Abstract
While most carriers of human T-cell leukemia virus type 1 (HTLV-1) remain asymptomatic throughout their lifetime, infection is associated with the development of adult T cell leukemia (ATL) and HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP). The exact parameters that determine these outcomes are unknown but are believed to include host genetic factors that control the immune response to infection. Host response to fellow retroviridae member HIV is influenced by the expression of members of the Killer Immunoglobulin Receptor (KIR) family including KIR3DS1. In this study we examined the association of KIR3DS1 with the outcome of HTLV-1 infection in three geographically distinct cohorts (Jamaican, Japanese and Brazilian). Despite increased prevalence of KIR3DS1 in the HAM/TSP patients of the Jamaican cohort, we found no evidence for a role of KIR3DS1 in influencing control of proviral load or disease outcome. This suggests that unlike HIV, KIR3DS1-mediated regulation of HTLV-1 infection does not occur, or is ineffective.
1. Introduction
Human T-cell leukemia virus type 1 (HTLV-1) is a human retrovirus, first discovered in the 1980s, that is believed to currently infect between 10 and 20 million people worldwide [1, 2]. HTLV-1 is endemic in many areas of the globe, including southwestern Japan, Africa, the Caribbean and Central and South America, particularly Brazil [3]. While most infected individuals remain asymptomatic (AS), between 1–5% globally develop an aggressive malignancy, adult T cell Leukemia (ATL), and a similar fraction develop the chronic neurological condition HAM/TSP (HTLV-1 associated myelopathy/tropical spastic paraparesis) [4].
HTLV-1 is found predominately as a cell associated virus, with little or no detectable cell-free virions in the plasma of infected individuals. The factors that control the various outcomes of HTLV-1 infection (AS, ATL, or HAM/TSP) are largely unknown, but variation in the virus sequence is not believed to play a dominant role. Mother to child transmission of the virus appears to be a risk factor for the development of ATL, whereas sexual transmission is more likely to give rise to HAM/TSP [4]. Although the sequence of viral isolates are remarkable consistent across populations [6], there are intriguing differences in the geographical distribution of disease outcomes, for example, ATL is much more common than HAM/TSP in Southwestern Japan [3]. These data suggest that in addition to route of transmission, host genetic factors, controlling perhaps the host immune response, may be critical in determining the final outcome of HTLV-1 infection and therefore development of associated conditions. Indeed, high proviral load is associated with the development of HAM/TSP – where the amount of provirus can be up to five times the levels seen in asymptomatic carriers [7]. Proviral load in turn is controlled by the host immune response with a dominant role for an effective cytotoxic T lymphocyte (CTL) response [8]. Consistent with the role of CTL in determining outcome, in Japanese cohorts both HLA-A*02 and HLA-C*08 are protective against the development of HAM/TSP, while the presence of HLA-B*54 is associated with higher levels of HAM/TSP. This is reportedly due to the elimination of infected cells displaying immunogenic HLA-A2 or -C*08 restricted HTLV-1 epitopes resulting in reduced proviral loads [9]. Recently, MacNamara et al have shown that the ability of HLA to present peptides derived from the viral protein HBZ to CD8+ T cells are most protective in HTLV-1 infection [10].
