Abstract
The biosynthesis of rubber is thought to take place on the surface of rubber particles in laticifers, highly specialized cells that are present in more than 40 plant families. The small rubber particle protein (SRPP) has been supposed to be involved in rubber biosynthesis, and recently five SRPPs (TbSRPP1–5) were identified in the rubber-producing dandelion species Taraxacum brevicorniculatum. Here, we demonstrate by immunogold labeling that TbSRPPs are localized to rubber particles, and that rubber particles mainly consist of TbSRPP3, 4 and 5 as shown by high-resolution two-dimensional gel electrophoresis and mass spectrometric analysis. We also carried out an RNA-interference approach in transgenic plants to address the function of TbSRPPs in rubber biosynthesis as well as rubber particle morphology and stability. TbSRPP-RNAi transgenic T. brevicorniculatum plants showed a 40–50% reduction in the dry rubber content, but neither the rubber weight average molecular mass nor the polydispersity of the rubber were affected. Although no phenotypical differences to wild-type particles could be observed in vivo, rubber particles from the TbSRPP-RNAi transgenic lines were less stable and tend to rapidly aggregate in expelling latex after wounding of laticifers. Our results prove that TbSRPPs are very crucial for rubber production in T. brevicorniculatum, probably by contributing to a most favourable and stable rubber particle architecture for efficient rubber biosynthesis and eventually storage.
Introduction
Approximately 2,500 plants produce natural rubber poly (cis-1,4-isoprene), which is stored in rubber particles dispersed throughout the cytosol of either laticifers or specialized parenchyma cells [1]. Natural rubber is synthesized by the successive addition of isopentenyl pyrophosphate (IPP) to an initiating allylic pyrophosphate, e.g. farnesyl pyrophosphate (FPP) [1]–[5]. This polymerization reaction is thought to take place on the surface of rubber particles, mediated by a particle bound cis-prenyltransferase (CPT) [1], [6]–[8]. Recently, we identified three rubber particle bound long-chain CPTs in Taraxacum brevicorniculatum (T. brevicorniculatum was erroneously labeled as T. koksaghyz in botanical gardens [9]) that appear to be the key enzymes for rubber biosynthesis in this species [8], [10]. Detailed analysis of rubber particles in different species revealed a conserved spherical structure with sizes ranging from 0.2–6.5 µm in Ficus carica and Ficus benghalensis, 1–2 µm in Parthenium argentatum, 0.08–2 µm in Hevea brasiliensis and 0.01–1 µm in T. brevicorniculatum [8], [11]–[13]. Rubber particles comprise a homogenous rubber core surrounded by a monolayer membrane containing lipids, proteins, and several minor compounds [12], [14], [15]. This composite monolayer membrane probably mediates the colloidal stability of rubber particles by providing a net negative surface charge that prevents their coalescence in the cytosol [12], [14]. In principle, colloidal stability depends on the balance between repulsion and attractive van-der-Waals forces, and electrostatic forces are often responsible for the repulsion of colloids in a suspension [16]–[18]. The net surface charge of colloids can be estimated by the determination of their zeta (ζ)-potential, which is the electrical potential difference at the interface of the suspending fluid and the stationary fluid attached to the colloid surface [16], [18]. In addition to repulsion through a similar (positive or negative) surface charge, repulsion of colloids can also be mediated by steric hindrance of adsorbed large molecules, such as proteins [17], [19]–[22]. Amongst others, two highly homologous proteins are associated with the surface of H. brasiliensis rubber particles, i.e. the small rubber particle protein (SRPP; here designated as HbSRPP) and the rubber elongation factor (REF; here designated as HbREF) [23]–[25]. SRPP homologs were also identified in P. argentatum (guayule homolog of SRPP, GHS [26]) and T. brevicorniculatum (TbSRPP1–5, previously designated as TkSRPP1–5 [27]). The functions of SRPP and REF are yet unclear. However, there is some evidence indicating a potential role in rubber biosynthesis: In vitro-experiments revealed that the addition of recombinant HbSRPP to H. brasiliensis rubber particles suspended in latex cytosol increased the incorporation of IPP, while addition of antibodies directed against HbSRPP and HbREF inhibited IPP incorporation [24], [25]. Moreover, the analysis of HbREF gene expression in H. brasiliensis accessions with low to high rubber yields revealed that HbREF expression correlates positively with the amount of rubber [28]. However, it has been proposed that the rubber-producing species F. carica and F. benghalensis do not possess a SRPP homolog, suggesting that SRPP is not a general and essential protein for rubber biosynthesis [13].
The recently identified SRPPs in T. brevicorniculatum (TbSRPP1–5) shared sequence identities of 41–66% to each other and 38–43% with HbSRPP. Except for TbSRPP2, all TbSRPPs are predominately expressed in latex and their spatiotemporal expression pattern correlates with latex and rubber production in T. brevicorniculatum [27]. To analyze the function of TbSRPPs in vivo, we generated transgenic T. brevicorniculatum plants with strongly reduced TbSRPP protein levels, caused by RNA-interference. We revealed that exuded latex as well as freeze-dried roots of transgenic plants contain significantly lower amounts of natural rubber compared to wild-type plants. Rubber particles in expelling latex of transgenic plants appeared to be less stable compared to wild-type, whereas neither the rubber weight average molecular mass nor the polydispersity were affected. Hence, we conclude that TbSRPPs probably support rubber biosynthesis by the stabilization of rubber particles.
Results
TbSRPPs are the major rubber particle proteins
We recently identified and cloned five genes (TbSRPP1–5) encoding small rubber particle proteins from T. brevicorniculatum [27]. As reported for other SRPPs [13], [25], we first set out to determine whether TbSRPP1–5 are associated with rubber particles by two-dimensional (2-D) gel electrophoresis followed by mass spectrometry. We revealed a cluster of highly abundant protein spots with isoelectric point (pI) values between pH 4 and pH 6 (Fig. 1A). Mass spectrometric (MS) analysis of these protein spots generated representative peptide sequences for TbSRPP2, TbSRPP3, TbSRPP4 and TbSRPP5, whereas no peptides were detected for TbSRPP1 (Tab. S1). The most abundant rubber particle protein was TbSRPP3, followed by TbSRPP4 and TbSRPP5, while TbSRPP2 was present only in small amounts (Fig. 1A, Tab. S1).
Figure 1. Association of TbSRPPs with rubber particles. A.
