Abstract
Several genome-wide association studies (GWAS) have been conducted to identify the common single nucleotide polymorphisms (SNPs) that influence the risk of prostate cancer. It was hypothesized that some prostate cancer-associated SNPs might relate to the clinical outcomes in patients treated for prostate cancer using androgen-deprivation therapy (ADT). A cohort of 601 patients who have received ADT for prostate cancer was genotyped for 29 SNPs that have been associated with prostate cancer in Cancer Genetic Markers of Susceptibility GWAS, and within the genes that have been implicated in cancer. Prognostic significance of these SNPs on the disease progression, prostate cancer-specific mortality (PCSM) and all-cause mortality (ACM) after ADT were assessed by Kaplan-Meier analysis and Cox regression model. Three SNPs, namely CASP3 rs4862396, BMP5 rs3734444 and IRS2 rs7986346, were found to be closely associated with the ACM (P≤0.042), and BMP5 rs3734444 and IRS2 rs7986346 were also noted to be significantly related to the PCSM (P≤0.032) after adjusting for the known clinicopathologic predictors. Moreover, patients carrying a greater number of unfavorable genotypes at the loci of interest had a shorter time to ACM and PCSM during ADT (P for trend <0.001). Our results suggest that CASP3 rs4862396, BMP5 rs3734444 and IRS2 rs7986346 may affect the survival in patients after ADT for prostate cancer, and the analysis of these SNPs can help identify patients at higher risk of poor outcome.
Introduction
Prostate cancer, the most frequently diagnosed cancer, is the second leading cause of cancer-related deaths among men in Western countries [1], [2]. Since it was first introduced more than 70 years ago, an androgen-deprivation therapy (ADT) has been the mainstay of treatment for advanced prostate cancer based on that the androgen signaling has been considered the main oncogenic driver in prostate carcinogenesis. The initial response rate of prostate cancer to ADT has been reported to be up to 80%, however, the disease in most of treated patients have progressed toward castration-resistant prostate cancer (CRPC). Unfortunately, CRPC is still incurable and the median survival for patients with CRPC is only 1–2 years. Nevertheless, poor understanding of the molecular mechanisms underlying CRPC might be the constraint on development of efficient therapy.
Genetic biomarkers have been demonstrated to have potential for enabling application of more effective personalized diagnosis, prognosis and treatment in clinics. Recently, millions of single nucleotide polymorphisms (SNPs) have been identified to be associated with the risk of prostate cancer in several genome-wide association studies, such as the Cancer Genetic Markers of Susceptibility (CGEMS) study [3]–[14]. Nonetheless, the prognostic value of the prostate cancer-associated variants has not been well documented.
The purpose of the current study was to investigate the prognostic significance of 29 SNPs that were associated with genes implicated in cancer progression and had low P values (P<0.01) in CGEMS (Table S1) for disease progression, prostate cancer-specific mortality (PCSM) and all-cause mortality (ACM) in a cohort of 601 patients treated with ADT for prostate cancer.
Methods
Patient recruitment and data collection
Patients with diagnosed and pathologically confirmed prostate cancer were actively recruited from three medical centers in Taiwan: Kaohsiung Medical University Hospital and Kaohsiung Veterans General Hospital in southern Taiwan, and National Taiwan University Hospital in northern Taiwan. Written informed consent was obtained from each participant and permission to conduct this study was provided by the Institutional Review Board of the three hospitals. Collection of the clinical data and patient characteristics described as previous study [15]–[19] are reported in Method S1.
Genotyping
Genomic DNA was extracted from the peripheral blood of each patient and genotyping was performed as described previously [20] using Sequenom iPLEX matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry technology at the National Center for Genome Medicine, Academia Sinica, Taiwan. Briefly, primers for locus-specific polymerase chain reaction (PCR) and allele-specific extension were designed by MassARRAY AssayDesign 3.0 software (Sequenom, San Diego, CA, USA). For primer sequences, see Table S2. The sample DNAs were amplified by primers flanking the targeted sequence, followed by dephosphorylation and allele-specific primer extension. The extension products were purified, loaded into a 384-format SpectroChip, and subjected to MALDI-TOF mass spectrometry. The resulting data were analyzed by the Sequenom MassARRAY TYPER software (Sequenom, San Diego, CA, USA). The average genotype call rate for these SNPs was 99.3% and the concordance rate was 99.8% among 55 blind duplicated quality control samples. Each of the SNPs was in Hardy-Weinberg equilibrium (P>0.01).