In addition to their role in presenting peptides to CTLs, HLA also acts as a ligand for members of the KIR family. The KIR (Killer Immunoglobulin-like Receptors) are a family of receptors expressed primarily on Natural Killer (NK) cells, which includes both inhibitory and activating members [11]. The genes encoding these receptors are very diverse – with variation among people in the presence or absence of individual genes as well as a high degree of allelic polymorphism of individual genes [12]. A member of this family, KIR3DS1, has been implicated in the response to HIV. Genetic association studies have suggested that HIV infected individuals who express both KIR3DS1 and HLABw4 (in particular Bw4 molecules with isoleucine at position 80, HLABw4-80I) have lower viral loads and progress more slowly towards AIDS [13]. There is also evidence that the KIR3DS1 expressing population of NK cells expands during acute HIV infection, and is capable of controlling viral replication in vitro [14, 15]. Interestingly, this putative interaction between KIR3DS1 and HLABw4-80I molecules has proved impossible to directly detect thus far, even using assays successful with other activating KIR [16, 17]. Moreover, while it has been suggested that presentation of an HIV-specific peptide by a HLABw4 molecule may be the critical factor in facilitating activation of KIR3DS1, repeated attempts to identify this peptide have failed [18]. An alternative hypothesis is that the activation of KIR3DS1+ NK cells may not be dependent on a HIV-specific signal but rather occur as a direct or indirect consequence of retroviral infection. If this is the case, it might be predicted that KIR3DS1 would play a role in the response to other retroviral infections such as HTLV-1. The ability of KIR3DS1 to suppress viral load in HIV infections suggests that, if it participates in anti-HTLV-1 responses, those patients that express this receptor may show lower viral loads and protection from the development of HAM/TSP. To address this possibility we initially assessed the role of carriage of KIR3DS1 on disease outcome in a Jamaican cohort of HTLV-1 patients. We then extended this analysis and compared the frequency of KIR3DS1, and its putative ligand HLABw4-80I, between HAM/TSP and asymptomatic carriers in two additional larger cohorts of HTLV-1 patients.
2. Methods
2.1 Study Subjects
Jamaican Cohort
This cohort consists of 56 asymptomatic carriers (AS), 34 adult T cell leukemia (ATL) and 21 HAM/TSP patients, identified through nationwide disease registries and referrals to the university clinic in Jamaica, or from participants in a nationwide serosurvey [19]. None of the samples are from biologically related individuals. Informed consent was obtained from all participants. Study protocols followed the human experimentation guidelines of the US Department of Health and Human Services and Institutional Review Board approvals at the National Cancer Institute and University of the West Indies.
Brazilian Cohort
This cohort consists of 145 asymptomatic carriers (AS), and 71 HAM/TSP patients. All Brazilian patients included in this study were followed at the Bahia School of Medicine and Public Health HTLV reference center in Salvador, located in northeastern Brazil, where HTLV-1 is prevalent [20, 21]. Informed consent was obtained from all participants. HAM/TSP diagnosis was made according established criteria [22]. Study protocols followed the human experimentation guidelines of the US Department of Health and Human Services and Institutional Review Board approvals at the National Cancer Institute and by both Institutional Review Board of the Oswaldo Cruz Foundation, and National Commission on Ethics in Research (CONEP record: 15271), Brazilian Ministry of Health.
Japanese Cohort
This cohort (N=402) consists of individuals recruited in Kagoshima Japan and has previously been described [23]. All individuals were of Japanese ethnic origin and resided in Kagoshima Prefecture, Japan. The cohort had 222 HAM/TSP patients and 180 asymptomatic carriers. HAM/TSP diagnosis was made according to World Health Organization criteria [24].
2.2 KIR and HLA Genotyping
HLA and KIR3DS1 genotyping was carried out as previously described [23, 25, 26].
2.3 Proviral Load Measurement
The proviral load (PVL) of the Brazilian cohort was measured as described [27]. In brief, SK110/SK111 primers were used to amplify a fragment of the pol gene and dual TaqMan probe was located at within the HTLV-1 reference sequence (HTLVATK). Albumin DNA was used as an endogenous reference. The value of HTLV-1 proviral load was reported as the [(HTLV-1 average copy number) / (albumin average copy number)] × 2 × 106 and expressed as the number of HTLV-1copies per 106 cells in PBMCs. The PVL of the Japanese cohort was measured as described [7]. In brief, a quantitative PCR reaction was performed by using an ABI 7700 sequence detector. A standard curve was generated by using the -actin gene from HTLV-1 negative PBMC and the Tax gene from TARL-2, a cell line containing a single copy of HTLV-I proviral DNA. The amount of HTLV-I proviral DNA was calculated as follows: copy number of HTLV-I (tax) per 104 PBMC = [copy number of tax/(copy number of β-actin/2)] × 104.
