Abstract
Stem cells sustain tissue regeneration by their remarkable ability to replenish the stem cell pool and to generate differentiating progeny. Signals from local microenvironments, or niches, control stem cell behavior. In the Drosophila testis, a group of somatic support cells called the hub creates a stem cell niche by locally activating the Janus Kinase-Signal Transducer and Activator of Transcription (JAK-STAT) pathway in two adjacent types of stem cells: germline stem cells (GSCs) and somatic cyst stem cells (CySCs). Here, we find that ken and barbie (ken) is autonomously required for the self-renewal of CySCs but not GSCs. Furthermore, Ken misexpression in the CySC lineage induces the cell-autonomous self-renewal of somatic cells as well as the nonautonomous self-renewal of germ cells outside the niche. Thus, Ken, like Stat92E and its targets ZFH1 (Leatherman and Dinardo, 2008) and Chinmo (Flaherty et al., 2010), is necessary and sufficient for CySC renewal. However, ken is not a JAK-STAT target in the testis, but instead acts in parallel to Stat92E to ensure CySC self-renewal. Ken represses a subset of Stat92E targets in the embryo (Arbouzova et al., 2006) suggesting that Ken maintains CySCs by repressing differentiation factors. In support of this hypothesis, we find that the global JAKSTAT inhibitor Protein tyrosine phosphatase 61F (Ptp61F) is a JAK-STAT target in the testis that is repressed by Ken. Together, our work demonstrates that Ken has an important role in the in the inhibition of CySC differentiation. Studies of ken may inform our understanding of its vertebrate orthologue B-Cell Lymphoma 6 (BCL6) and how misregulation of this oncogene leads to human lymphomas.
Keywords: ken and barbie, Drosophila, JAK-STAT, stem cell, niche
Introduction
Stem cells divide asymmetrically to give rise to one daughter that remains a stem cell (a process known as self-renewal) and another daughter that commits to differentiation. In this way, stem cells are able to provide a continuous source of differentiating cells for tissue regeneration while sustaining the original stem cell population. Signals from niches, or local microenvironments that regulate stem cell behavior, regulate the decision between stem cell fate and differentiation (Lin, 2002). Some of the best-characterized stem cell niches are found in the Drosophila gonads (Fuller and Spradling, 2007). The stem cells found in these tissues can be identified at single-cell resolution with markers that easily distinguish them from their differentiating progeny as well as from neighboring niche-generating cells. Furthermore, stem cells and their niches can be genetically manipulated in vivo in order to investigate the molecular requirements for stem cell maintenance (Morrison and Spradling, 2008). Studies using Drosophila spermatogenesis as a model system have shown that multiple conserved signaling pathways regulate stem cell self-renewal and differentiation in the testis niche (de Cuevas and Matunis, 2011).
Two populations of stem cells reside in the apex of the Drosophila testis: germline stem cells (GSCs) which produce sperm, and somatic stem cells known as cyst stem cells (CySCs) which produce support cells. Both types of stem cells are anchored around a cluster of somatic support cells known as the hub. The hub specifically expresses the secreted glycoprotein Upd, which activates the highly conserved Janus Kinase-Signal Transducer and Activator of Transcription (JAK-STAT) signaling pathway in adjacent stem cells via the transmembrane receptor Domeless. JAK-STAT signaling is required for the maintenance of both GSCs (Kiger et al., 2001; Tulina and Matunis, 2001) and CySCs (Issigonis et al., 2009; Leatherman and Dinardo, 2008). In CySCs, activation of JAK-STAT signaling leads to the expression of the Stat92E target Zinc finger homeodomain 1 (ZFH1), which is highly expressed in CySCs and quickly downregulated in cyst cell daughters (Leatherman and Dinardo, 2008). Similar to Stat92E, ZFH1 is required intrinsically for CySC maintenance; zfh1 or Stat92E mutant CySCs differentiate within 2-3 days (Issigonis et al., 2009; Leatherman and Dinardo, 2008). Furthermore, sustained activation of the JAK-STAT signaling pathway or its target zfh1 in the CySCs and cyst cells is sufficient to cause CySC-like cells to accumulate throughout the testis, far outside of the normal niche region. A striking consequence of this phenotype is that the excess CySCs nonautonomously promote the accumulation of GSCs throughout the testis. This is remarkable considering that ectopic activation of the JAK-STAT pathway throughout the germline is not sufficient to prevent differentiation of the germ cells (Leatherman and Dinardo, 2008). However, a yet unidentified signal from CySCs which activates the BMP pathway in neighboring GSCs may be partially responsible for the maintenance of GSCs in a GSC-like state (Leatherman and Dinardo, 2010). Therefore, the GSC niche is made up not only of hub cells, but CySCs as well.
GSCs and CySCs typically divide asymmetrically, such that one daughter cell remains adjacent to the hub while the other one gets pushed away from the niche (Cheng et al., 2011; Sheng and Matunis, 2011; Yamashita et al., 2003). Since Upd appears to act over a short distance, the GSC and CySC daughters (known as gonialblasts and cyst cells, respectively) that are displaced from the hub no longer receive the signals that specify stem cell identity and begin to differentiate. The gonialblast daughter undergoes four mitotic divisions with incomplete cytokinesis resulting in 16 interconnected spermatogonia, which further differentiate, undergoing meiosis and spermiogenesis to form sperm. Cyst cell daughters exit the mitotic cycle, but increase in size as they differentiate. Pairs of cyst cells continue to envelop each gonialblast and its descendants throughout spermatogenesis. In fact, encystment of the germline by the cyst cells is essential for their proper differentiation (Kiger et al., 2001; Matunis et al., 1997; Sarkar et al., 2007; Schulz et al., 2002; Tran et al., 2000).
Several negative regulators of the JAK-STAT pathway have been characterized. These include proteins of the Suppressor of Cytokine Signaling (SOCS) family; all contain an SH2 domain and a SOCS box (Hilton et al., 1998), and bind to phosphorylated tyrosines on receptors and/or JAKs to attenuate signaling by recruiting the proteasomal degradation machinery to these targets (Alexander, 2002; Alexander and Hilton, 2004). Socs36E, the best characterized Drosophila SOCS protein, is a known target of JAK-STAT signaling and behaves in a classic negative feedback loop to attenuate the pathway (Callus and Mathey-Prevot, 2002; Issigonis et al., 2009; Karsten et al., 2002). STAT itself can also be regulated by several different mechanisms. Phosphorylated STAT molecules can be dephosphorylated and thereby deactivated by protein tyrosine phosphatases (PTPases), leading to the global downregulation of STAT targets. Ptp61F is the Drosophila homologue of the human phosphotyrosine phosphatase B1 (PTPB1) and is one of 28 predicted PTPs in the fly genome. The expression pattern of Ptp61F during embryogenesis mirrors that of upd, suggesting that Ptp61F may be a target of JAK-STAT signaling (Baeg et al., 2005). Depletion of Ptp61F leads to increase JAK-STAT pathway activity. The precise mechanism of Ptp61F remains unclear but potentially involves the dephosphorylation of Stat92E (Baeg et al., 2005; Muller et al., 2005).
SOCS proteins and PTPases cause global downregulation of the JAK-STAT pathway by inhibition of the receptor/JAK complex in the cytoplasm or phosphorylated STATs in the nucleus, respectively. Recently, a JAK-STAT inhibitor was found in Drosophila that did not act in this global fashion. The ken & barbie (ken) gene was originally identified in a P-element mutagenesis screen for male sterility, and mutants of this gene lacked external genitalia (Castrillon et al., 1993). ken was later implicated to be a novel interactor of the JAK-STAT pathway (Arbouzova et al., 2006). In a genetic screen designed to uncover modifiers of the adult eye overgrowth phenotype caused by Upd overexpression in the developing eye imaginal disc, ken enhanced the eye overgrowth phenotype suggesting that, in this tissue, it normally inhibited the JAK-STAT signaling pathway (Arbouzova et al., 2006). Ken (and its vertebrate orthologue B-Cell Lymphoma 6, BCL6) is characterized by an N-terminal Broad complex, tramtrack, brica-brac (BTB) domain and C-terminal zinc finger (ZF) motifs, a domain structure shared by known transcriptional repressors. Ken was found to bind the sequence GAAA, which overlaps with a subset of Stat92E consensus binding sites (TTCNNNGAA). Furthermore, ectopic expression of Ken in the embryo inhibits the expression of known JAK-STAT target genes ventral veins lacking (vvl), trachealess (trh), and knirps (kni). In contrast, misexpression of Ken does not affect the expression of the JAK-STAT target Socs36E (Arbouzova et al., 2006). Therefore, Ken behaves as a selective inhibitor of a subset of JAK-STAT targets that contain DNA binding sites that accommodate both Stat92E and Ken (Stat92E+/Ken+) binding sites (TTCNNNGAAA).
Here, we investigate the role of Ken in the Drosophila testis niche. Although ken is expressed throughout the testis apex, it is cell-autonomously required in CySCs but not GSCs for their maintenance. Furthermore, expression of Ken in the CySC lineage is sufficient to cause CySCs as well as GSCs to self-renew outside of their normal niche.
