Abstract
Background
Some of the most frequent deficits seen in children with FASD and in animal models of FASD are spatial memory impairments and impaired executive functioning, which are likely related to alcohol-induced alterations of the hippocampus and prefrontal cortex (PFC), respectively. Choline, a nutrient supplement, has been shown in a rat model to ameliorate some of alcohol's teratogenic effects and this effect may be mediated through choline' effects on DNA methylation.
Methods
Alcohol was given by intragastric intubation to rat pups during the neonatal period (postnatal days 2–10) (ET group), which is equivalent to the third trimester in humans and a period of heightened vulnerability of the brain to alcohol exposure. Control groups included an intubated control group given the intubation procedure without alcohol (IC) and a non-treated control group (NC). Choline or saline was administered subcutaneously to each subject from postnatal day 2 to 20. On postnatal day 21, the brains of the subjects were removed and assayed for global DNA methylation patterning as measured by chemiluminescence using the cpGlobal assay in both the hippocampal region and PFC.
Results
Alcohol exposure caused hypermethylation in the hippocampus and PFC, which was significantly reduced after choline supplementation. In contrast, control animals showed increases in DNA methylation in both regions after choline supplementation, suggesting that choline supplementation has different effects depending upon the initial state of the brain.
Conclusions
This study is the first to show changes in global DNA methylation of the hippocampal region and PFC after neonatal alcohol exposure. Choline supplementation impacts global DNA methylation in these two brain regions in alcohol-exposed and control animals in a differential manner. The current findings suggest that both alcohol and choline have substantial impact on the epigenome in the prefrontal cortex and hippocampus and future studies will be needed to describe which gene families are impacted in such a way that function of the nervous system is changed.
Keywords: Fetal Alcohol Spectrum Disorder, Choline, DNA methylation, Hippocampus, Prefrontal Cortex
BACKGROUND
The teratogenic effects of alcohol exposure during development have been an intensive area of research for many years. Most recently, the term Fetal Alcohol Spectrum Disorders (FASD) has been used to describe the wide spectrum of teratogenic effects of alcohol on individuals who are exposed to the drug during fetal development. On the most severe end of this spectrum is Fetal Alcohol Syndrome (FAS), which is characterized by growth deficiencies, cranio-facial dysmorphology and central nervous system (CNS) dysfunction (Jones and Smith, 1973; Streissguth and O'Malley, 2000). Children with FASD may not show any physical abnormalities associated with developmental alcohol exposure or may show physical abnormalities that do not meet the requirements for a diagnosis of FAS. However, with or without the physical features, high levels of prenatal alcohol exposure may still lead to behavioral anomalies and intellectual deficits in newborns and young children (Landesman-Dwyer et al., 1978; Mattson et al., 1997). Because there is a significant amount of congruence between animals and humans in the effects of alcohol exposure (Driscoll et al., 1990; Hannigan, 1996; Hannigan and Abel, 1996), animal models have been used to further examine the effects of developmental alcohol exposure on brain and behavior.
In both clinical studies and studies using animal models of FASD, early ethanol exposure consistently leads to deficits in learning, memory, and executive functioning (Connor et al., 2000; Driscoll et al., 1990; Kodituwakku et al., 2001). Consistent with the behavioral effects, studies have shown teratogenic effects of alcohol exposure both on the hippocampus (Abel et al., 1983; Archibald et al., 2001; Autti-Ramo et al., 2002; Ba et al., 1996; Barnes and Walker, 1981; Berman and Hannigan, 2000; Davies and Smith, 1981; Diaz-Granados et al., 1993; Ferrer et al., 1988; Greene et al., 1992; Miller, 1995; Perez et al., 1991; Riikonen et al., 1999; Smith and Davies, 1990; Sowell et al., 2007; Tanaka, 1998; Tran and Kelly, 2003; Uecker and Nadel, 1998; West et al., 1981; West and Hodges-Savola, 1983; West and Pierce, 1984; Wigal and Amsel, 1990; Willoughby et al., 2008) and the prefrontal cortex (PFC) (Inomata et al., 1987; Nagahara and Handa, 1995; Sowell et al., 2002; Whitcher and Klintsova, 2008), which are structures importantly involved in learning, memory and executive functioning. Choline – a nutrient supplement – has been recently shown in a rat model to ameliorate some of the behavioral deficits (including deficits in learning, memory and executive functioning) observed in subjects pre- or neonatally exposed to alcohol (Ryan et al., 2008; Thomas et al., 2000; Thomas et al., 2004; Thomas et al., 2007; Thomas et al., 2009; Wagner & Hunt, 2006; Thomas & Tran, 2011).
Studies examining the impact of choline on brain development have suggested multiple mechanisms of action and have suggested that choline-induced changes in the epigenome may be a pivotal site of action (Niclulescu et al. 2004, Niculescu et al., 2006, Zeisel 2011). Research using rodent models has shown that choline supplementation during the gestational period critical for hippocampal development significantly impacts both hippocampal structure and functioning (Albright et al., 1999; Li et al., 2004; Williams et al., 1998). The long-term changes after choline supplementation have been suggested, in part, to be the result of alterations in the pattern of DNA methylation in the hippocampus (Davison et al., 2009; Jhaveri et al., 2001; Kovacheva et al., 2007; Mehedint et al., 2010; Niculescu et al., 2004; Niculescu et al., 2006; Pogribny et al., 2008; Waterland and Jirtle, 2003; Waterland et al., 2006). Importantly, Thomas and colleagues have shown that choline supplementation during early development alleviates spatial learning deficits in a rat model of developmental alcohol exposure and that this effect of choline is observed after choline treatment has ceased (Ryan et al., 2008). This long-lasting effect of choline suggests this nutrient may impact alcohol-induced effects on learning and memory through epigenetic changes in the alcohol-affected brain, specifically via changes in DNA methylation.
Only a few published studies have looked at possible global DNA methylation changes due to exposure to alcohol during the developmental period (Garro et al., 1991; Liu et al., 2009a) and none have focused on specific brain regions. The behavioral deficits and brain changes seen in FASD and animal models of FASD suggest that the changes in gene expression via alterations in the epigenome induced by alcohol are likely to be very complex and so as a first step, global methylation changes in brain regions were first examined in order to identify affected regions. In the present study, a rat model of FASD was used to investigate the underlying mechanism of alcohol-induced deficits by looking at changes in global DNA methylation in the prefrontal and hippocampal areas. In addition, this study also examined changes in DNA methylation when subjects were supplemented with choline during and after early neonatal alcohol exposure. Animals were exposed to alcohol during the third trimester equivalent of human pregnancy (Bayer et al., 1993) and then treated with choline supplementation from the early postnatal period (PD) 2 up to PD 20 (Ryan et al., 2008). It was hypothesized that alcohol exposure during the third trimester equivalent of human pregnancy will reduce global DNA methylation in the hippocampus and PFC areas. It was also expected that choline supplementation given to ethanol-exposed subjects will alter the levels of global DNA methylation in these two brain regions to that of control animals.
METHODS
Subjects
All animal procedures followed institutional guidelines outlined by the American Association of Laboratory Animal Care (AALAC) and were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of South Carolina. All subjects were born and bred in the animal colony at the Department of Psychology at the University of South Carolina. Housing conditions were maintained at 22°C with a 12-hr light-dark cycle, beginning at 0700 h. Female Long-Evans rats were housed with an experienced male over night and gestational day (GD) 1 was defined by the presence of sperm in the vaginal smear on the following morning. Food and water were provided ad libitum to all dams.
After birth, the pups in each litter were quasi-randomly assigned to one of three treatment groups – an ethanol-treated (ET) group, an intubated control (IC) group, and a non-treated control (NC) group. Each pup from each litter was also quasi-randomly assigned to either the choline-supplemented condition or the placebo (saline) condition. Quasi-random assignments in this study took into account the number of pups previously assigned to each experimental cell when assignments were done but was completely random in the selection of a pup from a particular litter for that cell. Thus, this study incorporated a 3 (group) X 2 (sex) X 2 (choline) design. There was no more than one animal from a litter assigned to a cell. Subject numbers are in Tables 1 and 2. Pups remained in the same cage as their dam until PD 21, after which they were sacrificed and their brains retrieved for further analyses.