2.4 Statistical analysis
We used logistic regression to investigate the impact of KIR3DS1 with or without HLA-HLABw4-80I on status with age and gender included in the model as covariates. We used linear regression to investigate the impact of KIR3DS1 on log proviral load, with age and gender included in the model as covariates. Statistical analysis was done using SAS or R statistical software package.
2.5 In vitro assays
MT-2, K562, UC 7296, and its transfectants were maintained in RPMI 1640 supplemented with 10% FCS. UC 7296 cell line transfectants were a kind gift from Dr Patrick Green, Ohio State University. Natural Killer cells were isolated from human peripheral blood using StemSep® Human NK Cell Enrichment Kit according to the manufacturer's instructions. Purified NK cells were plated at 1 × 106 cell/ml and stimulated with cell lines at a stimulator:responder ratio of 1:5. Anti-CD107a Ab was added at time of stimulation and 10μM monensin was added 1 h later. Cells were incubated at 37°C for an additional 5 h and then stained with anti-3DL1/S1 (clone Z27), and anti-CD56. KIR3DL1+ cells were identified as Z27hi cells, KIR3DS1+ cells were identified as Z27lo cells as described [16].
3. Results
To examine a possible role for KIR3DS1 in the outcome of HTLV-1 infection, we assessed the presence of this gene in cohorts of HTLV-1 infected patients (see Table 1). In a well-characterized Jamaican cohort [19], we examined 56 asymptomatic carriers (AS), 34 with ATL and 21 with HAM/TSP. In this limited cohort, we detected an apparent increased frequency of KIR3DS1 in patients with HAM/TSP (23.8% versus 3.6% for asymptomatic carriers and 8.8% for ATL patients; Table 2).
Table 1.
Summary of Population Demographics
| Population | n | Age Mean [rangel | Gender Male, female | Reference |
|---|---|---|---|---|
| Jamaican | 111 | - | - | 17 |
| Brazilian | 216 | 54 [10–89] | 68%, 32% | - |
| Japanese | 402 | 49 [15–80] | 38%, 62% | 22 |
Table 2.
KIR3DS1 Frequency in Jamaican HTLV-1 Cohort
| Group | N | 3DS1+ (%) |
|---|---|---|
| AS | 56 | 3.6 |
| ATL | 34 | 8.8 |
| HAM/TSP | 21 | 23.8 |
In light of the suggestion from the Jamaican cohort that KIR3DS1 may predispose HTLV-1 infected patients to the development of the neurological condition HAM/TSP, we extended our analysis to a larger cohort of patients. To this end, we examined two additional cohorts. The first, from Brazil consisted of 177 AS and 102 HAM/TSP, where we did not detect any increased frequency of KIR3DS1 in those who developed HAM/TSP. In this cohort the frequency of KIR3DS1 was 27.3% in asymptomatic carriers compared with 30.3% in the HAM/TSP group (p=0.59, Table 3).
Table 3.
Analysis of KIR3DS1 and HLA-Bw480I Influence on Disease Outcome in Brazilian HTLV-1 Cohort
|
|
||||||
|---|---|---|---|---|---|---|
| AS | HAM/TSP | |||||
|
|
||||||
| n | % | n | % | Odds Ratio [95% CI] | P Value | |
|
|
||||||
| 3DS1 | 145 | 27.3 | 71 | 30.3 | 1.18 [0.64–2.18] | 0.59 |
| Bw480I | 145 | 59.3 | 71 | 68.8 | 1.19 [0.63–2.23] | 0.59 |
| 3DS1 + Bw480I | 145 | 16.6 | 71 | 18.3 | 1.13 [0.60–2.15] | 0.7 |
The ligand for KIR3DS1 is believed to be the subgroup of HLA molecules that carry the Bw4 public epitope, specifically those with isoleucine at position 80 (HLABw4-80I). Due to the prominent role of HLA in the immune response and the known influence of HLA polymorphism on the outcome of a variety of diseases, we also examined the HLA genotypes of these patients. Carriage of Bw4-80I, either alone or in combination with KIR3DS1 did not show any correlation with disease outcome. Of asymptomatic carriers, 59.3% expressed Bw4-80I compared with 68.8% of HAM/TSP (p=0.59), while joint KIR3DS1+Bw4 was found in 16.6% of asymptomatic patients and 18.3% of HAM/TSP (p=0.59, Table 3). This result suggests that, at least in this Brazilian cohort, KIR3DS1 expression, even in the presence of the best candidate for its ligand, did not influence the development of HAM/TSP in HTLV-1 patients.