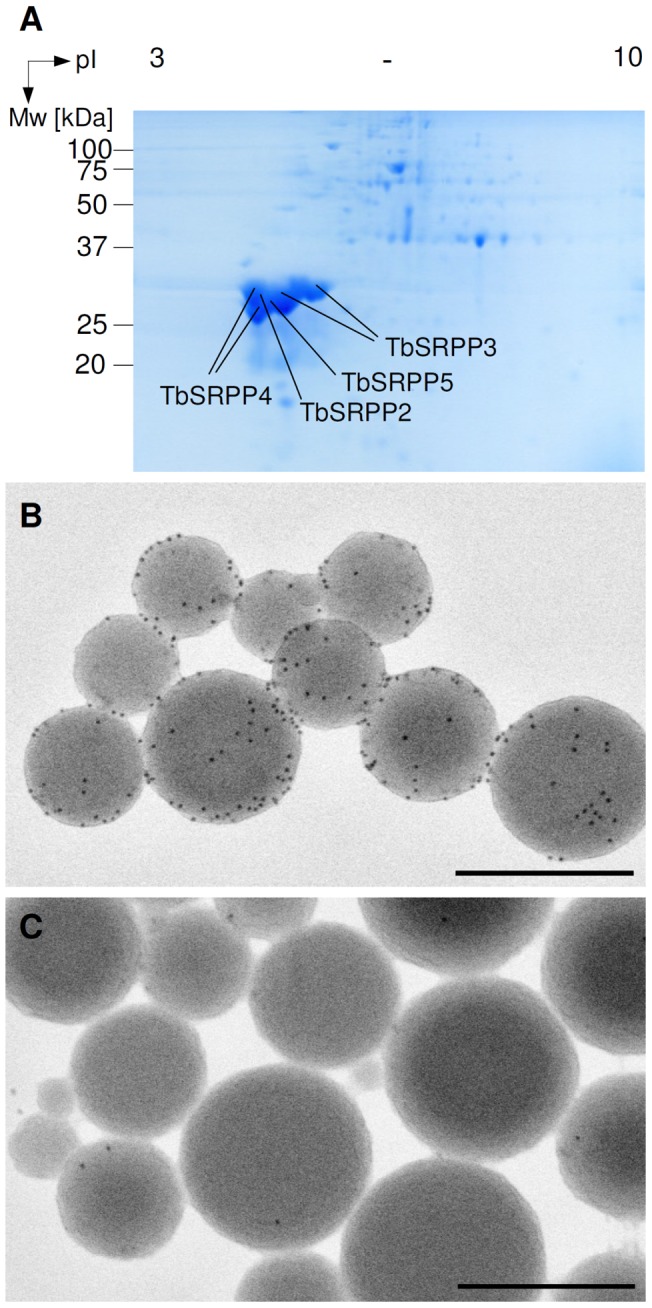
Fifty micrograms of purified rubber particle proteins were separated by isoelectric focussing (pH 3–10 IPG strips) and subsequent SDS-PAGE. The gel was stained with colloidal Coomassie Brilliant Blue. B. Backscattered electron imaging of rubber particles labeled with 10 nm gold particles by immunodetection using the TbSRPP-antibody. C. Gold labeling of rubber particles was not appreciably detected in backscattered electron images, when the corresponding pre-immune serum was used as primary antibody. Micrographs are shown as inverted images. Scale bars = 400 nm.
Next, the presence of TbSRPPs on rubber particles was investigated by scanning electron microscopy (SEM) using a polyclonal antibody raised against TbSRPP3. In Western blot experiments, this antibody recognized all TbSRPPs with the same efficiency, most probably due to the high amino acid sequence conservation between members of this protein family [27], and will therefore be referred to as anti-TbSRPP (Fig. S1, Text S1). Immunogold labeling of rubber particles using anti-TbSRPP and a gold-coupled secondary antibody with subsequent visualization by SEM directly confirmed the presence of TbSRPPs on the rubber particle surface (Fig. 1B). Control experiments with pre-immune serum did not show significant staining of rubber particles (Fig. 1C).
Depletion of TbSRPP1-5 by RNA-interference
To further address the function of TbSRPPs in rubber biosynthesis we developed transgenic T. brevicorniculatum plants with reduced TbSRPPs expression levels by RNA-interference (RNAi). Therefore, we introduced a TbSRPP3-RNAi construct into the genome of T. brevicorniculatum by Agrobacterium-mediated transformation. Out of ten independently regenerated transgenic plants, six lines displayed reduced transcript levels for all TbSRPP genes and were therefore designated as TbSRPP-RNAi transgenic plants (data not shown). Subsequently, three TbSRPP-RNAi transgenic plants, labeled S3, S76 and S85, were selected for further analysis of TbSRPP1–5 expression by quantitative RT-PCR using total RNA from latex. The expression of all TbSRPP genes was severely reduced by the TbSRPP3-RNAi construct when compared to wild-type plants (Fig. 2A). The most severe reduction in TbSRPPs expression was found in S3, where only residual amounts of TbSRPP5 are still visible.
Figure 2. Analysis of TbSRPP1–5 expression in TbSRPP-RNAi transgenic and wild-type plants. A.

TbSRPP transcription levels in the latex of three independent TbSRPP-RNAi transgenic lines (designated S3, S76 and S85) were quantified by RT-PCR. Relative transcription of TbSRPPs was normalized to the constitutively expressed control gene TbACTIN. The ratios of TbSRPPs/TbACTIN transcription levels in wild-type (wt) served as a reference and were set to the value of one. Values represent mean ± SE (n = 3). B, C. Fifty microgram of rubber particle proteins from wild-type (B) and TbSRPP-RNAi lines (C, exemplarily shown for line S3) was separated by isoelectric focussing (pH 3–6 IPG strips) and subsequent SDS-PAGE. Proteins were transferred to nitrocellulose membranes and TbSRPPs were detected using an anti-TbSRPP antibody and a secondary antibody conjugated with horse radish peroxidase. D, E. Ponceau-S staining of the nitrocellulose membranes confirmed equal loading of gels prepared for wild-type (D) and line S3 (E), by showing that proteins spots of the C-terminus of the Polyphenoloxidase-1 (TbPPO-1) were present in similar amounts [29], [30].
Next, the effect of the TbSRPP3-RNAi construct was analyzed on protein level by 2-D gel electrophoresis of rubber particle proteins followed by immunodetection using the anti-TbSRPP antibody. Equal loading of the gels was verified after Ponceau-S staining of the nitrocellulose membrane by comparing the intensities of the protein spots representing the C-terminus of the Polyphenoloxidase 1 (TbPPO-1), which is expressed consistently in T. brevicorniculatum and thus suitable as loading control [29], [30]. Compared to wild-type plants (Fig. 2B), the levels for TbSRPPs were strongly reduced as exemplarily shown for S3 (Fig. 2C). It should be noted that TbSRPP transcription (Fig. 2A) strongly correlates with the observed protein levels as only residual amounts were identified for TbSRPP5 by 2-D gel electrophoresis (Fig. 2C). The transgenic lines showed regular growth and development under standard greenhouse conditions.