Real-time reverse transcription-PCR (real-time RT-PCR)
The human prostate cell lines, namely LNCaP, DU 145 and PC-3, were purchased from Bioresource Collection and Research Center (Hsinchu, Taiwan) and maintained in RPMI 1640 with 10% fetal bovine serum, 100 U/ml penicillin and 100 U/ml streptomycin (Invitrogen, Carlsbad, CA, USA) at 37°C in a humidified 5% CO2-95% air atmosphere. Total RNA was extracted from LNCaP, DU 145 and PC-3 cells using Trizol (Invitrogen, Carlsbad, CA, USA). Reverse transcription with the High Capacity cDNA Reverse Transcription Kit (ABI, Foster city, CA, USA) and PCR amplifications with Smart Quant Green Master Mix (Protech, Taipei, Taiwan) were carried out on an MJ mini and MiniOpticon real-time PCR detection system (Bio-Rad, Hercules, CA, USA). PCR was performed as the following sequence: initial HotStart activation at 95°C for 15 min, and 40 cycles of denaturation at 95°C for 15 s, annealing and extension at 60°C for 1 min. Primer sequences were: caspase 3 (CASP3), sense 5′- GACATACTCCTTCCATCAA-3′ and antisense 5′- ATTCATAGCACAGCATCA-3′; bone morphogenetic protein 5 (BMP5), sense 5′- CCGTCTTCTGCTACATCA-3′ and antisense 5′- ACAACATCCTCACCGATT-3′; insulin receptor substrate 2 (IRS2), sense 5′- CAGTGTATTGACGCATAT-3′ and antisense 5′- AGCATATTATCATCTGTGTA-3′; Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), sense 5′-TCACCACCATGGAGAAGGC-3′ and antisense 5′-GCTAAGCAGTTGGTGGTGCA-3′. Quantification of each sample relative to the LNCaP sample was calculated using 2−ΔΔCT method [21]. The expected sizes and the absence of non-specific amplification products were confirmed by agarose gel electrophoresis and melting curve analysis.
Statistical analysis
Patient's clinicopathologic characteristics were summarized as number and percentage of patients, median, or interquartile range (IQR) of values. The continuous factors were dichotomized at the median value within the cohort, with the exception of prostate-specific antigen (PSA) nadir that was dichotomized at 0.2 ng/mL because of its correlation with disease progression and PCSM [22], [23]. The heterozygous and rare homozygous genotypes were collapsed in the analysis if the frequency of the rare homozygote was too low (<2%), or if the homozygous and heterozygous genotypes had the same direction of effect. The associations of 29 individual SNPs and clinicopathologic characteristics with time to progression, PCSM and ACM were assessed using the Kaplan-Meier analysis with log-rank test. Multivariate analyses to determine the interdependency of genotypes and other known prognostic factors, such as age at diagnosis, clinical stage, Gleason score, PSA at ADT initiation, PSA nadir and time to PSA nadir, were carried out using Cox proportional hazards regression model. As the 29 SNPs were tested, the false-discovery rates (q values) were calculated to determine the degree to which tests for association were prone to false-positives [24]. q values were estimated using R q value package (http://genomics.princeton.edu/storeylab/qvalue/) on the observed distribution of P values from the log-rank test for 29 SNPs. Statistical Package for the Social Sciences software version 16.0.1 (SPSS Inc., Chicago, IL) was used for other statistical analyses. A two-sided P value of ≤0.05 was considered statistically significant.
Results
The study population (N = 601) is derived from a previously described cohort of prostate cancer patients treated with ADT and its clinicopathologic characteristics are presented in Table S3. Clinical outcomes following ADT were measured by time to disease progression, PCSM and ACM. The clinical stage at diagnosis, PSA nadir and time to PSA nadir during ADT were significantly associated with all three clinical outcomes after ADT (P≤0.019). While age at diagnosis was only associated with ACM (P = 0.008), Gleason score at diagnosis and PSA level at ADT initiation were both associated with time to PCSM and ACM (P<0.001), but not with time to progression.
A total of 29 SNPs that were associated with genes implicated in cancer and had low P values (P<0.01) in CGEMS, were selected. Their associations with disease progression, PCSM and ACM after ADT were summarized in Table S1. Our primary log-rank tests revealed that BMP5 rs3734444, NCOR2 rs10846667, IRS2 rs7986346 and MAP2K6 rs1972933 significantly related to disease progression (nominal P≤0.043) with a false-discovery rate (q value) of 0.085 (Table 1). To evaluate the prognostic value of these SNPs beyond the currently used clinical factors, a multivariate Cox proportional hazards analysis adjusting for age, clinical stage, Gleason score at diagnosis, PSA nadir, time to PSA nadir and PSA at ADT initiation was performed. After adjusting for these predictors, no statistical association was observed between any SNPs and disease progression (P≥0.084, Table 1).
