Abstract
Heat shock factors (HSFs) are essential for all organisms to survive exposures to acute stress. They are best known as inducible transcriptional regulators of genes encoding molecular chaperones and other stress proteins. Four members of the HSF family are also important for normal development and lifespan-enhancing pathways, and the repertoire of HSF targets has thus expanded well beyond the heat shock genes. These unexpected observations have uncovered complex layers of post-translational regulation of HSFs that integrate the metabolic state of the cell with stress biology, and in doing so control fundamental aspects of the health of the proteome and ageing.
In the early 1960s, Ritossa made the seminal discovery of temperature-induced puffs in polytene chromosomes of Drosophila melanogaster larvae salivary glands1. A decade later, it was shown that the puffing pattern corresponded to a robust activation of genes encoding the heat shock proteins (HSPs), which function as molecular chaperones2. The heat shock response is a highly conserved mechanism in all organisms from yeast to humans that is induced by extreme proteotoxic insults such as heat, oxidative stress, heavy metals, toxins and bacterial infections. The conservation among different eukaryotes suggests that the heat shock response is essential for survival in a stressful environment.
The heat shock response is mediated at the transcriptional level by cis-acting sequences called heat shock elements (HSEs; BOX 1) that are present in multiple copies upstream of the HSP genes3. The first evidence for a specific transcriptional regulator, the heat shock factor (HSF) that can bind to the HSEs and induce HSP gene expression, was obtained through DNA–protein interaction studies on nuclei isolated from D. melanogaster cells4,5. Subsequent studies showed that, in contrast to a single HSF in invertebrates, multiple HSFs are expressed in plants and vertebrates6–8. The mammalian HSF family consists of four members: HSF1, HSF2, HSF3 and HSF4. Distinct HSFs possess unique and overlapping functions (FIG. 1), exhibit tissue-specific patterns of expression and have multiple post-translational modifications (PTMs) and interacting protein partners7,9,10. Functional crosstalk between HSF family members and PTMs facilitates the fine-tuning of HSF-mediated gene regulation. The identification of many targets has further extended the impact of HSFs beyond the heat shock response. Here, we present the recent discoveries of novel target genes and physiological functions of HSFs, which have changed the view that HSFs act solely in the heat shock response. Based on the current knowledge of small-molecule activators and inhibitors of HSFs, we also highlight the potential for pharmacologic modulation of HSF-mediated gene regulation.
Box 1. The heat shock element.
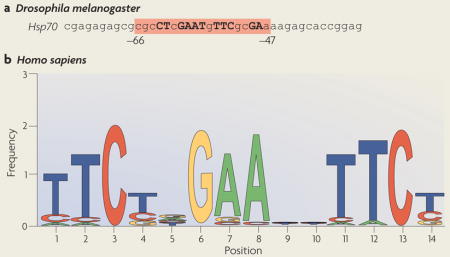
Heat shock factors (HSFs) act through a regulatory upstream promoter element, called the heat shock element (HSE). In the DNA-bound form of a HSF, each DNA-binding domain (DBD) recognizes the HSE in the major groove of the double helix6. The HSE was originally identified using S1 mapping of transcripts of the Drosophila melanogaster heat shock protein (HSP) genes3 (see the figure; part a). Residues –47 to –66 are necessary for heat inducibility. HSEs in HSP gene promoters are highly conserved and consist of inverted repeats of the pentameric sequence nGAAn132. The type of HSEs that can be found in the proximal promoter regions of HSP genes is composed of at least three contiguous inverted repeats: nTTCnnGAAnnTTCn132–134. The promoters of HSF target genes can also contain more than one HSE, thereby allowing the simultaneous binding of multiple HSFs. The binding of an HSF to an HSE occurs in a cooperative manner, whereby binding of an HSF trimer facilitates binding of the next one135. More recently, Trinklein and colleagues used chromatin immunoprecipitation to enrich sequences bound by HSF1 in heat-shocked human cells to define the HSE consensus sequence. They confirmed the original finding of Xiao and Lis, who identified guanines as the most conserved nucleotides in HSEs87,133 (see the figure; part b). Moreover, in a pair of inverted repeats, a TTC triplet 5′ of a GAA triplet is separated by a pyrimidine–purine dinucleotide, whereas the two nucleotides separating a GAA triplet 5′ from a TTC triplet is unconstrained87. The discovery of novel HSF target genes that are not involved in the heat shock response has rendered it possible that there may be HSEs in many genes other than the HSP genes. Although there are variations in these HSEs, the spacing and position of the guanines are invariable7. Therefore, both the nucleotides and the exact spacing of the repeated units are considered as key determinants for recognition by HSFs and transcriptional activation. Part b of the figure is modified, with permission, from REF. 87 © (2004) The American Society for Cell Biology.
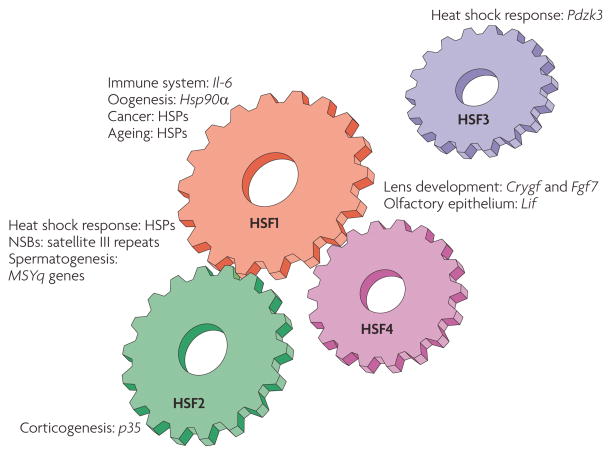
Figure 1. The mammalian HSF machinery.
An overview of the mammalian heat shock factor (HSF) family members and their biological functions. HSFs contribute to multiple normal physiological processes and pathologies through direct regulation of their target genes. The HSF target genes that have been identified in vivo are shown. HSF1 was originally recognized as the principal stress-responsive regulator of the heat shock response, but now HSF2 is known to modulate HSF1-mediated expression of heat shock protein (HSP) genes through heterocomplex formation. On heat shock, HSF1 and HSF2 accumulate into nuclear stress bodies (NSBs), where they bind to satellite III repeats. HSF1 is also a regulator of immune responses and cancer. So far, the regulation of HSP genes in ageing has most intensively been examined in Caenorhabditis elegans. Both HSF1 and HSF2 have been ascribed regulatory functions in several developmental processes, such as oogenesis, spermatogenesis and corticogenesis. HSF4 is involved in the development of different sensory organs in cooperation with HSF1, but has no role in the heat shock response. Murine HSF3 is the most recently identified mammalian HSF, which participates in the heat shock response by binding to the PDZ domain-containing 3 (Pdzk3) promoter10. Currently, HSF3 is not known to crosstalk with any member of the HSF family, and is therefore placed separately from the other HSFs. Crygf, crystallin γF; Fgf7, fibroblast growth factor 7; Il-6, interleukin-6; MSYq, male-specific long arm of the mouse Y chromosome.
HSFs as stress integrators
A hallmark of stressed cells and organisms is the increased synthesis of HSPs, which function as molecular chaperones to prevent protein misfolding and aggregation to maintain protein homeostasis, also called proteostasis11. The transcriptional activation of HSP genes is mediated by HSFs (FIG. 2a), of which HSF1 is the master regulator in vertebrates. Hsf1-knockout mouse and cell models have revealed that HSF1 is a prerequisite for the transactivation of HSP genes, maintenance of cellular integrity during stress and development of thermotolerance12–15. HSF1 is constitutively expressed in most tissues and cell types16, where it is kept inactive in the absence of stress stimuli. Thus, the DNA-binding and transactivation capacity of HSF1 are coordinately regulated through multiple PTMs, protein–protein interactions and subcellular localization. HSF1 also has an intrinsic stress-sensing capacity, as both D. melanogaster and mammalian HSF1 can be converted from a monomer to a homotrimer in vitro in response to thermal or oxidative stress17–19.
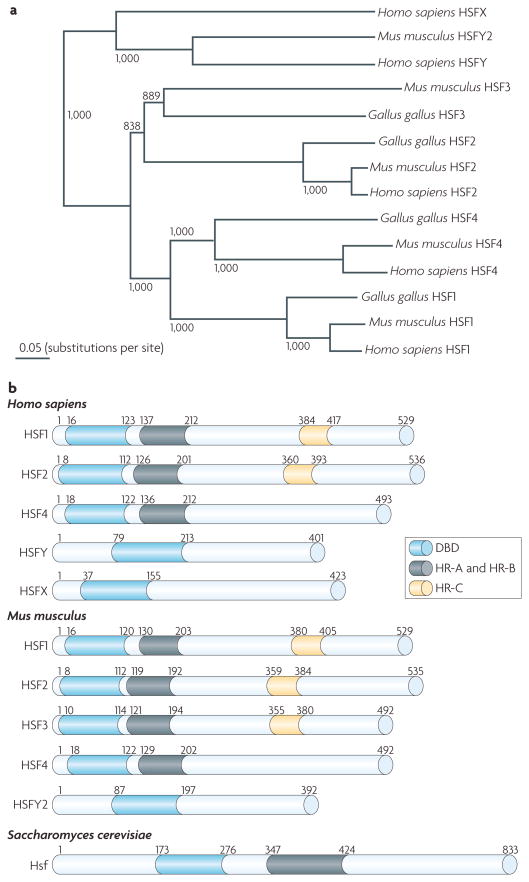
Figure 2. Members of the mammalian HSF family.
a | A phylogenetic tree showing the species-specific relationship of heat shock factors (HSFs) among higher eukaryotes. Two recently found, but still poorly characterized, family members are: HSFY, which is located on the human Y chromosome and on the murine chromosome 2 (HSFY2), and HSFX, which has only been found on the human X chromosome144–146. HSFY and HSFX exist in two identical copies on their respective chromosome. The phylogenetic tree was generated in CLUSTAL W147 and gaps were excluded from all phylogenetic analyses. The numbers represent bootstrap values (1000 bootstrap replicates were carried out). b | A schematic of the functional domains of the human and murine HSF family members. The conserved domains of distinct HSFs are indicated: the DNA-binding domain (DBD), the oligomerization domain (heptad repeat A (HR-A) and HR-B) and the carboxy-terminal HR-C. All HSFs contain the characteristic helix-loop-helix DBD. HSF1–HSF4 contain Leu zipper-like HR domains, which are required for homotrimerization or heterotrimerization. Yeast Hsf is included as a comparison. Image in part a is modified, with permission, from REF. 10 © (2010) The American Society for Cell Biology.