Table 1.
Mean body weight (g) and SEMs in male subjects across treatment and supplement.
| CHOLINE | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | PD 2 | PD 3 | PD 4 | PD 5 | PD 6 | PD 7 | PD 8 | PD 9 | PD 10 | PD 11 | |
| ET | 6 | 7.3 ± 0.2 | 7.9 ± 0.3 | 8.9 ± 0.4* | 9.9 ± 0.5* | 11.6 ± 0.6* | 13.2 ± 0.6* | 14.9 ± 0.7* | 17.0 ± 0.8* | 18.9 ± 0.9* | 21.1 ± 0.8* |
| IC | 3 | 7.1 ± 0.3 | 8.3 ± 0.4 | 9.6 ± 0.4 | 11.2 ± 0.5 | 12.9 ± 0.6 | 14.7 ± 0.7 | 16.5 ± 0.7 | 19.0 ± 0.8 | 21.0 ± 0.9 | 23.5 ± 1.0 |
| NC | 4 | 7.5 ± 0.2 | 8.6 ± 0.3 | 10.1 ± 0.3 | 11.7 ± 0.4 | 13.6 ± 0.5 | 15.5 ± 0.5 | 17.6 ± 0.5 | 19.8 ± 0.5 | 22.2 ± 0.5 | 25.0 ± 0.5 |
| PD 12 | PD 13 | PD 14 | PD 15 | PD 16 | PD 17 | PD 18 | PD 19 | PD 20 | PD 21 | ||
| ET | 6 | 23.2 ± 0.8* | 25.7 ± 0.8* | 28.5 ± 0.9* | 30.6 ± 0.9* | 32.9 ± 1.0* | 35.0 ± 1.1* | 37.0 ± 1.1* | 39.9 ± 1.4* | 44.1 ± 1.6* | 48.1 ± 1.9* |
| IC | 3 | 25.7 ± 1.0 | 28.0 ± 1.0 | 30.4 ± 1.0 | 32.2 ± 1.0 | 34.8 ± 1.0 | 36.8 ± 1.0 | 38.7 ± 1.1 | 41.3 ± 1.3 | 45.6 ± 1.5 | 50.1 ± 1.5 |
| NC | 4 | 27.0 ± 0.4 | 29.3 ± 0.4 | 31.9 ± 0.5 | 34.1 ± 0.6 | 36.2 ± 0.6 | 38.3 ± 0.7 | 40.5 ± 0.7 | 43.7 ± 0.8 | 48.2 ± 1.0 | 52.5 ± 1.4 |
| SALINE | |||||||||||
| N | PD 2 | PD 3 | PD 4 | PD 5 | PD 6 | PD 7 | PD 8 | PD 9 | PD 10 | PD 11 | |
| ET | 8 | 7.1 ± 0.2 | 7.9 ± 0.2 | 9.0 ± 0.2* | 10.0 ± 0.2* | 11.6 ± 0.3* | 13.3 ± 0.3* | 15.3 ± 0.4* | 17.4 ± 0.4* | 19.1 ± 0.4* | 21.5 ± 0.4* |
| IC | 4 | 7.1 ± 0.1 | 8.0 ± 0.3 | 9.3 ± 0.4 | 10.8 ± 0.6 | 12.4 ± 0.7 | 14.4 ± 0.7 | 16.2 ± 0.8 | 18.3 ± 1.0 | 20.4 ± 1.2 | 23.0 ± 1.2 |
| NC | 4 | 7.1 ± 0.2 | 8.4 ± 0.4 | 9.9 ± 0.4 | 11.5 ± 0.6 | 13.4 ± 0.7 | 15.1 ± 0.7 | 17.1 ± 0.7 | 19.4 ± 0.7 | 21.7 ± 0.9 | 24.5 ± 0.9 |
| PD 12 | PD 13 | PD 14 | PD 15 | PD 16 | PD 17 | PD 18 | PD 19 | PD 20 | PD 21 | ||
| ET | 8 | 23.5 ± 0.4* | 25.7 ± 0.5* | 28.3 ± 0.5* | 30.4 ± 0.6* | 32.7 ± 0.6* | 34.8 ± 0.7* | 36.6 ± 0.8* | 39.0 ± 0.9* | 42.7 ± 0.8* | 47.2 ± 0.9* |
| IC | 4 | 25.0 ± 1.2 | 27.0 ± 1.1 | 29.7 ± 1.2 | 31.6 ± 1.2 | 34.0 ± 1.2 | 36.0 ± 1.3 | 37.8 ± 1.3 | 40.7 ± 1.4 | 44.5 ± 1.5 | 48.9 ± 1.8 |
| NC | 4 | 26.5 ± 1.0 | 28.5 ± 1.0 | 31.1 ± 1.1 | 33.4 ± 1.2 | 35.5 ± 1.3 | 37.7 ± 1.4 | 39.3 ± 1.5 | 42.2 ± 1.5 | 46.8 ± 1.8 | 51.7 ± 2.0 |
The indicate significant difference from the NC group at p's < .05.
Table 2.
Mean body weight (g) and SEMs in female subjects across treatments and supplement.
| CHOLINE | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | PD 2 | PD 3 | PD 4 | PD 5 | PD 6 | PD 7 | PD 8 | PD 9 | PD 10 | PD 11 | |
| ET | 7 | 6.9 ± 0.1 | 7.7 ± 0.2 | 8.7 ± 0.2*° | 9.8 ± 0.3*° | 11.3 ± 0.4*° | 12.8 ± 0.4*° | 14.8 ± 0.5*° | 16.7 ± 0.5*° | 18.6 ± 0.6*° | 20.9 ± 0.6*° |
| IC | 4 | 6.8 ± 0.2 | 7.9 ± 0.2 | 9.3 ± 0.2 | 10.8 ± 0.3 | 12.6 ± 0.4 | 14.4 ± 0.5 | 16.3 ± 0.5 | 18.6 ± 0.6 | 20.7 ± 0.6 | 23.1 ± 0.7 |
| NC | 4 | 6.8 ± 0.2 | 7.8 ± 0.3 | 9.1 ± 0.3 | 10.5 ± 0.4 | 12.2 ± 0.5 | 14.1 ± 0.5 | 16.1 ± 0.5 | 18.3 ± 0.5 | 20.4 ± 0.5 | 23.1 ± 0.5 |
| PD 12 | PD 13 | PD 14 | PD 15 | PD 16 | PD 17 | PD 18 | PD 19 | PD 20 | PD 21 | ||
| ET | 7 | 23.0 ± 0.5*° | 25.1 ± 0.6*° | 21.5 ± 0.6*° | 29.6 ± 0.6* | 31.7 ± 0.8*° | 33.7 ± 0.8* | 35.3 ± 0.9* | 37.5 ± 1.0* | 41.7 ± 1.0*° | 45.8 ± 1.0* |
| IC | 4 | 24.9 ± 0.7 | 27.0 ± 0.7 | 29.4 ± 0.8 | 31.4 ± 0.9 | 33.8 ± 1.0 | 35.8 ± 1.1 | 37.4 ± 1.1 | 39.8 ± 1.2 | 44.8 ± 1.3 | 49.1 ± 1.4 |
| NC | 4 | 25.1 ± 0.5 | 27.2 ± 0.6 | 29.6 ± 0.5 | 31.9 ± 0.6 | 33.9 ± 0.7 | 35.9 ± 0.8 | 37.6 ± 0.8 | 40.3 ± 1.0 | 44.7 ± 1.1 | 48.3 ± 1.4 |
| SALINE | |||||||||||
| N | PD 2 | PD 3 | PD 4 | PD 5 | PD 6 | PD 7 | PD 8 | PD 9 | PD 10 | PD 11 | |
| ET | 8 | 6.7 ± 0.1 | 7.5 ± 0.2 | 8.6 ± 0.3*° | 9.6 ± 0.3*° | 11.1 ± 0.4*° | 12.8 ± 0.4*° | 14.6 ± 0.4*° | 16.4 ± 0.5*° | 18.3 ± 0.5*° | 20.6 ± 0.6*° |
| IC | 4 | 6.9 ± 0.3 | 8.0 ± 0.3 | 9.3 ± 0.4 | 10.9 ± 0.6 | 12.7 ± 0.6 | 14.4 ± 0.7 | 16.4 ± 0.8 | 18.6 ± 0.9 | 20.6 ± 0.9 | 23.1 ± 1.0 |
| NC | 4 | 6.9 ± 0.2 | 8.2 ± 0.2 | 9.6 ± 0.3 | 11.1 ± 0.5 | 12.8 ± 0.6 | 14.5 ± 0.6 | 16.6 ± 0.7 | 18.9 ± 0.8 | 21.1 ± 0.8 | 23.5 ± 0.8 |
| PD 12 | PD 13 | PD 14 | PD 15 | PD 16 | PD 17 | PD 18 | PD 19 | PD 20 | PD 21 | ||
| ET | 8 | 22.6 ± 0.5*° | 24.9 ± 0.6*° | 27.0 ± 0.6*° | 29.3 ± 0.8* | 31.1 ± 0.7*° | 33.2 ± 0.8* | 34.9 ± 0.8* | 37.5 ± 0.9* | 41.3 ± 1.0*° | 46.3 ± 1.2* |
| IC | 4 | 24.9 ± 1.0 | 26.9 ± 1.0 | 29.1 ± 1.0 | 31.3 ± 1.0 | 33.7 ± 1.0 | 35.5 ± 1.1 | 37.0 ± 1.1 | 40.0 ± 1.2 | 44.4 ± 1.3 | 48.8 ± 1.4 |
| NC | 4 | 25.8 ± 0.8 | 27.8 ± 0.9 | 30.1 ± 0.9 | 32.5 ± 1.0 | 34.6 ± 1.0 | 36.7 ± 1.1 | 38.5 ± 1.2 | 41.5 ± 1.4 | 45.8 ± 1.6 | 50.4 ± 1.6 |
The indicates significant differences from the NC group with p's < .05
the indicates significant differences from the IC group with p's < .05.