Table 1. Genotyping frequencies and the association of genotype with disease progression during ADT.
| Gene SNP | Genotype | No. of Patients | No. of Events | Median (months) | P * | q | HR (95% CI) | P † |
| BMP5 rs3734444 | AA+AG | 572 | 394 | 23 | 0.022 | 0.085 | 1.00 | |
| GG | 21 | 17 | 15 | 1.56 (0.94–2.60) | 0.087 | |||
| NCOR2 rs10846667 | CC | 395 | 266 | 24 | 0.011 | 0.085 | 1.00 | |
| CT+TT | 192 | 142 | 18 | 1.16 (0.93–1.43) | 0.184 | |||
| IRS2 rs7986346 | TT+TG | 512 | 355 | 21 | 0.040 | 0.085 | 1.00 | |
| GG | 71 | 48 | 32 | 0.76 (0.56–1.04) | 0.084 | |||
| MAP2K6 rs1972933 | GG | 208 | 141 | 25 | 0.043 | 0.085 | 1.00 | |
| GT+TT | 387 | 272 | 21 | 1.19 (0.97–1.48) | 0.102 |
Abbreviations: ADT, androgen-deprivation therapy; HR, hazard ratio; 95% CI, 95% confidence interval; PSA, prostate-specific antigen.
P values were calculated using the log-rank test.
HRs were adjusted for age, clinical stage, Gleason score, PSA at ADT initiation, PSA nadir, and time to PSA nadir.
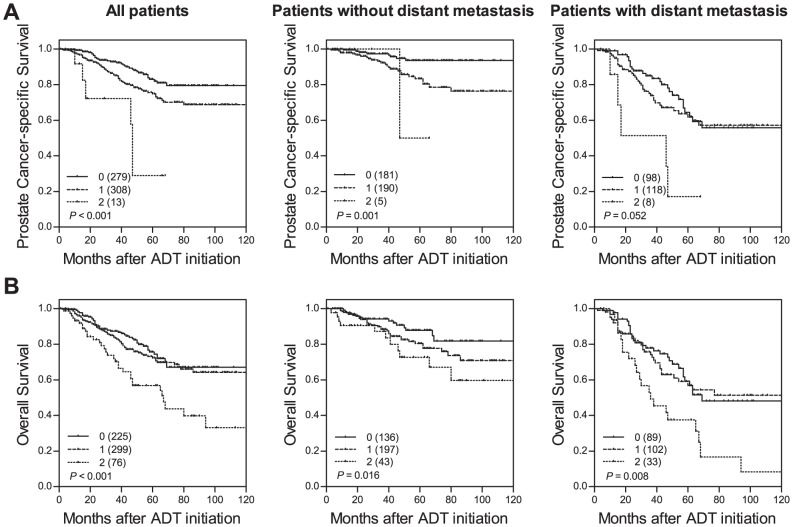
BMP5 rs373444, RXRA rs3118536, IRS2 rs7986346 and ERG rs2836370 were associated with PCSM with nominal P≤0.043 and q≤0.209 (Table 2). Six SNPs, namely CASP3 rs4862396, BMP5 rs373444, RXRA rs3118536, BMPR1A rs11597689, IRS2 rs7986346 and ERG rs2836370, also exhibited significant influence on ACM (nominal P≤0.037), and all had a q value of ≤0.179 (Table 3). After adjusting for clinical predictors, BMP5 rs3734444 and IRS2 rs7986346 remained significant association with both PCSM (adjusted P≤0.032, Table 2) and ACM (adjusted P≤0.034, Table 3). A strong gene-dosage effect on PCSM during ADT was noted when BMP5 rs3734444 and IRS2 rs7986346 were analyzed in combination (log-rank P<0.001, Table 2 and Figure 1A left), and the hazard ratios (HRs) increased as the number of unfavorable genotypes increased. In addition, the association of CASP3 rs4862396 with time to ACM also remained significant (adjusted P = 0.042, Table 3) when other risk factors were included in the multivariate analysis. Estimated time to ACM decreased progressively for patients with increasing number of unfavorable genotypes for BMP5 rs3734444, IRS2 rs7986346 and CASP3 rs4862396 (Table 3 and Figure 1B left). The risk of ACM increased in patients with 1 unfavorable genotype (HR 1.43, 95% confidence interval [CI] 0.97–2.11) and significantly increased for patients with >1 unfavorable genotypes (HR 2.48, 95% CI 1.54–3.98) when compared to patients without unfavorable genotypes (P for trend <0.001, Table 3).