Reduced TbSRPP protein levels had severe impacts on rubber production
To test if depleted TbSRPP levels affect rubber biosynthesis, the dry rubber content in exuding latex and freeze-dried roots was measured. Compared to wild-type, the latex of TbSRPP-RNAi transgenic plants contained significantly lower amounts of rubber (24–37% of wild-type, Tab. 1) and the same was evident for the rubber content of freeze-dried roots (Tab. 1). Additionally, a clear correlation between TbSRPP expression levels and rubber content was indicated (Tab. 1, see also Fig. 2A).
Table 1. Table 1. Content, molecular mass and polydispersity of rubber from exuded latex and freeze-dried roots and corresponding long-chain prenyltransferase activity.
| plant lines | latex dry rubber content | root dry rubber content | rubber molecular mass distribution a | [1-14C] IPP transferase activty | ||
| [mg ml−1] | [mg ml−1] | Μwb [106 Da] | Μnc [106 Da] | ΜwΜn−1 | [pmol mg−1 min−1] | |
| wt | 172.6 (±11.3) | 3.88 (±0.65) | 3.30 (±0.47) | 2.21 (±0.32) | 1.56 (±0.03) | 1.83 (±0.29) |
| S3 | 41.5 (±8.3) ** | 1.93 (±0.17)** | 2.74 (±0.23) | 1.97 (±0.15) | 1.39 (±0.04) | 1.09 (±0.04)* |
| S76 | 63.2 (±8.6) ** | 2.32 (±0.09)** | 2.69 (±0.84) | 1.86 (±0.11) | 1.45 (±0.04) | 0.96 (±0.09)** |
| S85 | 61.4 (±0.4) ** | 2.15 (±0.12)** | 3.14 (±0.27) | 1.99 (±0.34) | 1.62 (±0.23) | 1.29 (±0.20)* |
The values are given as the mean of at least three repititions ± SD.
Molecular mass distribution was measured by size exclusion chromatography of rubber from exuded latex.
W, weight average molecular mass.
N, number average molecular mass.
statistically significant difference compared to wild type control (P, 0.05).
statistically significant difference compared to wild type control (P, 0.01).
Next, we analyzed the weight average molecular mass (Μw) and number average molecular mass (Μn) of rubber obtained from exuding latex. Using size exclusion chromatography (SEC) no significant differences could be detected between rubber of wild-type and TbSRPP-RNAi transgenic plants indicating that polymer length and polydispersity of the residual rubber were not influenced by altered TbSRPPs expression (Tab. 1).
To figure out whether reduced TbSRPP levels affected the activity of the long chain prenyltransferase in latex, we carried out in vitro [1-14C] IPP incorporation assays using whole latex from TbSRPP-RNAi transgenic and wild-type plants as described by Post et al. [10]. After removal of the short- and medium-chain polyprenol diphosphates by n-butanol extraction, the long-chain products were obtained with a mixture of toluene and hexane and the incorporated radioactivity was measured. This experiment revealed that the activity of the long-chain prenyltransferase in latex of TbSRPP-RNAi transgenic plants was indeed significantly lower under this condition (Tab. 1).
Silencing of TbSRPPs reduces the stability of rubber particles in exuding latex
We set out to analyze if the depleted levels of TbSRPPs affected the architecture of rubber particles by ultrastructural investigations using transmission electron microscopy (TEM). TEM images of ultra-thin cross sections from roots of TbSRPP-RNAi transgenic (Fig. 3A) and wild-type plants (Fig. 3C) showed laticifers with electron dense material, organelle-like structures, and rubber particles. In the analyzed sections, rubber particles appeared spherical and did not show any obvious morphological differences in both, TbSRPP-RNAi transgenic (Fig. 3B) and wild-type (Fig. 3D) plants.
Figure 3. In situ analysis of the ultra-structure of rubber particles from TbSRPP-RNAi transgenic plants and wild-type plants by transmission electron microscopy.

A. Micrograph of ultra-thin cross sections of roots from TbSRPP-RNAi transgenic plant (S3) at low and B. higher magnification. C. Micrograph of ultra-thin cross sections of roots from a wild-type plant at low and D. higher magnification. Enlarged areas are indicated with dashed boxes. Key: RP, rubber particle; V, vacuole; CW, cell wall; PC, parenchyma cell. Scale bars = 1 µm.
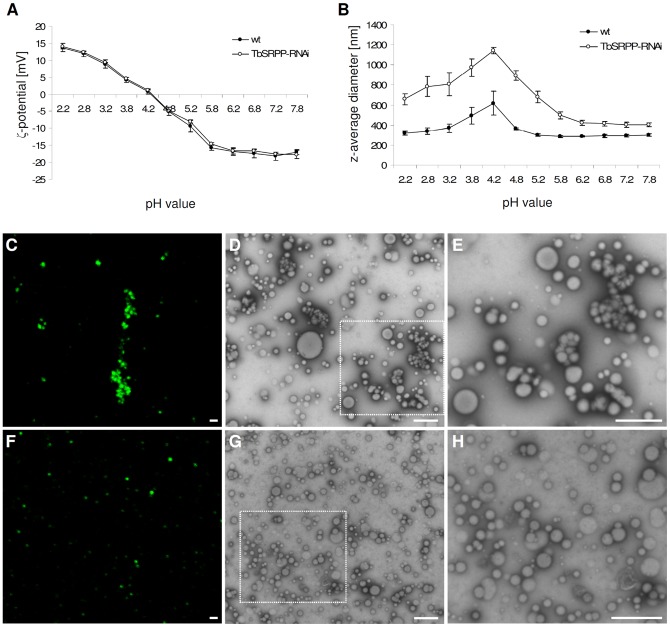
We further studied the colloidal stability of purified rubber particles from exuding latex of TbSRPP-RNAi transgenic and wild-type plants by measuring their ζ-potential at different pH values. Both types of rubber particles showed a similar ζ-potential ranging from +14 mV at pH 2.2 to −18 mV at pH 7.2 (Fig. 4A). The pI was approximately pH 4.2. In contrast to the ζ-potential, significant differences were obtained for their size. Rubber particles from wild-type latex revealed a z-average size of 295 nm at pH 7.2 (Fig. 4B) and showed an increased diameter at pH-values near their pI (pH 3.2–4.8, Fig. 4B). Rubber particles obtained from TbSRPP-RNAi transgenic plants, however, displayed with 406 nm at pH 7.2 a significantly higher z-average diameter compared to the wild-type control. A similar increase in z-average diameter was also evident at other pH values (Fig. 4B).
Figure 4. In vitro analysis of the colloidal stability of rubber particles from TbSRPP-RNAi transgenic plants and wild-type plants.