To confirm this finding, we examined an additional, ethnically distinct cohort of HTLV-1 infected patients. This cohort from Kagoshima Japan consists of 180 asymptomatic and 222 HAM/TSP patients. We genotyped these patients for the presence of KIR3DS1 and used this data in combination with the previously reported HLA information [23]. Similar to what we observed in the Brazilian cohort, we could not detect any role for KIR3DS1 in the development of HAM/TSP, with a KIR3DS1 frequency of 37.8% versus 36.5% (OR=1.19, p=0.5) in the asymptomatic carriers and the HAM/TSP patients respectively (Table 4). Expression of the proposed KIR3DS1 ligand Bw4-80I, either alone or in combination with KIR3DS1 was also not found to be higher in the HAM/TSP group.
Table 4.
Analysis of KIR3DS1 and HLA-Bw480I Influence on Disease Outcome in a Japanese HTLV-1 Cohort
|
|
||||||
|---|---|---|---|---|---|---|
| AS | HAM/TSP | |||||
|
|
||||||
| n | % | n | % | Odds Ratio [95% CI] | P Value | |
|
|
||||||
| 3DS1 | 180 | 37.8 | 222 | 36.5 | 1.19 [0.71–2.01] | 0.5 |
| Bw480I | 180 | 35.6 | 222 | 39.6 | 0.79 [0.47–1.32] | 0.37 |
| 3DS1+ Bw480I | 180 | 13.9 | 222 | 13.5 | 1.44 [0.71–2.9] | 0.3 |
Development of HAM/TSP is likely to be a complex process, dependent on a range of host, genetic and viral factors. The evidence that indicates KIR3DS1 does not play a role in the development of this condition does not preclude the possibility that KIR3DS1 is involved in the immune response to HTLV-1. To examine more broadly the potential role of KIR3DS1 in the control of this viral infection, we analyzed the effect of KIR3DS1 carriage on the proviral load of patients, both asymptomatic carriers and HAM/TSP, in the two larger cohorts. As shown in Table 5, we found no evidence for an effect of KIR3DS1 in proviral load levels, on its own or in the presence of its putative ligand, in either cohort.
Table 5.
Analysis of KIR3DS1 and HLA-Bw4 Influence on Proviral Load in HTLV-1 Cohorts
|
|
||||
|---|---|---|---|---|
| AS | HAM/TSP | |||
|
|
||||
| Slope | P value | Slope | P value | |
|
|
||||
| Japanese | ||||
| KIR3DS1 | −0.25 | 0.09 | −0.096 | 0.27 |
| KIR3DS1+Bw480I | −0.3 | 0.1 | −0.169 | 0.12 |
| Brazilian | ||||
| KIR3DS1 | −0.08 | 0.72 | 0.13 | 0.49 |
| KIR3DS1+Bw480I | −0.08 | 0.74 | 0.07 | 0.80 |
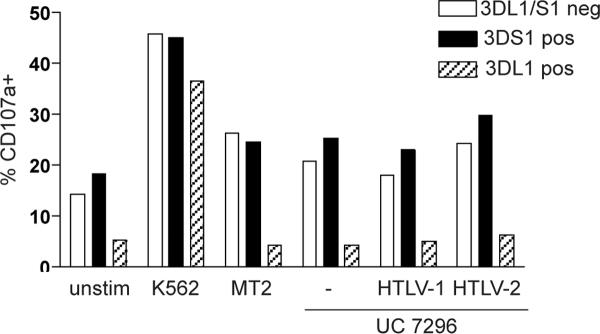
Lastly, we examined the possibility that KIR3DS1 might recognize a ligand on the surface of HTLV-1 infected cells, either a peptide presented by HLA-Bw4, a complex involving HLABw4, or an HLA-independent ligand. To do this, we used the well-characterized HTLV-1 positive MT-2 T cell line along with a panel of B cell (UC-7296) transfectants, expressing either HTLV-1 or HTLV-2 [28, 29]. We first verified that both MT-2 and UC-7296 cell lines express HLA-Bw4 by staining with an anti-Bw4 antibody (data not shown). We then used these cells as targets in an NK cell degranulation assay. By co-staining the cells with anti-KIR antibodies, we were able to identify cells expressing KIR3DS1 or KIR3DL1 or neither. KIR3DL1 is very similar to KIR3DS1, however it readily interacts with HLABw4 and delivers an inhibitory rather than an activating signal. As expected, all NK cell populations showed increased expression of the degranulation marker CD107a in response to the HLA class I negative K562 cell line (Figure 1). Similarly, there was an increase in CD107a expression in the KIR3DL1/S1 negative population in response to MT2 and UC-7296 cell lines. The KIR3DL1 expressing subset did not up-regulate CD107a due to the presence of HLABw4, a ligand for this inhibitory receptor. Importantly, there was no further increase in CD107a expression in the KIR3DS1 subset, either in comparison to KIR negative cells, or for the UC-7629 parental cell line in comparison to the HTLV-1 or -2 transfectants. These data suggest that there is no ligand for KIR3DS1 on these cells.
