dnj-10, dnj-15, and dnj-21) (Li et al., 2009) suggests that these interactions could expand the range of substrate interactions and, therefore, be important for regulated responses to diverse cellular or environmental stimuli. Likewise, C. elegans mitochondria express two ClpX ATPase orthologs, ClpX1 and ClpX2, which could interact with the proteolytic subunit, ClpP, to form the complete ClpXP protease essential for clearance of damaged mitochondrial proteins. It is not known whether both ClpX proteins can bind simultaneously to ClpP to form a mixed complex or whether interactions with ClpP is mutually exclusive and separately whether both have either a similar or different substrate spectrum. The amino acid sequence identity between ClpX1 and ClpX2 is less than 55%, which suggests the latter possibility. This speculation is further supported by the evidence that their N-termini, which are used as binding sites for either adaptor proteins or their direct interaction with substrates in Hsp100/AAA+ homologues, show the highest variation (Kirstein et al., 2009). No substrates for ClpXP and no potential adaptor proteins for either of the two ClpX proteins have been yet identified.
The differences in the chaperone and protease machines of the mitochondria relative to the cytosol lead to questions on folding specificity and capacity. This has been addressed initially by experiments in which a Huntingtin N-terminal fragment containing 73 glutamines was targeted to the mitochondria. Unexpectedly, despite having a polyQ expansion that causes cytoplasmic aggregation, this mitochondrial- targeted polyQ protein was soluble (Rousseau et al., 2004). This suggests that the subcellular compartment can have a strong consequence on the equilibrium between folding and misfolding, which leads to the question of whether protein quality control in a specific compartment can influence proteotoxicity beyond its organellar boundary. Consistent with this possibility, an RNAi study of modifiers of cytoplasmic-expressed polyQ identified mitochondrial Hsp70 (hsp-6) and several components of the respiration chain as modifiers of aggregation. Knock-down of hsp-6 enhances cytoplasmic polyQ aggregation, indicating that the mitochondrial protein quality control system contributes to the proteostasis and health of the whole cell (Nollen et al., 2004). How do chaperones in the mitochondria contribute to cellular proteostasis? hsp-6 was the only mitochondrial chaperone identified in this suppressor screen, which suggests either that Hsp70 specifically contributes to the cellular proteostasis network or that Hsp70, as the major mitochondrial chaperone, has the most dramatic effect on the mitochondrial folding environment. Because mitochondrial Hsp70 provides the driving force for translocation of proteins targeted to the mitochondria, a reduction in the levels of this essential protein should abolish import and could, therefore, lead to an accumulation of mitochondrial pre-proteins in the cytosol. One consequence could be an imbalance of the folding environment in the cytosol resulting in enhanced cytoplasmic aggregation of polyQ.
Studies of mitochondrial disorders provide additional support that the activity of specific chaperones is linked to neurological diseases. For example, of the 20 genes linked to spastic paraplegias, two of these are paraplegin (spg-7) and hsp-60 (Casari et al., 1998; Hansen et al., 2002; Fink, 2003a,b). Mutation of spg-7 leads to a deficient assembly and hence to a reduced activity of complex I of the respiratory chain and thus to a higher susceptibility to oxidative stress (Casari et al., 1998).
Importantly, all components of the mitochondrial proteostasis system are encoded by the nuclear genome. This leads to questions on the nature of the signaling mechanisms and regulation of these nuclear encoded components to meet the folding demands in the mitochondria.
SIGNALLING BETWEEN THE TWO COMPARTMENTS UPON PROTEOTOXIC CHALLENGES
mtUPR
The observation that perturbation of the folding environment in mitochondria up-regulates the expression of nuclear genes encoding mitochondrial chaperones led to the discovery of the mitochondrial unfolded protein response (UPRmt) (Zhao et al., 2002; Kuzmin et al., 2004; Yoneda et al., 2004). In mammalian cells, the expression of a truncated folding-deficient form of ornithine transcarbamylase (ΔOTC) specifically up-regulated the mitochondrial chaperones Hsp60/10, Hsp40, and the protease ClpP. This signaling pathway likely involves the sensing of mitochondrial stress in the mitochondrial matrix, transduction of the stress signal to the nucleus, and the specific expression of the genes encoding for mitochondrial chaperones. It has been demonstrated that the transcription factors CHOP and C/EBPβ regulate the expression of the mitochondrial chaperones in the nucleus (Zhao et al., 2002). However, CHOP is conserved only in vertebrates, which led to the suggestion that other signaling pathways are involved in the UPRmt (Yoneda et al., 2004).