Functional domains
HSFs, like other transcription factors, are composed of functional domains. These have been most thoroughly characterized for HSF1 and are schematically presented in FIG. 2b. The DNA-binding domain (DBD) is the best preserved domain in evolution and belongs to the family of winged helix-turn-helix DBDs20–22. The DBD forms a compact globular structure, except for a flexible wing or loop that is located between β-strands 3 and 4 (REF. 6). This loop generates a protein– protein interface between adjacent subunits of the HSF trimer that enhances high-affinity binding to DNA by cooperativity between different HSFs23. The DBD can also mediate interactions with other factors to modulate the transactivating capacity of HSFs24. Consequently, the DBD is considered as the signature domain of HSFs for target-gene recognition.
The trimerization of HSFs is mediated by arrays of hydrophobic heptad repeats (HR-A and HR-B) that form a coiled coil, which is characteristic for many Leu zippers6,25 (FIG. 2b). The trimeric assembly is unusual, as Leu zippers typically facilitate the formation of homodimers or heterodimers. Suppression of spontaneous HSF trimerization is mediated by yet another hydrophobic repeat, HR-C26–28. Human HSF4 lacks the HR-C, which could explain its constitutive trimerization and DNA-binding activity29. Positioned at the extreme carboxyl terminus of HSFs is the transactivation domain, which is shared among all HSFs6 except for yeast Hsf, which has transactivation domains in both the amino and C termini, and HSF4A, which completely lacks a transactivation domain29–31. In HSF1, the transactivation domain is composed of two modules — AD1 and AD2, which are rich in hydrophobic and acidic residues (FIG. 3a) — that together ensures a rapid and prolonged response to stress32,33. The transactivation domain was originally proposed to provide stress inducibility to HSF1 (REFS 34,35), but it soon became evident that an intact regulatory domain, located between the HR-A and HR-B and the transactivation domain, is essential for the responsiveness to stress stimuli32,33,36,37. Because several amino acids that are known targets for different PTMs reside in the regulatory domain33,38–42, the structure and function of this domain are under intensive investigation.
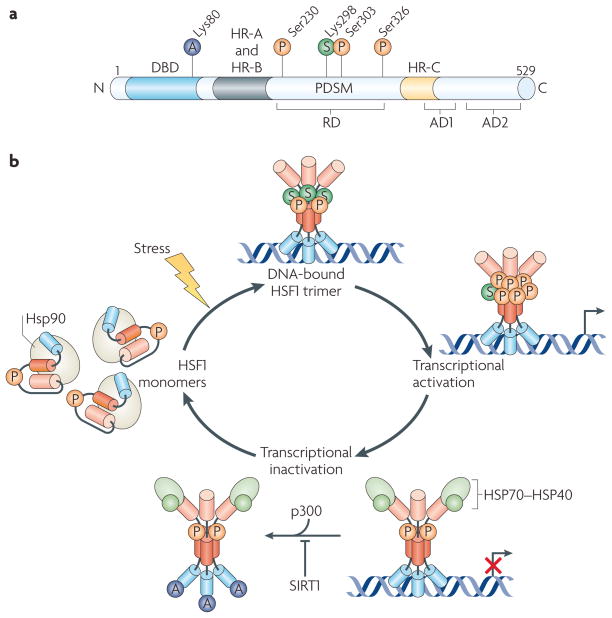
Figure 3. HSF1 undergoes multiple PTMs on activation.
a | An overview of heat shock factor 1 (HSF1)-related post-translational modifications (PTMs). Some of the identified sites for acetylation (A), phosphorylation (P) and sumoylation (S) are indicated, as well as the phosphorylation-dependent sumoylation motif (PDSM). The DNA-binding domain (DBD) and the heptad repeats (HR-A and HR-B, and HR-C) are indicated as in FIG. 2, as well as the regulatory domain (RD) and activation domains (AD1 and AD2). b | The HSF1 activation and attenuation cycle, involving trimerization, multiple PTMs and feedback from heat shock proteins (HSPs). In the resting state, HSF1 is a monomer in both the cytoplasm and nucleus. Monomeric HSF1 is already a phosphoprotein under non-stress conditions and it interacts with HSP90. On stress, HSF1 dissociates from the HSP90 complex, allowing HSF1 to trimerize and bind to the heat shock elements (HSEs) in HSP genes. Several PTMs, such as phosphorylation and sumoylation, are involved in regulating the transactivation capacity of HSF1. HSF1 acquires transcriptional activity, which is abrogated during the attenuation phase. Attenuation involves two regulatory steps: negative feedback from HSPs, which represses the transactivation of DNA-bound HSF1, and inhibition of DNA binding by the acetylation of Lys80 in the DBD of HSF1. The sirtuin SIRT1 regulates the attenuation phase of the heat shock response by preventing HSF1 acetylation42.
Regulation of the HSF1 activation–attenuation cycle
The conversion of the inactive monomeric HSF1 to high-affinity DNA-binding trimers is the initial step in the multistep activation process and is a common feature of all eukaryotic HSFs43,44 (FIG. 3b). There is compelling evidence for HSF1 interacting with multiple HSPs at different phases of its activation cycle. For example, monomeric HSF1 interacts weakly with HSP90 and, on stress, HSF1 dissociates from the complex, allowing HSF1 trimerization45,46 (FIG. 3b). Trimeric HSF1 can be kept inactive when its regulatory domain is bound by a multi-chaperone complex of HSP90, co-chaperone p23 (also known as PTGES3) and immunophilin FK506-binding protein 5 (FKBP52; also known as FKBP4)46–51. Elevated levels of both HSP90 and HSP70 negatively regulate HSF1 and prevent trimer formation on heat shock52. Activated HSF1 trimers also interact with HSP70 and the co-chaperone HSP40 (also known as DNAJB1), but instead of suppressing the DNA-binding activity of HSF1, this interaction inhibits its transactivation capacity52–54. Although the inhibitory mechanism is still unknown, the negative feedback from the end products of HSF1-dependent transcription (the HSPs) provides an important control step in adjusting the duration and intensity of HSF1 activation according to the levels of chaperones and presumably the levels of nascent and misfolded peptides.
A ribonucleoprotein complex containing eukaryotic elongation factor 1A (eEF1A) and a non-coding RNA, heat shock RNA-1 (HSR-1), has been reported to possess a thermosensing capacity. According to the proposed model, HSR-1 undergoes a conformational change in response to heat stress and together with eEF1A facilitates trimerization of HSF1 (REF. 55). How this activation mode relates to the other regulatory mechanisms associated with HSFs remains to be elucidated.
Throughout the activation–attenuation cycle, HSF1 undergoes extensive PTMs, including acetylation, phosphorylation and sumoylation (FIG. 3). HSF1 is also a phosphoprotein under non-stress conditions, and the results from mass spectrometry (MS) analyses combined with phosphopeptide mapping experiments indicate that at least 12 Ser residues are phosphorylated41,56–59. Among these sites, stress-inducible phosphorylation of Ser230 and Ser326 in the regulatory domain contributes to the transactivation function of HSF1 (REFS 38,41). Phosphorylation-mediated sumoylation on a single Lys residue in the regulatory domain occurs rapidly and transiently on exposure to heat shock; Ser303 needs to be phosphorylated before a small ubiquitin-related modifier (SUMO) can be conjugated to Lys298 (REF. 39). The extended consensus sequence ΨKxExxSP has been named the phosphorylation-dependent sumoylation motif (PDSM; FIG. 3)40. The PDSM was initially discovered in HSF1 and subsequently found in many other proteins, especially transcriptional regulators such as HSF4, GATA1, myocyte-specific enhancer factor 2A (MEF2A) and SP3, which are substrates for both SUMO conjugation and Pro-directed kinases40,60–62.
Recently, Mohideen and colleagues showed that a conserved basic patch on the surface of the SUMO-conjugating enzyme ubiquitin carrier protein 9 (UBC9; also known as UBE2I) discriminates between the phosphorylated and non-phosphorylated PDSM of HSF1 (REF. 63). Future studies will be directed at elucidating the molecular mechanisms for dynamic phosphorylation and UBC9-dependent SUMO conjugation in response to stress stimuli and establishing the roles of kinases, phosphatases and desumoylating enzymes in the heat shock response. The kinetics of phosphorylation-dependent sumoylation of HSF1 correlates inversely with the severity of heat stress, and, as the transactivation capacity of HSF1 is impaired by sumoylation and this PTM is removed when maximal HSF1 activity is required40, sumoylation could modulate HSF1 activity under moderate stress conditions. The mechanisms by which SUMO modification represses the transactivating capacity of HSF1, and the functional relationship of this PTM with other modifications that HSF1 is subjected to, will be investigated with endogenous substrate proteins.
Phosphorylation and sumoylation of HSF1 occur rapidly on heat shock, whereas the kinetics of acetylation are delayed and coincide with the attenuation phase of the HSF1 activation cycle. Stress-inducible acetylation of HSF1 is regulated by the balance of acetylation by p300–CBP (CREB-binding protein) and deacetylation by the NAD+-dependent sirtuin, SIRT1. Increased expression and activity of SIRT1 enhances and prolongs the DNA-binding activity of HSF1 at the human HSP70.1 promoter, whereas downregulation of SIRT1 enhances the acetylation of HSF1 and the attenuation of DNA-binding without affecting the formation of HSF1 trimers42. This finding led to the discovery of a novel regulatory mechanism of HSF1 activity, whereby SIRT1 maintains HSF1 in a state that is competent for DNA binding by counteracting acetylation (FIG. 3). In the light of current knowledge, the attenuation phase of the HSF1 cycle is regulated by a dual mechanism: a dependency on the levels of HSPs that feed back directly by weak interactions with HSF1, and a parallel step that involves the SIRT1-dependent control of the DNA-binding activity of HSF1. Because SIRT1 has been implicated in caloric restriction and ageing, the age-dependent loss of SIRT1 and impaired HSF1 activity correlate with an impairment of the heat shock response and proteostasis in senescent cells, connecting the heat shock response to nutrition and ageing (see below).