Postnatal Treatment
Dams gave birth on gestational day 23, and this day was designated as postnatal (PD) 1 for the pups. All dams and their pups were left undisturbed on this day and on PD 2, litters were culled to 10 pups (5 males and 5 females) when possible. Treatments occurred from PD 2 through PD 10, equaling the third trimester of human pregnancy (Bayer et al., 1993; Dobbing and Sands, 1979).
For the ET group, ethanol was administered via intragastric intubation from PD 2 through 10. ET pups received a single daily dose of ethanol (3.0 g/kg) in a volume of 27.8 ml/kg of milk solution (West et al., 1984) which produces peak blood alcohol concentrations (BACs) between 300–400 mg/dl (Marino et al., 2002). Intubation involved placing PE10 Intramedic tubing, attached to a syringe and dipped in corn oil for lubrication, down the esophagus of the pup and injecting the solution directly into the stomach. Two hours after ethanol administration, the ET pups were intubated a second time, but with 27.8 ml/kg of milk solution only to compensate for any reduction in milk intake from the dam due to intoxication (Tran and Kelly, 2003). The IC pups received the same treatment procedure, twice per day, but were not given milk alone or ethanol in milk. The NC pups were weighed daily. On PD 7, all pups were permanently marked on one or more of their paws for identification using the AIMS Animal Tattoo Identification kit (Serial # NEO9001000). Each paw to be marked was sterilized, and then tattooed using a standard tip tattoo needle dipped in India ink.
Postnatal Choline Supplementation
From PD 2 to 20, each pup in each experimental group (ET, IC and NC) was quasi-randomly assigned to one of two supplement conditions. Animals in the choline-supplement condition received subcutaneous (s.c.) injections of 100 mg/kg/day choline chloride in a volume of 6.66 ml/kg of saline, while the animals in the saline-supplement condition received s.c. injections of saline vehicle. Subjects from all groups were weighed daily throughout the days of supplementation.
Blood Ethanol Concentrations (BECs)
On PD 10, samples of 10 μl of blood from each pup in the ET and IC conditions were collected via a nick to the tail, 2 hours after ethanol administration, in order to assess maximum BAC levels (Marino et al., 2002). No blood was taken from the NC pups. Blood samples from the ET group were assayed for blood ethanol content using a colorimetric assay (Dudek and Abbot, 1984).
Tissue Collection
On PD 21, subjects were sacrificed via decapitation. Immediately after removal, the brain of each subject was divided sagittally into left and right hemispheres. The PFC and entire hippocampus were dissected from one hemisphere, left and right were balanced within each treatment and supplemental condition, and immediately frozen in a container of isopentane, which was kept chilled with dry ice. After freezing, the tissue was placed in labeled tubes and kept frozen at −80 °C until time of assays.
DNA Methylation
Genomic DNA (gDNA) was isolated from approximately 30 mg of rat brain tissue using the Wizard® genomic DNA purification kit (Promega # A1120) following the protocol of the manufacturer. The quantity and quality of the gDNA samples were assessed by spectrophotometry using the NanoDrop 8000, (Thermo Scientific) and agarose gel electrophoresis, respectively. To assess global methylation status, methods were similar to that of Anisowicz et al., 2008 with the following modifications: Samples were processed in triplicate with a no enzyme, buffer only control and DNA was digested with HpaII (methylation sensitive/dependent) and MspI (methylation insensitive) restriction enzymes. First, aliquots of 150 ng gDNA for each sample were placed into 9 wells of a 96 well MaxiSorp™ plate (Nunc #436110). DNA in the first three wells digested with 1 unit of HpaII (NEB # R0171S), the second three wells was digested with1 unit of MspI (NEB # R0106S), and no enzyme added to the last three wells (buffer only control (NEBuffer1)). The total volume of the reactions was 30 μl and the plate was then incubated in a 37 °C dry incubator for 1 hour.
After DNA digestion, 20 ul of the end fill reaction mix containing 0.1 μM Biotin-11-dCTP (PerkinElmer # NEL538001EA), 0.1 μM Biotin-11-dGTP (PerkinElmer # NEL541001EA), and 0.1 units Sequenase (USB 70775Y) was added to each well. The plate was then incubated at 37 °C for 2 hours.
After end-fill with biotinylated nucleotides, 100 μl of Reacti Bind DNA coating solution (Pierce # 17250) was added to each well of the plate and incubated overnight at room temperature with shaking at 150 RPM. After coating, the plate was washed two times with TBS (10 mM Tris-HCL pH 8.0, 150 mM NaCl), and then 200 μl of the Detector Block solution (KPL # 71–83-00) was added to each well and incubated at room temperature for 1 hour. 150 μl of Detector Block containing 2 μg/ml of HRP Neutravidin (1: 500 dilution) (Pierce # 31030) was added to each well after removing the Detector Block solution. The plate was again incubated at room temperature for 2 hours.
The Dector Block/HRP Neutravidin solution was removed from the plate and 1X Biotin wash solution (KPL # 50–63-06) was used to wash the plate 2 times. Finally, 150 μl of LumiGlo chemiluminescence substrate (KPL # 54–61-00) was added to each well, and after 4 minutes, net luminescence was measured by a GENios microplate reader (Tecan) at 440 nM. To determine the net luminescence, the average of the 3 values obtained from the no-enzyme (buffer) control was subtracted from the average of the 3 values obtained from the HPaII and MSP I enzyme values of each subject. To calculate the global DNA methylation index GDMI, the average net luminescence value obtained for the methylation sensitive enzyme (HPAII) was divided by the average net luminescence value obtained for the methylation insensitive enzyme (Msp I) of each subject. The GDMI values were the dependent measure analyzed and the value of GDMI is inversely related to the amount of methylation.