Materials and methods
Fly stocks and culture
Flies were raised on standard yeast/molasses medium at 25°C unless otherwise stated. The following stocks were used: y w (wild-type), ken alleles (which also serve as enhancer trap lines): ken1 (hypomorphic allele; Bloomington Stock Center), ken02970 (hypomorphic allele; Bloomington Stock Center), kenk11035 (strongest hypomorphic allele; M. Zeidler), UAS-ken (M. Zeidler), UAS-zfh1 (Bloomington Stock Center), UAS-hopTumL (D. Harrison), UAS-stat92E-RNAi (Transformants 43866 and 106980; Vienna Drosophila RNAi Center), UAS-zfh1-RNAi (Transformants 42856 and 42857; Vienna Drosophila RNAi Center), c587-GAL4 (A. Spradling), nanos-GAL4, hs-upd (D. Harrison), and hs-ken (Drosophila Genetic Resource Center, Kyoto).
Induction of ectopic ken, zfh1, hopTumL, upd, and RNAi constructs
Ectopic Ken, Zfh1, or HopTumL was induced in c587-GAL4/Y;; UAS-ken/tub-GAL80ts, c587-GAL4/Y; UAS-zfh1/+; UAS/tub-GAL80ts/+, or c587-GAL4/Y; UAS-hopTumL +; UAS/tub-GAL80ts/+ males by setting up crosses at 18°C to permit survival until adulthood. Newly eclosed males were then shifted to 31°C (for Ken) or 29°C (for Zfh1 and HopTumL) for two weeks before dissection. Stat92E-RNAi and Zfh1-RNAi were induced in c587-GAL4/Y; UAS-stat92ERNAi/+; tub-GAL80ts/+ or c587-GAL4/Y; UAS-zfh1-RNAi/+; tub-GAL80ts/+ males by shifting newly eclosed males raised at 18°C to 31°C for one week before dissection. For in situ hybridization and qPCR experiments, newly eclosed hs-upd or hs-ken males were heat-shocked for 45 minutes at 37°C and then allowed to recover for 1 hour at 25°C.
Mosaic analysis
ken mutant alleles ken1, ken02970, and kenk11035 were recombined onto FRT42B chromosomes and crossed to FRT42B Ubi-GFP::nls; hsFLP flies. The FLP-mediated mitotic recombination technique (Xu and Rubin, 1993) was used to generate negatively marked ken homozygous mutant GSC and/or CySC clones. Newly eclosed males of the genotype +/Y; P{FRT(w[hs])}G13 ken*/P{FRT(w[hs])}G13 P{GFP::nls}; MKRS, P[hsFLP]/+ (experimental) and +/Y; P{FRT(w[hs])}G13/P{FRT(w[hs])}G13 P{GFP::nls}; MKRS, P[hsFLP]/+ (control) were heat-shocked 3 times for 30 minutes at 37°C, then dissected 2, 6, 10, and 14 days after clone induction (ACI). Negatively marked GSC clones were identified by their absence of GFP and the somatic markers ZFH1 or Traffic jam (Tj) and by their position adjacent to the hub. Negatively marked CySC clones were identified by their absence of GFP, presence of ZFH1 or Tj, and position within 2 cell diameters from the hub. Statistical analysis on percentage testes with clones (normalized to basal clone induction rates obtained from non heat-shocked controls) was performed using the Fisher Exact or Chi-Squared tests.
In situ hybridization
To generate probes for in situ hybridization, cDNAs for ken (GH12495, Berkley Drosophila Genome Project [BDGP]) and Ptp61F (LP02164, BDGP) were PCR amplified with primers that contained restriction enzyme sites XbaI and EcoRI at the 5′ ends to allow for subsequent cloning. PCR amplified products were digested with XbaI and EcoRI, and then ligated into the pBluescript II KS(+) vector (Stratagene). Digoxigenin-labeled anti-sense RNA probes were transcribed in vitro using T3 RNA polymerase according to the manufacturer’s instructions (Roche) from plasmid templates linearized with XbaI. Control sense probes were transcribed with T7 RNA polymerase from plasmids linearized with EcoRI. In situ hybridizations were performed as described (Terry et al., 2006) and visualized with an Olympus BX51 microscope.
Immunostaining
Testes were dissected from newly eclosed flies (less than 3 days old unless otherwise stated) and were fixed and immunostained as previously described (Matunis et al., 1997). To visualize ken expression in the ken enhancer trap lines, tyramide signal amplification was used to increase sensitivity of the anti- -galactosidase ( -GAL) staining according to the manufacturer’s instructions (Molecular Probes/Invitrogen). Antibodies used were rabbit anti-Vasa (d-260) (Santa Cruz Biotechnology; 1:400), rabbit anti-GFP (Torrey Pines Biolabs; 1:10,000), mouse anti- -GAL (Promega; 1:1000), affinity-purified rabbit anti-Stat92E (E. Bach; 1:400) (Flaherty et al., 2010), guinea pig anti-ZFH1 (J. Skeath; 1:1000), mouse monoclonal antibody 1B1 (Developmental Studies Hybridoma Bank; 1:50), rabbit anti-phospho-Histone H3 (Upstate/Millipore; 1:200). Alexa 488- and Alexa 568-conjugated secondary antibodies were used (Molecular Probes/Invitrogen; 1:400 and 1:200, respectively). DNA was counterstained with 4,6-diamidino-2-phenylindole (DAPI) (Sigma; 1 g/ml). Confocal images were acquired with a Zeiss LSM 5 Pascal microscope and figures were assembled with Adobe Photoshop CS3 and Adobe Illustrator CS3.
Antibody generation and Western blotting
Rabbit polyclonal antiserum was raised to the following Ken peptide: DRKHLLEAQRNRAQSPE (Open Biosystems). Western blots were performed using standard methods (Harlow and Lane, 1988). Protein extracts were made from 20 pairs of testes or 10 adult males for each genotype. Testis protein extracts were prepared as previously described (Chen et al., 2005). Adult flies were homogenized in 50 l 2X SDS loading buffer, boiled for 5 minutes, and then centrifuged at 13,200 rpm for 1 minute. 5 l of the protein lysate (equivalent to one fly) was then loaded onto a 4-12% Bis-Tris gel (NuPage/Invitrogen). Following SDS-PAGE. proteins were transferred to nitrocellulose membranes and immunoblotting was performed using Anti-Ken antisera diluted 1:1000 in 5% dry milk in 1X PBS-Tween (0.1% Tween-20), peroxidase-conjugated anti-rabbit IgG secondary antisera diluted 1:10,000 (Jackson Immunoresearch Laboratories), and ECL Plus Western blotting detection reagents according to the manufacturer’s instructions (Amersham/GE Healthcare).
In silico identification of potential Stat92E- and Ken-binding sites
We searched the promoter proximal regions of Socs36E and Ptp61F for Stat92E-binding sites TTC[N]3GAA or TTC[N]4GAA that were +/− 5 kb from the transcription start site in sequences obtained from the UCSC Drosophila melanogaster Genome Browser (April 2006 assembly). Stat92E+ sites that were immediately followed by an extra A (TTC[N]3GAAA or TTC[N]4GAAA) represented potential Stat92E+/Ken+-binding sites.
Quantitative Real-Time PCR analysis
Fifty pairs of testes were dissected from 0-3 day old males of the appropriate genotype and separated from other reproductive tissues such as the seminal vesicles and accessory glands. RNA was extracted with TRIzol® Reagent (Invitrogen) and RNA cleanup was performed using QIAgen’s RNeasy kit followed by treatment with DNase I, Amplification Grade (Invitrogen). cDNAs were synthesized from total RNA primed with oligo(dT) using SuperScript® III First-Strand Synthesis (Invitrogen). qPCR was performed with SYBR Green Supermix (Bio-Rad) and a CFX96 Real-Time PCR detection thermal cycler (Bio-Rad) using primers specific for Socs36E, ken, Ptp61F, and Gapdh2.
Socs36E (forward): GCCAACTAGCCAAAAGTAACG
Socs36E (reverse): TGCTGAGAACTTGCTAAGGTG
ken (forward): GCGAGAACAAAGTAAAGCTGC
ken (reverse): AAGTAGGTGGCATTCACGTC
Ptp61F (forward): ATCGATCCAATTCCAGGCC
Ptp61F (reverse): CTGTTTGTCCTCGTTCTCCC
Gapdh2 (forward): GAGTTTTCGCCCATAGAAAGC
Gapdh2 (reverse): CGATGCGACCAAATCCATTG
All reactions were performed in triplicate and the relative expression levels of each target gene were normalized to that of Gapdh2. qPCR analysis was performed with Excel and graphing was performed using Prism software (Graphpad). One representative experiment is shown (n = 3 or 4). P values were obtained using two-tailed Student’s t test.