Figure 1. KIR3DS1+ NK cells are not preferentially activated by HTLV-1 expressing cells.
Freshly isolated NK cells were stimulated for 6 h with the indicated cell lines at a ratio of 5:1. Cells were analyzed by flow cytometry and KIR3DS1 and KIR3DL1 positive NK cell populations were identified. Representative data are shown from one of two independent experiments.
4. Discussion
The role of KIR3DS1 in retroviral infections remains an exciting but elusive prospect. The genetic data that support a protective role for KIR3DS1 in HIV infection continues to accumulate and is supported by studies of in vitro interaction of KIR3DS1+ cells with autologous HIV-infected targets and evidence of in vivo expansion [13–15]. In light of the recent emergence of HIV, specific activation of KIR3DS1 during HIV infection cannot have been the driving force behind the evolution of this KIR. In contrast, HTLV-1 has been present in the human population for a much longer period of time [30, 31]. It is tempting to speculate that KIR3DS1 may have arisen to recognize changes induced upon infection with HTLV-1 (e.g. a peptide associated with the virus, or the cellular response to the infection), which now also results in recognition of HIV-infected cells. This hypothesis predicts that KIR3DS1 would play a role in modulating the immune response to HTLV-1. In this study we examined the frequency of KIR3DS1 in HTLV-1 patients but did not detect any consistent role for KIR3DS1 in determining the outcome of the disease or in controlling viral load.
Our initial study focused on a cohort of Jamaican patients, where we saw a low frequency of KIR3DS1 in the population. This is likely to be a reflection of African admixture in this population, as the frequency of KIR3DS1 is known to be low in those of African descent [32]. In this cohort we saw an increased frequency of KIR3DS1 in HAM/TSP patients. We next expanded our study to include two (larger) additional cohorts with Brazilian and Japanese patients. In both of these populations the frequency of KIR3DS1 was higher, and in line with reported frequencies for these populations (www.allelefrequencies.net). The frequency of HLABw4-80I was higher in the Brazilian cohort than in the Japanese, but did not differ dramatically between disease groups. However in these two cohorts, we found no association of KIR3DS1, either alone or together with HLABw4-80I, with disease outcome. This is in agreement with the study of Talledo et al which found no difference in the presence of the KIR3DS1 gene in HAM/TSP patients relative to ACs in a cohort of HTLV-1 patients in Peru [33]. Our initial finding in the Jamacian cohort may be a reflection of the small size of this group or a reflection of bias in the selection of the AS group which were identified from a nationwide serosurvey rather than disease registries and clinics. In addition, the protective role of KIR3DS1 in HIV has been reported in Caucasian populations, and there may be confounding population differences.