RESULTS
The statistical significance level was set at α = .05 and data analyses were performed using the Statistical Package for the Social Sciences (SPSS) 16.0 for Windows. Greenhouse-Geisser degrees of freedom were used for analyses of repeated measures and body weight data were analyzed for each sex separately because of a priori expectations of a sex difference. Tukey's HSD post hoc tests were used where appropriate.
Male Body Weights
A repeated measures ANOVA, with ethanol treatment and choline supplement as between factors and Day (PD 2–21) as the repeated measure, was performed on the body weights across days. Tests of within-subjects effects showed a significant main effect of Day F (19, 1026) = 4810, p < .01, and a significant Day × Treatment interaction F (38, 1026) = 3.2, p < .05. Post-hoc analyses revealed the ET group weighed significantly less than the NC group from PD 4 to 21 (p's < .05), and had a non-significant tendency to weigh less than the IC group only on PDs 11 and 12 (p = .067 and .054, respectively). This suggests that stress (specifically of the administration procedure) may account for some of the effects seen. Body weight data for the males are shown in Table 1. Importantly, there was no impact of choline supplement on body weight in males.
Female Body Weights
A repeated measures ANOVA, with ethanol treatment and choline supplement as between factors and Day from PD 2 to 21 as the repeated measures, was performed on body weight across days. Tests of within-subjects effects showed a significant main effect of Day, F (19, 1026) = 6272, p < .01, on body weight, and a significant Day × Treatment interaction, F (38, 1026) = 4.0, p < .01. Post-hoc analyses revealed the ET groups weighed less than the NC groups from PD 4 to 21 (p's < .05), and the IC groups from PD 4 to 14, PD 16 and PD 20 (p's < .05). Body weight data for the females are shown in Table 2. There was no impact of supplement on body weight in females.
Blood Ethanol Concentrations (BECs)
A 2-way ANOVA (Supplement × Sex) was performed to determine whether these two factors have any effect on BEC levels. There were no significant differences found in the average BEC levels between male and female subjects or between subjects receiving the choline supplement and those receiving saline. These data are shown in Table 3.
Table 3.
. Blood Ethanol Concentrations (BECs) (mg/dl) and SEMs in ethanol-exposed subjects
| Treatment | Supplement | Sex | BEC |
|---|---|---|---|
| ET | Saline | Male | 358.2 ± 18.1 |
| ET | Saline | Female | 365.0 ± 22.5 |
| ET | Choline | Male | 367.6 ± 17.6 |
| ET | Choline | Female | 365.3 ± 26.0 |
DNA Methylation
Initial 3-way ANOVAs (Treatment × Supplement × Sex) were performed on the GDMI for all treatment and supplement groups from each brain region separately and found no significant differences between the sexes or the two control groups. Further analyses were performed with sex and control groups combined.
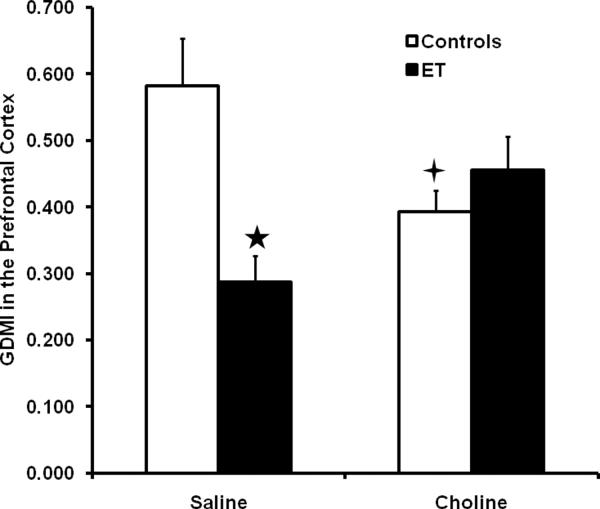
Analysis of the PFC GDMI with a 2 × 2 ANOVA (Treatment × Supplement) revealed a significant Treatment × Supplement effect in the PFC, F(1, 55) = 10.7, p < .01. Tukey's HSD post hoc analyses showed that the saline-supplemented ET group had a lower GDMI than the saline-supplemented control groups, indicating a hypermethylation in this region (p's < 0.05). Choline supplementation reduced the GDMI in the control group and increased the GDMI in the ET group compared to the saline-supplemented group given the same treatment (p's < 0.05). The changes in GDMI indicate that ethanol treatment caused a hypermethylation in this region and this was partially ameliorated by choline treatment. Choline treatment in the control subjects caused a hypermethylation in this region, an effect opposite to the effect in the ET subjects. These data are shown in Figure 1.
Figure 1.
Average Global DNA Methylation Index (GDMI) values in the PFC. Data are collapsed across sex and control group. Error bars represent the SEM. The GDMI is inversely related to the amount of DNA methylation. The five-pointed star indicates a significant difference from both the saline-supplemented control group and the choline supplemented ET group. The four-pointed star indicates a significant difference from the saline-supplemented control group.
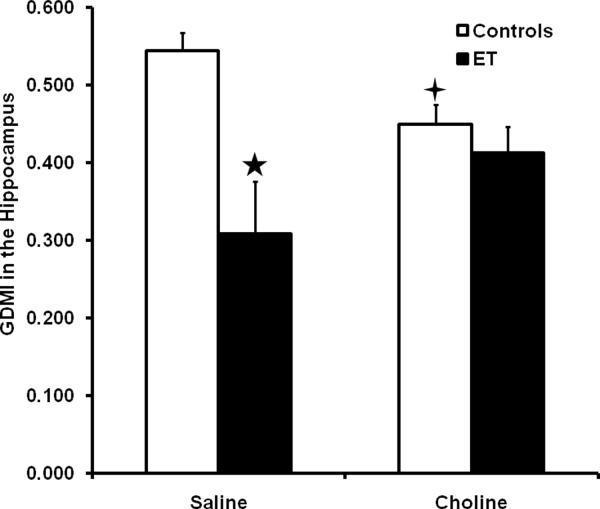
Analysis of the hippocampal GDMI with a 2 × 2 ANOVA (Treatment × Supplement) revealed a significant Treatment × Supplement interaction, F(1, 55) = 5.7, p < .05. Tukey's HSD post hoc analyses showed a pattern of findings in the hippocampal region similar to that in the PFC region. The saline-supplemented ET group had a lower GDMI than the saline-supplemented control group, indicating a hypermethylation in this region (p's < 0.05). Choline supplementation reduced the GDMI in the control group and increased the GDMI in the ET group compared to the saline-supplemented group given the same treatment (p's < 0.05). These data are shown in Figure 2.
Figure 2.
Average Global DNA Methylation Index (GDMI) values in the hippocampus. Data are collapsed across sex and control group. Error bars represent the SEM. The GDMI is inversely related to the amount of DNA methylation. The five-pointed star indicates a significant difference from both the saline-supplemented control group and the choline supplemented ET group. The four-pointed star indicates a significant difference from the saline-supplemented control group.
DISCUSSION
Temporal and spatial gene expression during development is an important and complex process that is regulated by epigenetic marks that are crucial for the maintenance of normal cellular homeostasis (Robertson 2005). It has been demonstrated that aberrant DNA methylation patterns are associated with a growing number of diseases (Robertson 2005). This study is among the first to show that alcohol exposure during the third trimester equivalent of human pregnancy in the rat causes changes in global DNA methylation in the PFC and hippocampus, and these changes are evident in the early adolescent stage. Developmental alcohol exposure led to hypermethylation in both these brain areas compared to control subjects. This study also shows significant changes in global DNA methylation in ethanol-exposed animals after choline supplementation. Choline given to ethanol-exposed subjects from PD 2 to 20 led to a decrease in methylation, while control animals showed a state of increased methylation after choline supplementation in both brain regions. The effects of choline were dependent on the prior developmental treatment of the subjects.