Results
ken is expressed in the Drosophila testis apex
The expression pattern of ken mRNA during Drosophila development is very dynamic and is present in many of the tissues where JAK-STAT signaling occurs (Arbouzova et al., 2006). To determine if this is also the case in the adult Drosophila testis niche, we generated a polyclonal antiserum to Ken to visualize the ken expression pattern in the testis. However, we could not detect endogenous Ken protein above background levels by immunofluorescence on whole testes (data not shown) or by Western analysis on extracts from testes or whole adult males (Supplemental Figure 1). Nevertheless, by Western analysis, this antiserum recognizes a specific band at approximately 67 kDa within 30 minutes of global ectopic Ken induction in transgenic adult males carrying ken wild-type cDNA driven by the hsp70 promoter (hs-ken) (Supplemental Figure 1). Similarly, ken mRNA is undetectable by in situ hybridization in wild-type testes but is readily detected in testes with ectopic ken expression (data not shown). Taken together, these results indicate that ken is not expressed at high levels in adults or in testes. Although endogenous ken mRNA is undetectable by in situ hybridization, recent RNA-Sequencing (RNASeq) studies (Gan et al., 2010) have shown that the ken gene is expressed in Drosophila testes, which we have verified by performing our own real-time quantitative PCR (qPCR) of wild-type testes (data not shown). Therefore, ken is expressed in the Drosophila testis, albeit at low levels.
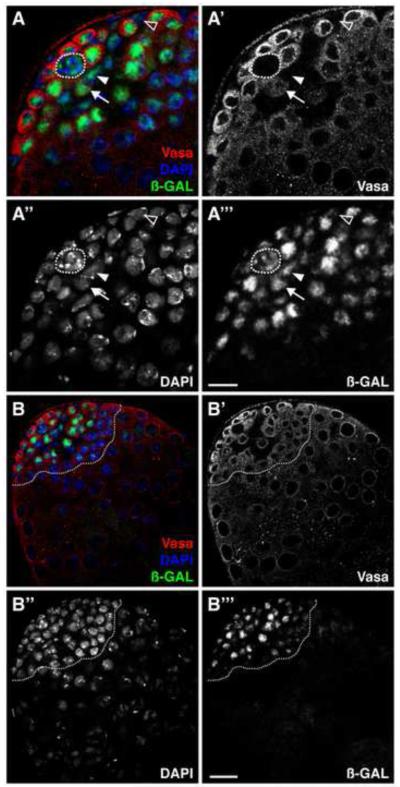
Since ken expression is not readily detectable by in situ hybridization or immunofluorescence, we used three independent enhancer detector lines inserted in the ken locus as tools to obtain further clues about the spatial distribution of ken expression in the testis (Figure 1). All three enhancer traps are expressed in this tissue with expression patterns restricted to the testis apex. In ken1 heterozygous flies, -GAL staining is detected in both the germline and somatic lineages (Figure 1A). The highest levels are detected in the hub, in GSCs, and in all spermatogonial stages (Figure 1A) with an abrupt decrease in expression at the spermatogonialto-spermatocyte transition (Figure 1B, dotted line). Expression is detectable in CySCs and cyst cells as well (Figure 1A). ken02970 and kenk11035 heterozygous flies also express LacZ in hub cells, GSCs and early spermatogonia, as well as CySCs and cyst cells, albeit at lower levels than ken1 flies (data not shown). Taken together, these results indicate that ken is expressed at low levels in the testis apex, in the hub as well as in both stem cell populations and their early progeny. Even though the enhancer trap lines might not reflect the full expression pattern of ken, their expression patterns are restricted to the testis apex, which suggests that ken could be functioning in the testis niche.
Figure 1. ken is expressed in the Drosophila testis apex.
(A, B) ken1 heterozygous testes immunostained with anti-Vasa (germline, red), anti- -galactosidase ( -GAL)(Ken, green), and DAPI (DNA, blue). (A) Ken is expressed in the hub (outlined), the CySCs (one indicated, arrowhead), the GSCs (one indicated, arrow), and their differentiating cyst cell (one indicated, open arrowhead) and germline daughters. Scale bar, 10 m. (B) Ken is expressed in the testis apex (outlined), but not in highly differentiated germ cells or somatic cells. Scale bar, 20 m.
ken is required cell-autonomously for CySC but not GSC self-renewal
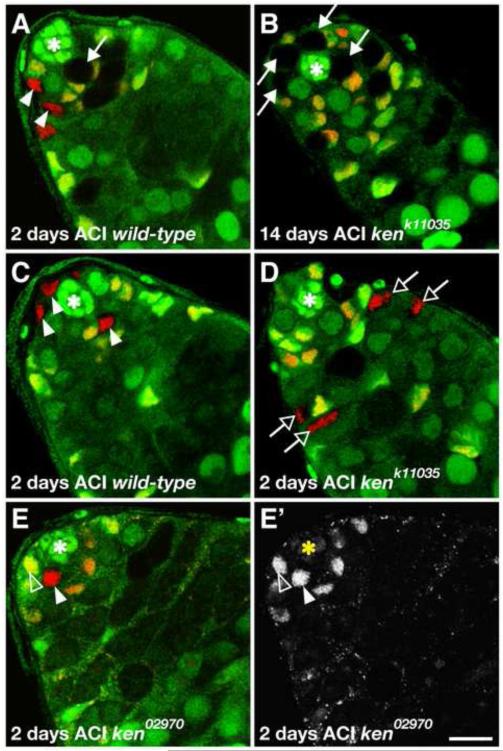
Since ken is expressed in both stem cell populations in the testis, we used mosaic analysis to determine whether ken is required in the GSCs and/or CySCs. The Flipase (FLP)-mediated mitotic recombination technique (Xu and Rubin, 1993) was used to generate ken mutant clones of three loss-of-function alleles in the testis. By counting the proportion of testes with mutant GSCs or CySCs at 2, 6, 10, and 14 days after clone induction (ACI), we found that ken mutant GSC clones are recovered as efficiently as wild-type clones (Table 1, Figure 2A) and are maintained at a rate comparable with wild-type clones over time (Table 1, Figure 2B). In contrast, even though a similar number of ken mutant and wild-type CySCs were initially induced (2 days ACI, Table 1), ken mutant CySCs are lost at a faster rate (Table 1, Compare Figure 2C to 2D). As the number of ken mutant CySCs diminishes over time, ken mutant cyst cells are still detected for up to two weeks (Table 1). These cyst cells are not likely to arise during the initial clonal induction event, since the entire process of spermatogenesis is complete in 10 days (Fuller, 1993). Instead, it is likely that ken mutant CySCs are able to generate cyst cell daughters. This suggests that ken mutant CySCs are lost from the tissue via differentiation, though we have not ruled out the possibility that apoptosis may play a role as well. Taken together, these data indicate that ken does not play a cell-autonomous role in GSCs for their maintenance or differentiation, but is required cell-autonomously in CySCs for their maintenance.
Table 1. ken is necessary for the maintenance of CySCs, but not GSCs.
The percentage of testes with at least one clone is indicated. The total number of testes scored is shown in parentheses.
| Days ACI | ken allele | Number of testes with one or more clones/total testes |
|||
|---|---|---|---|---|---|
| GSC | CySC | SGa | CCb | ||
| no hs control | Wild-type | 6% (36) | 0% (36) | 8% (36) | 0% (36) |
| 1 | 0% (17) | 0% (17) | 0% (17) | 0% (17) | |
| 02970 | 0% (20) | 0% (20) | 0% (20) | 0% (20) | |
| k11035 | 0% (17) | 0% (17) | 0% (17) | 0% (17) | |
| n.s | n.s | ||||
|
| |||||
| 2 | Wild-type | 59% (51) | 33% (51) | 86% (51) | 57% (51) |
| 1 | 49% (45) | 49% (45) | 71% (45) | 76% (45) | |
| 02970 | 57% (49) | 53% (49) | 80% (49) | 71% (49) | |
| k11035 | 34% (47) | 55% (47) | 60% (47) | 60% (47) | |
| n.s | n.s | ||||
|
| |||||
| 6 | Wild-type | 54% (57) | 30% (57) | 56% (57) | 33% (57) |
| 1 | 39% (44) | 5% (44) | 41% (44) | 11% (44) | |
| 02970 | 48% (31) | 16% (31) | 55% (31) | 19% (31) | |
| k11035 | 56% (25) | 8% (25) | 60% (25) | 16% (25) | |
| n.s | P < 0.01 | ||||
|
| |||||
| 10 | Wild-type | 53% (43) | 26% (43) | 53% (43) | 30% (43) |
| 1 | 39% (31) | 0% (31) | 39% (31) | 0% (31) | |
| 02970 | 54% (26) | 15% (26) | 54% (26) | 15% (26) | |
| k11035 | 25% (44) | 2% (44) | 25% (44) | 2% (44) | |
| n.s | P < 0.001 | ||||
|
| |||||
| 14 | Wild-type | 39% (79) | 14% (79) | 39% (79) | 19% (79) |
| 1 | 33% (73) | 4% (73) | 34% (73) | 10% (73) | |
| 02970 | 37% (63) | 5% (63) | 38% (63) | 5% (63) | |
| k11035 | 26% (38) | 3% (38) | 26% (38) | 3% (38) | |
| n.s | P < 0.05 | ||||
SG, spermatogonial clone;
CC, cyst cell clone; n.s., not significant (Chi square and Fisher’s exact test).