The lack of any detectable influence of KIR3DS1 on disease outcome is potentially due to a number of reasons. We had predicted that carriage of KIR3DS1 would lead to a reduction in viral load (as seen in HIV), which would in turn protect against the development of HAM/TSP. However, it is possible that KIR3DS1 plays a conflicting role in HAM/TSP and, despite early control of viral load, may actually contribute to disease progression and clinical manifestation by supporting the inflammatory conditions seen in the spinal cord lesions [34]. This idea is challenged however by the proviral load data, which suggest that KIR3DS1 is not effective in reducing viral replication and/or expansion of HTLV-I infected clones.
By examining three different cohorts of HTLV-1 patients we were able to exclude a prominent role for KIR3DS1 in controlling viral load or influencing disease outcome. In contrast to HIV, KIR3DS1 expressing NK cells do not appear to recognize or respond to HTLV-1 infection. This suggests that there are unique conditions present in HIV infection, not seen in HTLV-1, that control KIR3DS1 activation. Candidates are numerous and include expression of a specific HIV peptide and the presence of free viral particles and intermediates from viral replication such as RNA, known to activate NK cells, among others. As we continue to expand our understanding of KIR3DS1 function, and the types of pathogenic conditions in which it is involved, we grow closer to understanding its role and mechanism of action.
Acknowledgements
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. This research was supported in part by funding from the Wellcome Trust and Medical Research Council. This research was supported in part by the Brazilian National Research Council (CNPq).
We would like to acknowledge the kind assistance of K Usuku, M Osame, and CRM Bangham.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Hlela C, Shepperd S, Khumalo NP, Taylor GP. The prevalence of human T-cell lymphotropic virus type 1 in the general population is unknown. AIDS Rev. 2009;11:205–14. [PubMed] [Google Scholar]
- [2].Watanabe T. Current status of HTLV-1 infection. Int J Hematol. 2011;94:430–4. doi: 10.1007/s12185-011-0934-4. [DOI] [PubMed] [Google Scholar]
- [3].Verdonck K, Gonzalez E, Van Dooren S, Vandamme AM, Vanham G, Gotuzzo E. Human T-lymphotropic virus 1: recent knowledge about an ancient infection. Lancet Infect Dis. 2007;7:266–81. doi: 10.1016/S1473-3099(07)70081-6. [DOI] [PubMed] [Google Scholar]
- [4].Proietti FA, Carneiro-Proietti AB, Catalan-Soares BC, Murphy EL. Global epidemiology of HTLV-I infection and associated diseases. Oncogene. 2005;24:6058–68. doi: 10.1038/sj.onc.1208968. [DOI] [PubMed] [Google Scholar]
- [5].Slattery JP, Franchini G, Gessain A. Genomic evolution, patterns of global dissemination, and interspecies transmission of human and simian T-cell leukemia/lymphotropic viruses. Genome Res. 1999;9:525–40. [PubMed] [Google Scholar]
- [6].Manns A, Hisada M, La Grenade L. Human T-lymphotropic virus type I infection. Lancet. 1999;353:1951–8. doi: 10.1016/s0140-6736(98)09460-4. [DOI] [PubMed] [Google Scholar]
- [7].Nagai M, Usuku K, Matsumoto W, Kodama D, Takenouchi N, Moritoyo T, Hashiguchi S, Ichinose M, Bangham CR, Izumo S, Osame M. Analysis of HTLV-I proviral load in 202 HAM/TSP patients and 243 asymptomatic HTLV-I carriers: high proviral load strongly predisposes to HAM/TSP. J Neurovirol. 1998;4:586–93. doi: 10.3109/13550289809114225. [DOI] [PubMed] [Google Scholar]
- [8].Bangham CR, Osame M. Cellular immune response to HTLV-1. Oncogene. 2005;24:6035–46. doi: 10.1038/sj.onc.1208970. [DOI] [PubMed] [Google Scholar]