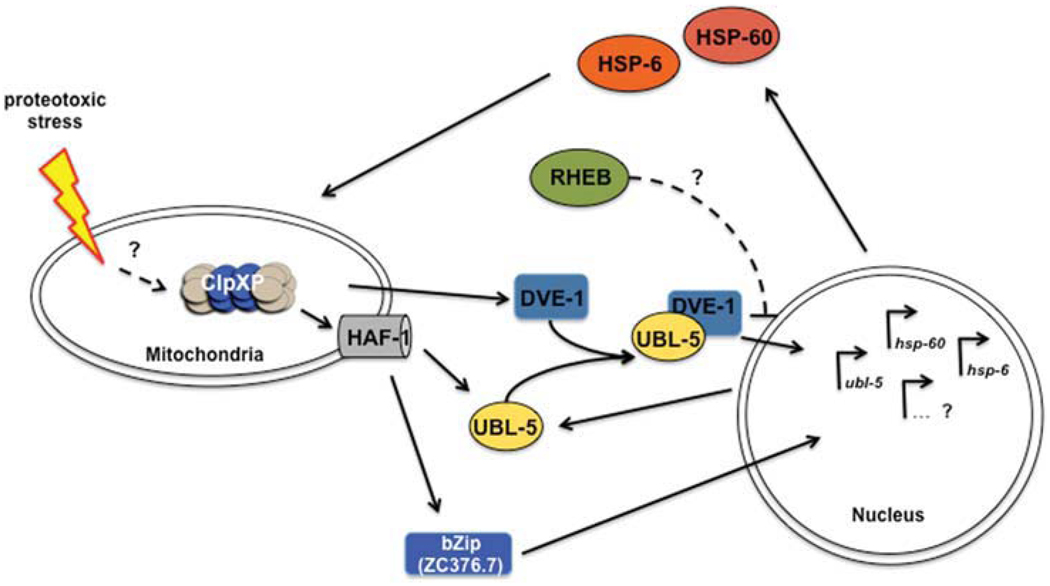
To identify the signaling components of the UPRmt, an RNAi screen of C. elegans chromosome I was performed to identify genes required for activation of the hsp-6 (Hsp70) and hsp-60 transcriptional GFP fusions. The genes, whose knock-down resulted in up-regulation of chaperone reporters were either encoding for proteins directly involved in the mitochondrial biogenesis, processing, and folding, or were components of multimeric complexes (Yoneda et al., 2004). The modifiers that contribute to the folding environment would be expected to activate molecular chaperones. By comparison, knock-down of components of multisubunit protein complexes would be expected to cause an imbalance of subunits that could result in misfolding and aggregation of the partner proteins and thus increasing the number of chaperone client proteins. In subsequent genome-wide screens, eight components of the UPRmt signaling pathway were identified: (1) ubl-5, a ubiquitin-like protein, (2) dve-1, a homeobox containing transcription factor, (3) clpp-1, encoding the proteolytic component of the mitochondrial Clp protease, (4 and 5) the AAA+ partner Clp-ATPases, ClpX1 (K07A3.3) and ClpX2 (D2030.2), (6) haf-1, encoding an ABC transporter, (7) the bZip transcription factor ZC376.7, and (8) F54C8.5, a GTPase homologous to Rheb (Benedetti et al., 2006; Haynes et al., 2007, 2010; see Fig. 3). Induction of mitochondrial stress promotes the accumulation of UBL-5 in nuclei, indicating that UBL-5 acts at a nuclear-localized step of the UPRmt (Benedetti et al, 2006). DVE-1 displays a nuclear redistribution upon mt stress induction and directly binds to the promoter of the chaperone genes, hsp-6 and hsp-60. In addition, DVE-1 interacts with UBL-5, indicating that both act together in the transcriptional activation of the mitochondrial chaperone genes. UBL-5 appears to activate its own expression, leading to a feed forward regulation of the signal transduction. The function of ClpXP can be placed upstream of DVE-1 and UBL-5 since knock-down of clpp-1 (ClpP), K07A3.3 (ClpX1), and D2030.2 (ClpX2) attenuates the induction of ubl-5, hsp-6, and hsp-60 and also prevents the redistribution of DVE-1 and its binding to the promoter regions of hsp-6 and hsp-60 (Haynes et al., 2007, 2010). It was proposed that the peptides generated by the proteolytic activity of ClpXP are then transported into the cytosol by the ABC transporter HAF-1 to transmit the UPRmt signal to the nucleus. Since the degradation of mitochondrial proteins results in an efflux of peptides in yeast by a homologous transporter (Mdl1p), it was speculated that a potential peptide efflux by HAF-1 could play a role in the UPRmt (Young et al., 2001; Haynes et al., 2010). Mitochondria isolated from C. elegans, indeed, exhibited an ATP-dependent release of peptides, which was abolished upon knock-down of either clpp-1 or haf-1. Down-regulation of haf-1 also attenuated ubl-5 expression upon mt stress, supporting the role of haf-1 in the signaling of UPRmt (Haynes et al., 2010). Notably, haf-1 had no influence on the nuclear redistribution of DVE-1, emphasizing that there are other yet unidentified regulators of the UPRmt. Additional screens have also revealed, ZC376.7, a bZip transcription factor as a regulator of the UPRmt. Upon mt stress, ZC376.7 relocated from the cytosol to the nucleus, which was attenuated