Table 2. Genotyping frequencies and the association of genotype with PCSM during ADT.
| Gene SNP | Genotype | No. of Patients | No. of Events | Estimated Mean (months) | P * | q | HR (95% CI) | P † |
| BMP5 rs3734444 | AA+AG | 575 | 93 | 135 | 0.002 | 0.039 | 1.00 | |
| GG | 21 | 8 | 96 | 2.28 (1.08–4.82) | 0.032 | |||
| RXRA rs3118536 | CC | 392 | 75 | 130 | 0.043 | 0.209 | 1.00 | |
| CA+AA | 204 | 25 | 148 | 0.74 (0.47–1.16) | 0.189 | |||
| IRS2 rs7986346 | TT | 272 | 35 | 147 | 0.007 | 0.068 | 1.00 | |
| TG+GG | 313 | 65 | 122 | 1.90 (1.25–2.90) | 0.003 | |||
| ERG rs2836370 | TT+TC | 506 | 92 | 128 | 0.035 | 0.209 | 1.00 | |
| CC | 85 | 7 | 157 | 0.50 (0.23–1.08) | 0.079 | |||
| No. of Unfavorable Genotypes Present‡ | ||||||||
| 0 | 279 | 34 | 143 | <0.001 | 1.00 | |||
| 1 | 308 | 61 | 132 | 1.94 (1.26–2.99) | 0.003 | |||
| 2 | 13 | 6 | 44 | 3.90 (1.60–9.52) | 0.003 | |||
| P-trend | <0.001 | |||||||
Abbreviations: ADT, androgen-deprivation therapy; HR, hazard ratio; 95% CI, 95% confidence interval; PSA, prostate-specific antigen.
P values were calculated using the log-rank test.
HRs were adjusted for age, clinical stage, Gleason score, PSA at ADT initiation, PSA nadir, and time to PSA nadir.
Unfavorable genotypes refer to GG in BMP5 rs3734444 and TG+GG in IRS2 rs7986346.
P≤0.05 are in boldface.
Table 3. Genotyping frequencies and the association of genotype with ACM during ADT.
| Gene SNP | Genotype | No. of Patients | No. of Events | Estimated Mean (months) | P * | q | HR (95% CI) | P † |
| CASP3 rs4862396 | TT | 475 | 105 | 127 | 0.037 | 0.179 | 1.00 | |
| TC+CC | 123 | 39 | 102 | 1.49 (1.01–2.19) | 0.042 | |||
| BMP5 rs3734444 | AA+AG | 575 | 136 | 120 | 0.014 | 0.138 | 1.00 | |
| GG | 21 | 9 | 80 | 2.13 (1.06–4.28) | 0.034 | |||
| RXRA rs3118536 | CC | 392 | 108 | 114 | 0.016 | 0.138 | 1.00 | |
| CA+AA | 204 | 36 | 134 | 0.68 (0.47–1.01) | 0.054 | |||
| BMPR1A rs11597689 | GG | 523 | 134 | 116 | 0.028 | 0.162 | 1.00 | |
| GA+AA | 76 | 11 | 146 | 0.64 (0.34–1.21) | 0.172 | |||
| IRS2 rs7986346 | TT | 272 | 54 | 133 | 0.017 | 0.138 | 1.00 | |
| TG+GG | 313 | 87 | 109 | 1.63 (1.15–2.32) | 0.006 | |||
| ERG rs2836370 | TT+TC | 506 | 132 | 123 | 0.019 | 0.138 | 1.00 | |
| CC | 85 | 11 | 149 | 0.55 (0.30–1.03) | 0.061 | |||
| No. of Unfavorable Genotypes Present‡ | ||||||||
| 0 | 225 | 45 | 128 | <0.001 | 1.00 | |||
| 1 | 299 | 68 | 126 | 1.43 (0.97–2.11) | 0.069 | |||
| >1 | 76 | 32 | 84 | 2.48 (1.54–3.98) | <0.001 | |||
| P-trend | <0.001 | |||||||
Abbreviations: ADT, androgen-deprivation therapy; HR, hazard ratio; 95% CI, 95% confidence interval; PSA, prostate-specific antigen.
P values were calculated using the log-rank test.
HRs were adjusted for age, clinical stage, Gleason score, PSA at ADT initiation, PSA nadir, and time to PSA nadir.