A. Measurement of the zeta (ζ)-potential of rubber particles from wild-type plants (filled circles) and TbSRPP-RNAi transgenic plants (open circles) suspended in modified rubber extraction buffer (REB) at given pH values. Values represent mean ± SD of three independent plant lines (S3, S76 and S85) respectively of six wild-type plants (wt). No significant differences were detected among the plants using Students t-test (P, 0.05). B. Measurement of the z-average diameter of rubber particles from wild-type plants (filled circles) and TbSRPP-RNAi transgenic plants (open circles) suspended in modified REB with the indicated pH values. Values represent mean ± SD of three independent plant lines (S3, S76 and S85) respectively of six wild-type plants (wt). Significant differences have been detected among TbSRPP-RNAi lines and wild-type plants using Students t-test (P, 0.01). C, F. Confocal laser scanning microscopic analysis of isolated rubber particles from TbSRPP-RNAi transgenic plants (C) and wild-type plants (F) that were stained with nile red. D, E and G, H. Transmission electron microscopic analysis of isolated rubber particles from TbSRPP-RNAi transgenic and wild-type plants. D. Micrograph of isolated rubber particles from TbSRPP-RNAi transgenic plant line S3 suspended in modified rubber extraction buffer (REB) pH 7.2 at low and E. higher magnification. G. Micrograph of rubber particles from wild-type suspended in modified REB pH 7.2 at low and H. higher magnification. Enlarged areas are indicated with dashed boxes. Scale bars = 1 µm.
The increase of the z-average diameter could indicate an aggregation of rubber particles, although such structures have not been observed in situ (Fig. 3). To proof if the differences in z-average diameter point toward the formation of rubber particle aggregates, we stained the rubber particles with the lipophilic fluorescent dye nile red [31] and analyzed the samples by confocal laser scanning microscopy (CLSM, Fig. 4C, F). While the rubber particles of TbSRPP-RNAi transgenic plants formed large aggregates at pH 7.2, their wild-type counterparts appeared to be evenly distributed in the suspending buffer. For more detailed observations we again carried out TEM-analysis and confirmed that rubber particles fromTbSRPP-RNAi transgenic plants indeed tend to aggregate at pH 7.2 and in some cases are also partially fused (Fig. 4D, E). No similar observations were made for rubber particles of wild-type latex (Fig. 4G, H).
Discussion
The biosynthesis of rubber at the surface of rubber particles is only little understood. Recently, CPTs were identified as one important component of the polymer elongation process in T. brevicorniculatum [10], however other proteinaceous and non-proteinaceous factors appear to be necessary for efficient rubber production.
In this study we showed that the three SRPP homologs TbSRPP3, 4 and 5 are the major rubber particle proteins in T. brevicorniculatum and confirmed their association with rubber particles by immunogold labeling and SEM (Fig. 1, Tab. S1). While we were also able to detect small amounts of TbSRPP2 at the rubber particle surface, TbSRPP1 seems to be not or only in non-detectable amounts (by 2D-gel electrophoresis) associated with the rubber particle surface, since no peptide sequences for TbSRPP1 could be detected. This could result from the washing steps during the purification procedure applied to the rubber particles by which more loosely associated proteins might detach from the rubber particle surface (Fig. 1A, Tab. S1). In H. brasiliensis, the closely related proteins HbREF and HbSRPP are also associated with rubber particles, whereby HbREF is the most abundant rubber particle protein followed by HbSRPP [13], [24], [25]. HbREF and HbSRPP as well as their homolog GHS from P. argentatum have been supposed to positively influence rubber biosynthesis: It has been shown that heterologously expressed GST-HbSRPP as well as His-GHS fusion proteins enhance IPP incorporation activity of rubber particles in vitro, while immunoinhibition by appropriate HbREF and HbSRPP antibodies reduces the IPP incorporation [24]–[26]. Therefore, it has been suggested that HbSRPP is part of the rubber transferase complex or that it is involved in the synthesis or recruitment of substrates for rubber biosynthesis initiation and/or rubber chain elongation [25]. However, Singh and co-workers speculated that SRPP is not crucial for rubber biosynthesis since its detection on F. carica and F. benghalensis rubber particles by immunogold labeling with an HbSRPP antibody failed [13]. As SRPP homologs have been detected in species that produce rubber of high molecular mass (H. brasiliensis and P. argentatum) but not in species possessing only low molecular mass rubber (F. carica and F. benghalensis), it has been supposed that HbSRPP and GHS might be important factors for determination of the rubber molecular mass by assisting CPT function [26]. Due to the lack of in vivo evidence, the roles of SRPP, REF, and GHS in rubber biosynthesis have yet not been sufficiently proven.
To elucidate the role of TbSRPPs in rubber biosynthesis in vivo, we generated transgenic plants expressing an RNAi construct directed against TbSRPPs. Quantitative RT-PCR and 2-D gel electrophoresis with subsequent immunodetection using the anti-TbSRPP antibody confirmed that expression of all TbSRPPs was severely silenced in the transgenic lines (Fig. 2). By determination of the dry rubber content from exuded latex and freeze-dried roots we were able to show that the TbSRPP-RNAi transgenic plants contained significantly lower amounts of natural rubber compared to wild-type controls (Tab. 1) indicating an impaired rubber biosynthesis in the TbSRPP-RNAi transgenic lines. In contrast, SEC-analysis of the rubber samples revealed a wild-type like rubber chain elongation since neither the weight average molecular mass nor the polydispersity was affected by reduced TbSRPP protein levels (Tab. 1). Therefore, we propose that TbSRPPs are not mandatory for the formation of high molecular mass rubber. Instead, the rubber biosynthesis seems to be less effective in TbSRPP-RNAi transgenic plants, probably resulting from an inefficient enzymatic activity of the long-chain prenyltransferase (TbCPT1–3) [10], as also suggested by the data obtained from the in vitro prenyltransferase assay (Tab. 1).
Rubber particles are not only required for the storage of rubber molecules, but also to provide a suitable and stable platform for rubber biosynthesis. Although one could expect that the depletion of major integral rubber particle proteins might have a dramatic effect on the morphology of rubber particles, TEM-analysis of ultra-thin cross sections of laticifers revealed no obvious differences in the architecture of rubber particles from TbSRPP-RNAi transgenic and wild-type plants (Fig. 3). In addition, TbSRPPs appear not to contribute to the surface charge and subsequently to the electrostatic repulsion of rubber particles since both, rubber particles from TbSRPP-RNAi transgenic and wild-type plants showed a nearly identical ζ-potential (Fig. 4A). This is in consistence with results obtained for H. brasiliensis rubber particles, whose negative surface charge is mainly derived from adsorbed long chain fatty acids and only little from proteins [32].