Given the initial literature (Garro et al., 1991) that reported a state of hypomethylation after exposure to gestational alcohol, and that decreases in methylation have been implicated in reductions in learning and memory abilities (Liu et al., 2009b), it was hypothesized that developmental alcohol during the third trimester equivalent would lead to global DNA hypomethylation. However, the present findings of alcohol-induced DNA hypermethylation corroborate a more recent report showing that developmental alcohol (GD 0.5 to 8.5 - equivalent to the first trimester of human pregnancy) increases the probability of transcriptional silencing (hypermethylation) of the allele, Agouti viable yellow (Avy), in the mouse (Kaminen-Ahola et al., 2010). Others have shown that alcohol exposure during development can cause both hypo- and hypermethylation of specific genes (Zhou et al., 2011). Since both hypo- and hypermethylation can lead to deficits in brain function and behavior (Jiang et al., 2004; Robertson, 2005), the deficits induced by alcohol exposure during development may be mediated by either direction of change in methylation. Suffice it to say, our findings of global DNA hypermethylation in the PFC and hippocampus provide only a piece of the puzzle in determining possible molecular mechanisms of developmental alcohol's effects, with the next necessary step being to examine methylation changes in specific cells and genes in these two brain areas. Importantly, the findings of changes in global DNA methylation in the current study suggest that there are many genes affected by alcohol and choline and it will be necessary to examine methylation changes in functionally-related genes to fully understand the relationship of dysfunction in the prefrontal cortex and hippocampus to changes in the epigenome.
Given the complexity of effects of alcohol exposure during development, it seems likely that methylation changes will vary according to not only timing of exposure, degree of exposure, brain regions examined and individual variation, but also to specific tissues, cells, and genes. Indeed, using a whole embryo culture, Liu and colleagues (2009a) showed that there was increased DNA methylation on chromosomes 10 and X due to alcohol exposure but the degree of alcohol-induced effect on the methylation was a function of whether the embryo showed a neural tube deficit caused by the alcohol exposure. Zhou et al. (2011) has followed up this study and has shown that the hypermethylation in the embryos with alcohol-induced neural tube deficits is associated with reduced expression of genes involved in neural specification and neural growth factors. Some of the gene-specific changes were not seen in alcohol-exposed embryos without neural tube deficits.
Moreover, as clearly shown by Zhou and colleagues (2011), even with global hypermethylation in the embryos, methylation of specific genes/promoter regions can either be increased or decreased depending upon the specific gene and/or tissue. Thus, it will be necessary to investigate the regulators of the epigenome and the methylation status of specific genes to fully describe the impact of alcohol on the epigenome. The current study was aimed at investigating neural regions that have been shown to be impacted by alcohol exposure during development with the eventual goal of targeting the investigation on specific genes involved in known alcohol-related effects, such as changes in the dendritic tree of neurons in the PFC and hippocampus or genes involved in neural plasticity in these regions. The eventual link of alcohol's impact on the epigenome to effects on neural tissue needs further investigation. Nevertheless, the current findings suggest that the overall impact of alcohol on the epigenome will be skewed towards hypermethylation and thus, gene suppression. Examination of regulators of the epigenome such as methyl-CpG-binding 2 (MeCP2) protein (Moretti et al., 2006; Pogribny et al., 2008) and DNMT would provide more specific information on how alcohol is affecting gene expression. Importantly, the current findings suggest that the changes in DNA methylation after alcohol exposure and choline supplementation are very substantial since a global measure was changed. In order to fully explain how epigenetic changes induced by alcohol and/or choline are related to function of the nervous system, examination of regulation of functional gene families rather than single genes will be necessary.
The methylation changes observed after choline supplementation in ET subjects led to a significant decrease in the alcohol-induced hypermethylation state of the DNA, a change that was not significantly different from the methylation levels of control subjects that were also given choline. These choline-induced changes corroborate past research showing that methylation changes do occur in the brains of animals treated with varying levels of choline availability. However, the manner in which choline impacts methylation has varied across studies. Some studies examining methylation changes after choline supplementation have found hypomethylation of the DNA (Kovacheva et al., 2007). Choline-deficient diets have been shown to cause DNA hypomethylation (Niculescu et al., 2006), DNA hypermethylation (Mehedint et al., 2010) and no effect on DNA methylation (Kovacheva et al., 2007). The variability of DNA methylation changes due to choline availability may be the result of the initial state of the animals as was observed in the present study where choline supplementation led to opposite changes in DNA methylation in ethanol-exposed and control subjects. This finding may be explained by observations that DNA methylation is regulated by a number of factors such as DNMT enzymes and binding proteins (Robertson, 2005), shows co-activity with other epigenetic processes such as histone methylation (Pogribny et al., 2008), which in turn are regulated by multiple factors (Davison et al., 2009). Thus, the disruption of this complex process by agents such as alcohol and choline can have cascading effects throughout the epigenome. One of the major findings of the present study is that choline alters the epigenome in ethanol-exposed subjects, although an important and still unanswered question is whether choline is reversing gene-specific effects of alcohol or whether the impacted genes differ. This information will help determine whether choline's beneficial effects on behavior in rat models of FASD are due to reversing the ethanol effects or simply compensating for them.
Given the variability of DNA methylation among different tissue regions, the variability of and regional specificity of alcohol's teratogenic effects, and the variability of the effects of choline availability on DNA methylation, it is difficult to generalize ethanol's effects on the epigenome. Moreover, given the likely complexity of alcohol's impact and choline's impact on the epigenome, careful programmatic research will be necessary to determine time course effects, tissue effects, and most important gene-specific effects and their relationship to brain deficits seen in animal models of FASD. The impact of choline availability on DNA methylation is dependent on the time period and the length of choline supplementation, the organ or brain regions of interest, and the original state of the individual or organism (e.g. Pogribny et al., 2006; 2008).
Choline may also have additional effects in addition to DNA methylation, such as its involvement in acetylcholine neurotransmission (Albuquerque et al., 1998; Alkondon et al., 1997; Alkondon et al., 1999; Cermak et al., 1998; Uteshev et al., 2003), NMDA-receptor mediated transmission (Montoya and Swartzwelder, 2000), growth factors (Napoli et al., 2008; Sandstrom et al., 2002; Wong-Goodrich et al., 2008a; 2008b), and even cell membrane integrity (Blusztajn, 1998; Zeisel and Blusztajn, 1994; Zeisel & Niculescu, 2006). In fact, ethanol exposure during the 3rd trimester equivalent leads to an upregulation of M2/4 muscarinic receptors, an effect mitigated with perinatal choline supplementation (Monk et al., 2011).
Along with the Garro et al. (1991), Kaminen-Ahola et al. (2010) and Liu et al. (2009a) studies, the present finding of an alcohol-induced change in DNA methylation points towards a central role of epigenetics in the etiology of FASD and an understanding of alcohol's teratology at the molecular level. As asserted by both Haycock (2009) and Zeisel (2011), examining the effects of developmental alcohol from an epigenetic viewpoint will help to explain the several behavioral and physical deficits definitive of FASD. This implication is further supported by the observation of a significant change in the alcohol-induced methylation state after choline supplementation, a nutrient shown to attenuate some of the behavioral learning and memory deficits caused by developmental alcohol (Ryan et al., 2008; Thomas et al., 2000; 2004; 2007; Thomas and Tran, 2011). It remains to be seen whether the impact of choline on the alcohol-exposed brain is reversing the impact of alcohol on the methylation state of specific genes or whether the impact of choline is on the methylation of a different set of genes. Either way, it is known that choline can reverse some of the behavioral changes in an animal model of FASD and it is possible that the mechanism of this treatment is through modification of the epigenome. The next step in this promising line of research is to examine the methylation of specific gene families after ethanol exposure and choline supplementation to begin to target the global mechanisms underlying the effects of both these substances. The current findings suggest that the changes in the epigenetic regulation of genes will be widespread in both the hippocampus and prefrontal cortex after alcohol exposure and choline supplementation and that the changes induced by alcohol exposure are long-lasting.
Acknowledgements
The project described was supported by Award Number RO1AA011566 and RO1AA12446 from the National Institute on Alcohol Abuse and Alcoholism. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We would like to thank Amy Perkins and Dr. R. Charles Lawrence for their helpful feedback on the manuscript.