HSF dynamics on the HSP70 promoter
For decades, the binding of HSF to the HSP70.1 gene has served as a model system for inducible transcription in eukaryotes. In D. melanogaster, HSF is constitutively nuclear and low levels of HSF are associated with the HSP70 promoter before heat shock64–66. The uninduced HSP70 promoter is primed for transcription by a transcriptionally engaged paused RNA polymerase II (RNAP II)67,68. RNAP II pausing is greatly enhanced by nucleosome formation in vitro, implying that chromatin remodelling is crucial for the release of paused RNAP II69. It has been proposed that distinct hydrophobic residues in the transactivation domain of human HSF1 can stimulate RNAP II release and directly interact with BRG1, the ATPase subunit of the chromatin remodelling complex SWI/SNF70,71. Upon heat shock, RNAP II is released from its paused state, leading to the synthesis of a full-length transcript. Rapid disruption of nucleosomes occurs across the entire HSP70 gene, at a rate that is faster than RNAP II-mediated transcription72. The nucleosome displacement occurs simultaneously with HSF recruitment to the promoter in D. melanogaster. Downregulation of HSF abrogates the loss of nucleosomes, indicating that HSF provides a signal for chromatin rearrangement, which is required for HSP70 nucleosome displacement. Within seconds of heat shock, the amount of HSF at the promoter increases drastically and HSF translocates from the nucleoplasm to several native loci, including HSP genes. Interestingly, the levels of HSF occupying the HSP70 promoter reach saturation soon after just one minute65,73.
HSF recruits the co-activating mediator complex to the heat shock loci, which acts as a bridge to transmit activating signals from transcription factors to the basal transcription machinery. The mediator complex is recruited by a direct interaction with HSF: the transactivation domain of D. melanogaster HSF binds to TRAP80 (also known as MED17), a subunit of the mediator complex74. HSF probably has other macromolecular contacts with the preinitiation complex as it binds to TATA-binding protein (TBP) and the general transcription factor TFIIB in vitro75,76. In contrast to the rapid recruitment and elongation of RNAP II on heat shock, activated HSF exchanges very slowly at the HSP70 promoter. HSF stays stably bound to DNA in vivo and no turnover or disassembly of transcription activator is required for successive rounds of HSP70 transcription65,68.
Functional interplay between HSFs
Although HSF1 is the principal regulator of the heat shock response, HSF2 also binds to the promoters of HSP genes. In light of our current knowledge, HSF2 strictly depends on HSF1 for its stress-related functions as it is recruited to HSP gene promoters only in the presence of HSF1 and this cooperation requires an intact HSF1 DBD77. Nevertheless, HSF2 modulates, both positively and negatively, the HSF1-mediated inducible expression of HSP genes, indicating that HSF2 can actively participate in the transcriptional regulation of the heat shock response. Coincident with the stress-induced transcription of HSP genes, HSF1 and HSF2 colocalize and accumulate rapidly on stress into nuclear stress bodies (NSBs; BOX 2), where they bind to a subclass of satellite III repeats, predominantly in the human chromosome 9q12 (REFS 78-80). Consequently, large and stable non-coding satellite III transcripts are synthesized in an HSF1-dependent manner in NSBs81,82. The function of these transcripts and their relationship with other HSF1 targets, and the heat shock response in general, remain to be elucidated.
Box 2. Nuclear stress bodies.
The cell nucleus is highly compartmentalized and dynamic. Many nuclear factors are diffusely distributed throughout the nucleoplasm, but they can also accumulate in distinct subnuclear compartments, such as nucleoli, speckles, Cajal bodies and promyelocytic leukaemia (PML) bodies136. Nuclear stress bodies (NSBs) are different from any other known nuclear bodies137,138. Although NSBs were initially thought to contain aggregates of denatured proteins and be markers of heat-shocked cells, their formation can be elicited by various stresses, such as heavy metals and proteasome inhibitors137. NSBs are large structures, 0.3–3 μm in diameter, and are usually located close to the nucleoli or nuclear envelope137,138. NSBs consist of two populations: small, brightly stained bodies and large, clustered and ring-like structures137.
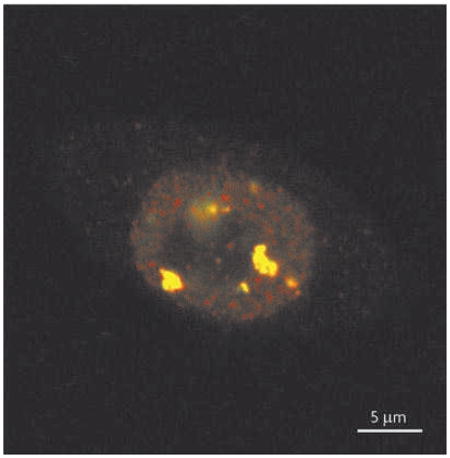
NSBs appear transiently and are the main site of heat shock factor 1 (HSF1) and HSF2 accumulation in stressed human cells80. HSF1 and HSF2 form a physically interacting complex and colocalize into small and barely detectable NSBs after only five minutes of heat shock, but the intensity and size of NSBs increase after hours of continuous heat shock. HSF1 and HSF2 colocalize in HeLa cells that have been exposed to heat shock for one hour at 42°C (see the figure; confocal microscopy image with HSF1–green fluorescent protein in green and endogenous HSF2 in red). NSBs form on specific chromosomal loci, mainly on q12 of human chromosome 9, where HSFs bind to a subclass of satellite III repeats78,79,83. Stress-inducible and HSF1-dependent transcription of satellite III repeats has been shown to produce non-coding RNA molecules, called satellite III transcripts81,82. The 9q12 locus consists of pericentromeric heterochromatin, and the satellite III repeats provide scaffolds for docking components, such as splicing factors and other RNA-processing proteins139–143.
HSF2 also modulates the heat shock response through the formation of heterotrimers with HSF1 in the NSBs when bound to the satellite III repeats83 (FIG. 4). Studies on the functional significance of heterotrimerization indicate that HSF1 depletion prevents localization of HSF2 to NSBs and abolishes the stress-induced synthesis of satellite III transcripts. By contrast, increased expression of HSF2 leads to its own activation and the localization of both HSF1 and HSF2 to NSBs, where transcription is spontaneously induced in the absence of stress stimuli. These results suggest that HSF2 can incorporate HSF1 into a transcriptionally competent heterotrimer83. It is possible that the amounts of HSF2 available for heterotrimerization with HSF1 influence stress-inducible transcription, and that HSF1–HSF2 heterotrimers regulate transcription in a temporal manner. During the acute phase of heat shock, HSF1 is activated and HSF1–HSF2 heterotrimers are formed, whereas upon prolonged exposures to heat stress the levels of HSF2 are diminished, thereby limiting heterotrimerization83. Intriguingly, in specific developmental processes such as corticogenesis and spermatogenesis, the expression of HSF2 increases spatiotemporarily, leading to its spontaneous activation. Therefore, it has been proposed that HSF-mediated transactivation can be modulated by the levels of HSF2 to provide a switch that integrates the responses to stress and developmental stimuli83 (FIG. 4). Functional relationships between different HSFs are emerging, and the synergy of DNA-binding activities among HSF family members offers an efficient way to control gene expression in a cell- and stimulus-specific manner to orchestrate the differential upstream signalling and target-gene networks.
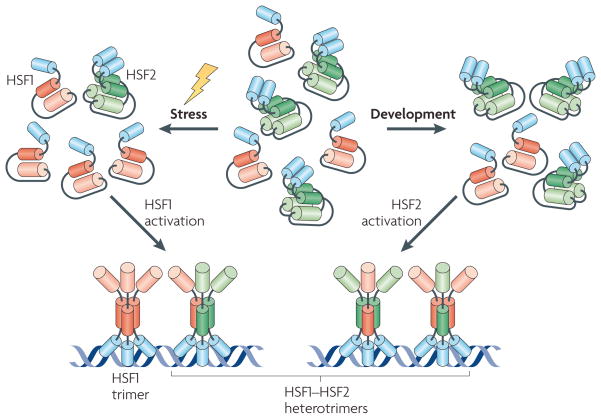
Figure 4. Interactions between different HSFs provide distinct functional modes in transcriptional regulation.
On stress, heat shock factor 1 (HSF1) is activated and HSF1–HSF2 heterotrimers are formed. Heat shock stress diminishes the levels of HSF2 and restricts heterotrimerization by limiting the availability of HSF2. Biochemical characterization of HSF2 has revealed that, unlike HSF1, which undergoes a monomer-to-trimer transition, HSF2 is mainly converted from a dimer to a trimer on activation148. In certain developmental processes, such as corticogenesis and spermatogenesis, HSF2 levels are elevated in specific cell types and tissues, leading to activation of HSF2. Increased HSF2 expression then induces the formation of heterotrimers with HSF1. It has therefore been suggested that HSF1–HSF2 heterotrimerization provides a switch that integrates the transcriptional activation in response to specific stimuli83.
A new member of the mammalian HSF family, mouse HSF3, was recently identified10. Avian HSF3 was shown to be activated at higher temperatures and with different kinetics than HSF1 (REF. 84), whereas in mice, heat shock induces the nuclear translocation of HSF3 and activation of stress-responsive genes other than HSP genes10. Future experiments will determine whether HSF3 is capable of interacting with other HSFs, potentially through heterocomplex formation. HSF4 has not been implicated in the heat shock response, but it competes with HSF1 for common target genes in mouse lens epithelial cells85, which will be discussed below. It is important to elucidate whether the formation of homotrimers or hetero trimers between different family members is a common theme in HSF-mediated transcriptional regulation.
HSFs as developmental regulators
Evidence is accumulating that HSFs are highly versatile transcription factors that, in addition to protecting cells against proteotoxic stress, are vital for many physioogical functions, especially during development. The initial observations using deletion experiments of the D. melanogaster Hsf gene revealed defective oogenesis and larvae development86. These effects were not caused by obvious changes in HSP gene expression patterns, which is consistent with the subsequent studies showing that basal expression of HSP genes during mouse embryogenesis is not affected by the lack of HSF1 (REF. 13). These results are further supported by genome-wide gene expression studies revealing that numerous genes, not classified as HSP genes or molecular chaperones, are under HSF1-dependent control87,88.