Figure 2. ken is required cell-autonomously for CySC but not GSC maintenance.
(A-E) Confocal sections through the testis apex. Hubs denoted by asterisks. (A) Wild-type GSC clone (arrow) adjacent to the hub identified by absence of GFP (green) and Tj (red) at 2 days after clone induction (ACI). Wild-type CySC clones (arrowheads) identified by absence of GFP (green) and presence of Tj (red) are visible in this testis as well. (B) kenk11035 mutant GSC clones (arrows) are still present 14 days ACI and producing normal differentiating spermatogonia whereas there are virtually no remaining kenk11035 CySC clones by this time point. (C) Wild-type CySC clones (arrowheads) surrounding the hub at 2 days after clone induction (ACI). (D) Differentiating kenk11035 mutant cyst cell clones (open arrows) identified by absence of GFP (green), presence of Tj (red), and distance from the hub. There are no kenk11035 CySCs surrounding the hub indicating that these cyst cells originated from CySC clones generated 2 days prior. (E) ken02970 mutant CySC clone (arrowhead) 2 days ACI identified by the absence of GFP (green) and presence of ZFH1 (red) has similar ZFH1 levels relative to neighboring wild-type CySCs (one indicated, open arrowhead). (E’) ZFH1 channel alone. Scale bar, 10 m.
Since ken mutant CySCs are likely lost to differentiation, we analyzed the expression of ZFH1, a known JAK-STAT target required for CySC self-renewal (Leatherman and Dinardo, 2008), in ken CySC clones. ZFH1 is highly expressed in CySCs and is quickly downregulated in their daughters (cyst cells) (Leatherman and Dinardo, 2008). When we examined testes with ken1, ken02970, or kenk11035 mutant CySC clones, we found that there is no discernible decrease in ZFH1 expression in ken mutant CySCs compared to neighboring wild-type CySCs (example in Figure 2E). Taken together, these data indicate that ken is required in CySCs for their self-renewal and ken mutant CySCs appropriately express ZFH1 prior to differentiating into cyst cells.
Ectopic ken expression in the CySC lineage causes an accumulation of somatic and germ cells that retain stem cell-like properties
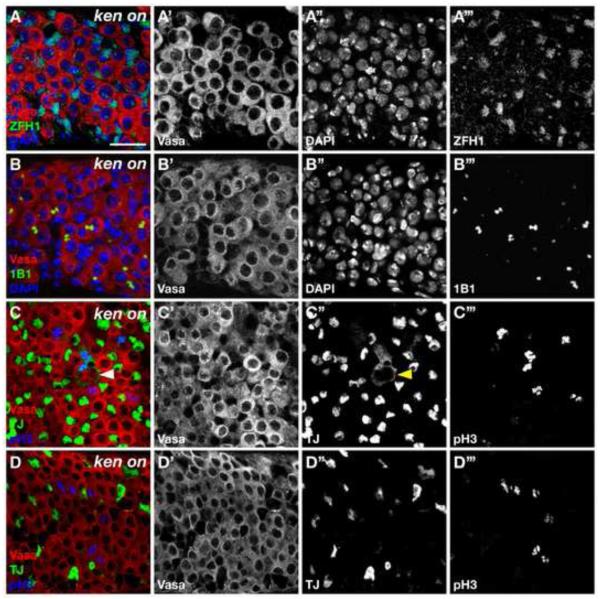
Since we observed that CySCs autonomously require Ken for their maintenance, we speculated whether ken is sufficient to maintain CySC fate. To address this, we used the binary GAL4/UAS system (Brand and Perrimon, 1993) combined with a temperature-sensitive GAL80 (McGuire et al., 2004) to overexpress Ken in the CySCs and their daughters in newly eclosed males. This is sufficient to cause a dramatic accumulation of ZFH1-positive early somatic cells as well as early germ cells throughout the testis (Figure 3A). This is reminiscent of the phenotype seen when the JAK-STAT targets ZFH1 or Chinmo are overexpressed in the CySC lineage (Flaherty et al., 2010; Leatherman and Dinardo, 2008). Furthermore, overexpression of Ken in the germline (nanos>ken) does not result in any phenotypes (data not shown). Therefore, ken overexpression in CySCs, but not GSCs, results in the accumulation of GSC- and CySC-like cells. Taken together, these data are consistent with the emerging model that CySCs behave as a niche for GSCs, and under certain conditions, the somatic lineage can cause GSC-like cells to accumulate throughout the testis (Flaherty et al., 2010; Leatherman and Dinardo, 2008).
Figure 3. Misexpression of Ken in the CySC lineage causes an accumulation of CySC- and GSC-like cells.
(A-D) Confocal section through Ken misexpressing testis. (A) Testis immunostained with anti-Vasa (red), anti-ZFH1 (green), and DAPI (blue). Ken misexpression in the CySC lineage leads to excess ZFH1-positive somatic cells, which are displaced from the hub. (B) Testis immunostained with anti-Vasa (red), anti-1B1 (green), and DAPI (blue). Ken misexpression in the CySC lineage causes early germ cells to accumulate throughout the testis. These germ cells are characterized by a dot or dumbbell-shaped fusome usually found only in GSCs and GBs in wild-type testes. (C, D) Testes immunostained with anti-Vasa (red), anti-Tj (green), and pH3 (blue). (C) Ectopic somatic cells cycle as single cells (Tj+/pH3+) displaced far from the hub. Note the cytoplasmic projections that a cycling CySC (arrowhead) forms around a GSC-GB pair. Breakdown of the nuclear membrane during mitosis in this cell causes the nuclear localization of Tj to accumulate in the cytoplasm. (D) Ectopic early germ cells only cycle as single cells (reminiscent of GSCs or GBs) or as GSC-GB-like pairs. Scale bar, 20 m.
To further characterize the effects of ectopic Ken expression on the testis stem cells, we examined these testes for additional evidence of CySC identity. In wild-type testes CySCs undergo mitosis, but their daughters (cyst cells) exit the cell cycle. Sustained Ken expression in the cyst cell lineage causes somatic cells displaced far from the hub to undergo mitosis as single cells (Figure 3C). These data, as well as the expression of the CySC self-renewal factor ZFH1 throughout the testis, indicate that ectopic Ken is sufficient to promote CySC identity.
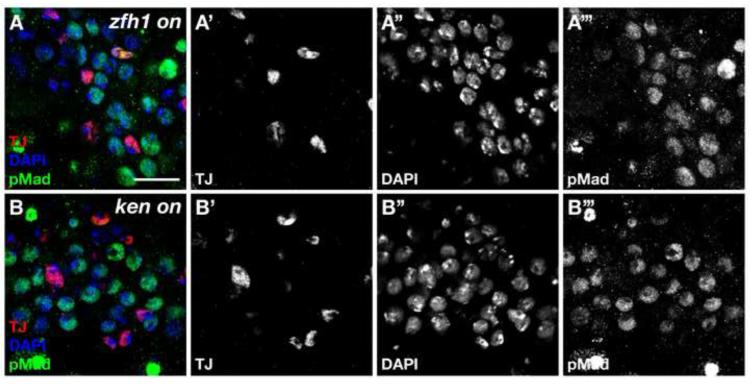
In testes ectopically expressing Ken, the germ cells intermingled with ZFH1-positive cells typically appear to be single cells or two interconnected cells, suggesting that they are GSCs or GSC-GB pairs. Thus, we assayed for different features of GSCs or GBs, which distinguish them from differentiating spermatogonia. First, we looked for the presence of spherical or dumbbell-shaped fusomes by 1B1 staining, a hallmark of GSCs or GSC-GB pairs. We found that most germ cells are found in pairs containing a dumbbell-shaped fusome (Figure 3B). Furthermore, despite being far removed from the hub, these germ cells undergo mitosis as single cells or in pairs, much like GSCs or GSC-GB pairs, as shown by phospho-Histone H3 staining (Figure 3D). In wild-type testes, only GSCs and GBs cycle as single cells while differentiating spermatogonia divide synchronously. Finally, GSCs self-renewing far from the niche in testes ectopically expressing Ken display elevated levels of the BMP pathway activation indicator pMad (Compare Figure 4A to 4B). Together, these data indicate that expression of Ken in the somatic lineage causes an expansion of both germline and somatic stem cell populations in a manner very similar to that seen with ectopic expression of the Stat92E or its target ZFH1. This led us to speculate that ken could be acting either together with the Upd/JAK-STAT signaling pathway and its target ZFH1, or in a parallel pathway.
Figure 4. The BMP pathway is highly activated in ectopic GSCs caused by misexpression of Ken in the CySC lineage.