- [9].Jeffery KJ, Siddiqui AA, Bunce M, Lloyd AL, Vine AM, Witkover AD, Izumo S, Usuku K, Welsh KI, Osame M, Bangham CR. The influence of HLA class I alleles and heterozygosity on the outcome of human T cell lymphotropic virus type I infection. J Immunol. 2000;165:7278–84. doi: 10.4049/jimmunol.165.12.7278. [DOI] [PubMed] [Google Scholar]
- [10].Macnamara A, Rowan A, Hilburn S, Kadolsky U, Fujiwara H, Suemori K, Yasukawa M, Taylor G, Bangham CR, Asquith B. HLA class I binding of HBZ determines outcome in HTLV-1 infection. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gardiner CM. Killer cell immunoglobulin-like receptors on NK cells: the how, where and why. Int J Immunogenet. 2008;35:1–8. doi: 10.1111/j.1744-313X.2007.00739.x. [DOI] [PubMed] [Google Scholar]
- [12].Parham P. The genetic and evolutionary balances in human NK cell receptor diversity. Semin Immunol. 2008;20:311–6. doi: 10.1016/j.smim.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O'Brien SJ, Carrington M. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–34. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- [14].Alter G, Rihn S, Walter K, Nolting A, Martin M, Rosenberg ES, Miller JS, Carrington M, Altfeld M. HLA class I subtype-dependent expansion of KIR3DS1+ and KIR3DL1+ NK cells during acute human immunodeficiency virus type 1 infection. J Virol. 2009;83:6798–805. doi: 10.1128/JVI.00256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, Streeck H, Waring M, Meier A, Brander C, Lifson JD, Allen TM, Carrington M, Altfeld M. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204:3027–36. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].O'Connor GM, Guinan KJ, Cunningham RT, Middleton D, Parham P, Gardiner CM. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol. 2007;178:235–41. doi: 10.4049/jimmunol.178.1.235. [DOI] [PubMed] [Google Scholar]
- [17].Carr WH, Rosen DB, Arase H, Nixon DF, Michaelsson J, Lanier LL. Cutting Edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12-associated receptor expressed on NK cells that triggers NK cell activation. J Immunol. 2007;178:647–51. doi: 10.4049/jimmunol.178.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gillespie GM, Bashirova A, Dong T, McVicar DW, Rowland-Jones SL, Carrington M. Lack of KIR3DS1 binding to MHC class I Bw4 tetramers in complex with CD8+ T cell epitopes. AIDS Res Hum Retroviruses. 2007;23:451–5. doi: 10.1089/aid.2006.0165. [DOI] [PubMed] [Google Scholar]
- [19].Goedert JJ, Li HC, Gao XJ, Chatterjee N, Sonoda S, Biggar RJ, Cranston B, Kim N, Carrington M, Morgan O, Hanchard B, Hisada M. Risk of human T-lymphotropic virus type I-associated diseases in Jamaica with common HLA types. Int J Cancer. 2007;121:1092–7. doi: 10.1002/ijc.22767. [DOI] [PubMed] [Google Scholar]
- [20].Galvao-Castro B, Loures L, Rodriques LG, Sereno A, Ferreira OC, Junior, Franco LG, Muller M, Sampaio DA, Santana A, Passos LM, Proietti F. Distribution of human T-lymphotropic virus type I among blood donors: a nationwide Brazilian study. Transfusion. 1997;37:242–3. doi: 10.1046/j.1537-2995.1997.37297203532.x. [DOI] [PubMed] [Google Scholar]
- [21].Dourado I, Alcantara LC, Barreto ML, da Gloria Teixeira M, Galvao-Castro B. HTLV-I in the general population of Salvador, Brazil: a city with African ethnic and sociodemographic characteristics. J Acquir Immune Defic Syndr. 2003;34:527–31. doi: 10.1097/00126334-200312150-00013. [DOI] [PubMed] [Google Scholar]