Although mice lacking HSF1 can survive to adulthood, they exhibit multiple defects, such as increased prenatal lethality, growth retardation and female infertility13. Fertilized oocytes do not develop past the zygotic stage when HSF1-deficient female mice are mated with wild-type male mice, indicating that HSF1 is a maternal factor that is essential for early post-fertilization development89. Recently, it was shown that HSF1 is abundantly expressed in maturing oocytes, where it regulates specifically Hsp90α transcription90. The HSF1-deficient oocytes are devoid of HSP90α and exhibit a blockage of meiotic maturation, including delayed G2–M transition or germinal vesicle breakdown and defective asymmetrical division90. Moreover, intra-ovarian HSF1-depleted oocytes contain dysfunctional mitochondria and are sensitive to oxidative stress, leading to reduced survival91. The complex phenotype of Hsf1-knockout mice also demonstrates the involvement of HSF1 in placenta formation, placode development and the immune system15,85,92,93, further strengthening the evidence for a protective function of HSF1 in development and survival.
Both HSF1 and HSF2 are key regulators in the developing brain and in maintaining proteostasis in the central nervous system. Disruption of Hsf1 results in enlarged ventricles, accompanied by astrogliosis, neurodegeneration, progressive myelin loss and accumulation of ubiquitylated proteins in specific regions of the postnatal brain under non-stressed conditions94,95. The expression of HSP25 (also known as HSPB1) and α-crystallin B chain (CRYAB), which are known to protect cells against stress-induced protein damage and cell death, is dramatically decreased in brains lacking HSF1 (REF. 13). In contrast to HSF1, HSF2 is already at peak levels during early brain development in mice and is predominantly expressed in the proliferative neuronal progenitors of the ventricular zone and post-mitotic neurons of the cortical plate96–99. HSF2-deficient mice have enlarged ventricles and defects in cortical lamination owing to abnormal neuronal migration97–99. Incorrect positioning of superficial neurons during cortex formation in HSF2-deficient embryos is caused by decreased expression of the cyclin-dependent kinase 5 (CDK5) activator p35, which is a crucial regulator of the cortical migration signalling pathway100,101. The p35 gene was identified as the first direct target of HSF2 in cortex development99. As correct cortical migration requires the coordination of multiple signalling molecules, it is likely that HSF2, either directly or indirectly, also regulates other components of the same pathway.
Cooperativity of HSFs in development
In adult mice, HSF2 is most abundantly expressed in certain cell types of testes, specifically pachytene spermatocytes and round spermatids102. The cell-specific expression of HSF2 in testes is regulated by a microRNA, miR-18, that directly binds to the 3′ untranslated region (UTR) of HSF2 (J.K. Björk, A. Sandqvist, A.N. Elsing, N. Kotaja and L.S., unpublished observations). Targeting of HSF2 in spermatogenesis reveals the first physiological role for miR-18, which belongs to the oncomir-1 cluster associated mainly with tumour progression103. In accordance with the expression pattern during the maturation of male germ cells, HSF2-null male mice display several abnormal features in spermatogenesis, ranging from smaller testis size and increased apoptosis at the pachytene stage to a reduced amount of sperm and abnormal sperm head shape97,98,104. A genome-wide search for HSF2 target promoters in mouse testis revealed the occupancy of HSF2 on the sex chromosomal multi-copy genes spermiogenesis specific transcript on the Y 2 (Ssty2), Sycp3-like Y-linked (Sly) and Sycp3-like X-linked (Slx), which are important for sperm quality104. Compared with the Hsf2-knockout phenotype, disruption of both Hsf1 and Hsf2 results in a more pronounced phenotype, including larger vacuolar structures, more widely spread apoptosis and a complete lack of mature spermatozoa and male sterility105. The hypo thesis that the activities of HSF1 and HSF2 are intertwined and essential for spermatogenesis is further supported by our results that HSF1 and HSF2 synergistically regulate the sex chromosomal multi-copy genes in post-meiotic round spermatids (M.Å., A. Vihervaara, E.S. Christians, E. Henriksson and L.S., unpublished observations). Given that the sex chromatin mostly remains silent after meiosis, HSF1 and HSF2 are currently the only known transcriptional regulators during post-meiotic repression. These results, together with the earlier findings that HSF2 can also form heterotrimers with HSF1 in testes83, strongly suggest that HSF1 and HSF2 act in a heterocomplex and fine-tune transcription of their common target genes during the maturation of male germ cells.
HSF1 and HSF4 are required for the maintenance of sensory organs, especially when the organs are exposed to environmental stimuli for the first time after birth85,88. During the early postnatal period, Hsf1-knockout mice display severe atrophy of the olfactory epithelium, increased accumulation of mucus and death of olfactory sensory neurons88. Although lens development in HSF4-deficient mouse embryos is normal, severe abnormalities, including inclusion-like structures in lens fibre cells, appear soon after birth and the mice develop cataracts85,106,107. Intriguingly, inherited severe cataracts occurring in Chinese and Danish families have been associated with a mutation in the DBD of HSF4 (REF. 108). In addition to the established target genes, Hsp25, Hsp70 and Hsp90, several new targets for HSF1 and HSF4, such as crystallin γF (Crygf), fibroblast growth factor 7 (Fgf7) and leukaemia inhibitory factor (Lif) have been found to be crucial for sensory organs85,88. Furthermore, binding of either HSF1 or HSF4 to the Fgf7 promoter shows opposite effects on gene expression, suggesting competitive functions between the two family members85. In addition to the proximal promoters, HSF1, HSF2 and HSF4 bind to other genomic regions (that is, introns and distal parts of protein-coding genes in mouse lens), and there is also evidence for either synergistic interplay or competition between distinct HSFs occupying the target-gene promoters109. It is possible that the different HSFs are able to compensate for each other to some extent. Thus, the identification of novel functions and target genes for HSFs has been a considerable step forward in understanding their regulatory mechanisms in development.
HSFs and lifespan
The lifespan of an organism is directly linked to the health of its tissues, which is a consequence of the stability of the proteome and functionality of its molecular machineries. During its lifetime, an organism constantly encounters environmental and physiological stress and requires an efficient surveillance of protein quality to prevent the accumulation of protein damage and the disruption of proteostasis. Proteotoxic insults contribute to cellular ageing, and numerous pathophysiological conditions, associated with impaired protein quality control, increase prominently with age11. From studies on the molecular basis of ageing, in which a wide range of different model systems and experimental strategies have been used, the insulin and insulin-like growth factor 1 receptor (IGF1R) signalling pathway, which involves the phosphoinositide 3-kinase (PI3K) and AKT kinases and the Forkhead box protein O (FOXO) transcription factors (such as DAF-16 in Caenorhabditis elegans), has emerged as a key process. The downregulation of HSF reduces the lifespan and accelerates the formation of protein aggregates in C. elegans carrying mutations in different components of the IGF1R-mediated pathway. Conversely, inhibition of IGF1R signalling results in HSF activation and promotes longevity by maintaining proteostasis110,111. These results have prompted many laboratories that use other model organisms to investigate the functional relationship between HSFs and the IGF1R signalling pathway.
The impact of HSFs on the lifespan of whole organisms is further emphasized by a recent study, in which proteome stability was examined during C. elegans ageing112. The age-dependent misfolding and downregulation of distinct metastable proteins, which display temperature-sensitive missense mutations, was examined in different tissues. Widespread failure in proteostasis occurred rapidly at an early stage of adulthood, coinciding with the severely impaired heat shock response and unfolded protein response112. The age-dependent collapse of proteostasis could be restored by overexpression of HSF and DAF-16, strengthening the evidence for the unique roles of these stress-responsive transcription factors to prevent global instability of the proteome.
Limited food intake or caloric restriction is another process that is associated with an enhancement of lifespan. In addition to promoting longevity, caloric restriction slows down the progression of age-related diseases such as cancer, cardiovascular diseases and metabolic disorders, stimulates metabolic and motor activities, and increases resistance to environmental stress stimuli113. To this end, the dynamic regulation of HSF1 by the NAD+-dependent protein deacetylase SIRT1, a mammalian orthologue of the yeast transcriptional regulator Sir2, which is activated by caloric restriction and stress, is of particular interest. Indeed, SIRT1 directly deacetylates HSF1 and keeps it in a state that is competent for DNA binding. During ageing, the DNA-binding activity of HSF1 and the amount of SIRT1 are reduced. Consequently, a decrease in SIRT1 levels was shown to inhibit HSF1 DNA-binding activity in a cell-based model of ageing and senescence42. Furthermore, an age-related decrease in the HSF1 DNA-binding activity is reversed in cells exposed to caloric restriction114. These results indicate that HSF1 and SIRT1 function together to protect cells from stress insults, thereby promoting survival and extending lifespan. Impaired proteostasis during ageing may at least partly reflect the compromised HSF1 activity due to lowered SIRT1 expression.
Impact of HSFs in disease
The heat shock response is thought to be initiated by the presence of misfolded and damaged proteins, and is thus a cell-autonomous response. When exposed to heat, cells in culture, unicellular organisms, and cells in a multicellular organism can all trigger a heat shock response autonomously115–117. However, it has been proposed that multicellular organisms sense stress differently to isolated cells. For example, the stress response is not properly induced even if damaged proteins are accumulated in neurodegenerative diseases like Huntington’s disease and Parkinson’s disease, suggesting that there is an additional control of the heat shock response at the organismal level118. Uncoordinated activation of the heat shock response in cells in a multicellular organism could cause severe disturbances of interactions between cells and tissues. In C. elegans, a pair of thermosensory neurons called AFDs, which sense and respond to temperature, regulate the heat shock response in somatic tissues by controlling HSF activity119,120. Moreover, the heat shock response in C. elegans is influenced by the metabolic state of the organism and is reduced under conditions that are unfavourable for growth and reproduction121. Neuronal control may therefore allow organisms to coordinate the stress response of individual cells with the varying metabolic requirements in different tissues and developmental stages. These observations are probably relevant to diseases of protein misfolding that are highly tissue-specific despite the often ubiquitous expression of damaged proteins and the heat shock response.