(A, B) Confocal section through testis immunostained with anti-Tj (red), anti-pMad (green), and DAPI (blue). High levels of pMad indicative of BMP pathway activation accumulate in ectopic GSCs in ZFH1 misexpressing (A) and Ken misexpressing (B) testes. Scale bar, 20 m.
ken-induced CySC and GSC self-renewal is not due to ectopic JAK-STAT pathway activation
To determine whether the phenotype that we observed with Ken overexpression in the CySC lineage is due to the ectopic activation of the JAK-STAT pathway ligand Upd, we examined the expression of upd in testes with ectopic Ken expression by in situ hybridization. We found that levels of upd are not altered in Ken overexpressing testes (Compare Supplemental Figure 2A to 2B). We next asked whether ectopic Ken expression promotes the stabilization of Stat92E in the CySC- and GSC-like cells accumulating outside of the niche in these testes. However, unlike testes overexpressing HopTumL (Figure 5A), which are known to contain high levels of Stat92E in early germline and somatic cells far from the niche (Leatherman and Dinardo, 2008), Ken overexpressing testes do not express Stat92E in CySC-like cells far removed from the hub (Figure 5B). These data indicate that Ken overexpression is not sufficient to induce ectopic Upd or Stat92E activation outside of their normal domain. However, Ken overexpression is sufficient to induce high levels of ZFH1 expression, raising the possibility that Ken may induce ZFH1 in a Stat92E-independent manner.
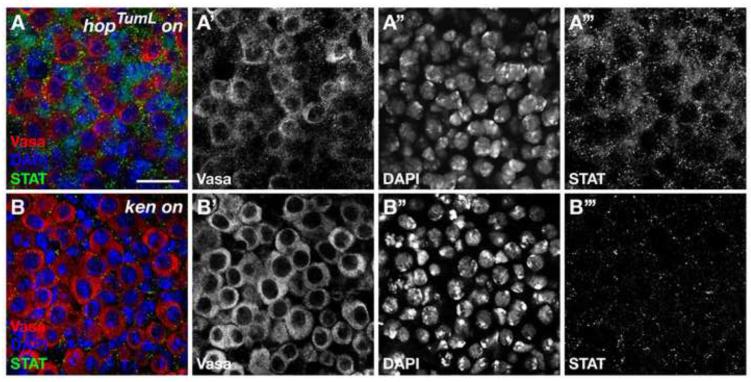
Figure 5. ken-mediated CySC and GSC self-renewal is not due to ectopic JAK-STAT pathway activation.
(A, B) Confocal sections through testes immunostained with anti-Stat92E (STAT, green), anti-Vasa (red), and DAPI (blue). (A) In positive control testes with sustained hopTumL expression in the CySC lineage, Vasa-negative somatic cells far away from the hub (not shown) express elevated levels of Stat92E due to ectopic JAK-STAT pathway activation. (B) In testes misexpressing ken in the CySC lineage, Stat92E expression is not detected above background levels in ectopic CySCs or GSCs. Scale bar, 20 m.
To further explore the epistatic relationship between ken, stat92E, and zfh1, we asked whether overexpression of Ken could rescue the loss of CySCs caused by RNA interference (RNAi) of stat92E or zfh1. Expression of stat92E-RNAi in the CySC lineage causes a significant loss of CySCs, which in turrn leads to a loss of germ cells as well (Compare Supplemental Figure 3A to 3C and 3C’). Co-expression of Ken and stat92E (or zfh1) RNAi partially rescued the CySC loss phenotype (Supplemental Figure 3D’ and 3E). Furthermore, CySCs in testes concomitantly overexpressing Ken and stat92E-RNAi in the CySC lineage continued to express ZFH1 (Supplemental Figure 3D’). While we cannot rule out that the presence of ZFH1 staining in these testes is partly due to incomplete knockdown of stat92E, this finding, along with our data above, suggest that ZFH1 expression in Ken overexpressing testes may not be Stat92E-dependent (Figure 5). This is consistent with data indicating that there may be additional inputs to ZFH1 expression other than Stat92E (Leatherman and Dinardo, 2008). Ken becomes a reasonable candidate for such an input.
ken is not a Stat92E target in the Drosophila testis
If Ken constitutes part of a JAK-STAT independent input promoting ZFH1 expression, stat92E should not be required for ken expression in the testis. To determine if ken expression is influenced by JAK-STAT signaling, we crossed the ken enhancer trap lines into transgenic flies carrying upd cDNA driven by the hsp70 promoter (hs-upd) and then examined the expression pattern of ken before and after heat-shock induced activation of the JAK-STAT pathway. However, we did not observe any appreciable differences in the expression pattern of ken with and without ectopic JAK-STAT signaling (data not shown). Consistent with these results, we also did not detect any changes in ken mRNA expression levels by qPCR in wild-type versus heat-shocked hs-upd testes (Figure 6A, left). However, these conditions are sufficient to significantly up-regulate the expression of a known Stat92E target, Socs36E (Issigonis et al., 2009) (Figure 6A, middle). Therefore, ken is not a Stat92E target in the testis. This distinguishes ken from the other known CySC maintenance factors, zfh1 and chinmo, which are Stat92E targets in the testis (Flaherty et al., 2010; Leatherman and Dinardo, 2008).
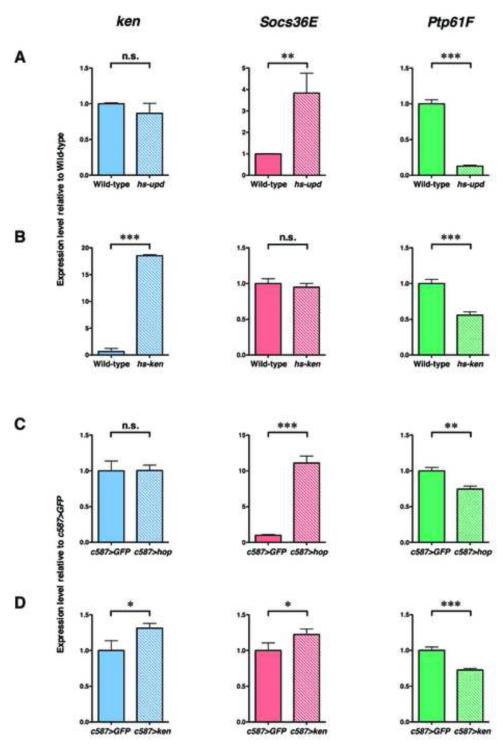
Figure 6. Stat92E and Ken both negatively regulate Ptp61F expression.
(A) Quantitative RT-PCR analysis of ken, Socs36E, and Ptp61F transcript levels in wild-type and heat-shocked hs-upd testes. (A, left) ken expression does not appreciably change in response to ectopic JAK-STAT pathway activation. (A, middle) Socs36E expression levels increase 5-fold in response to ectopic JAK-STAT signaling. (A, right) Ptp61F levels significantly decrease due to JAK-STAT pathway activation. (B) qPCR analysis of ken, Socs36E, and Ptp61F transcript levels in wild-type and heat-shocked hs-ken (ken misexpressing) testes. (B, left) A single heat-shock pulse significantly upregulates ken in hs-ken testes. (B, middle) Socs36E expression is not influenced by ectopic ken expression. (B, right) Ken negatively regulates Ptp61F expression levels. For comparison, expression levels of each gene were normalized to wild-type (set to 1). (A) qPCR analysis of ken, Socs36E, and Ptp61F transcript levels in c587>GFP and c587>hopTumL testes from flies that have been shifted to 31°C for one week. (C, left) ken expression does not appreciably change in response to ectopic JAK-STAT pathway activation. (C, middle) Socs36E expression levels increase more than 10-fold in response to ectopic JAK-STAT signaling. (C, right) Ptp61F expression significantly decreases in response to JAK-STAT pathway activation. (D) qPCR analysis of ken, Socs36E, and Ptp61F transcript levels in c587>GFP and c587>ken testes. (D, left) Shifting c587>ken flies to 31°C for one week causes a significant increase in ken expression. (D, middle) Socs36E expression is not influenced by ectopic ken expression. (D, right) Ken negatively regulates Ptp61F expression levels specifically in the CySC lineage. For comparison, expression levels of each gene were normalized to c587>GFP controls (set to 1). Error bars represent standard deviations of the mean. n.s., not significant; **, P < 0.01; ***, P < 0.001 (Student’s t test).
Both Stat92E and Ken affect the expression of Ptp61F
All our data indicate that ken positively regulates JAK-STAT signaling in the testis niche. Similar to Stat92E, ken is autonomously required in CySCs to prevent CySC differentiation, and ectopic Ken expression in the CySC lineage leads to ectopic CySCs and GSCs. Our results are surprising, since previous studies have shown that Ken behaves as a selective inhibitor of JAKSTAT signaling by negatively regulating the expression of a subset of JAK-STAT targets in the embryo (Arbouzova et al., 2006). Therefore, ken may maintain CySCs either by activating genes required for CySC maintenance (e.g. JAK-STAT signaling) or by repressing an inhibitor of the pathway. Since Ken is known to behave as a transcriptional repressor, we hypothesized that it could be acting on Socs36E or Protein tyrosine phosphatase 61 (Ptp61F), two known JAKSTAT inhibitors. Socs36E is expressed in the testis niche and is an induced antagonist of the JAK-STAT pathway (Callus and Mathey-Prevot, 2002; Issigonis et al., 2009; Karsten et al., 2002; Singh et al., 2010). However, previous results have demonstrated that Socs36E does not respond to Ken in the embryo (Arbouzova et al., 2006), and quantitative real-time PCR (qPCR) analysis of Socs36E in wild-type testes versus testes with ectopic JAK-STAT signaling revealed this to be the case in the testis as well (Figure 6B, middle). Therefore, we focused on the effects of Ken on the candidate JAK-STAT target and inhibitor Ptp61F. According to RNA-Seq data, Ptp61F is expressed in the testis (Gan et al., 2010) and has also been shown to be a JAK-STAT target in Drosophila (Baeg et al., 2005; Muller et al., 2005). Furthermore, an in silico search for Stat92E-binding sites (TTC[N]3GAA and TTC[N]4GAA) in the promoter proximal region of Ptp61F revealed a high number of Stat92E-binding sites, many of which are also potential Ken-binding sites (42/76 Stat92E+/Ken+ sites ).