by knock-down of clpP-1 and haf-1, thus placing ZC376.7 downstream of the proteolytic signaling cascade (Haynes et al., 2010). Little is known about the activity of the eighth and last component of the UPRmt, Rheb, but it seems to attenuate the UPRmt and might gain a role to shut off the UPRmt with cessation of the stress conditions. Knock-down by RNAi of F54C8.5 (RHEB) promotes the nuclear redistribution of DVE-1, the induction of ubl-5, and the complex formation of DVE-1 and UBL-5 (Haynes et al., 2007).
Fig. 3.
Model of the mitochondrial unfolded protein response (UPRmt) in C. elegans. Proteotoxic stress triggers a response of ClpXP, whose proteolytic activity is required to transmit the signal to the cytoplasm presumably via peptide translocation by HAF-1. Here, the transcription factors, UBL-5 and DVE-1, which form a complex, and ZC376.7 redistribute to the nucleus. DVE-1 binds to the promoter region of the mitochondrial chaperone genes, hsp-6 and hsp-60. The expression of ubl-5 is also up-regulated and probably acts as a feed-forward regulation to enhance the signal. This activation of the UPRmt results in an accumulation of HSP-6 (mt Hsp70) and HSP-60 (mt chaperonin) in the mitochondria. RHEB is thought to act as a suppressor of the UPRmt, probably with cessation of the stress conditions. Open questions of the regulation of the UPRmt are indicated with question marks and are further discussed in the text.
While there has been much recent progress, numerous questions remain: (1) on the initial sensing of mt stress: Does proteotoxic stress result in general misfolding of proteins, which are recognized by ClpXP or is there a specific substrate?, (2) characterization of the signal transduction events from the mitochondria to the nucleus: are there receptors for mitochondrial-specific peptides/components released by ClpXP-HAF-1 in the cytosol/nucleus? How are these peptides protected from cytosolic peptidases?, (3) identification of the specific targets of ZC376.7: Do the number and specificity differ between ZC376.7 and UBL-5/DVE-1?, and (4) the role of Rheb. A model for the signal transduction pathway of the UPRmt is summarized in Figure 3.
CONCLUSION AND PERSPECTIVE
C. elegans has proven to be an excellent model organism to study inter-compartmental proteostasis and has contributed greatly to our understanding of the role of mitochondria and its integration into the proteostasis network for the health of the cell and organism. The benefits of using C. elegans are its versatility to address specific questions on the sub-cellular, tissue-specific, and organismal level. This, combined with genetic and proteomic studies, has provided insights into the molecular composition of the chaperones and proteases in the mitochondria and into the biological role of mitochondria within the compartment and beyond the organellar boundaries. The function of mitochondria affects all developmental processes and the lifespan of the organism. Dysfunction of mitochondria can lead to oxidative stress, accelerated aging, apoptotic cell death, the development of neurodegenerative diseases, or the enhancement of its progression. Thus, the balance of the mitochondrial folding capacity, which maintains the correct fold and function of the mitochondrial proteome and its integrity into the cellular proteostasis network, is of vital importance for the health of the organism.