Depletion of the rubber particle surface charge by suspending rubber particles in a buffer with a pH-value close to their isoelectric point led to an increase of their z-average diameter, indicating the formation of aggregates (Fig. 4B). This suggests that electrostatic repulsion contributes to a great extent to the colloidal stability of T. brevicorniculatum rubber particles. Nevertheless, even at pH 7.2 the rubber particle ζ-potential of approximately −18 mV remains insufficient for electrostatic force to solely provide particle stabilization, which requires a ζ-potential of at least ±30 mV [18], [33]. Therefore, the colloidal stability of T. brevicorniculatum rubber particles is not exclusively mediated by electrostatic repulsion. This is strongly supported by the fact that the depletion of TbSRPPs, which have no impact on the rubber particle surface charge, led to a significant increase of the z-average diameter of rubber particles from TbSRPP-RNAi transgenic plants compared to wild-type (Fig. 4A, B). Microscopic analysis of purified rubber particles using CLSM and TEM confirmed that even at pH 7.2, where the surface charge of the rubber particles is considerably high, rubber particles from TbSRPP-RNAi transgenic plants aggregated intensely, while wild-type particles remained solitary (Fig. 4A, C–H). Therefore, we suppose that in addition to the electrostatic repulsion of rubber particles (most probably provided by their membrane lipids) TbSRPPs contribute to their stabilization by steric hindrance. This is supported by the fact that at low surface charges rubber particles from TbSRPP-RNAi transgenic plants showed an even more intensive aggregation compared to wild-type particles, suggesting an additive effect of electrostatic repulsion and steric repulsion mediated by TbSRPPs as also considered for other colloidal systems (Fig. 4A, B) [19]–[22]. Accordingly, the monolayer-membrane surrounded seed oil bodies are sterically stabilized by oleosin, their major surface protein [34], [35]. The assumption that proteins on the surface of rubber particles contribute to their stability is also supported by the fact that H. brasiliensis rubber particles become unstable when treated with proteolytic enzymes [36].
In summary, our data obtained by photon correlation spectroscopy of rubber particles suggest that TbSRPPs are involved in the stabilization of the rubber particle membrane. As a consequence, TbSRPPs-depleted rubber particles may not provide a suitable environment for the CPT and subsequently efficient IPP-polymerization. To gain deeper insight into the functional role of individual SRPPs in rubber biosynthesis, it will be necessary to generate transcript specific knock-down plants and to analyze their respective effect on rubber biosynthesis and rubber particle biogenesis as well as rubber particle architecture and stability.
Materials and Methods
Plant material and cultivation conditions
T. brevicorniculatum plants and seeds were obtained from the Botanical Gardens Karlsruhe (Germany) and cultivated in a greenhouse at 18°C and a 16 h photoperiod with 20 kilolux light intensity.
Preparation of T. brevicorniculatum rubber particles
Latex from T. brevicorniculatum was harvested as described previously [8]. The latex suspension was separated into three phases by centrifugation at 4°C for 5 min and 8,000×g. The rubber phase and the soluble phase were transferred to a new tube and centrifuged at 4°C for 15 min at 10,000xg. The soluble phase was isolated using a syringe and afterwards the remaining rubber particle suspension, if not stated otherwise, was diluted with an appropriate volume rubber extraction buffer (REB) [8]. All experiments described were carried out with freshly prepared rubber particles.
Two-dimensional gel electrophoresis of rubber particle proteins
Fifty microliter of rubber particle suspension was used for protein extraction using the ZOOM® 2D Protein Solubilizer 1 according to the manufacturer's recommendations (Invitrogen, Karlsruhe, Germany) with the following modification: Before rehydration, acetone precipitation was carried out to concentrate proteins. After solubilising proteins in rehydration buffer for at least 4 h, 50 µg protein (determined according to Bradford) [37] was loaded on pH 3–10 7 cm or pH 3–6 7 cm ReadyStripTM IPG strips (Biorad, Munich, Germany). The strips were rehydrated overnight and isoelectric focussing was carried out at 20°C and a maximum current of 50 µA using the following program: 300 V for 30 min, 1000 V for 30 min, 5000 V until 12 kVh were reached. Proteins were either stained with colloidal Coomassie Brilliant Blue (Fermentas, St. Leon-Rot, Germany) or transferred to nitrocellulose membranes, which were stained with 0.1% Ponceau-S in 5% acetic acid to confirm equal loading of proteins. Membranes were incubated with the anti-TbSRPP antibody (1∶250) for 1 h and a secondary mouse anti-rabbit IgG conjugated to horse radish peroxidase according to manufacturer's recommendation (Sigma, Schnelldorf, Germany). TbSRPPs were visualized on X-ray films by chemiluminescence detection.
Protein identification by mass spectrometrical analysis
Protein bands were excised from gels and subjected to in-gel trypsin digestion following the descriptions in Shevchenko et al. [38]. Subsequent LC-MS/MS was accomplished using an Ultimate 3000 system (Dionex, Sunnyvale, CA, USA) for Nano-LC and a LTQ Orbitrap XL mass spectrometer (Thermo, Bremen, Germany) [39]. The five most abundant precursor ions in the first MS were used for MS/MS fragmentations. MS/MS data was searched against a database created from all protein data available at the public NCBI non-redundant (nr) database (http://www.ncbi.nlm.nih.gov/, October 2009 status) using the OMSSA software (version 2.1.4) [40]. Mass tolerances were set to 0.02 Da for the precursor ions and to 0.5 Da for the product ions. An estimation of true protein identification hits was achieved by searching a concatenated database of target and decoy peptide mass information and defining an adaptive E-value threshold for all hits originating from one sample in such a way that the estimated false positive rate was ≤1% and by a mass accuracy requirement of 5 ppm.
Construction of expression vector for heterologous expression of TbSRPP3 and generation of antibodies against TbSRPPs
The full length cDNA encoding TbSRPP3 was amplified using the primer combination srpp3-EcoRI and srpp3-rev_NotI (Tab. S1). The cDNA was subsequently inserted into the pCRII-TOPO® vector (Invitrogen, Karlsruhe, Germany), sequenced and then transferred into the expression vector pGEX4-T1, using the EcoRI and NotI restriction sites (GE Healthcare Europe GmbH, Freiburg, Germany). This allowed the expression of TbSRPP3 in C-terminal fusion to glutathione-S-transferase (GST) for heterologous expression in Escherichia coli BL21-cells and subsequent antibody generation. The TbSRPP3 clone in pGEX4-T1 was expressed in 300-ml cultures of E. coli strain BL21, induced with 1 mM isopropyl-beta-D-thiogalactopyranoside (IPTG). The protein band containing heterologously expressed TbSRPP3 was excised from the gel, the eluted protein was sequenced by MALDI-MS and send to EUROGENTEC (Seraing, Belgium) for antibody production. The pre-immune and antibody sera were tested by western blot analysis against recombinant TbSRPP1-5 proteins.