To examine the expression pattern of Ptp61F in the Drosophila testis, we performed in situ hybridization to Ptp61F mRNA and found that it is expressed at low levels in the testis apex (including the hub, CySCs, GSCs, and their daughters) and is slightly upregulated in late spermatocytes and in cyst cells (Figure 7). Since previous data have shown that, similar to Socs36E, Ptp61F is an induced antagonist of the JAK-STAT signaling pathway (Baeg et al., 2005; Muller et al., 2005), we asked whether Ptp61F expression is also controlled by JAK-STAT signaling in the testis. To do this, we performed quantitative real-time PCR (qPCR) analysis of Ptp61F in wild-type testes versus testes with ectopic JAK-STAT signaling. Surprisingly, Ptp61F expression is significantly downregulated in response to JAK-STAT pathway activation (Figure 6A, right). Taken together, these data suggest that Ptp61F is a target of JAK-STAT signaling and that Stat92E differentially regulates distinct targets, either by upregulating (i.e. Socs36E) or downregulating (i.e. Ptp61F) gene expression.
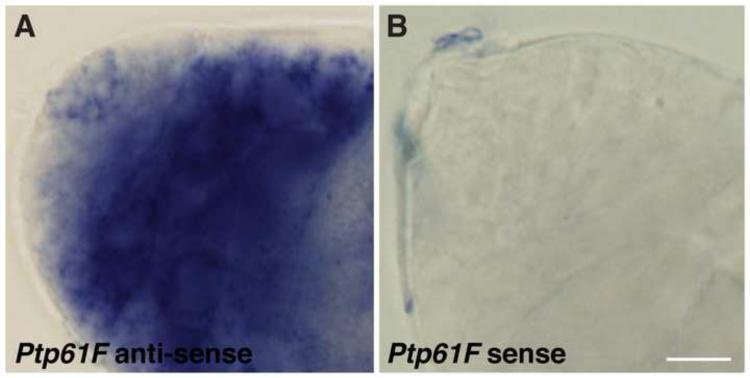
Figure 7. Ptp61F expression in the Drosophila testis apex.
(A, B) In situ hybridization on wild-type testes. (A) Ptp61F mRNA is expressed at low levels around the niche (in the hub, GSCs, and CySCs), and is greatly upregulated in differentiating germ cells and somatic cells. (B) Ptp61F sense probe is shown as a negative control. Scale bar, 10 m.
To test whether Ken can also modulate the expression of Ptp61F, we performed qPCR analysis of Ptp61F in wild-type versus Ken overexpressing testes. Since misexpression of both Upd and Ken lead to the same phenotype (ectopic CySCs and GSCs), we hypothesized that Ptp61F expression would decrease in testes with ectopic Ken (as it does in testes with ectopic Upd). We found that Ptp61F expression is significantly downregulated in Ken overexpressing testes (Figure 6B, right). However, not all Stat92E targets are similarly affected; Socs36E expression is unaffected by ectopic Ken expression (Figure 6B, middle). We conclude that Ptp61F, but not Socs36E, is a target of the transcriptional repressor Ken in the testis, and that global ectopic expression of either Upd or Ken is sufficient to downregulate the expression of Ptp61F.
Although global induction of either JAK-STAT signaling or Ken throughout the testis is sufficient to reduce the levels of Ptp61F expression (Figure 6C and 6D), Ken is required specifically in the CySC lineage (Figure 2). Therefore, we sought to determine whether ectopic expression of Ken or Hop TumL specifically in the CySC lineage is sufficient to reduce PTP61F expression as detected via RT-PCR. Testes from c587>hopTumL and c587>ken flies that have been shifted for 1 week at 31°C are wild-type in appearance (data not shown). However, induction of ectopic HopTumL in the CySC lineage is sufficient to yield a significant increase in JAK-STAT pathway activity as evidenced by an increase in Socs36E expression (Figure 6C, middle). Testes misexpressing Ken in the CySC lineage alone also exhibit a significant decrease in Ptp61F expression (Figure 6D, right). These data indicate that ectopic expression of either the JAK-STAT pathway or Ken specifically in the CySCs lineage is sufficient to downregulate the expression of Ptp61F in these cells.
Discussion
Here, we demonstrate that ken, the orthologue of the human oncogene BCL6, plays a novel and crucial role in adult stem cell maintenance. Furthermore, our data show that ken is sufficient to promote the self-renewal of CySCs outside of their normal niche, which in turn drives the nonautonomous self-renewal of GSCs. This is consistent with previous studies, which have shown that hyperactivation of JAK-STAT signaling or misexpression of the Stat92E targets ZFH1 or Chinmo are sufficient to induce ectopic CySCs and GSCs (Flaherty et al., 2010; Leatherman and Dinardo, 2008, 2010). This work also reveals a previously unappreciated role for Stat92E in the Drosophila testis-transcriptional repression of target genes.
Transcriptional repressors are essential for CySC self-renewal
This study demonstrates the importance of ken in maintaining CySC fate. The only three genes other than Stat92E currently known to be necessary and sufficient for CySC self-renewal are ken, zfh1, and chinmo. Remarkably, all three genes are known to behave as transcriptional repressors. Furthermore, both ken and chinmo encode proteins that share the same overall domain structure: an N-terminal BTB domain and C-terminal DNA-binding zinc fingers. The Drosophila genome encodes 32 BTB-ZF proteins, so it would be interesting to see whether other BTB-ZF proteins are also sufficient to induce ectopic CySCs and GSCs when expressed in the CySC lineage. BTB-ZF proteins regulate many important biological processes such as cell survival and differentiation and generally behave as transcriptional repressors (Beaulieu and Sant’Angelo, 2011; Perez-Torrado et al., 2006). Therefore, it is clear that transcriptional repression plays a critical role in regulating CySC fate (Figure 8).
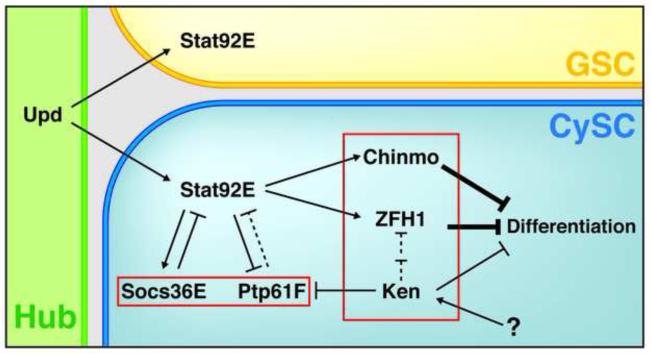
Figure 8. A model for the cell-autonomous requirement of ZFH1, Chinmo, and Ken in CySCs to prevent differentiation.
Upd from the hub activates the JAK-STAT signaling pathway in adjacent GSCs and CySCs. In CySCs, activation of Stat92E leads to expression of its targets ZFH1 and Chinmo, which are both required for CySCs self-renewal. Similarly, Ken is cell-autonomously required for CySC self-renewal in a Stat92E-independent manner. Stat92E activates Socs36E and represses Ptp61F expression. Whether Ptp61F behaves as an inhibitor of Stat92E in the testis remains unknown (dashed line). See text for details.
It will be interesting to learn whether Ken, ZFH1, and Chinmo each control a distinct set of genes, or whether some of their targets are co-regulated. Both ZFH1 and BCL6, the mammalian homolog of Ken, are known to interact with the corepressor CtBP (C-terminal binding protein) (Mendez et al., 2008; Postigo and Dean). Furthermore, heterodimerization between different BTB-ZF family members has been shown to occur (Davies et al., 1999; Dhordain et al., 2000; Korutla et al., 2009; Phan et al., 2005). Since the transcriptional repressors Ken, ZFH1, and Chinmo have similar loss-of-function phenotypes (CySC loss) and gain-of-function phenotypes (ectopic CySCs), it seems likely that identifying their common targets will lead to identification of key effectors required to promote CySC self-renewal. An important parallel can be drawn to studies on embryonic stem (ES) cells which demonstrate that the main ES cell self-renewal factors OCT4, SOX2, and NANOG promote stem cell fate by transcriptionally repressing genes required for differentiation. Interestingly, OCT4, SOX2, and NANOG have been shown to co-occupy a number of target genes (Boyer et al., 2005). Mapping Ken as well as ZFH1 and Chinmo to their binding sites within CySCs will reveal how these transcriptional regulators behave to promote self-renewal and block differentiation.