This work was supported by NIAAA RO1 11566 to S.J.K. and NIAAA RO1 12446 to J.D.T.
REFERENCES
- Abel EL, Jacobson S, Sherwin BT. In utero alcohol exposure: functional and structural brain damage. Neurobehav Toxicol Teratol. 1983;5:363–366. [PubMed] [Google Scholar]
- Albright CD, Friedrich CB, Brown EC, Mar MH, Zeisel SH. Maternal dietary choline availability alters mitosis, apoptosis and the localization of TOAD-64 protein in the developing fetal rat septum. Brain Res Dev Brain Res. 1999;115:123–129. doi: 10.1016/s0165-3806(99)00057-7. [DOI] [PubMed] [Google Scholar]
- Albuquerque EX, Pereira EF, Braga MF, Alkondon M. Contribution of nicotinic receptors to the function of synapses in the central nervous system: the action of choline as a selective agonist of α7 receptors. J Physiol Paris. 1998;92:309–316. doi: 10.1016/s0928-4257(98)80039-9. [DOI] [PubMed] [Google Scholar]
- Alkondon M, Pereira EF, Cortes WS, Maelicke A, Albuquerque EX. Choline is a selective agonist of α7 nicotinic acetylcholine receptors in the rat brain neurons. Eur J Neurosci. 1997;9:2734–2742. doi: 10.1111/j.1460-9568.1997.tb01702.x. [DOI] [PubMed] [Google Scholar]
- Alkondon M, Pereira EFR, Eisenberg HM, Albuquerque EX. Choline and selective antagonists identify two subtypes of nicotinic actylcholine receptors that modulate GABA release from CA1 interneurons in rat hippocampal slices. J Neurosci. 1999;19:2693–2705. doi: 10.1523/JNEUROSCI.19-07-02693.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisowicz A, Huang H, Braunschweiger K, Liu Z, Giese H, Wang H, Mamaev S, Olejnik J, Massion P, Mastro R. A high-throughput and sensitive method to measure global DNA methylation: Application in lung cancer. BMC Cancer. 2008;8:222. doi: 10.1186/1471-2407-8-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald SL, Fennema-Notestine C, Gamst A, Riley EP, Mattson SN, Jernigan TL. Brain dysmorphology in individuals with severe prenatal alcohol exposure. Dev Med Child Neurol. 2001;43:148–154. [PubMed] [Google Scholar]
- Autti-Ramo I, Autti T, Korkman M, Kettunen S, Salonen O, Valanne L. MRI findings in children with school problems who had been exposed prenatally to alcohol. Dev Med Child Neurol. 2002;44:98–106. doi: 10.1017/s0012162201001748. [DOI] [PubMed] [Google Scholar]
- Ba A, Seri BV, Han SH. Thiamine administration during chronic alcohol intake in pregnant and lactating rats: effects on the offspring neurobehavioural development. Alcohol Alcohol. 1996;31:27–40. doi: 10.1093/oxfordjournals.alcalc.a008113. [DOI] [PubMed] [Google Scholar]
- Barnes DE, Walker DW. Prenatal ethanol exposure permanently reduces the number of pyramidal neurons in rat hippocampus. Brain Res. 1981;227:333–340. doi: 10.1016/0165-3806(81)90071-7. [DOI] [PubMed] [Google Scholar]
- Bayer SA, Altman J, Russo RJ, Zhang X. Timtables of neurogenesis in the human brain based on experimentally determined patterns in the rat. Neurotoxicology. 1993;14:83–144. [PubMed] [Google Scholar]
- Berman RF, Hannigan JH. Effects of prenatal alcohol exposure on the hippocampus: spatial behavior, electrophysiology, and neuroanatomy. Hippocampus. 2000;10:94–110. doi: 10.1002/(SICI)1098-1063(2000)10:1<94::AID-HIPO11>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Blusztajn JK. Choline, a vital amine. Science. 1998;281:794–795. doi: 10.1126/science.281.5378.794. [DOI] [PubMed] [Google Scholar]
- Cermak JM, Holler T, Jackson DA, Blusztajn JK. Prenatal availability of choline modifies development of the hippocampal cholinergic system. FASEB J. 1998;12:349–357. doi: 10.1096/fasebj.12.3.349. [DOI] [PubMed] [Google Scholar]
- Connor PD, Sampson PD, Bookstein FL, Barr HM, Streissguth AP. Direct and indirect effects of prenatal alcohol damage on executive function. Dev Neuropsychol. 2000;18:331–354. doi: 10.1207/S1532694204Connor. [DOI] [PubMed] [Google Scholar]
- Davies DL, Smith DE. A golgi study of mouse hippocampal CA1 pyramidal neurons following perinatal alcohol exposure. Neurosci Lett. 1981;26:49–54. doi: 10.1016/0304-3940(81)90424-9. [DOI] [PubMed] [Google Scholar]
- Davison JM, Mellott TJ, Kovacheva VP, Blusztajn JK. Gestational choline supply regulates methylation of histone H3, expression of histone methyltransferases G9a (Kmt1c) and Suv39h1 (Kmt1a), and DNA methylation of their genes in rat fetal liver and brain. J Biol Chem. 2009;284:1982–1989. doi: 10.1074/jbc.M807651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Granados JL, Greene PL, Amsel A. Mitigating effects of combined prenatal and postnatal exposure to ethanol on learned persistence in the weanling rat: a replication under high-peak conditions. Behav Neurosci. 1993;107:1059–1066. doi: 10.1037//0735-7044.107.6.1059. [DOI] [PubMed] [Google Scholar]
- Dobbing J, Sands J. Comparative aspects of the brain growth spurt. Early Hum Dev. 1979;3:79–83. doi: 10.1016/0378-3782(79)90022-7. [DOI] [PubMed] [Google Scholar]
- Driscoll CD, Streissguth AP, Riley EP. Prenatal alcohol exposure: comparability of effects on humans and animal models. Neurotoxicol Teratol. 1990;12:231–238. doi: 10.1016/0892-0362(90)90094-s. [DOI] [PubMed] [Google Scholar]
- Dudek BC, Abbott ME. A biometrical genetic analysis of ethanol response in selectively bred long-sleep and short-sleep mice. Behavioral Genetics. 1984;14:1–19. doi: 10.1007/BF01066065. [DOI] [PubMed] [Google Scholar]
- Ferrer I, Galofre F, Lopez-Tejero D, Llobera M. Morphological recovery of hippocampal pyramidal neurons in the adult rat exposed in utero to ethanol. Toxicology. 1988;48:191–197. doi: 10.1016/0300-483x(88)90100-x. [DOI] [PubMed] [Google Scholar]
- Garro AJ, McBeth DL, Lima V, Lieber CS. Ethanol consumption inhibits DNA methylation in mice: implication for the fetal alcohol syndrome. Alcohol Clin Exp Res. 1991;15:395–398. doi: 10.1111/j.1530-0277.1991.tb00536.x. [DOI] [PubMed] [Google Scholar]
- Greene PL, Diaz-Granados JL, Amsel A. Blood ethanol concentration from early postnatal exposure: effects on memory-based learning and hippocampal neuroanatomy in infant and adult Rats. Behav Neurosci. 1992;106:51–61. doi: 10.1037//0735-7044.106.1.51. [DOI] [PubMed] [Google Scholar]
- Hannigan JH. What research with animals is telling us about alcohol-related neurodevelopmental disorder. Pharmacol Biochem Behav. 1996;55:489–499. doi: 10.1016/s0091-3057(96)00251-1. [DOI] [PubMed] [Google Scholar]
- Hannigan JH, Abel EL. In: Animal models for the study of alcohol-related birth defects, in Alcohol Pregnancy and Child Development. Spohr H-L, Steinhaussen H-C, editors. Cambridge University Press; Cambridge: 1996. pp. 77–102. [Google Scholar]
- Haycock PC. Fetal alcohol spectrum disorders: the epigenetic perspective. Biol Reprod. 2009;81:607–617. doi: 10.1095/biolreprod.108.074690. [DOI] [PubMed] [Google Scholar]
- Inomata K, Nasu F, Tanaka H. Decreased density of synaptic formation in the frontal cortex of neonatal rats exposed to ethanol in utero. Int J Dev Neurosci. 1987;5:455–460. doi: 10.1016/0736-5748(87)90023-2. [DOI] [PubMed] [Google Scholar]