Elevated levels of HSF1 have been detected in several types of human cancer, such as breast cancer and prostate cancer122,123. Mice deficient in HSF1 exhibit a lower incidence of tumours and increased survival than their wild-type counterparts in a classical chemical skin carcinogenesis model and in a genetic model expressing an oncogenic mutation of p53. Similar results have been obtained in human cancer cells lines, in which HSF1 was depleted using an RNA interference strategy124. HSF1 expression is likely to be crucial for non-oncogene addiction and the stress phenotype of cancer cells, which are attributes given to many cancer cells owing to their high intrinsic level of proteotoxic and oxidative stress, frequent spontaneous DNA damage and aneuploidy125. Each of these features may disrupt proteostasis, raising the need for efficient chaperone and proteasome activities. Accordingly, HSF1 would be essential for the survival of cancer cells that experience constant stress and develop non-oncogene addiction.
HSFs as therapeutic targets
Given the unique role of HSF1 in stress biology and proteostasis, enhanced activity of this principal regulator during development and early adulthood is important for the stability of the proteome and the health of the cell. However, HSF1 is a potent modifier of tumorigenesis and, therefore, a potential target for cancer therapeutics125. In addition to modulating the expression of HSF1, the various PTMs of HSF1 that regulate its activity should be considered from a clinical perspective. As many human, age-related pathologies are associated with stress and misfolded proteins, several HSF-based therapeutic strategies have been proposed126. In many academic and industrial laboratories, small molecule regulators of HSF1 are actively being searched for (see Supplementary information S1 (table)). For example, celastrol, which has antioxidant properties and is a natural compound derived from the Celastreace family of plants, activates HSF1 and induces HSP expression with similar kinetics to heat shock, and could therefore be a potential candidate molecule for treating neurodegenerative diseases127,128. In a yeast-based screen, a small-molecule activator of human HSF1 was found and named HSF1A129. HSF1A, which is structurally distinct from the other known activators, activates HSF1 and enhances chaperone expression, thereby counteracting protein misfolding and cell death in polyQ-expressing neuronal precursor cells129. Triptolide, also from the Celastreace family of plants, is a potent inhibitor of the transactivating capacity of HSF1 and has been shown to have beneficial effects in treatments of pancreatic cancer xenografts130,131. These examples of small-molecule regulators of HSF1 are promising candidates for drug discovery and development. However, the existence of multiple mammalian HSFs and their functional interplay should also be taken into consideration when planning future HSF-targeted therapies.
Concluding remarks and future perspectives
HSFs were originally identified as specific heat shock-inducible transcriptional regulators of HSP genes, but now there is unambiguous evidence for a wide variety of HSF target genes that extends beyond the molecular chaperones. The known functions governed by HSFs span from the heat shock response to development, metabolism, lifespan and disease, thereby integrating pathways that were earlier strictly divided into either cellular stress responses or normal physiology.
Although the extensive efforts from many laboratories focusing on HSF biology have provided a richness of understanding of the complex regulatory mechanisms of the HSF family of transcription factors, several key questions remain. For example, what are the initial molecular events (that is, what is the ‘thermometer’) leading to the multistep activation of HSFs? The chromatin-based interaction between HSFs and the basic transcription machinery needs further investigation before the exact interaction partners at the chromatin level can be established. The activation and attenuation mechanisms of HSFs require additional mechanistic insights, and the roles of the multiple signal transduction pathways involved in post-translational regulation of HSFs are only now being discovered and are clearly more complex than anticipated. Although still lacking sufficient evidence, the PTMs probably serve as rheostats to allow distinct forms of HSF-mediated regulation in different tissues during development. Further emphasis should therefore be placed on understanding the PTMs of HSFs during development, ageing and different protein folding diseases. Likewise, the subcellular distribution of HSF molecules, including the mechanism by which HSFs shuttle between the cytoplasm and the nucleus, remains enigmatic, as do the movements of HSF molecules in different nuclear compartments such as NSBs.
Most studies on the impact of HSFs in lifespan and disease have been conducted with model organisms such as D. melanogaster and C. elegans, which express a single HSF. The existence of multiple members of the HSF family in mammals warrants further investigation of their specific and overlapping functions, including their extended repertoire of target genes. The existence of multiple HSFs in higher eukaryotes with different expression patterns suggests that they may have functions that are triggered by distinct stimuli, leading to activation of specific target genes. The impact of the HSF family in the adaptation to diverse biological environments is still poorly understood, and future studies are likely to broaden the prevailing view of HSFs being solely stress-inducible factors. To this end, the crosstalk between distinct HSFs that has only recently been uncovered raises obvious questions about the stoichiometry between the components in different complexes residing in different cellular compartments, and the mechanisms by which the factors interact with each other. Interaction between distinct HSF family members could generate new opportunities in designing therapeutics for protein-folding diseases, metabolic disorders and cancer.
Acknowledgments
We apologize to our colleagues whose original work could only be cited indirectly owing to space limitations. Members of our laboratories are acknowledged for valuable comments on the manuscript. Our own work is supported by The Academy of Finland, The Sigrid Jusélius Foundation, The Finnish Cancer Organizations and Åbo Akademi University. The image in box 2 is courtesy of A. Sandqvist, Department of Biosciences, Åbo Akademi University, Turku, Finland.
Glossary
- Polytene chromosome
A chromosome that undergoes multiple rounds of DNA replication, without cell division, and produces many sister chromatids that remain synapsed together;for example in larval salivary glands of D. melanogaster
- Proteostasis
Also called protein homeostasis, this refers to the control of the concentration, three-dimensional structure, binding interactions and cellular location of individual proteins making up the proteome
- S1 mapping
A method for mapping precursor or mature mRNAs that correspond to particular DNA sequences using the S1 nuclease enzyme
- Coiled coil
A structural motif in proteins, in which α-helices are coiled together like the strands of a rope, most commonly as dimers and trimers
- Leu zipper
A common three-dimensional structural motif in regulatory proteins that functions as an oligomerization domain and generates adhesion forces in parallel α-helices
- ΨKxExxSP
Many SUMO substrates contain this extended consensus sequence (called the PDSM), in which Ψ denotes a branched hydrophobic amino acid, the Lys is the SUMO acceptor and x is any amino acid
- Sirtuin
A class of proteins that posses either histone deacetylase or monoribosyltransferase activity. SIRT1 is activated by resveratrol, which is a phytoalexin produced naturally by several plants when under attack by pathogens, and has been suggested to trigger mechanisms that counteract ageing-related effects in animals
- Paused RNA polymerase II
An RNA polymerase II molecule that is engaged in transcription but has arrested after synthesizing ~25 nucleotides; for example on the HSP70 promoter under non-heat-shock conditions
- Mediator complex
A multiprotein complex that functions as a transcriptional co-activator and binds to the C-terminal domain of the RNA polymerase II holoenzyme, acting as a bridge between this enzyme and transcription factors
- Preinitiation complex
A large protein complex that is necessary for the transcription of protein-coding genes in eukaryotes and typically consists of six general transcription factors; TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH
- Satellite III repeat
A highly repetitive DNA element that is so called because repetitions of a short DNA sequence tend to produce a different frequency of the nucleotides adenine, cytosine, guanine and thymine compared with bulk DNA
- Germinal vesicle breakdown
The process whereby a greatly enlarged nucleus of an egg in prophase of the first meiotic division breaks down, permitting entry into metaphase I
- Cortical plate
The middle layer of the cerebral cortex. The cerebral cortex is formed during development, and neurons migrate from the ventricular zone to form layers. The cortical plate will form the deep layers of the mature cortex
- Ventricle
A brain structure that contains cerebrospinal fluid. There are four cerebral ventricles: the paired lateral ventricles that are large and C shaped, and the third and fourth ventricles in the midline
- Cortical lamination
The characteristic distribution of neuronal cell types and their connections in the six main layers of the cerebral cortex
- Neuronal migration
The process whereby neurons migrate from their place of origin or birth to their final position in the brain by, for example, radial migration or tangential migration
- Pachytene spermatocyte
A male gametocyte that derives from a spermatogonium, resides in the seminiferous tubules of the testis and is in the developmental stage of spermatogenesis, before which meiosis occurs
- Round spermatid
A haploid male gamete derived from spermatocyte division. As a result of meiosis, each round spermatid contains only half of the genetic material present in the original spermatocyte
- Sex chromosomal multi-copy gene
A large amplified region that comprises palindromic or tandem segmental duplications, contains multi-copy gene families and resides in the X and Y chromosomes
- Sex chromatin
The complex combination of DNA and proteins that makes up chromatin of the X and Y chromosomes during spermatogenesis
- Cataract
Clouding that develops in the crystalline lens of the eye or in its envelope, varying in degree from slight to complete opacity and obstructing the passage of light
- Unfolded protein response
A cellular stress response that is activated by an accumulation of unfolded or misfolded proteins in the lumen of the endoplasmic reticulum
- Non-oncogene addiction
The increased dependence of cancer cells on the normal cellular functions of certain genes, which are not classical cancer genes or oncogenes but, if targeted, they could be equally effective at treating cancer
- Aneuploidy
An abnormal number of chromosomes generated during cell division when the chromosomes do not separate properly between the two daughter cells. This is a characteristic of cancer cells
- PolyQ
A protein sequence that consists of multiple (typically tens to hundreds) Glu repeats. Several inheritable neurodegenerative disorders are polyQ diseases, characterized by a mutation that extends a CAG (which encodes Glu) repeat in a specific gene (for example, huntingtin (HTT) in Huntington’s disease) beyond a certain length
- Rheostat
An adjustment device (a term borrowed from electronics)
Footnotes
Competing interests statement
The authors declare competing financial interests: see web version for details.