Total RNA extraction from T. brevicorniculatum latex and cDNA synthesis
Extraction of total RNA from T. brevicorniculatum latex and cDNA synthesis was carried out as previously described [27], whereas random hexamer primer were used to initiate the reverse transcription reaction (Fermentas, St. Leon-Rot, Germany).
Quantitative real time PCR experiments
Quantitative real time PCR was carried out using an iCycler realtime PCR system (Biorad, Munich, Germany). 7.5 µl of serial cDNA dilutions was added to 12.5 µl IQTMSYBRGreen Supermix and 5 µl Primermix (50 pmol each). The primers used for the amplification of the TbSRPP transcripts are listed in table S2. The transcript of the housekeeping gene TbACTIN was used as a reference (Tab. S2). Melting curves were analyzed to exclude false amplifications. After determination of the Ct-values, the relative expression of the genes was calculated using the REST-MCS software [41].
Cloning of TbSRPP3-RNAi construct
For cloning of the TbSRPP3-RNAi construct, a 363 bp-fragment specific for all TbSRPP genes was obtained from TbSRPP3 cDNA (coordinates 270–632 of cDNA) by PCR using the primers TbSRPP3 RNAi KpnI fw and TbSRPP3 RNAi XhoI rev (Tab. S2). To avoid off-target suppression of genes, we validated that the chosen cDNA fragment gave no hits for other than TbSRPP1–5 homologs in the public available sequences and especially in EST datasets for the closely related plant species T. koksaghyz and T. officinale using the Basic Local Alignment Search Tool (BLAST; http://www.ncbi.nlm.nih.gov). The cDNA fragment was inserted into the KpnI and XhoI sites of the Gateway® pENTRTM4 and transferred into the pFGC5941 (www.chromDB.org) using the Gateway®LR Clonase enzyme mix (Invitrogen, Karlsruhe, Germany). The integrity of the construct was verified by sequencing.
Agrobacterium-mediated transformation of T. brevicorniculatum
Agrobacterium-mediated transformation was done as previously described [29] using the Agrobacterium tumefaciens strain EHA105 that has been transformed with the binary plasmid.
Determination of dry rubber content
To determine the dry rubber content in exuding latex, roots of T. brevicorniculatum plants were dissected with a razor blade and 20 µl of the expelling latex was transferred to 100 µl REB supplemented with 5 mM DTT. The samples were centrifuged at 4°C and 12000×g and overlaid with 100 µl methanol to coagulate rubber particles. The rubber coagulum was air-dried for 24 h and weighted. To determine the dry rubber content of whole roots, the freeze-dried entire roots were ground into a fine powder using a coffee grinder. For extraction, 200 mg root powder and 4 ml toluene were mixed thoroughly and incubated at 70°C for 18 h. The organic phase was transferred to a new glass vial and toluene was evaporated. The remaining material was dissolved in 300 µl toluene for 30 min and then 600 µl methanol was added to precipitate the rubber. After 30 min the samples were centrifuged at RT and 16,000×g for 2 min. The precipitated rubber was washed with water and acetone by vigorous shaking for 30 min at RT to remove proteins and resins. Subsequently the rubber samples were dried and weighted.
Size exclusion chromatography of T. brevicorniculatum rubber
Toluene was added to air-dried rubber samples to a final concentration of 0.75 mg ml−1 and dissolved at 60°C for 16 h. The samples were filtered with Cameo 3 N syringe (Nylon, 0.22 micron) filters (Roth, Karlsruhe, Germany) and transferred to new glass vials. Size exclusion chromatography (SEC)-analysis was carried out using a SECcurity GPC system with a column combination including a PSS SDV 20 µ 8.0×50 mm precolumn, a PSS SDV Linear XL 20 µ 8×300 nm analytical column and a PSS SDV 100 Å 20 µ 8×300 mm analytical column (Polymer Standards Service, Mainz, Germany).
[1-14C] IPP incorporation assay
The [1-14C] IPP incorporation rate of crude latex was determined as described previously [10].
Determination of rubber particle ζ-potential and z-average diameter
20 µl of rubber particle suspension was diluted in 1180 µl modified REB (350 mM Sorbitol, 5 mM MgCl, 50 mM NaCl, 40 mM NaH2PO4 titrated with 20 mM Citrate to the desired pH) to reach a minimal derived count rate of 250 kcps. One milliliter of the suspension was injected into a Malvern Clear Zeta Potential Cell. The Derjaguin-Landau-Verwey-Overbeek theory allows the quantification of the electrostatic stability of lyophobic colloids by measuring their ζ-potential (Verwey, 1947). The electrophoretic mobility of rubber particles was measured by Laser Doppler Velocimetry and was converted to ζ-potential by the Dispersion Technology Software (DTS) using Henrýs equation and the Smoluchowski approximation. The intensity weighted mean hydrodynamic diameter (z-average diameter) was measured by dynamic light scattering. The ζ-potential and z-average diameter of rubber particles at different pH-values were determined using a Zetasizer Nano ZS and DTS (Malvern Instruments Ltd., Worcestershire, United Kingdom).
Scanning electron microscopy
Rubber particles were fixed with 2.5% glutaraldehyde in 0.05 M sodium cacodylate buffer for 1 hour at room temperature. After 3 times of washing with 0.05 M sodium cacodylate buffer pH 7.3 (at 5,000×g), they were postfixed with 1% osmium tetroxide for 1 hour. Finally the particles were washed 3 times with 0.05 M sodium cacodylate buffer and subsequently 3 times with PBS (pH 7.4), each for 5 minutes.
For immunogold labeling, 10 µl aliquots of the suspensions were placed on a freshly glow-discharged carbon-layer (thickness ∼10 nm) onto a Pioloform film supported by a Cu-mesh grid, let sediment for 5 minutes, and washed with PBS (pH 7.4) for 10 minutes. The grids were then incubated in PBS buffer containing 5% bovine serum albumin (BSA) (pH 7.4) for 15 minutes, followed by 3 times washing with PBS (pH 7.4) each for 5 minutes. After this, the grids were incubated for 3 hours either with the primary anti-TbSRPP antibody (1∶5 in PBS, pH 7.4) or with the pre-immunserum (1∶5 in PBS, pH 7.4) as a negative control. Subsequently, grids were washed 5 times for 5 minutes with PBS (pH 7.7) and then 5 minutes with Tris-BSA-buffer (pH 8.2). After this the samples were incubated with a secondary goat-anti rabbit IgG antibody coupled to 10 nm colloidal gold particles (Sigma-Aldrich, Schnelldorf, Germany) diluted 1∶20 in Tris-BSA-buffer (pH 8.2) for 2 hours. Finally, samples were washed with Tris-BSA-buffer (pH 8.2) 6 times for 5 minutes and once with distilled water for 5 minutes. After air drying, the sample was covered by 2 nm platinum/carbon. Images were recorded by a field emission SEM S-5000 (Hitachi, Japan) at electron energy of 20 kV; both, secondary and backscattered electron signals were recorded.