- Jhaveri MS, Wagner C, Trepel JB. Impact of extracellular folate levels on global gene expression. Mol Pharmacol. 2001;60:1288–1295. doi: 10.1124/mol.60.6.1288. [DOI] [PubMed] [Google Scholar]
- Jiang Y-h, Bressler J, Beaudet AL. Epigenetics and human disease. Annu Rev Genomics Hum Genet. 2004;5:479–510. doi: 10.1146/annurev.genom.5.061903.180014. [DOI] [PubMed] [Google Scholar]
- Jones KL, Smith DW. Recognition of fetal alcohol syndrome in early infancy. Lancet. 1973;302:999–1001. doi: 10.1016/s0140-6736(73)91092-1. [DOI] [PubMed] [Google Scholar]
- Kaminen-Ahola N, Ahola A, Maga M, Mallitt K-A, Fahey P, Cox TC, Whitelaw E, Chong S. Maternal ethanol consumption alters the epigenotype and the phenotype of offspring in a mouse model. PLoS Genet. 2010;6:e1000811. doi: 10.1371/journal.pgen.1000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodituwakku PW, Kalberg W, May PA. The effects of prenatal alcohol exposure on executive functioning. Alcohol Res Health. 2001;25:192–198. [PMC free article] [PubMed] [Google Scholar]
- Kovacheva VP, Mellott TJ, Davison JM, Wagner N, Lopez-Coviella I, Schnitzler AC, Blusztajn JK. Gestational choline deficiency causes global and Igf2 gene DNA hypermethylation by up-regulation of Dnmt1 expression. J Biol Chem. 2007;282:31777–31788. doi: 10.1074/jbc.M705539200. [DOI] [PubMed] [Google Scholar]
- Landesman-Dwyer S, Keller LS, Streissguth AP. Naturalistic observations of newborns: effects of maternal alcohol intake. Alcohol Clin Exp Res. 1978;2:171–177. doi: 10.1111/j.1530-0277.1978.tb04718.x. [DOI] [PubMed] [Google Scholar]
- Li Q, Guo-Ross S, Lewis DV, Turner D, White AM, Wilson WA, Swartzwelder HS. Dietary prenatal choline supplementation alters postnatal hippocampal structure and function. J Neurophysiol. 2004;91:1545–55. doi: 10.1152/jn.00785.2003. [DOI] [PubMed] [Google Scholar]
- Liu Y, Balaraman Y, Wang G, Nephew KP, Zhou FC. Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation. Epigenetics. 2009a;4:500–511. doi: 10.4161/epi.4.7.9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, van Groen T, Kadish I, Tollefsbol TO. DNA methylation impacts on learning and memory in aging. Neurobiol Aging. 2009b;30:549–560. doi: 10.1016/j.neurobiolaging.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadev K, Vemuri MC. Effect of ethanol on chromatin and nonhistone nuclear proteins in rat brain. Neurochem Res. 1998;23:1179–1184. doi: 10.1023/a:1020778018149. [DOI] [PubMed] [Google Scholar]
- Marino MD, Cronise K, Lugo J, Kelly SJ. Ultrasonic vocalizations and maternal infant interactions in a rat model of fetal alcohol syndrome. Dev Psychobiol. 2002;41:341–51. doi: 10.1002/dev.10077. [DOI] [PubMed] [Google Scholar]
- Mattson SN, Riley EP, Gramling L, Delis DC, Jones KL. Heavy prenatal alcohol exposure with or without physical features of fetal alcohol syndrome leads to IQ deficits. J Pediatr. 1997;131:718–721. doi: 10.1016/s0022-3476(97)70099-4. [DOI] [PubMed] [Google Scholar]
- Mehedint MG, Niculescu MD, Craciunescu CN, Zeisel SH. Choline deficiency alters global histone methylation and epigenetic marking at the Rel site of the calbindin 1 gene. FASEB J. 2010;24:184–195. doi: 10.1096/fj.09-140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MW. Generation of neurons in the rat dentate gyrus and hippocampus: effects of prenatal and postnatal treatment with ethanol. Alcohol Clin Exp Res. 1995;19:1500–1509. doi: 10.1111/j.1530-0277.1995.tb01014.x. [DOI] [PubMed] [Google Scholar]
- Monk BR, Leslie FM, Thomas JD. The effects of perinatal choline supplementation on Hippocampal cholinergic development in rats exposed to alcohol during the brain growth spurt. Hippocampus. 2011 doi: 10.1002/hipo.22009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya D, Swartzwelder HS. Prenatal choline supplementation alters hippocampal N methyl-D-aspartate receptor-mediated neurotransmission in adult rats. Neurosci Lett. 2000;296:85–88. doi: 10.1016/s0304-3940(00)01660-8. [DOI] [PubMed] [Google Scholar]
- Moretti P, Levenson JM, Battaglia F, Atkinson R, Teague R, Antalffy B, Armstrong D, Arancio O, Sweatt JD, Zoghbi HY. Learning and memory and synaptic plasticity are impaired in a mouse model of rett syndrome. J Neurosci. 2006;26:319–327. doi: 10.1523/JNEUROSCI.2623-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahara AH, Handa RJ. Fetal alcohol exposure alters the induction of immediate early gene mRNA in the rat prefrontal cortex after an alternation task. Alcohol Clin Exp Res. 1995;19:1389–1397. doi: 10.1111/j.1530-0277.1995.tb00997.x. [DOI] [PubMed] [Google Scholar]
- Napoli I, Blusztajn JK, Mellott TJ. Prenatal choline supplementation in rats increases the expression of IGF2 and its receptor IGF2R and enhances IGF2-induced acetylcholine release in hippocampus and frontal cortex. Brain Res. 2008;1237:124–135. doi: 10.1016/j.brainres.2008.08.046. [DOI] [PubMed] [Google Scholar]
- Niculescu MD, Craciunescu CN, Zeisel SH. Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. FASEB J. 2006;20:43–49. doi: 10.1096/fj.05-4707com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu MD, Yamamuro Y, Zeisel SH. Choline availability modulates human neuroblastoma cell proliferation and alters the methylation of the promoter region of the cyclin dependent kinase inhibitor gene. J Neurochem. 2004;89:1252–1259. doi: 10.1111/j.1471-4159.2004.02414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez HD, Villanueva JE, Salas JM. Behavioral and hippocampal morphological changes induced by ethanol administration to pregnant rats. Annals NY Acad Sci. 1991:300–304. doi: 10.1111/j.1749-6632.1991.tb33855.x. [DOI] [PubMed] [Google Scholar]
- Pogribny IP, Karpf AR, James SR, Melnyk S, Han T, Tryndyak VP. Epigenetic alterations in the brain of Fisher 344 rats induced by long-term administration of folate/methyl-deficient diet. Brain Res. 2008;1237:25–34. doi: 10.1016/j.brainres.2008.07.077. [DOI] [PubMed] [Google Scholar]
- Pogribny IP, Ross SA, Wise C, Pogribna M, Jones EA, Tryndyak VP, James SJ, Dragan YP, Poirier LA. Irreversible global DNA hypomethylation as a key step in hepatocarcinogenesis induced by dietary methyl deficiency. Mutat Res. 2006;593:80–87. doi: 10.1016/j.mrfmmm.2005.06.028. [DOI] [PubMed] [Google Scholar]
- Riikonen R, Salonen I, Partanen K, Verho S. Brain perfusion SPECT and MRI in foetal alcohol syndrome. Dev Med Child Neurol. 1999;41:652–659. doi: 10.1017/s0012162299001358. [DOI] [PubMed] [Google Scholar]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Ryan SH, Williams JK, Thomas JD. Choline supplementation attenuates learning deficits associated with neonatal alcohol exposure in the rat: effects of varying the timing of choline administration. Brain Res. 2008;1237:91–100. doi: 10.1016/j.brainres.2008.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandstrom NJ, Loy R, Williams CL. Prenatal choline supplementation increases NGF levels in the hippocampus and frontal cortex of young and adult rats. Brain Res. 2002;947:9–16. doi: 10.1016/s0006-8993(02)02900-1. [DOI] [PubMed] [Google Scholar]