References
- 1.Ritossa F. A new puffing pattern induced by temperature shock and DNP in Drosophila. Experimentia. 1962;18:571–573. [Google Scholar]
- 2.Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- 3.Pelham HRB. A regulatory upstream promoter element in the Drosophila hsp70 heat-shock gene. Cell. 1982;30:517–528. doi: 10.1016/0092-8674(82)90249-5. [DOI] [PubMed] [Google Scholar]
- 4.Wu C. Activating protein factor binds in vitro to upstream control sequences in heat shock gene chromatin. Nature. 1984;311:81–84. doi: 10.1038/311081a0. [DOI] [PubMed] [Google Scholar]
- 5.Parker CS, Topol J. A Drosophila RNA polymerase II transcription factor binds to the regulatory site of an hsp70 gene. Cell. 1984;37:273–283. doi: 10.1016/0092-8674(84)90323-4. [DOI] [PubMed] [Google Scholar]
- 6.Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 7.Åkerfelt M, Trouillet D, Mezger V, Sistonen L. Heat shock factors at a crossroad between stress and development. Ann N Y Acad Sci. 2007;1113:15–27. doi: 10.1196/annals.1391.005. [DOI] [PubMed] [Google Scholar]
- 8.Nover L, et al. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones. 2001;6:177–189. doi: 10.1379/1466-1268(2001)006<0177:aathst>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anckar J, Sistonen L. Heat shock factor 1 as a coordinator of stress and developmental pathways. Adv Exp Med Biol. 2007;594:78–88. doi: 10.1007/978-0-387-39975-1_8. [DOI] [PubMed] [Google Scholar]
- 10.Fujimoto M, et al. A novel mouse HSF3 has the potential to activate nonclassical heat-shock genes during heat shock. Mol Biol Cell. 2010;21:106–116. doi: 10.1091/mbc.E09-07-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 12.McMillan DR, Xiao X, Shao L, Graves K, Benjamin IJ. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J Biol Chem. 1998;273:7523–7528. doi: 10.1074/jbc.273.13.7523. [DOI] [PubMed] [Google Scholar]
- 13.Xiao X, et al. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 1999;18:5943–5952. doi: 10.1093/emboj/18.21.5943. Generation of the first HSF-knockout mouse, Hsf1−/−, revealing the physiological function of HSF1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pirkkala L, Alastalo TP, Zuo X, Benjamin IJ, Sistonen L. Disruption of heat shock factor 1 reveals an essential role in the ubiquitin proteolytic pathway. Mol Cell Biol. 2000;20:2670–2675. doi: 10.1128/mcb.20.8.2670-2675.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Huang L, Zhang J, Moskophidis D, Mivechi NF. Targeted disruption of hsf1 leads to lack of thermotolerance and defines tissue-specific regulation for stress-inducible Hsp molecular chaperones. J Cell Biochem. 2002;86:376–393. doi: 10.1002/jcb.10232. [DOI] [PubMed] [Google Scholar]
- 16.Fiorenza MT, Farkas T, Dissing M, Kolding D, Zimarino V. Complex expression of murine heat shock transcription factors. Nucleic Acids Res. 1995;23:467–474. doi: 10.1093/nar/23.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodson ML, Sarge KD. Heat-inducible DNA binding of purified heat shock transcription factor 1. J Biol Chem. 1995;270:2447–2450. doi: 10.1074/jbc.270.6.2447. [DOI] [PubMed] [Google Scholar]
- 18.Larson JS, Schuetz TJ, Kingston RE. In vitro activation of purified human heat shock factor by heat. Biochemistry. 1995;34:1902–1911. doi: 10.1021/bi00006a011. [DOI] [PubMed] [Google Scholar]
- 19.Zhong M, Orosz A, Wu C. Direct sensing of heat and oxidation by Drosophila heat shock transcription factor. Mol Cell. 1998;2:101–108. doi: 10.1016/s1097-2765(00)80118-5. [DOI] [PubMed] [Google Scholar]
- 20.Damberger FF, Pelton JG, Harrison CJ, Nelson HCM, Wemmer DE. Solution structure of the DNA-binding domain of the heat shock transcription factor determined by multidimensional heteronuclear magnetic resonance spectroscopy. Protein Sci. 1994;3:1806–1821. doi: 10.1002/pro.5560031020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison CJ, Bohm AA, Nelson HCM. Crystal structure of the DNA binding domain of the heat shock transcription factor. Science. 1994;263:224–227. doi: 10.1126/science.8284672. [DOI] [PubMed] [Google Scholar]
- 22.Vuister GW, et al. Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor. Nature Struct Biol. 1994;1:605–614. [PubMed] [Google Scholar]
- 23.Littlefield O, Nelson HCM. A new use for the ‘wing’ of the ‘winged’ helix-turn-helix motif in the HSF-DNA cocrystal. Nature Struct Biol. 1999;6:464–470. doi: 10.1038/8269. [DOI] [PubMed] [Google Scholar]
- 24.Bulman AL, Hubl ST, Nelson HCM. The DNA-binding domain of yeast heat shock transcription factor independently regulates both the N- and C-terminal activation domains. J Biol Chem. 2001;276:40254–40262. doi: 10.1074/jbc.M106301200. [DOI] [PubMed] [Google Scholar]
- 25.Sorger PK, Nelson HCM. Trimerization of a yeast transcriptional activator via a coiled-coil motif. Cell. 1989;59:807–813. doi: 10.1016/0092-8674(89)90604-1. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Barlev NA, Westergaard O, Jakobsen BK. Identification of the C-terminal activator domain in yeast heat shock factor: independent control of transient and sustained transcriptional activity. EMBO J. 1993;12:5007–5018. doi: 10.1002/j.1460-2075.1993.tb06194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabindran SK, Haroun RI, Clos J, Wisniewski J, Wu C. Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science. 1993;259:230–234. doi: 10.1126/science.8421783. [DOI] [PubMed] [Google Scholar]
- 28.Nakai A, et al. HSF4, a new member of the human heat shock factor family which lacks properties of a transcriptional activator. Mol Cell Biol. 1997;17:469–481. doi: 10.1128/mcb.17.1.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanabe M, et al. The mammalian HSF4 gene generates both an activator and a repressor of heat shock genes by alternative splicing. J Biol Chem. 1999;274:27845–27856. doi: 10.1074/jbc.274.39.27845. [DOI] [PubMed] [Google Scholar]
- 30.Nieto-Sotelo J, Wiederrecht G, Okuda A, Parker CS. The yeast heat shock transcription factor contains a transcriptional activation domain whose activity is repressed under nonshock conditions. Cell. 1990;62:807–817. doi: 10.1016/0092-8674(90)90124-w. [DOI] [PubMed] [Google Scholar]
- 31.Sorger PK. Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell. 1990;62:793–805. doi: 10.1016/0092-8674(90)90123-v. [DOI] [PubMed] [Google Scholar]
- 32.Green M, Schuetz TJ, Sullivan EK, Kingston RE. A heat shock-responsive domain of human HSF1 that regulates transcription activation domain function. Mol Cell Biol. 1995;15:3354–3362. doi: 10.1128/mcb.15.6.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newton EM, Knauf U, Green M, Kingston RE. The regulatory domain of human heat shock factor 1 is sufficient to sense heat stress. Mol Cell Biol. 1996;16:839–846. doi: 10.1128/mcb.16.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hensold JO, Hunt CR, Calderwood SK, Housman DE, Kingston RE. DNA binding of heat shock factor to the heat shock element is insufficient for transcriptional activation in murine erythroleukemia cells. Mol Cell Biol. 1990;10:1600–1608. doi: 10.1128/mcb.10.4.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jurivich DA, Sistonen L, Kroes RA, Morimoto RI. Effect of sodium salicylate on the human heat shock response. Science. 1992;255:1243–1245. doi: 10.1126/science.1546322. [DOI] [PubMed] [Google Scholar]
- 36.Shi Y, Kroeger PE, Morimoto RI. The carboxyl-terminal transactivation domain of heat shock factor 1 is negatively regulated and stress responsive. Mol Cell Biol. 1995;15:4309–4318. doi: 10.1128/mcb.15.8.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuo J, Rungger D, Voellmy R. Multiple layers of regulation of human heat shock transcription factor 1. Mol Cell Biol. 1995;15:4319–4330. doi: 10.1128/mcb.15.8.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holmberg CI, et al. Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. EMBO J. 2001;20:3800–3810. doi: 10.1093/emboj/20.14.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hietakangas V, et al. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol Cell Biol. 2003;23:2953–2968. doi: 10.1128/MCB.23.8.2953-2968.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hietakangas V, et al. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci USA. 2006;103:45–50. doi: 10.1073/pnas.0503698102. Discovery of a motif for phosphorylation-dependent sumoylation, called PDSM, that is present in the regulatory domain of HSF1 and HSF4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guettouche T, Boellmann F, Lane WS, Voellmy R. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem. 2005;6:4. doi: 10.1186/1471-2091-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–1066. doi: 10.1126/science.1165946. Describes the mechanism of SIRT1-mediated deacetylation of HSF1, which maintains HSF1 in a state that is competent for DNA binding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baler R, Dahl G, Voellmy R. Activation of human heat shock genes is accompanied by oligomerization, modification, and rapid translocation of heat shock transcription factor HSF1. Mol Cell Biol. 1993;13:2486–2496. doi: 10.1128/mcb.13.4.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarge KD, Murphy SP, Morimoto RI. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ali A, Bharadwaj S, O’Carroll R, Ovsenek N. HSP90 interacts with and regulates the activity of heat shock factor 1 in Xenopus oocytes. Mol Cell Biol. 1998;18:4949–4960. doi: 10.1128/mcb.18.9.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- 47.Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 48.Duina AA, Kalton HM, Gaber RF. Requirement for Hsp90 and a CyP-40-type cyclophilin in negative regulation of the heat shock response. J Biol Chem. 1998;273:18974–18978. doi: 10.1074/jbc.273.30.18974. [DOI] [PubMed] [Google Scholar]
- 49.Zou J, Salminen WF, Roberts SM, Voellmy R. Correlation between glutathione oxidation and trimerization of heat shock factor 1, an early step in stress induction of the Hsp response. Cell Stress Chaperones. 1998;3:130–141. doi: 10.1379/1466-1268(1998)003<0130:cbgoat>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bharadwaj S, Ali A, Ovsenek N. Multiple components of the HSP90 chaperone complex function in regulation of heat shock factor 1 in vivo. Mol Cell Biol. 1999;19:8033–8041. doi: 10.1128/mcb.19.12.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo Y, et al. Evidence for a mechanism of repression of heat shock factor 1 transcriptional activity by a multichaperone complex. J Biol Chem. 2001;276:45791–45799. doi: 10.1074/jbc.M105931200. [DOI] [PubMed] [Google Scholar]