The functional emphasis of mitochondria differs between tissues and varies from a biosynthetic role (e.g., intestine, liver) to a primarily energy-providing organelle (heart and skeletal muscles). Differential gene expression of the nuclear-encoded mitochondrial genes and varying the mitochondrial content enables cells to meet tissue-specific demands of mitochondrial activity (Veltri et al., 1990). Consequently, different tissues exhibit distinct mitochondrial proteomes (Johnson et al., 2007a,b). How do cells adjust to this variable folding load? Do the expression levels and/or composition of the mitochondrial chaperones and proteases differ in various tissues? How does the cellular proteases network adjust to changes in the compartmental folding machineries?
ACKNOWLEDGMENTS
Work in the lab of R.M. is supported by grants from the National Institute of Health (National Institute of General Medical Sciences and National Institute on Aging) and the Daniel F. and Ada L. Rice Foundation. J.K.-M. is supported by a long-term postdoctoral fellowship of the Human Frontier Science Program (HFSP).
REFERENCES
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Artal-Sanz M, Tavernarakis N. Prohibitin couples diapause signalling to mitochondrial metabolism during ageing in C. elegans. Nature. 2009;461:793–797. doi: 10.1038/nature08466. [DOI] [PubMed] [Google Scholar]
- Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci USA. 2009;106:14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 2006;174:229–239. doi: 10.1534/genetics.106.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- Brignull HR, Moore FE, Tang SJ, Morimoto RI. Polyglutamine proteins at the pathogenic threshold display neuron-specific aggregation in a panneuronal Caenorhabditis elegans model. J Neurosci. 2006a;26:7597–7606. doi: 10.1523/JNEUROSCI.0990-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brignull HR, Morley JF, Garcia SM, Morimoto RI. Modeling polyglutamine pathogenesis in C. elegans. Methods Enzymol. 2006b;412:256–282. doi: 10.1016/S0076-6879(06)12016-9. [DOI] [PubMed] [Google Scholar]
- Casari G, De Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, De Michele G, Filla A, Cocozza S, Marconi R, Durr A, Fontaine B, Ballabio A. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell. 1998;93:973–983. doi: 10.1016/s0092-8674(00)81203-9. [DOI] [PubMed] [Google Scholar]
- Condo I, Ventura N, Malisan F, Tomassini B, Testi R. A pool of extramitochondrial frataxin that promotes cell survival. J Biol Chem. 2006;281:16750–16756. doi: 10.1074/jbc.M511960200. [DOI] [PubMed] [Google Scholar]
- Curran SP, Leverich EP, Koehler CM, Larsen PL. Defective mitochondrial protein translocation precludes normal Caenorhabditis elegans development. J Biol Chem. 2004;279:54655–54662. doi: 10.1074/jbc.M409618200. [DOI] [PubMed] [Google Scholar]
- Dodson MW, Guo M. Pink1, Parkin, DJ-1 and mitochondrial dysfunction in Parkinson’s disease. Curr Opin Neurobiol. 2007;17:331–337. doi: 10.1016/j.conb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Fink JK. Advances in the hereditary spastic paraplegias. Exp Neurol. 2003a;184(Suppl 1):S106–S110. doi: 10.1016/j.expneurol.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Fink JK. The hereditary spastic paraplegias: nine genes and counting. Arch Neurol. 2003b;60:1045–1049. doi: 10.1001/archneur.60.8.1045. [DOI] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Fornai F, Schluter OM, Lenzi P, Gesi M, Ruffoli R, Ferrucci M, Lazzeri G, Busceti CL, Pontarelli F, Battaglia G, Pellegrini A, Nicoletti F, Ruggieri S, Paparelli A, Sudhof TC. Parkinson-like syndrome induced by continuous MPTP infusion: convergent roles of the ubiquitin-proteasome system and alpha-synuclein. Proc Natl Acad Sci U S A. 2005;102:3413–3418. doi: 10.1073/pnas.0409713102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- Gidalevitz T, Krupinski T, Garcia S, Morimoto RI. Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS Genet. 2009;5:e1000399. doi: 10.1371/journal.pgen.1000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves LC, Turnbull DM. Mitochondrial DNA mutations and ageing. Biochim Biophys Acta. 2009;1790:1015–1020. doi: 10.1016/j.bbagen.2009.04.018. [DOI] [PubMed] [Google Scholar]