Transmission electron microscopy
TEM of ultra-thin root cross sections was carried out as described previously [10].
Confocal laser scanning microscopy
Fifty microliters of freshly prepared rubber particles were supplemented with 1 µl nile red (15 µM solubilised in DMSO) and fluorescence was visualized by CLSM (excitation 514 nm, emission 509–620 nm, Leica DMIRE2, Wetzlar, Germany).
Supporting Information
Anti-TbSRPP antibody detects heterologously expressed TbSRPP1–5. A. Protein extracts from pellet and supernatant from E. coli cultures heterologously expressing TbSRPP1–5 proteins were separated by SDS-PAGE and subsequently stained with Coomassie Brilliant Blue. B. Protein extracts from pellet and supernatant from E. coli cultures heterologously expressing TbSRPP1–5 proteins were separated by SDS-PAGE and subsequently transferred to a nitrocellulose membrane. TbSRPP1–5 were detected using the anti-TbSRPP antibody and a secondary antibody coupled with alkaline phosphatase (1, TbSRPP1; 2, TbSRPP2; 3, TbSRPP3; 4, TbSRPP4; 5, TbSRPP5; P, pellet; S, supernatant).
(TIF)
Identification of TbSRPP2, TbSRPP3, TbSRPP4, and TbSRPP5 on T. brevicorniculatum rubber particles by mass spectrometric analysis.
(DOC)
Oligonucleotide sequences and respective assignments.
(DOC)
Materials and Methods section regarding experiments performed for Figure S1.
(DOC)
Acknowledgments
The technical assistance of Sandra Ponanta and Daniela Ahlert (Institute of Plant Biology and Biotechnology, Münster), Ulrike Keller and Ursula Malkus (Institute for Medical Physics and Biophysics, Münster) as well as Raphael Soeur (Fraunhofer Institute for Molecular Biology and Applied Ecology, Aachen) is kindly acknowledged. We thank Fabrice Brunel and Francisco M. Goycoolea (Institute of Plant Biology and Biotechnology, Münster) for their support concerning dynamic light scattering and size exclusion chromatography experiments.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a grant from the European Union Framework Program 7 (project no. 212827, EU-PEARLS), the Deutsche Bundesstiftung Umwelt (AZ13255) and the Ministry of Science and Education of Germany (grant no. FKZ 0315971C). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cornish K. Similarities and differences in rubber biochemistry among plant species. Phytochemistry. 2001;57:1123–1134. doi: 10.1016/s0031-9422(01)00097-8. [DOI] [PubMed] [Google Scholar]
- 2.Poulter CD, Rilling HC. Prenyltransferase – mechanism of reaction. Biochemistry. 1976;15:1079–1083. doi: 10.1021/bi00650a019. [DOI] [PubMed] [Google Scholar]
- 3.Poulter CD, Rilling HC. The prenyl transfer-reaction. Enzymatic and mechanistic studies of 1′-4 coupling reaction in the terpene biosynthetic-pathway. Accounts Chem Res. 1978;11:307–313. [Google Scholar]
- 4.Cornish K, Backhaus RA. Rubber transferase activity in rubber particles of guayule. Phytochemistry. 1990;29:3809–3813. [Google Scholar]
- 5.Kharel Y, Koyama T. Molecular analysis of cis-prenyl chain elongating enzymes. Nat Prod Rep. 2003;20:111–118. doi: 10.1039/b108934j. [DOI] [PubMed] [Google Scholar]
- 6.Archer BL, Audley BG, Cockbain EG, McSweeny GP. The biosynthesis of rubber – incorporation of mevalonate and isopentenyl pyrophosphate into rubber by Hevea brasiliensis -latex fractions. Biochem J. 1963;89:565–574. doi: 10.1042/bj0890565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siler DJ, Cornish K. A protein from Ficus elastica rubber particles is related to proteins from Hevea brasiliensis and Parthenium argentatum. Phytochemistry. 1993;32:1097–1102. [Google Scholar]
- 8.Schmidt T, Lenders M, Hillebrand A, van Deenen N, Munt O, et al. Characterization of rubber particles and rubber chain elongation in Taraxacum koksaghyz. BMC Biochem. 2010;11:11. doi: 10.1186/1471-2091-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirschner J, Štěpánek J, Černý T, De Heer P, van Dijk PJ. Available ex situ germplasm of the potential rubber crop Taraxacum koksaghyz belongs to a poor rubber producer, T. brevicorniculatum (Compositae-Crepidinae). Genet Resour Crop Evol. 2012. DOI: 10.1007/s10722-012-9848-0.
- 10.Post J, van Deenen N, Fricke J, Kowalski N, Wurbs D, et al. Laticifer-Specific cis-Prenyltransferase Silencing Affects the Rubber, Triterpene, and Inulin Content of Taraxacum brevicorniculatum. Plant Physiol. 2012;158:1406–1417. doi: 10.1104/pp.111.187880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomez JB, Hamzah S. Particle size distribution in Hevea latex – some observations on the electron microscopic method. J Nat Rubber Res. 1989;4:204–211. [Google Scholar]
- 12.Cornish K, Wood DF, Windle JJ. Rubber particles from four different species, examined by transmission electron microscopy and electronparamagnetic-resonance spin labeling, are found to consist of a homogeneous rubber core enclosed by a contiguous, monolayer biomembrane. Planta. 1999;210:85–96. doi: 10.1007/s004250050657. [DOI] [PubMed] [Google Scholar]
- 13.Singh AP, Wi SG, Chung GC, Kim YS, Kang H. The micromorphology and protein characterization of rubber particles in Ficus carica, Ficus benghalensis and Hevea brasiliensis. J Exp Bot. 2003;54:985–992. doi: 10.1093/jxb/erg107. [DOI] [PubMed] [Google Scholar]
- 14.Siler DJ, Goodrich-Tanrikulu M, Cornish K, Stafford AE, McKeon TA. Composition of rubber particles of Hevea brasiliensis, Parthenium argentatum, Ficus elastica, and Euphorbia lactiflua indicates unconventional surface structure. Plant Physiol Bioch. 1997;35:881–889. [Google Scholar]
- 15.Wood DF, Cornish K. Microstructure of purified rubber particles. Int J Plant Sci. 2000;161:435–445. doi: 10.1086/314269. [DOI] [PubMed] [Google Scholar]
- 16.Verwey EJW. Theory of the stability of lyophobic colloids. J Phys Chem. 1947;51:631–636. doi: 10.1021/j150453a001. [DOI] [PubMed] [Google Scholar]
- 17.Heller W, Pugh TL. “Steric” Stabilization of Colloidal Solutions by Adsorption of Flexible Macromolecules. J Polym Sci. 1960;47:203–217. [Google Scholar]
- 18.Riddick TM. Zeta-Meter Manual. Zeter Meter inc. New York. 1968.