- Smith DE, Davies DL. Effect of perinatal administration of ethanol on the CA1 pyramidal cell of the hippocampus and Purkinje cell of the cerebellum: an ultrastructural survey. J Neurocytol. 1990;19:708–717. doi: 10.1007/BF01188039. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Lu LH, O'Hare ED, McCourt ST, Mattson SN, O'Connor MJ, Bookheimer SY. Functional magnetic resonance imaging of verbal learning in children with heavy prenatal alcohol exposure. Neuroreport. 2007;18:635–639. doi: 10.1097/WNR.0b013e3280bad8dc. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Thompson PM, Mattson SN, Tessner KD, Jernigan TL, Riley EP, Toga AW. Regional brain shape abnormalities persist into adolescence after heavy prenatal alcohol exposure. Cereb Cortex. 2002;12:856–865. doi: 10.1093/cercor/12.8.856. [DOI] [PubMed] [Google Scholar]
- Streissguth AP, O'Malley K. Neuropsychiatric implications and long-term consequences of fetal alcohol spectrum disorders. Semin Clin Neuropsychiatry. 2000;5:177–190. doi: 10.1053/scnp.2000.6729. [DOI] [PubMed] [Google Scholar]
- Tanaka H. Fetal alcohol syndrome: a Japanese perspective. Ann Med. 1998;30:21–26. doi: 10.3109/07853899808999381. [DOI] [PubMed] [Google Scholar]
- Thomas JD, Abou EJ, Dominguez HD. Prenatal choline supplementation mitigates the adverse effects of prenatal alcohol exposure on development in rats. Neurotoxicol Teratol. 2009;31:303–311. doi: 10.1016/j.ntt.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JD, Biane JS, O'Bryan KA, O'Neill TM, Dominguez HD. Choline supplementation following third-trimester-equivalent alcohol exposure attenuates behavioral alterations in rats. Behav Neurosci. 2007;121:120–130. doi: 10.1037/0735-7044.121.1.120. [DOI] [PubMed] [Google Scholar]
- Thomas JD, Garrison M, O'Neill TM. Perinatal choline supplementation attenuates behavioral alterations associated with neonatal alcohol exposure in rats. Neurotoxicol Teratol. 2004;26:35–45. doi: 10.1016/j.ntt.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Thomas JD, La Fiette MH, Quinn VRE, Riley EP. Neonatal choline supplementation ameliorates the effects of prenatal alcohol exposure on a discrimination learning task in rats. Neurotoxicol Teratol. 2000;22:703–711. doi: 10.1016/s0892-0362(00)00097-0. [DOI] [PubMed] [Google Scholar]
- Thomas JD, Tran TD. Choline supplementation mitigates trace, but not delay, eyeblink conditioning deficits in rats exposed to alcohol during development. Hippocampus. doi: 10.1002/hipo.20925. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TD, Kelly SJ. Critical periods for ethanol-induced cell loss in the hippocampal formation. Neurotoxicol Teratol. 2003;25:519–528. doi: 10.1016/s0892-0362(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Uecker A, Nadel L. Spatial but not object memory impairments in children with fetal alcohol syndrome. Am J Ment Retard. 1998;103:12–18. doi: 10.1352/0895-8017(1998)103<0012:SBNOMI>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Uteshev VV, Meyer EM, Papke RL. Regulation of neuronal function by choline and 4OH GTS-21 through α7 nicotinic receptors. J Neurophysiol. 2003;89:1797–1806. doi: 10.1152/jn.00943.2002. [DOI] [PubMed] [Google Scholar]
- Wagner AF, Hunt PS. Impaired trace fear conditioning following neonatal ethanol: reversal by choline. 2006;120:482–487. doi: 10.1037/0735-7044.120.2.482. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Dolinoy DC, Lin J-R, Smith CA, Shi X, Tahiliani KG. Maternal methyl supplements increase offspring DNA methylation at axin fused. Genesis. 2006;44:401–40. doi: 10.1002/dvg.20230. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JR, Hamre KM, Pierce DR. Delay in brain growth induced by alcohol in artificially reared rat pups. Alcohol. 1984;1:213–222. doi: 10.1016/0741-8329(84)90101-0. [DOI] [PubMed] [Google Scholar]
- West JR, Hodges CA, Black AC. Prenatal exposure to ethanol alters the organization of hippocampal mossy fibers in rats. Science. 1981;211:957–959. doi: 10.1126/science.7466371. [DOI] [PubMed] [Google Scholar]
- West JR, Hodges-Savola CA. Permanent hippocampal mossy fiber hyperdevelopment following prenatal ethanol exposure. Neurobehav Toxicol Teratol. 1983;5:139–150. [PubMed] [Google Scholar]
- West JR, Pierce DR. The effect of in utero ethanol exposure on hippocampal mossy fibers: an HRP study. Brain Res. 1984;317:275–279. doi: 10.1016/0165-3806(84)90104-4. [DOI] [PubMed] [Google Scholar]
- Whitcher LT, Klintsova AY. Postnatal binge-like alcohol exposure reduces spine density without affecting dendritic morphology in rat mPFC. Synapse. 2008;62:566–573. doi: 10.1002/syn.20532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigal T, Amsel A. Behavioral and neuroanatomical effects of prenatal, postnatal, or combined exposure to ethanol in weanling rats. Behav Neurosci. 1990;104:116–126. doi: 10.1037//0735-7044.104.1.116. [DOI] [PubMed] [Google Scholar]
- Williams CL, Meck WH, Heyer DD, Loy R. Hypertrophy of basal forebrain neurons and enhanced visuospatial memory in perinatally choline supplemented rats. Brain Res. 1998;794:225–238. doi: 10.1016/s0006-8993(98)00229-7. [DOI] [PubMed] [Google Scholar]
- Willoughby KA, Sheard ED, Nash K, Rovet J. Effects of prenatal alcohol exposure on hippocampal volume, verbal learning, and verbal and spatial recall in late childhood. J Int Neuropsychol Soc. 2008;14:1022–1033. doi: 10.1017/S1355617708081368. [DOI] [PubMed] [Google Scholar]
- Wong-Goodrich SJE, Mellott TJ, Glenn MJ, Blusztajn JK, Williams CL. Prenatal choline supplementation attenuates neuropathological response to status epilepticus in the adult rat hippocampus. Neurobiol Dis. 2008a;30:255–269. doi: 10.1016/j.nbd.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong-Goodrich SJE, Glenn MJ, Mellott TJ, Blusztajn JK, Meck WH, Williams CL. Spatial memory and hippocampal plasticity are differentially sensitive to the availability of choline in adulthood as a function of choline supply in utero. Brain Res. 2008b;1237:153–166. doi: 10.1016/j.brainres.2008.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel SH. What choline metabolism can tell us about the underlying mechanisms of Fetal Alcohol Spectrum Disorders. Mol Neurobiol. 2011;44:185–191. doi: 10.1007/s12035-011-8165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel SH, Blusztajn JK. Choline and human nutrition. Annu Rev Nutr. 1994;14:269–96. doi: 10.1146/annurev.nu.14.070194.001413. [DOI] [PubMed] [Google Scholar]
- Zeisel SH, Niculescu MD. Perinatal choline influences brain structure and function. Nutr Rev. 2006;64:197–203. doi: 10.1111/j.1753-4887.2006.tb00202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FC, Zhao Q, Liu Y, Goodlett CR, Lian T, McClintick JN, Edenberg HJ, Li L. Alteration of gene expression by alcohol exposure at early neurulation. BMC Genomics. 2011;21 doi: 10.1186/1471-2164-12-124. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]