Unfavorable genotypes refer to TC+CC in CASP3 rs4862396, GG in BMP5 rs3734444, and TG+GG in IRS2 rs7986346.
P≤0.05 are in boldface.
Figure 1. The influence of the genetic loci of interest on prostate cancer prognosis.
Kaplan-Meier curves of (A) time to PCSM during ADT for patients with 0, 1, or 2 unfavorable genotypes at the 2 genetic loci of interest, and (B) time to ACM during ADT for patients with 0, 1, or >1 unfavorable genotypes at the 3 genetic loci of interest; measured in all patients (left), in patients without distant metastasis (middle), or in patients with distant metastases (right). Numbers in parentheses indicate the number of patients.
Since patients with distant metastasis are considered high risk, patients were further stratified based on their metastatic status at the initiation of ADT for evaluating the clinical relevance of these SNPs. The combined genotypes still had effects on PCSM and ACM in patients with or without distant metastasis, respectively (P≤0.052, Figure 1 middle and right). These results support that these SNPs could be the independent survival predictors following ADT, along with the current clinicopathologic prognostic markers. Integration of these SNPs with the known predictors may improve risk stratification and help making treatment decisions.
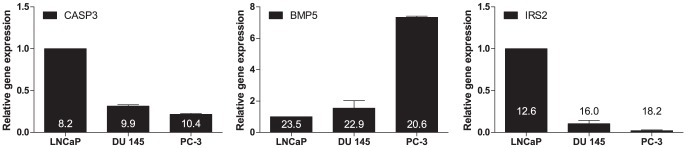
To gain an initial indication of these candidate genes in prostate cancer, the expression levels of CASP3, BMP5 and IRS2 in three most commonly used human prostate cancer cell lines, namely LNCaP, DU 145 and PC-3, were examined using real-time RT-PCR (Figure 2). It has been well established that LNCaP cells express androgen receptor (AR) and are androgen-sensitive with markedly less invasive potential [25]. DU 145 and PC-3 cells, on the other hand, do not express AR and are androgen-insensitive with highly aggressive behavior [26]. mRNA for all three genes were able to be detected in all prostate cancer cell lines. CASP3 is one of the main executors of apoptosis. The expression of endogenous CASP3 transcripts was inversely correlated to the aggressiveness in human prostate cancer cell lines, showing that it was higher in LNCaP than in DU 145 and PC-3. Elevated expression of BMPs has been implicated in prostate cancer, particularly in the disease-specific bone-metastasis [27]. A positive correlation was observed between the metastatic potential of these prostate cancer cell lines and BMP5 expression. Although IRS2 has been positively associated with aggressive tumor behavior [28], it has also been identified as a primary target gene of AR [29]. The endogenous IRS2 expressed higher levels in AR-positive LNCaP cells than in AR-negative DU 145 and PC-3 cells. All of these independent lines of evidence support the biological plausibility of our suggestion that the risk variants of prostate cancer may also affect the survival in patients treated with ADT for prostate cancer.
Figure 2. The mRNA expressions of endogenous CASP3, BMP5 and IRS2 in human prostate cancer cell lines.
Total RNAs were prepared from LNCaP, DU 145 and PC-3 cells, and gene expressions were analyzed using real-time RT-PCR. Relative gene expression represents the fold changes in gene expression relative to LNCaP cells set at 1.0. Numbers on the bars represent the difference in threshold cycles between genes of interest and internal control gene. Data are expressed as the mean ± SE of three independent experiments.
Discussion
This study aimed to investigate the prognostic effect of 29 prostate cancer-associated SNPs selected from CGEMS in patients receiving ADT for prostate cancer. Our results revealed that CASP3 rs4862396, BMP5 rs3734444 and IRS2 rs7986346 were significantly associated with ACM, and BMP5 rs3734444 and IRS2 rs7986346 were also significantly related to PCSM. In addition, multivariate analysis showed that these SNPs were independent prognostic factors after adjusting for the known clinicopathologic predictors, including age, clinical stage, Gleason score at diagnosis, PSA nadir, time to PSA nadir and PSA at ADT initiation. It suggested that CASP3 rs4862396, BMP5 rs3734444 and IRS2 rs7986346 may be useful markers for predicting the survival in patients receiving ADT for prostate cancer. To the best of our knowledge, this is the first study to determine whether the genetic variants in CASP3, BMP5 and IRS genes have prognostic effects on the clinical outcomes of patients with prostate cancer.