- [22].De Castro-Costa CM, Araujo AQ, Barreto MM, Takayanagui OM, Sohler MP, da Silva EL, de Paula SM, Ishak R, Ribas JG, Rovirosa LC, Carton H, Gotuzzo E, Hall WW, Montano S, Murphy EL, Oger J, Remondegui C, Taylor GP. Proposal for diagnostic criteria of tropical spastic paraparesis/HTLV-I-associated myelopathy (TSP/HAM) AIDS Res Hum Retroviruses. 2006;22:931–5. doi: 10.1089/aid.2006.22.931. [DOI] [PubMed] [Google Scholar]
- [23].Jeffery KJ, Usuku K, Hall SE, Matsumoto W, Taylor GP, Procter J, Bunce M, Ogg GS, Welsh KI, Weber JN, Lloyd AL, Nowak MA, Nagai M, Kodama D, Izumo S, Osame M, Bangham CR. HLA alleles determine human T-lymphotropic virus-I (HTLV-I) proviral load and the risk of HTLV-I-associated myelopathy. Proc Natl Acad Sci U S A. 1999;96:3848–53. doi: 10.1073/pnas.96.7.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Osame M. Review of WHO Kagoshima meeting and diagnostic guidelines for HAM/TSP. In: Blattner WA, editor. Human retrovirology. 1990. pp. 191–7. [Google Scholar]
- [25].John Hansen e. Joint report: 13th IHWS, Section A5: Sequence based HLA typing. Proceedings of the 13th International Histocompatibility Workshop and Conference.pp. 304–416. [Google Scholar]
- [26].Kulkarni S, Martin MP, Carrington M. KIR genotyping by multiplex PCR-SSP. Methods Mol Biol. 2010;612:365–75. doi: 10.1007/978-1-60761-362-6_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dehee A, Cesaire R, Desire N, Lezin A, Bourdonne O, Bera O, Plumelle Y, Smadja D, Nicolas JC. Quantitation of HTLV-I proviral load by a TaqMan real-time PCR assay. J Virol Methods. 2002;102:37–51. doi: 10.1016/s0166-0934(01)00445-1. [DOI] [PubMed] [Google Scholar]
- [28].Anderson M, Green PL. Growth and manipulation of a human T-cell leukemia virus type 2 full-length molecular clone. Methods Mol Biol. 2005;304:409–21. doi: 10.1385/1-59259-907-9:409. [DOI] [PubMed] [Google Scholar]
- [29].Anderson MD, Ye J, Xie L, Green PL. Transformation studies with a human T-cell leukemia virus type 1 molecular clone. J Virol Methods. 2004;116:195–202. doi: 10.1016/j.jviromet.2003.11.016. [DOI] [PubMed] [Google Scholar]
- [30].Coulthart MB, Posada D, Crandall KA, Dekaban GA. On the phylogenetic placement of human T cell leukemia virus type 1 sequences associated with an Andean mummy. Infect Genet Evol. 2006;6:91–6. doi: 10.1016/j.meegid.2005.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li HC, Fujiyoshi T, Lou H, Yashiki S, Sonoda S, Cartier L, Nunez L, Munoz I, Horai S, Tajima K. The presence of ancient human T-cell lymphotropic virus type I provirus DNA in an Andean mummy. Nat Med. 1999;5:1428–32. doi: 10.1038/71006. [DOI] [PubMed] [Google Scholar]
- [32].Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, Bruno D, Carrington CV, Chandanayingyong D, Chang YH, Crespi C, Saruhan-Direskeneli G, Fraser PA, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Mila J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Verity DH, Vaughan RW, Tyan D, Davis RW, Riley EM, Ronaghi M, Parham P. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39:1092–9. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- [33].Talledo M, Lopez G, Huyghe JR, Verdonck K, Gonzalez E, Clark D, Senitzer D, Vanham G, Gotuzzo E, Van Camp G, Van Laer L. Role of killer cell immunoglobulin-like receptor gene content and human leukocyte antigen-C group in susceptibility to human T-lymphotropic virus 1-associated myelopathy/tropical spastic paraparesis in Peru. Hum Immunol. 2010;71:804–8. doi: 10.1016/j.humimm.2010.05.011. [DOI] [PubMed] [Google Scholar]
- [34].Matsuura E, Yamano Y, Jacobson S. Neuroimmunity of HTLV-I Infection. J Neuroimmune Pharmacol. 2010;5:310–25. doi: 10.1007/s11481-010-9216-9. [DOI] [PMC free article] [PubMed] [Google Scholar]