- Hamamichi S, Rivas RN, Knight AL, Cao S, Caldwell KA, Caldwell GA. Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson’s disease model. Proc Natl Acad Sci USA. 2008;105:728–733. doi: 10.1073/pnas.0711018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JJ, Durr A, Cournu-Rebeix I, Georgopoulos C, Ang D, Nielsen MN, Davoine CS, Brice A, Fontaine B, Gregersen N, Bross P. Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am J Hum Genet. 2002;70:1328–1332. doi: 10.1086/339935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Neupert W. Protein sorting to mitochondria: evolutionary conservations of folding and assembly. Science. 1990;247:930–938. doi: 10.1126/science.2406905. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev Cell. 2007;13:467–480. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell. 2010;37:529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heschl MF, Baillie DL. Characterization of the hsp70 multigene family of Caenorhabditis elegans. DNA. 1989;8:233–243. doi: 10.1089/dna.1.1989.8.233. [DOI] [PubMed] [Google Scholar]
- Hoffmann M, Bellance N, Rossignol R, Koopman WJ, Willems PH, Mayatepek E, Bossinger O, Distelmaier F. C. elegans ATAD-3 is essential for mitochondrial activity and development. PLoS One. 2009;4:e7644. doi: 10.1371/journal.pone.0007644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Jenkins BG, Koroshetz WJ, Beal MF, Rosen BR. Evidence for impairment of energy metabolism in vivo in Huntington’s disease using localized 1H NMR spectroscopy. Neurology. 1993;43:2689–2695. doi: 10.1212/wnl.43.12.2689. [DOI] [PubMed] [Google Scholar]
- Johnson DT, Harris RA, Blair PV, Balaban RS. Functional consequences of mitochondrial proteome heterogeneity. Am J Physiol Cell Physiol. 2007a;292:C698–C707. doi: 10.1152/ajpcell.00109.2006. [DOI] [PubMed] [Google Scholar]
- Johnson DT, Harris RA, French S, Blair PV, You J, Bemis KG, Wang M, Balaban RS. Tissue heterogeneity of the mammalian mitochondrial proteome. Am J Physiol Cell Physiol. 2007b;292:C689–C697. doi: 10.1152/ajpcell.00108.2006. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ. Molecular chaperones and the heat shock response. Sponsored by Cold Spring Harbor Laboratory, 6–10 May 1998. Biochim Biophys Acta. 1999;1423:R13–R27. doi: 10.1016/s0304-419x(98)00029-8. [DOI] [PubMed] [Google Scholar]
- Kimura K, Tanaka N, Nakamura N, Takano S, Ohkuma S. Knockdown of mitochondrial heat shock protein 70 promotes progeria-like phenotypes in Caenorhabditis elegans. J Biol Chem. 2007;282:5910–5918. doi: 10.1074/jbc.M609025200. [DOI] [PubMed] [Google Scholar]
- Kirstein J, Moliere N, Dougan DA, Turgay K. Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat Rev Microbiol. 2009;7:589–599. doi: 10.1038/nrmicro2185. [DOI] [PubMed] [Google Scholar]
- Kuwahara T, Koyama A, Koyama S, Yoshina S, Ren CH, Kato T, Mitani S, Iwatsubo T. A systematic RNAi screen reveals involvement of endocytic pathway in neuronal dysfunction in alpha-synuclein transgenic C. elegans. Hum Mol Genet. 2008;17:2997–3009. doi: 10.1093/hmg/ddn198. [DOI] [PubMed] [Google Scholar]
- Kuzmin EV, Karpova OV, Elthon TE, Newton KJ. Mitochondrial respiratory deficiencies signal up-regulation of genes for heat shock proteins. J Biol Chem. 2004;279:20672–20677. doi: 10.1074/jbc.M400640200. [DOI] [PubMed] [Google Scholar]
- Lemire BD, Behrendt M, DeCorby A, Gaskova D. C. elegans longevity pathways converge to decrease mitochondrial membrane potential. Mech Ageing Dev. 2009;130:461–465. doi: 10.1016/j.mad.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Lettre G, Hengartner MO. Developmental apoptosis in C. elegans: a complex CEDnario. Nat Rev Mol Cell Biol. 2006;7:97–108. doi: 10.1038/nrm1836. [DOI] [PubMed] [Google Scholar]