Previous studies have uncovered the dependence of the germ cells on CySCs for their self-renewal (Flaherty et al., 2010; Leatherman and Dinardo, 2008, 2010) and on cyst cells for their proper differentiation (Kiger et al., 2001; Matunis et al., 1997; Sarkar et al., 2007; Tran et al., 2000). However, further investigation is required to elucidate the mechanisms by which ectopic CySCs are induced, and how this consequently leads to GSC self-renewal. It is unknown whether blocking differentiation in CySCs is sufficient to stall GSCs in an undifferentiated state or whether CySCs send a signal to neighboring germ cells causing them to self-renew. This work and previous studies have begun to uncover the regulatory network comprised of transcription factors (Flaherty et al., 2010; Leatherman and Dinardo, 2008, 2010) and chromatin remodelers (Cherry and Matunis, 2010) in CySCs. In order to understand how these transcriptional regulatory networks control the decision between stem cell fate versus differentiation in CySCs, and how CySC self-renewal promotes GSC identity, one must identify the downstream target genes of these critical transcriptional regulators.
Global and specific JAK-STAT pathway inhibition is critical for stem cell maintenance
Previous work from several labs has shown the importance of JAK-STAT activity for the maintenance of both CySCs and GSCs (Kiger et al., 2001; Leatherman and Dinardo, 2008; Tulina and Matunis, 2001). In CySCs, JAK-STAT signaling promotes stem cell identity by activating the transcription of self-renewal factors, and in GSCs, pathway activation primarily regulates their adhesion to the hub (de Cuevas and Matunis, 2011; Leatherman and Dinardo, 2010). However, attenuation of JAK-STAT signaling is critical as well; expression of the Stat92E target Socs36E in CySCs is necessary to create a negative feedback loop that prevents CySCs from activating Stat92E at aberrantly high levels and consequently outcompeting neighboring GSCs (Issigonis et al., 2009). Therefore, differentially fine-tuning the overall global levels of JAK-STAT pathway activation in the two stem cell types is essential. But how do the stem cells precisely regulate which JAK-STAT targets are activated in the appropriate cell lineage? For example, even though the JAK-STAT pathway is activated in both CySCs and GSCs (albeit at different levels), the target genes zfh1 and Socs36E are expressed in the CySCs but not the GSCs (Figure 8). It is possible that distinct STAT targets respond to different thresholds of STAT activation. Furthermore, certain co-activators or co-repressors may be uniquely expressed or may function exclusively in one cell lineage and not the other. For example, ZFH1 is only expressed in CySCs and is required for their maintenance. On the other hand, Chinmo is expressed in both GSCs and CySCs, but functions solely in the latter stem cell population for their maintenance. Ken is enriched in the testis apex, and similar to the transcriptional repressors ZFH1 and Chinmo, is required in CySCs, but not GSCs. However, in the testis, ken is not a target of the JAK-STAT pathway, unlike zfh1 and chinmo. It is worth noting that although their loss-of-function phenotypes are similar, ken mutant CySC clones are lost more slowly than stat92E, zfh1, or chinmo mutant CySCs (which are depleted within ~2 days of clonal induction) (Flaherty et al., 2010; Issigonis et al., 2009; Leatherman and Dinardo, 2008). One reason for this difference may be attributed to the fact that the available ken alleles are not null. However, it is also possible that genes such as zfh1 and chinmo may have stronger loss-of-function phenotypes because they play a primary role in CySC maintenance whereas Ken may perform secondary functions such as fine-tuning the transcriptional output of the JAKSTAT pathway. The Drosophila testis niche presents a unique opportunity to study how a single signaling pathway regulates two different stem cell populations within a niche via (1) differential regulation of global antagonists (i.e. Socs36E), (2) activation of a distinct set of target genes exclusively in one stem cell type (i.e. zfh1), and (3) differential regulation by transcriptional repressors (i.e. ken and chinmo) (Figure 8).
Stat92E as a transcriptional repressor
An interesting discovery from this study is that Stat92E represses the expression of Ptp61F (Figure 8). STATs were originally discovered as activators of gene transcription in response to interferons (Schindler and Darnell, 1995; Shuai et al., 1993). Recently, however, increasing evidence indicates that in addition to their more familiar and well-documented role as transcriptional activators, STATs can also behave as functional repressors in an indirect manner (via STAT-induced activation of a repressor) or directly (through interactions with DNA methyltransferases, histone deacetylases, or heterochromatin proteins) (Shi et al., 2008; Tran et al., 2010; Walker et al., 2007; Zhang et al., 2005).
In Drosophila, JAK-STAT pathway activation is known to upregulate the transcription of some targets, while repressing others (Flaherty et al., 2009). However, how a transcription factor such as Stat92E can stimulate the expression of individual genes while inhibiting others that have potentially conflicting roles is not well understood. The Drosophila testis provides a good model system to study this problem; Stat92E is required for the self-renewal of CySCs, presumably by positively regulating genes required for stem-cell identity while repressing those which would lead to opposite fates (i.e. differentiation). Our results indicate that Ptp61F is negatively regulated by JAK-STAT signaling in the testis since the activation of JAK-STAT leads to a dramatic decrease in Ptp61F expression (Figure 8). Since Ptp61F expression was quickly downregulated in hs-upd testes after a single heat-shock pulse, we think that Stat92E may be directly repressing Ptp61F transcription instead of activating the expression of a Ptp61F repressor. Support for this comes from work performed in an ex vivo system using Drosophila haemocyte-like cells to identify JAK-STAT targets (Bina et al., 2010). Upd or HopTumL stimulation of these haemocyte-like cells leads to a significant increase in the transcript levels of the “immediate-early” JAK-STAT target Socs36E, which responds within two hours of pathway activation (Bina et al., 2010). We were able to recapitulate these observations in vivo as we observe a robust increase in Socs36E expression levels in response to our heat-shocking protocol in hs-upd testes. Similarly, the rapid response seen in Ptp61F expression levels upon JAKSTAT pathway activation may reflect a direct repression of this target as opposed to a secondary effect. Future studies will address the mechanism by which Stat92E represses the JAK-STAT inhibitor Ptp61F to promote CySC self-renewal.
Ken and its mammalian orthologue BCL6
While the mechanism by which Ken represses JAK-STAT targets is currently unknown, clues to how Ken may be behaving can be drawn from its orthologue BCL6, which interacts with chromatin modifiers such as SMRT, mSIN3A, N-CoR, BcoR, and histone deacetylases (HDACs) (Dhordain et al., 1997; Dhordain et al., 1998; Huynh and Bardwell, 1998; Huynh et al., 2000; Lemercier et al., 2002; Wong and Privalsky, 1998). This suggests that Ken may be acting through these partners to block transcriptional activation through chromatin modification. Another possibility is that Ken directly blocks Stat92E from binding to and transcriptionally activating expression of target genes. Furthermore, since Stat92E can either activate or repress expression of targets, it is also possible that Ken behaves as a Stat92E co-repressor. Any of these non-exclusive possibilities will further our understanding of how a signaling pathway is able to transcriptionally activate different target genes in different cell types and stages of development as opposed to eliciting the indiscriminate activation of all possible target genes at once.
Chromosomal rearrangements and point mutations that lead to the misregulation of BCL6 occur frequently in human lymphomas (Baron et al., 1993; Kerckaert et al., 1993; Ye et al., 1993). Furthermore, constitutive overexpression of BCL6 in mice promotes the development of lymphomas (Baron et al., 2004; Cattoretti et al., 2005). BCL6 has been shown to repress differentiation of B-cells and mammary cells (Dent et al., 1997; Fukuda et al., 1997; Logarajah et al., 2003; Ye et al., 1997). In this study, we find that Ken plays an analogous role in repressing differentiation of CySCs in the Drosophila testis. Future studies on Drosophila Ken and its targets will further our understanding of the mammalian oncogene BCL6.
Supplementary Material
Supplemental Figure 1. Western blot analysis of Ken. Western blot probed with anti-Ken antibody of protein lysates prepared from hs-ken adults that were not heat-shocked (Lane 1) or heat-shocked for 45 minutes at 37°C and then allowed to recover for the time indicated (Lanes 2-8). Ken is transiently detected as a ~67 kDa band. Note the presence of a smaller non-specific band in all lanes (serves as a loading control).
Supplemental Figure 2. Ken overexpression does not cause ectopic Upd expression. (A, B) In situ hybridization with upd antisense probe on whole testes. (A) In wild-type, upd mRNA is expressed specifically in the hub. (B) In heat-shocked hs-ken testes, upd mRNA is present only in the hub, its normal expression domain. Scale bar, 10 m.