CASP3 encodes a member of the caspase family, which is responsible for apoptosis execution. Once CASP3 is proteolytically activated by the initiator caspases during death receptor- or DNA-damage-induced apoptosis, it cleaves a large set of substrates and results in the disassembly of the cell. Although the molecular mechanisms by which prostate cancer cells survive under ADT are unknown, dysregulation of survival/apoptosis proteins has been suspected to render cancer cells less prone to cell death induced by androgen deprivation [30]. rs4862396 resides 5-kb downstream of the CASP3 gene. Although the function predicted for rs4862396 has not been known, several untyped CASP3 SNPs are tightly linked (r 2>0.9) with rs4862396. According to the prediction of SNP Function Portal [31], many of the linked SNPs are able to alter binding sites for transcription factors (i.e. rs7675251 in hepatocyte nuclear factor 1 binding site), target sites for microRNAs (i.e. rs8549 in the target site of hsa-miR-196a), and amino acid sequence (i.e. rs1049210 leading to E189D). Therefore, further fine mapping of CASP3 gene might help the identification of the strongest markers and improve our understanding of the contribution of CASP3 rs4862396 to prostate cancer progression.
BMP5 encodes a member of the BMP family which belongs to the transforming growth factor-β superfamily. Since BMPs are potent regulators for bone homeostasis and prostate cancer is most likely to metastasize to the bone, there is an increasing interest to investigate the roles of BMPs in prostate cancer bone metastasis. The expression levels of several BMPs in prostate cancer have been linked with the acquisition of osteogenic characteristics and the tumor progression to bone [32]–[34]. Furthermore, amplifications of the BMP5 gene loci appeared to be more prevalent in tumors with a higher Gleason score and occurred in 50% of prostate tumors, which might account for the abnormal gene expression patterns of BMP5 in prostate cancer [27]. rs3734444 is located in the exon 1 of the BMP5 gene. Although rs3734444 is a synonymous SNP and does not affect amino acid sequence, it is predicted to alter binding sites for the transcription factor CP2 and myocyte enhancer factor 2A. In addition, rs3734444 is also predicted to locate at the binding sites for exonic splicing silencer and enhancer, which play important roles in constitutive and alternative splicing. Therefore, it is possible that rs3734444 might directly or indirectly, through other linked SNPs, influence BMP5 expression, interaction between prostate cancer cells and the bone microenvironment, and ultimately progression to bone metastasis.
IRS2 encodes a member of a family of intracellular signaling adaptor proteins that coordinate numerous biologically key extracellular signals within the cell, including insulin, insulin-like growth factor 1 (IGF1), hormones, cytokines and integrins. Alterations in IRS expression have been linked to not only metabolic diseases but also many types of cancer [35]–[37]. Upon ligand-binding, insulin receptor and IGF1 receptor are autophosphorylated and present docking sites for IRSs, which are phosphorylated by the receptor tyrosine kinase. The phospho-IRSs recruit and activate many downstream pathways: the two best studied being the phosphatidylinositol 3-kinase (PI3K)/phosphatase and tensin homolog (PTEN) and the extracellular signal-regulated kinase pathways. Deletion of IRS2 has been found to suppress prostate cancer cell growth, proliferation and invasion in PTEN +/− mice by decreasing MYC expression and DNA synthesis [28]. While ADT controls the prostate cancer growth, it is also associated with a pattern of metabolic alterations, such as hyperinsulinaemia [38]. These high circulating levels of serum insulin has been shown to act directly on CRPC cells and to enhance de novo androgen synthesis through upregulation of IRS2 and several downstream steroidogenic enzymes [39]. rs7986346 located at 15-kb upstream of the IRS2 gene was predicted to alter CCAAT/enhancer binding protein γ by SNP Function Portal. Therefore, rs7986346 might regulate IRS2 expression and affect prostate cancer progression to castrate resistance.
In conclusion, this study lights up several pathways to influence the survival after ADT, such as CASP3 in apoptosis, BMP5 in bone homeostasis and IRS2 in steroidogenesis, and the cell survival. CASP3 rs4862396, BMP5 rs3734444 and IRS2 rs7986346 were found to be the independent prognostic markers for patients receiving ADT for prostate cancer. It suggest that, in addition to the currently used clinicopathologic predictors, test of the three SNPs might help identify the patient subgroups at higher risk for poor outcome, thus assisting in tailoring individual therapeutic interventions for advanced prostate cancer. However, this study is still limited by sample size in subset analyses and multiple comparisons. In addition, our homogeneous Chinese Han population may make our findings less generalizable to other ethnic groups. Further functional analyses and large independent studies in other ethnic populations are necessary to validate the relevance of the observed associations to the efficacy of ADT for prostate cancer.
Supporting Information
Patient recruitment and data collection.