- Levine RL, Stadtman ER. Oxidative modification of proteins during aging. Exp Gerontol. 2001;36:1495–1502. doi: 10.1016/s0531-5565(01)00135-8. [DOI] [PubMed] [Google Scholar]
- Li J, Cai T, Wu P, Cui Z, Chen X, Hou J, Xie Z, Xue P, Shi L, Liu P, Yates JR, 3rd, Yang F. Proteomic analysis of mitochondria from Caenorhabditis elegans. Proteomics. 2009;9:4539–4553. doi: 10.1002/pmic.200900101. [DOI] [PubMed] [Google Scholar]
- Mann VM, Cooper JM, Javoy-Agid F, Agid Y, Jenner P, Schapira AH. Mitochondrial function and parental sex effect in Huntington’s disease. Lancet. 1990;336:749. doi: 10.1016/0140-6736(90)92242-a. [DOI] [PubMed] [Google Scholar]
- Martin J, Mahlke K, Pfanner N. Role of an energized inner membrane in mitochondrial protein import. Delta psi drives the movement of presequences. J Biol Chem. 1991;266:18051–18057. [PubMed] [Google Scholar]
- Martin LJ, Pan Y, Price AC, Sterling W, Copeland NG, Jenkins NA, Price DL, Lee MK. Parkinson’s disease alpha-synuclein transgenic mice develop neuronal mitochondrial degeneration and cell death. J Neurosci. 2006;26:41–50. doi: 10.1523/JNEUROSCI.4308-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzstein MM, Stanfield GM, Horvitz HR. Genetics of programmed cell death in C. elegans: past, present and future. Trends Genet. 1998;14:410–416. doi: 10.1016/s0168-9525(98)01573-x. [DOI] [PubMed] [Google Scholar]
- Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollen EA, Garcia SM, van Haaften G, Kim S, Chavez A, Morimoto RI, Plasterk RH. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci USA. 2004;101:6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Merkwirth C, Langer T. Prohibitins and the functional compartmentalization of mitochondrial membranes. J Cell Sci. 2009;122:3823–3830. doi: 10.1242/jcs.037655. [DOI] [PubMed] [Google Scholar]
- Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, Greenamyre JT. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- Pastore A, Tozzi G, Gaeta LM, Bertini E, Serafini V, Di Cesare S, Bonetto V, Casoni F, Carrozzo R, Federici G, Piemonte F. Actin glutathionylation increases in fibroblasts of patients with Friedreich’s ataxia: a potential role in the pathogenesis of the disease. J Biol Chem. 2003;278:42588–42595. doi: 10.1074/jbc.M301872200. [DOI] [PubMed] [Google Scholar]
- Raha S, Robinson BH. Mitochondria, oxygen free radicals, disease and ageing. Trends Biochem Sci. 2000;25:502–508. doi: 10.1016/s0968-0004(00)01674-1. [DOI] [PubMed] [Google Scholar]
- Rea SL, Ventura N, Johnson TE. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rotig A, de Lonlay P, Chretien D, Foury F, Koenig M, Sidi D, Munnich A, Rustin P. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat Genet. 1997;17:215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- Rousseau E, Dehay B, Ben-Haiem L, Trottier Y, Morange M, Bertolotti A. Targeting expression of expanded polyglutamine proteins to the endoplasmic reticulum or mitochondria prevents their aggregation. Proc Natl Acad Sci USA. 2004;101:9648–9653. doi: 10.1073/pnas.0403015101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyal SH, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer JM, Morimoto RI. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2000;97:5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira AH, Cooper JM, Dexter D, Jenner P, Clark JB, Marsden CD. Mitochondrial complex I deficiency in Parkinson’s disease. Lancet. 1989;1:1269. doi: 10.1016/s0140-6736(89)92366-0. [DOI] [PubMed] [Google Scholar]
- Schmidt E, Seifert M, Baumeister R. Caenorhabditis elegans as a model system for Parkinson’s disease. Neurodegener Dis. 2007;4:199–217. doi: 10.1159/000101845. [DOI] [PubMed] [Google Scholar]