- 19.Jucker BA, Zehnder AJB, Harms H. Quantification of Polymer Interactions in Bacterial Adhesion. Environ Sci Technol. 1998;32:2909–2915. [Google Scholar]
- 20.Giasson S, Kuhl TL, Israelachvili JN. Adsorption and Interaction Forces of Micellar and Microemulsion Solutions in Ultrathin Films. Langmuir. 1998;14:891–898. [Google Scholar]
- 21.Freitas AM, Sharma MM. Detachment of Particles from Surfaces: An AFM Study. J Colloid Interface Sci. 2001;233:73–82. doi: 10.1006/jcis.2000.7218. [DOI] [PubMed] [Google Scholar]
- 22.Grasso D, Subramaniam K, Butkus M, Strevett K, Bergendahl J. A review of non-DLVO interactions in environmental colloidal systems. Rev Environ Sci Biotechnol. 2002;1:17–38. [Google Scholar]
- 23.Dennis MS, Henzel WJ, Bell J, Kohr W, Light DR. Amino acid sequence of rubber elongation factor protein associated with rubber particles in Hevea Latex. J Biol Chem. 1989;264:18618–18626. [PubMed] [Google Scholar]
- 24.Dennis MS, Light DR. Rubber elongation factor from Hevea brasiliensis. J Biol Chem. 1989;264:18608–1861. [PubMed] [Google Scholar]
- 25.Oh SK, Kang H, Shin DH, Yang J, Chow K-S, et al. Isolation, Characterization, and Functional Analysis of a Novel cDNA Clone Encoding a Small Rubber Particle Protein from Hevea brasiliensis. J Biol Chem. 1999;274:17132–17138. doi: 10.1074/jbc.274.24.17132. [DOI] [PubMed] [Google Scholar]
- 26.Kim IJ, Ryu SB, Kwak YS, Kang H. A novel cDNA from Parthenium argentatum Gray enhances the rubber biosynthetic activity in vitro. J Exp Bot. 2004;55:377–385. doi: 10.1093/jxb/erh039. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt T, Hillebrand A, Wurbs D, Wahler D, Lenders M, et al. Molecular cloning and characterization of rubber biosynthetic genes from Taraxacum koksaghyz. Plant Mol Biol Rep. 2010;28:277–284. [Google Scholar]
- 28.Priya P, Venkatachalan P, Thulaseedharan A. Differential expression pattern of rubber elongation factor (REF) mRNA transcripts from high and low yielding clones of rubber tree (Hevea brasiliensis Muell. Arg.). Plant Cell Rep. 2007;26:1833–1838. doi: 10.1007/s00299-007-0402-z. [DOI] [PubMed] [Google Scholar]
- 29.Wahler D, Schulze Gronover C, Richter C, Foucu F, Twyman RM, et al. Polyphenoloxidase Silencing Affects Latex Coagulation in Taraxacum Species. Plant Physiol. 2009;151:334–346. doi: 10.1104/pp.109.138743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wahler D, Colby T, Kowalski NA, Harzen A, Wotzka SY, et al. Proteomic analysis of latex from the rubber-producing plant Taraxacum brevicorniculatum. Proteomics. 2012;12:901–905. doi: 10.1002/pmic.201000778. [DOI] [PubMed] [Google Scholar]
- 31.Sando T, Hayashi T, Takeda T, Akiyama Y, Nakazawa Y, et al. Histochemical study of detailed laticifer structure and rubber biosynthesis-related protein localization in Hevea brasiliensis using spectral confocal laser scanning microscopy. Planta. 2009;230:215–225. doi: 10.1007/s00425-009-0936-0. [DOI] [PubMed] [Google Scholar]
- 32.Ho CC. Changes in electrokinetic properties of natural rubber latex after surface chemical modifications. Colloid Polym Sci. 1989;267:643–647. [Google Scholar]
- 33.Genovese DB, Lozano JE. The effect of hydrocolloids on the stability and viscosity of cloudy apple juices. Food Hydrocolloid. 2001;15:1–7. [Google Scholar]
- 34.Tzen JTC, Huang AHC. Surface Structure and Properties of Plant Seed Oil Bodies. J Cell Biol. 1992;117:327–335. doi: 10.1083/jcb.117.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng C-C, Lin I-P, Lin C-K, Tzen JTC. Size and Stability of Reconstituted Sesame Oil Bodies. Biotechnol. 2003;Progr19:1623–1626. doi: 10.1021/bp034129z. [DOI] [PubMed] [Google Scholar]
- 36.Sansatsadeekul J, Sakdapipanich J, Rojruthai P. Characterization of associated proteins and phospholipids in natural rubber latex. J Biosci Bioeng. 2011;111:628–634. doi: 10.1016/j.jbiosc.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 37.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. 1976;Biochem72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 38.Shevchenko A, Tomas H, Havli J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2007;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 39.Naumann B, Stauber EJ, Busch A, Sommer F, Hippler M. N-terminal processing of Lhca3 is a key step in remodeling of the photosystem I-light-harvesting complex under iron deficiency in Chlamydomonas reinhardtii. J Biol Chem. 2005;280:20431–20441. doi: 10.1074/jbc.M414486200. [DOI] [PubMed] [Google Scholar]
- 40.Geer LY, Markey SP, Kowalak J A, Wagner L, Xu M, et al. Open mass spectrometry search algorithm. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 41.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acid Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Anti-TbSRPP antibody detects heterologously expressed TbSRPP1–5. A. Protein extracts from pellet and supernatant from E. coli cultures heterologously expressing TbSRPP1–5 proteins were separated by SDS-PAGE and subsequently stained with Coomassie Brilliant Blue. B. Protein extracts from pellet and supernatant from E. coli cultures heterologously expressing TbSRPP1–5 proteins were separated by SDS-PAGE and subsequently transferred to a nitrocellulose membrane. TbSRPP1–5 were detected using the anti-TbSRPP antibody and a secondary antibody coupled with alkaline phosphatase (1, TbSRPP1; 2, TbSRPP2; 3, TbSRPP3; 4, TbSRPP4; 5, TbSRPP5; P, pellet; S, supernatant).
(TIF)
Identification of TbSRPP2, TbSRPP3, TbSRPP4, and TbSRPP5 on T. brevicorniculatum rubber particles by mass spectrometric analysis.
(DOC)
Oligonucleotide sequences and respective assignments.
(DOC)
Materials and Methods section regarding experiments performed for Figure S1.
(DOC)