(DOC)
Genotyped SNPs and the P values of their association with time to progression, PCSM, ACM during ADT.
(DOC)
Oligonucleotides used for genotyping analysis.
(DOC)
Distribution of clinicopathologic characteristics and their associations with disease progression, PCSM, and ACM in prostate cancer patients receiving ADT.
(DOC)
Acknowledgments
The authors gratefully acknowledge the technical support from the National Center for Genome Medicine, National Science Council (NSC) of Taiwan.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the NSC grants NSC-98-2320-B-039-019-MY3, NSC-99-2314-B-037-018-MY3, NSC-100-2314-B-039-009-MY3, and China Medical University grant CMU99-COL-13. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.La Vecchia C, Bosetti C, Lucchini F, Bertuccio P, Negri E, et al. Cancer mortality in Europe, 2000–2004, and an overview of trends since 1975. Ann Oncol. 2010;21:1323–1360. doi: 10.1093/annonc/mdp530. [DOI] [PubMed] [Google Scholar]
- 3.Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet. 2009;41:1058–1060. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 4.Eeles RA, Kote-Jarai Z, Al Olama AA, Giles GG, Guy M, et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2009;41:1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 6.Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 8.Gudmundsson J, Sulem P, Rafnar T, Bergthorsson JT, Manolescu A, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008;40:281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haiman CA, Chen GK, Blot WJ, Strom SS, Berndt SI, et al. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat Genet. 2011;43:570–573. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kote-Jarai Z, Olama AA, Giles GG, Severi G, Schleutker J, et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet. 2011;43:785–791. doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takata R, Akamatsu S, Kubo M, Takahashi A, Hosono N, et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat Genet. 2010;42:751–754. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- 12.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 13.Yeager M, Chatterjee N, Ciampa J, Jacobs KB, Gonzalez-Bosquet J, et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet. 2009;41:1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 15.Bao BY, Pao JB, Huang CN, Pu YS, Chang TY, et al. Significant associations of prostate cancer susceptibility variants with survival in patients treated with androgen-deprivation therapy. Int J Cancer. 2012;130:876–884. doi: 10.1002/ijc.26091. [DOI] [PubMed] [Google Scholar]
- 16.Bao BY, Pao JB, Huang CN, Pu YS, Chang TY, et al. Polymorphisms inside microRNAs and microRNA target sites predict clinical outcomes in prostate cancer patients receiving androgen-deprivation therapy. Clin Cancer Res. 2011;17:928–936. doi: 10.1158/1078-0432.CCR-10-2648. [DOI] [PubMed] [Google Scholar]
- 17.Huang CN, Huang SP, Pao JB, Chang TY, Lan YH, et al. Genetic polymorphisms in androgen receptor-binding sites predict survival in prostate cancer patients receiving androgen-deprivation therapy. Ann Oncol. 2012;23:707–713. doi: 10.1093/annonc/mdr264. [DOI] [PubMed] [Google Scholar]
- 18.Huang CN, Huang SP, Pao JB, Hour TC, Chang TY, et al. Genetic polymorphisms in oestrogen receptor-binding sites affect clinical outcomes in patients with prostate cancer receiving androgen-deprivation therapy. J Intern Med. 2012;271:499–509. doi: 10.1111/j.1365-2796.2011.02449.x. [DOI] [PubMed] [Google Scholar]
- 19.Pao JB, Yang YP, Huang CN, Huang SP, Hour TC, et al. Vitamin D receptor gene variants and clinical outcomes after androgen-deprivation therapy for prostate cancer. World J Urol 10.1007/s00345-011-0813-x. 2011. [DOI] [PubMed]
- 20.Huang SP, Huang LC, Ting WC, Chen LM, Chang TY, et al. Prognostic significance of prostate cancer susceptibility variants on prostate-specific antigen recurrence after radical prostatectomy. Cancer Epidemiol Biomarkers Prev. 2009;18:3068–3074. doi: 10.1158/1055-9965.EPI-09-0665. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Hussain M, Tangen CM, Higano C, Schelhammer PF, Faulkner J, et al. Absolute prostate-specific antigen value after androgen deprivation is a strong independent predictor of survival in new metastatic prostate cancer: data from Southwest Oncology Group Trial 9346 (INT-0162). J Clin Oncol. 2006;24:3984–3990. doi: 10.1200/JCO.2006.06.4246. [DOI] [PubMed] [Google Scholar]