- 52.Shi Y, Mosser DD, Morimoto RI. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abravaya K, Myers MP, Murphy SP, Morimoto RI. The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev. 1992;6:1153–1164. doi: 10.1101/gad.6.7.1153. [DOI] [PubMed] [Google Scholar]
- 54.Baler R, Welch WJ, Voellmy R. Heat shock gene regulation by nascent polypeptides and denatured proteins: hsp70 as a potential autoregulatory factor. J Cell Biol. 1992;117:1151–1159. doi: 10.1083/jcb.117.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 56.Chu B, Soncin F, Price BD, Stevenson MA, Calderwood SK. Sequential phosphorylation by mitogen-activated protein kinase and glycogen synthase kinase 3 represses transcriptional activation by heat shock factor-1. J Biol Chem. 1996;271:30847–30857. doi: 10.1074/jbc.271.48.30847. [DOI] [PubMed] [Google Scholar]
- 57.Cotto JJ, Kline M, Morimoto RI. Activation of heat shock factor 1 DNA binding precedes stress-induced serine phosphorylation. Evidence for a multistep pathway of regulation. J Biol Chem. 1996;271:3355–3358. doi: 10.1074/jbc.271.7.3355. [DOI] [PubMed] [Google Scholar]
- 58.Kline MP, Morimoto RI. Repression of the heat shock factor 1 transcriptional activation domain is modulated by constitutive phosphorylation. Mol Cell Biol. 1997;17:2107–2115. doi: 10.1128/mcb.17.4.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xia W, Guo Y, Vilaboa N, Zuo J, Voellmy R. Transcriptional activation of heat shock factor HSF1 probed by phosphopeptide analysis of factor 32P-labeled in vivo. J Biol Chem. 1998;273:8749–8755. doi: 10.1074/jbc.273.15.8749. [DOI] [PubMed] [Google Scholar]
- 60.Collavin L, et al. Modification of the erythroid transcription factor GATA-1 by SUMO-1. Proc Natl Acad Sci USA. 2004;101:8870–8875. doi: 10.1073/pnas.0308605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Riquelme C, Barthel KK, Liu X. SUMO-1 modification of MEF2A regulates its transcriptional activity. J Cell Mol Med. 2006;10:132–144. doi: 10.1111/j.1582-4934.2006.tb00295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sapetschnig A, et al. Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J. 2002;21:5206–5215. doi: 10.1093/emboj/cdf510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mohideen F, et al. A molecular basis for phosphorylation-dependent SUMO conjugation by the E2 UBC9. Nature Struct Mol Biol. 2009;16:945–952. doi: 10.1038/nsmb.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andrulis ED, Guzman E, Doring P, Werner J, Lis JT. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yao J, Munson KM, Webb WW, Lis JT. Dynamics of heat shock factor association with native gene loci in living cells. Nature. 2006;442:1050–1053. doi: 10.1038/nature05025. Multi-photon microscopy imaging of polytene nuclei in living D. melanogaster salivary glands, allowing real-time analysis of HSF recruitment to the Hsp70 gene. [DOI] [PubMed] [Google Scholar]
- 66.Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT. Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol Cell. 2007;28:978–990. doi: 10.1016/j.molcel.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 67.Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 68.Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- 69.Brown SA, Imbalzano AN, Kingston RE. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- 70.Brown SA, Weirich CS, Newton EM, Kingston RE. Transcriptional activation domains stimulate initiation and elongation at different times and via different residues. EMBO J. 1998;17:3146–3154. doi: 10.1093/emboj/17.11.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sullivan EK, Weirich CS, Guyon JR, Sif S, Kingston RE. Transcriptional activation domains of human heat shock factor 1 recruit human SWI/SNF. Mol Cell Biol. 2001;21:5826–5837. doi: 10.1128/MCB.21.17.5826-5837.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Park JM, Werner J, Kim JM, Lis JT, Kim YJ. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol Cell. 2001;8:9–19. doi: 10.1016/s1097-2765(01)00296-9. [DOI] [PubMed] [Google Scholar]
- 75.Mason PB, Jr, Lis JT. Cooperative and competitive protein interactions at the Hsp70 promoter. J Biol Chem. 1997;272:33227–33233. doi: 10.1074/jbc.272.52.33227. [DOI] [PubMed] [Google Scholar]
- 76.Yuan CX, Gurley WB. Potential targets for HSF1 within the preinitiation complex. Cell Stress Chaperones. 2000;5:229–242. doi: 10.1379/1466-1268(2000)005<0229:ptfhwt>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Östling P, Björk JK, Roos-Mattjus P, Mezger V, Sistonen L. Heat shock factor 2 (HSF2) contributes to inducible expression of hsp genes through interplay with HSF1. J Biol Chem. 2007;282:7077–7086. doi: 10.1074/jbc.M607556200. [DOI] [PubMed] [Google Scholar]
- 78.Denegri M, et al. Human chromosomes 9, 12, and 15 contain the nucleation sites of stress-induced nuclear bodies. Mol Biol Cell. 2002;13:2069–2079. doi: 10.1091/mbc.01-12-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jolly C, et al. In vivo binding of active heat shock transcription factor 1 to human chromosome 9 heterochromatin during stress. J Cell Biol. 2002;156:775–781. doi: 10.1083/jcb.200109018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alastalo TP, et al. Formation of nuclear stress granules involves HSF2 and coincides with the nucleolar localization of Hsp70. J Cell Sci. 2003;116:3557–3570. doi: 10.1242/jcs.00671. [DOI] [PubMed] [Google Scholar]
- 81.Jolly C, et al. Stress-induced transcription of satellite III repeats. J Cell Biol. 2004;164:25–33. doi: 10.1083/jcb.200306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rizzi N, et al. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell. 2004;15:543–551. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sandqvist A, et al. Heterotrimerization of heat-shock factors 1 and 2 provides a transcriptional switch in response to distinct stimuli. Mol Biol Cell. 2009;20:1340–1347. doi: 10.1091/mbc.E08-08-0864. Provides the first evidence of a functional interplay between HSFs through heterotrimerization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tanabe M, Nakai A, Kawazoe Y, Nagata K. Different thresholds in the responses of two heat shock transcription factors, HSF1 and HSF3. J Biol Chem. 1997;272:15389–15395. doi: 10.1074/jbc.272.24.15389. [DOI] [PubMed] [Google Scholar]
- 85.Fujimoto M, et al. HSF4 is required for normal cell growth and differentiation during mouse lens development. EMBO J. 2004;23:4297–4306. doi: 10.1038/sj.emboj.7600435. Shows that there is competition between distinct HSF family members for regulating FGF expression in the lens. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jedlicka P, Mortin MA, Wu C. Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J. 1997;16:2452–2462. doi: 10.1093/emboj/16.9.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Trinklein ND, Murray JI, Hartman SJ, Botstein D, Myers RM. The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response. Mol Biol Cell. 2004;15:1254–1261. doi: 10.1091/mbc.E03-10-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Takaki E, et al. Maintenance of olfactory neurogenesis requires HSF1, a major heat shock transcription factor in mice. J Biol Chem. 2006;281:4931–4937. doi: 10.1074/jbc.M506911200. [DOI] [PubMed] [Google Scholar]
- 89.Christians E, Davis AA, Thomas SD, Benjamin IJ. Maternal effect of Hsf1 on reproductive success. Nature. 2000;407:693–694. doi: 10.1038/35037669. [DOI] [PubMed] [Google Scholar]
- 90.Metchat A, et al. Mammalian heat shock factor 1 is essential for oocyte meiosis and directly regulates Hsp90α expression. J Biol Chem. 2009;284:9521–9528. doi: 10.1074/jbc.M808819200. Establishes a role for the maternal transcription factor HSF1 in the normal progression of meiosis in oocytes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bierkamp C, et al. Lack of maternal heat shock factor 1 results in multiple cellular and developmental defects, including mitochondrial damage and altered redox homeostasis, and leads to reduced survival of mammalian oocytes and embryos. Dev Biol. 2010;339:338–353. doi: 10.1016/j.ydbio.2009.12.037. [DOI] [PubMed] [Google Scholar]
- 92.Inouye S, et al. Impaired IgG production in mice deficient for heat shock transcription factor 1. J Biol Chem. 2004;279:38701–38709. doi: 10.1074/jbc.M405986200. [DOI] [PubMed] [Google Scholar]
- 93.Takii R, et al. Heat shock transcription factor 1 inhibits expression of IL-6 through activating transcription factor 3. J Immunol. 2010;184:1041–1048. doi: 10.4049/jimmunol.0902579. [DOI] [PubMed] [Google Scholar]
- 94.Santos SD, Saraiva MJ. Enlarged ventricles, astrogliosis and neurodegeneration in heat shock factor 1 null mouse brain. Neuroscience. 2004;126:657–663. doi: 10.1016/j.neuroscience.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 95.Homma S, et al. Demyelination, astrogliosis, and accumulation of ubiquitinated proteins, hallmarks of CNS disease in hsf1-deficient mice. J Neurosci. 2007;27:7974–7986. doi: 10.1523/JNEUROSCI.0006-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rallu M, et al. Function and regulation of heat shock factor 2 during mouse embryogenesis. Proc Natl Acad Sci USA. 1997;94:2392–2397. doi: 10.1073/pnas.94.6.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kallio M, et al. Brain abnormalities, defective meiotic chromosome synapsis and female subfertility in HSF2 null mice. EMBO J. 2002;21:2591–2601. doi: 10.1093/emboj/21.11.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang G, Zhang J, Moskophidis D, Mivechi NF. Targeted disruption of the heat shock transcription factor (hsf)-2 gene results in increased embryonic lethality, neuronal defects, and reduced spermatogenesis. Genesis. 2003;36:48–61. doi: 10.1002/gene.10200. [DOI] [PubMed] [Google Scholar]
- 99.Chang Y, et al. Role of heat-shock factor 2 in cerebral cortex formation and as a regulator of p35 expression. Genes Dev. 2006;20:836–847. doi: 10.1101/gad.366906. The first identification of a direct target gene of HSFs in development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tsai LH, Delalle I, Caviness VS, Jr, Chae T, Harlow E. p35 is a neural-specific regulatory subunit of cyclin-dependent kinase 5. Nature. 1994;371:419–423. doi: 10.1038/371419a0. [DOI] [PubMed] [Google Scholar]