- Shi Y. Apoptosome assembly. Methods Enzymol. 2008;442:141–156. doi: 10.1016/S0076-6879(08)01407-9. [DOI] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, Fraser AG, Ahringer J, Plasterk RH. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song DD, Shults CW, Sisk A, Rockenstein E, Masliah E. Enhanced substantia nigra mitochondrial pathology in human alpha-synuclein transgenic mice after treatment with MPTP. Exp Neurol. 2004;186:158–172. doi: 10.1016/S0014-4886(03)00342-X. [DOI] [PubMed] [Google Scholar]
- Tan FJ, Fire AZ, Hill RB. Regulation of apoptosis by C. elegans CED-9 in the absence of the C-terminal transmembrane domain. Cell Death Differ. 2007;14:1925–1935. doi: 10.1038/sj.cdd.4402215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ham TJ, Thijssen KL, Breitling R, Hofstra RM, Plasterk RH, Nollen EA. C. elegans model identifies genetic modifiers of alpha-synuclein inclusion formation during aging. PLoS Genet. 2008;4:e1000027. doi: 10.1371/journal.pgen.1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000361. doi: 10.1371/journal.pgen.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ved R, Saha S, Westlund B, Perier C, Burnam L, Sluder A, Hoener M, Rodrigues CM, Alfonso A, Steer C, Liu L, Przedborski S, Wolozin B. Similar patterns of mitochondrial vulnerability and rescue induced by genetic modification of alpha-synuclein, parkin, and DJ-1 in Caenorhabditis elegans. J Biol Chem. 2005;280:42655–42668. doi: 10.1074/jbc.M505910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltri KL, Espiritu M, Singh G. Distinct genomic copy number in mitochondria of different mammalian organs. J Cell Physiol. 1990;143:160–164. doi: 10.1002/jcp.1041430122. [DOI] [PubMed] [Google Scholar]
- Ventura N, Rea S, Henderson ST, Condo I, Johnson TE, Testi R. Reduced expression of frataxin extends the lifespan of Caenorhabditis elegans. Aging Cell. 2005;4:109–112. doi: 10.1111/j.1474-9726.2005.00149.x. [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, Magrane J, Moore DJ, Dawson VL, Grailhe R, Dawson TM, Li C, Tieu K, Przedborski S. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W. Mitochondrial protein homeostasis: the cooperative roles of chaperones and proteases. Res Microbiol. 2009;160:718–725. doi: 10.1016/j.resmic.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Voos W, Rottgers K. Molecular chaperones as essential mediators of mitochondrial biogenesis. Biochim Biophys Acta. 2002;1592:51–62. doi: 10.1016/s0167-4889(02)00264-1. [DOI] [PubMed] [Google Scholar]
- Voos W, Martin H, Krimmer T, Pfanner N. Mechanisms of protein translocation into mitochondria. Biochim Biophys Acta. 1999;1422:235–254. doi: 10.1016/s0304-4157(99)00007-6. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lim PJ, Karbowski M, Monteiro MJ. Effects of overexpression of huntingtin proteins on mitochondrial integrity. Hum Mol Genet. 2009;18:737–752. doi: 10.1093/hmg/ddn404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway: a mitochondrial quality control system? J Bioenerg Biomembr. 2009;41:499–503. doi: 10.1007/s10863-009-9253-3. [DOI] [PubMed] [Google Scholar]
- Wolozin B, Saha S, Guillily M, Ferree A, Riley M. Investigating convergent actions of genes linked to familial Parkinson’s disease. Neurodegener Dis. 2008;5:182–185. doi: 10.1159/000113697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Xu Y, Shi Y. 2:1 Stoichiometry of the CED-4-CED-9 complex and the tetrameric CED-4: insights into the regulation of CED-3 activation. Cell Cycle. 2006;5:31–34. doi: 10.4161/cc.5.1.2263. [DOI] [PubMed] [Google Scholar]
- Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- Young L, Leonhard K, Tatsuta T, Trowsdale J, Langer T. Role of the ABC transporter Mdl1 in peptide export from mitochondria. Science. 2001;291:2135–2138. doi: 10.1126/science.1056957. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]