- 23.Stewart AJ, Scher HI, Chen MH, McLeod DG, Carroll PR, et al. Prostate-specific antigen nadir and cancer-specific mortality following hormonal therapy for prostate-specific antigen failure. J Clin Oncol. 2005;23:6556–6560. doi: 10.1200/JCO.2005.20.966. [DOI] [PubMed] [Google Scholar]
- 24.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horoszewicz JS, Leong SS, Kawinski E, Karr JP, Rosenthal H, et al. LNCaP model of human prostatic carcinoma. Cancer Res. 1983;43:1809–1818. [PubMed] [Google Scholar]
- 26.Kaighn ME, Narayan KS, Ohnuki Y, Lechner JF, Jones LW. Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Invest Urol. 1979;17:16–23. [PubMed] [Google Scholar]
- 27.Doak SH, Jenkins SA, Hurle RA, Varma M, Hawizy A, et al. Bone morphogenic factor gene dosage abnormalities in prostatic intraepithelial neoplasia and prostate cancer. Cancer Genet Cytogenet. 2007;176:161–165. doi: 10.1016/j.cancergencyto.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 28.Szabolcs M, Keniry M, Simpson L, Reid LJ, Koujak S, et al. Irs2 inactivation suppresses tumor progression in Pten+/− mice. Am J Pathol. 2009;174:276–286. doi: 10.2353/ajpath.2009.080086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, et al. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 2007;21:2005–2017. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang M, Latham DE, Delaney MA, Chakravarti A. Survivin mediates resistance to antiandrogen therapy in prostate cancer. Oncogene. 2005;24:2474–2482. doi: 10.1038/sj.onc.1208490. [DOI] [PubMed] [Google Scholar]
- 31.Wang P, Dai M, Xuan W, McEachin RC, Jackson AU, et al. SNP Function Portal: a web database for exploring the function implication of SNP alleles. Bioinformatics. 2006;22:e523–529. doi: 10.1093/bioinformatics/btl241. [DOI] [PubMed] [Google Scholar]
- 32.Bentley H, Hamdy FC, Hart KA, Seid JM, Williams JL, et al. Expression of bone morphogenetic proteins in human prostatic adenocarcinoma and benign prostatic hyperplasia. Br J Cancer. 1992;66:1159–1163. doi: 10.1038/bjc.1992.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris SE, Harris MA, Mahy P, Wozney J, Feng JQ, et al. Expression of bone morphogenetic protein messenger RNAs by normal rat and human prostate and prostate cancer cells. Prostate. 1994;24:204–211. doi: 10.1002/pros.2990240406. [DOI] [PubMed] [Google Scholar]
- 34.Masuda H, Fukabori Y, Nakano K, Takezawa Y, CSuzuki T, et al. Increased expression of bone morphogenetic protein-7 in bone metastatic prostate cancer. Prostate. 2003;54:268–274. doi: 10.1002/pros.10193. [DOI] [PubMed] [Google Scholar]
- 35.Bergmann U, Funatomi H, Kornmann M, Beger HG, Korc M. Increased expression of insulin receptor substrate-1 in human pancreatic cancer. Biochem Biophys Res Commun. 1996;220:886–890. doi: 10.1006/bbrc.1996.0500. [DOI] [PubMed] [Google Scholar]
- 36.Chang Q, Li Y, White MF, Fletcher JA, Xiao S. Constitutive activation of insulin receptor substrate 1 is a frequent event in human tumors: therapeutic implications. Cancer Res. 2002;62:6035–6038. [PubMed] [Google Scholar]
- 37.Tanaka S, Wands JR. A carboxy-terminal truncated insulin receptor substrate-1 dominant negative protein reverses the human hepatocellular carcinoma malignant phenotype. J Clin Invest. 1996;98:2100–2108. doi: 10.1172/JCI119016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith MR, Lee H, Nathan DM. Insulin sensitivity during combined androgen blockade for prostate cancer. J Clin Endocrinol Metab. 2006;91:1305–1308. doi: 10.1210/jc.2005-2507. [DOI] [PubMed] [Google Scholar]
- 39.Lubik AA, Gunter JH, Hendy SC, Locke JA, Adomat HH, et al. Insulin increases de novo steroidogenesis in prostate cancer cells. Cancer Res. 2011;71:5754–5764. doi: 10.1158/0008-5472.CAN-10-2470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Patient recruitment and data collection.
(DOC)
Genotyped SNPs and the P values of their association with time to progression, PCSM, ACM during ADT.
(DOC)
Oligonucleotides used for genotyping analysis.
(DOC)
Distribution of clinicopathologic characteristics and their associations with disease progression, PCSM, and ACM in prostate cancer patients receiving ADT.
(DOC)