- 101.Chae T, et al. Mice lacking p35, a neuronal specific activator of Cdk5, display cortical lamination defects, seizures, and adult lethality. Neuron. 1997;18:29–42. doi: 10.1016/s0896-6273(01)80044-1. [DOI] [PubMed] [Google Scholar]
- 102.Sarge KD, Park-Sarge OK, Kirby JD, Mayo KE, Morimoto RI. Expression of heat shock factor 2 in mouse testis: potential role as a regulator of heat-shock protein gene expression during spermatogenesis. Biol Reprod. 1994;50:1334–1343. doi: 10.1095/biolreprod50.6.1334. [DOI] [PubMed] [Google Scholar]
- 103.Mendell JT. miRiad roles for the miR-17–92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Åkerfelt M, et al. Promoter ChIP-chip analysis in mouse testis reveals Y chromosome occupancy by HSF2. Proc Natl Acad Sci USA. 2008;105:11224–11229. doi: 10.1073/pnas.0800620105. Provides a global description of HSF2 target genes in mouse testis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang G, et al. Essential requirement for both hsf1 and hsf2 transcriptional activity in spermatogenesis and male fertility. Genesis. 2004;38:66–80. doi: 10.1002/gene.20005. [DOI] [PubMed] [Google Scholar]
- 106.Min JN, Zhang Y, Moskophidis D, Mivechi NF. Unique contribution of heat shock transcription factor 4 in ocular lens development and fiber cell differentiation. Genesis. 2004;40:205–217. doi: 10.1002/gene.20087. [DOI] [PubMed] [Google Scholar]
- 107.Shi X, et al. Removal of Hsf4 leads to cataract development in mice through down-regulation of γS-crystallin and Bfsp expression. BMC Mol Biol. 2009;10:10. doi: 10.1186/1471-2199-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bu L, et al. Mutant DNA-binding domain of HSF4 is associated with autosomal dominant lamellar and Marner cataract. Nature Genet. 2002;31:276–278. doi: 10.1038/ng921. The first study to link a mutation of an HSF gene to a particular disease. [DOI] [PubMed] [Google Scholar]
- 109.Fujimoto M, et al. Analysis of HSF4 binding regions reveals its necessity for gene regulation during development and heat shock response in mouse lenses. J Biol Chem. 2008;283:29961–29970. doi: 10.1074/jbc.M804629200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. Establishes the link between HSF and the insulin-like signalling pathway in the regulation of C. elegans lifespan. [DOI] [PubMed] [Google Scholar]
- 111.Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. Provides the first evidence for the important role of HSF and molecular chaperone networks in ageing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci USA. 2009;106:14914–14919. doi: 10.1073/pnas.0902882106. Shows that failure in proteostasis occurs at an early stage of adulthood and coincides with a severely reduced activation of the heat shock response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nature Rev Genet. 2007;8:835–844. doi: 10.1038/nrg2188. [DOI] [PubMed] [Google Scholar]
- 114.Heydari AR, et al. Effect of caloric restriction on the expression of heat shock protein 70 and the activation of heat shock transcription factor 1. Dev Genet. 1996;18:114–124. doi: 10.1002/(SICI)1520-6408(1996)18:2<114::AID-DVG4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 115.Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 116.Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 117.Stringham EG, Candido EP. Targeted single-cell induction of gene products in Caenorhabditis elegans: a new tool for developmental studies. J Exp Zool. 1993;266:227–233. doi: 10.1002/jez.1402660309. [DOI] [PubMed] [Google Scholar]
- 118.Prahlad V, Morimoto RI. Integrating the stress response: lessons for neurodegenerative diseases from C. elegans. Trends Cell Biol. 2009;19:52–61. doi: 10.1016/j.tcb.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Clark DA, Gabel CV, Gabel H, Samuel AD. Temporal activity patterns in thermosensory neurons of freely moving Caenorhabditis elegans encode spatial thermal gradients. J Neurosci. 2007;27:6083–6090. doi: 10.1523/JNEUROSCI.1032-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mori I. Genetics of chemotaxis and thermotaxis in the nematode Caenorhabditis elegans. Annu Rev Genet. 1999;33:399–422. doi: 10.1146/annurev.genet.33.1.399. [DOI] [PubMed] [Google Scholar]
- 121.Prahlad V, Cornelius T, Morimoto RI. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 2008;320:811–814. doi: 10.1126/science.1156093. Introduces the concept that the heat shock response in somatic cells is not cell-autonomous, but depends on thermosensory neurons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tang D, et al. Expression of heat shock proteins and heat shock protein messenger ribonucleic acid in human prostate carcinoma in vitro and in tumors in vivo. Cell Stress Chaperones. 2005;10:46–58. doi: 10.1379/CSC-44R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Khaleque MA, et al. Heat shock factor 1 represses estrogen-dependent transcription through association with MTA1. Oncogene. 2008;27:1886–1893. doi: 10.1038/sj.onc.1210834. [DOI] [PubMed] [Google Scholar]
- 124.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–1018. doi: 10.1016/j.cell.2007.07.020. Describes HSF1 as a non-oncogene that is required for tumour initiation and maintenance in various cancer models. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Whitesell L, Lindquist S. Inhibiting the transcription factor HSF1 as an anticancer strategy. Expert Opin Ther Targets. 2009;13:469–478. doi: 10.1517/14728220902832697. [DOI] [PubMed] [Google Scholar]
- 126.Westerheide SD, Morimoto RI. Heat shock response modulators as therapeutic tools for diseases of protein conformation. J Biol Chem. 2005;280:33097–33100. doi: 10.1074/jbc.R500010200. [DOI] [PubMed] [Google Scholar]
- 127.Westerheide SD, et al. Celastrols as inducers of the heat shock response and cytoprotection. J Biol Chem. 2004;279:56053–56060. doi: 10.1074/jbc.M409267200. [DOI] [PubMed] [Google Scholar]
- 128.Trott A, et al. Activation of heat shock and antioxidant responses by the natural product celastrol: transcriptional signatures of a thiol-targeted molecule. Mol Biol Cell. 2008;19:1104–1112. doi: 10.1091/mbc.E07-10-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Neef DW, Turski ML, Thiele DJ. Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PLoS Biol. 2010;8:e1000291. doi: 10.1371/journal.pbio.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Westerheide SD, Kawahara TL, Orton K, Morimoto RI. Triptolide, an inhibitor of the human heat shock response that enhances stress-induced cell death. J Biol Chem. 2006;281:9616–9622. doi: 10.1074/jbc.M512044200. [DOI] [PubMed] [Google Scholar]
- 131.Phillips PA, et al. Triptolide induces pancreatic cancer cell death via inhibition of heat shock protein 70. Cancer Res. 2007;67:9407–9416. doi: 10.1158/0008-5472.CAN-07-1077. [DOI] [PubMed] [Google Scholar]
- 132.Amin J, Ananthan J, Voellmy R. Key features of heat shock regulatory elements. Mol Cell Biol. 1988;8:3761–3769. doi: 10.1128/mcb.8.9.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xiao H, Lis JT. Germline transformation used to define key features of heat-shock response elements. Science. 1988;239:1139–1142. doi: 10.1126/science.3125608. [DOI] [PubMed] [Google Scholar]
- 134.Perisic O, Xiao H, Lis JT. Stable binding of Drosophila heat shock factor to head-to-head and tail-to-tail repeats of a conserved 5 bp recognition unit. Cell. 1989;59:797–806. doi: 10.1016/0092-8674(89)90603-x. [DOI] [PubMed] [Google Scholar]
- 135.Xiao H, Perisic O, Lis JT. Cooperative binding of Drosophila heat shock factor to arrays of a conserved 5 bp unit. Cell. 1991;64:585–593. doi: 10.1016/0092-8674(91)90242-q. [DOI] [PubMed] [Google Scholar]
- 136.Gorisch SM, Lichter P, Rippe K. Mobility of multi-subunit complexes in the nucleus: accessibility and dynamics of chromatin subcompartments. Histochem Cell Biol. 2005;123:217–228. doi: 10.1007/s00418-005-0752-y. [DOI] [PubMed] [Google Scholar]
- 137.Cotto J, Fox S, Morimoto R. HSF1 granules: a novel stress-induced nuclear compartment of human cells. J Cell Sci. 1997;110:2925–2934. doi: 10.1242/jcs.110.23.2925. [DOI] [PubMed] [Google Scholar]
- 138.Chiodi I, et al. Structure and dynamics of hnRNP-labelled nuclear bodies induced by stress treatments. J Cell Sci. 2000;113:4043–4053. doi: 10.1242/jcs.113.22.4043. [DOI] [PubMed] [Google Scholar]
- 139.Denegri M, et al. Stress-induced nuclear bodies are sites of accumulation of pre-mRNA processing factors. Mol Biol Cell. 2001;12:3502–3514. doi: 10.1091/mbc.12.11.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Metz A, Soret J, Vourc’h C, Tazi J, Jolly C. A key role for stress-induced satellite III transcripts in the relocalization of splicing factors into nuclear stress granules. J Cell Sci. 2004;117:4551–4558. doi: 10.1242/jcs.01329. [DOI] [PubMed] [Google Scholar]
- 141.Weighardt F, et al. A novel hnRNP protein (HAP/SAF-B) enters a subset of hnRNP complexes and relocates in nuclear granules in response to heat shock. J Cell Sci. 1999;112:1465–1476. doi: 10.1242/jcs.112.10.1465. [DOI] [PubMed] [Google Scholar]
- 142.Chiodi I, et al. RNA recognition motif 2 directs the recruitment of SF2/ASF to nuclear stress bodies. Nucleic Acids Res. 2004;32:4127–4136. doi: 10.1093/nar/gkh759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Valgardsdottir R, et al. Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol Biol Cell. 2005;16:2597–2604. doi: 10.1091/mbc.E04-12-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shinka T, et al. Molecular characterization of heat shock-like factor encoded on the human Y chromosome, and implications for male infertility. Biol Reprod. 2004;71:297–306. doi: 10.1095/biolreprod.103.023580. [DOI] [PubMed] [Google Scholar]
- 145.Tessari A, et al. Characterization of HSFY, a novel AZFb gene on the Y chromosome with a possible role in human spermatogenesis. Mol Hum Reprod. 2004;10:253–258. doi: 10.1093/molehr/gah036. [DOI] [PubMed] [Google Scholar]
- 146.Bhowmick BK, Takahata N, Watanabe M, Satta Y. Comparative analysis of human masculinity. Genet Mol Res. 2006;5:696–712. [PubMed] [Google Scholar]
- 147.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sistonen L, Sarge KD, Morimoto RI. Human heat shock factors 1 and 2 are differentially activated and can synergistically induce hsp70 gene transcription. Mol Cell Biol. 1994;14:2087–2099. doi: 10.1128/mcb.14.3.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]