Supplemental Figure 3. Ken overexpression partially rescues CySC loss induced by Stat92E- or Zfh1-RNAi. (A-D) Testes immunostained with anti-Vasa (red), anti-ZFH1 (green), and DAPI (blue). Hubs are outlined. Scale bar, 10 m. All testes are from flies that were raised at the indicated temperature for 1 week. (A) Control c587>stat92E-RNAi testes raised at the restrictive temperature (18°C) are wild-type in appearance. (B) Control testes from c587>stat92E-RNAi flies that also contain a UAS-Ken construct appear wild-type. (C, C’) When shifted to the permissive temperature (31°C), c587>stat92E-RNAi induces loss of CySCs and their daughters which also leads to subsequent loss of their associated germ cells. (D, D’) Ken overexpression in the CySC lineage partially rescues CySC (and corresponding germ cell) loss induced by c587>stat92E-RNAi. (E) Graph depicting CySC (and early germ cell) loss caused by stat92E- or zfh1-RNAi. Simultaneous Ken overexpression partially rescues the RNAi-mediated phenotypes.
Acknowledgements
We thank members of the fly community for their generosity including Dr. Martin Zeidler for the kenk11035 and UAS-ken stocks; Dr. Doug Harrison for the UAS-hopTumL and hs-upd stocks; Dr. Allan Spradling for the c587-GAL4 stock; and Dr. Erika Bach for the Stat92E antibody. We also thank the Bloomington Stock Center; the Developmental Studies Hybridoma Bank; members of the Matunis lab for helpful discussion; Judy Qiu for generating FRT, ken recombinants; Rachel Stine for assistance with fly work; and Dr. Margaret de Cuevas and Rachel Stine for critical reading of the manuscript. This work was supported by NIH grant RO1HD40307 (E.M.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander WS. Suppressors of cytokine signalling (SOCS) in the immune system. Nat Rev Immunol. 2002;2:410–416. doi: 10.1038/nri818. [DOI] [PubMed] [Google Scholar]
- Alexander WS, Hilton DJ. The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu Rev Immunol. 2004;22:503–529. doi: 10.1146/annurev.immunol.22.091003.090312. [DOI] [PubMed] [Google Scholar]
- Arbouzova NI, Bach EA, Zeidler MP. Ken & barbie selectively regulates the expression of a subset of Jak/STAT pathway target genes. Curr Biol. 2006;16:80–88. doi: 10.1016/j.cub.2005.11.033. [DOI] [PubMed] [Google Scholar]
- Baeg GH, Zhou R, Perrimon N. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev. 2005;19:1861–1870. doi: 10.1101/gad.1320705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron BW, Anastasi J, Montag A, Huo D, Baron RM, Karrison T, Thirman MJ, Subudhi SK, Chin RK, Felsher DW, Fu YX, McKeithan TW, Baron JM. The human BCL6 transgene promotes the development of lymphomas in the mouse. Proc Natl Acad Sci U S A. 2004;101:14198–14203. doi: 10.1073/pnas.0406138101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron BW, Nucifora G, McCabe N, Espinosa R, 3rd, Le Beau MM, McKeithan TW. Identification of the gene associated with the recurring chromosomal translocations t(3;14)(q27;q32) and t(3;22)(q27;q11) in B-cell lymphomas. Proc Natl Acad Sci U S A. 1993;90:5262–5266. doi: 10.1073/pnas.90.11.5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu AM, Sant’Angelo DB. The BTB-ZF family of transcription factors: key regulators of lineage commitment and effector function development in the immune system. J Immunol. 2011;187:2841–2847. doi: 10.4049/jimmunol.1004006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina S, Wright VM, Fisher KH, Milo M, Zeidler MP. Transcriptional targets of Drosophila JAK/STAT pathway signalling as effectors of haematopoietic tumour formation. EMBO reports. 2010;11:201–207. doi: 10.1038/embor.2010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Callus BA, Mathey-Prevot B. SOCS36E, a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene. 2002;21:4812–4821. doi: 10.1038/sj.onc.1205618. [DOI] [PubMed] [Google Scholar]
- Castrillon DH, Gonczy P, Alexander S, Rawson R, Eberhart CG, Viswanathan S, DiNardo S, Wasserman SA. Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single P element mutagenesis. Genetics. 1993;135:489–505. doi: 10.1093/genetics/135.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoretti G, Pasqualucci L, Ballon G, Tam W, Nandula SV, Shen Q, Mo T, Murty VV, Dalla-Favera R. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Chen X, Hiller M, Sancak Y, Fuller MT. Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science. 2005;310:869–872. doi: 10.1126/science.1118101. [DOI] [PubMed] [Google Scholar]
- Cheng J, Tiyaboonchai A, Yamashita YM, Hunt AJ. Asymmetric division of cyst stem cells in Drosophila testis is ensured by anaphase spindle repositioning. Development. 2011;138:831–837. doi: 10.1242/dev.057901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry CM, Matunis EL. Epigenetic regulation of stem cell maintenance in the Drosophila testis via the nucleosome-remodeling factor NURF. Cell Stem Cell. 2010;6:557–567. doi: 10.1016/j.stem.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JM, Hawe N, Kabarowski J, Huang QH, Zhu J, Brand NJ, Leprince D, Dhordain P, Cook M, Morriss-Kay G, Zelent A. Novel BTB/POZ domain zinc-finger protein, LRF, is a potential target of the LAZ-3/BCL-6 oncogene. Oncogene. 1999;18:365–375. doi: 10.1038/sj.onc.1202332. [DOI] [PubMed] [Google Scholar]
- de Cuevas M, Matunis EL. The stem cell niche: lessons from the Drosophila testis. Development. 2011;138:2861–2869. doi: 10.1242/dev.056242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- Dhordain P, Albagli O, Honore N, Guidez F, Lantoine D, Schmid M, The HD, Zelent A, Koken MH. Colocalization and heteromerization between the two human oncogene POZ/zinc finger proteins, LAZ3 (BCL6) and PLZF. Oncogene. 2000;19:6240–6250. doi: 10.1038/sj.onc.1203976. [DOI] [PubMed] [Google Scholar]
- Dhordain P, Albagli O, Lin RJ, Ansieau S, Quief S, Leutz A, Kerckaert JP, Evans RM, Leprince D. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc Natl Acad Sci U S A. 1997;94:10762–10767. doi: 10.1073/pnas.94.20.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhordain P, Lin RJ, Quief S, Lantoine D, Kerckaert JP, Evans RM, Albagli O. The LAZ3(BCL-6) oncoprotein recruits a SMRT/mSIN3A/histone deacetylase containing complex to mediate transcriptional repression. Nucleic Acids Res. 1998;26:4645–4651. doi: 10.1093/nar/26.20.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty MS, Salis P, Evans CJ, Ekas LA, Marouf A, Zavadil J, Banerjee U, Bach EA. chinmo is a functional effector of the JAK/STAT pathway that regulates eye development, tumor formation, and stem cell self-renewal in Drosophila. Dev Cell. 2010;18:556–568. doi: 10.1016/j.devcel.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty MS, Zavadil J, Ekas LA, Bach EA. Genome-wide expression profiling in the Drosophila eye reveals unexpected repression of notch signaling by the JAK/STAT pathway. Dev Dyn. 2009;238:2235–2253. doi: 10.1002/dvdy.21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Yoshida T, Okada S, Hatano M, Miki T, Ishibashi K, Okabe S, Koseki H, Hirosawa S, Taniguchi M, Miyasaka N, Tokuhisa T. Disruption of the Bcl6 gene results in an impaired germinal center formation. J Exp Med. 1997;186:439–448. doi: 10.1084/jem.186.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller MT. Spermatogenesis. In: Bate M, Arias A. Martinez, editors. The Development of Drosphila melanogaster. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. pp. 71–147. [Google Scholar]
- Fuller MT, Spradling AC. Male and female Drosophila germline stem cells: two versions of immortality. Science. 2007;316:402–404. doi: 10.1126/science.1140861. [DOI] [PubMed] [Google Scholar]
- Gan Q, Schones DE, Ho Eun S, Wei G, Cui K, Zhao K, Chen X. Monovalent and unpoised status of most genes in undifferentiated cell-enriched Drosophila testis. Genome Biol. 2010;11:R42. doi: 10.1186/gb-2010-11-4-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1988. [Google Scholar]
- Hilton DJ, Richardson RT, Alexander WS, Viney EM, Willson TA, Sprigg NS, Starr R, Nicholson SE, Metcalf D, Nicola NA. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proc Natl Acad Sci U S A. 1998;95:114–119. doi: 10.1073/pnas.95.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh KD, Bardwell VJ. The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene. 1998;17:2473–2484. doi: 10.1038/sj.onc.1202197. [DOI] [PubMed] [Google Scholar]
- Huynh KD, Fischle W, Verdin E, Bardwell VJ. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000;14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- Issigonis M, Tulina N, de Cuevas M, Brawley C, Sandler L, Matunis E. JAKSTAT signal inhibition regulates competition in the Drosophila testis stem cell niche. Science. 2009;326:153–156. doi: 10.1126/science.1176817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten P, Hader S, Zeidler MP. Cloning and expression of Drosophila SOCS36E and its potential regulation by the JAK/STAT pathway. Mech Dev. 2002;117:343–346. doi: 10.1016/s0925-4773(02)00216-2. [DOI] [PubMed] [Google Scholar]
- Kerckaert JP, Deweindt C, Tilly H, Quief S, Lecocq G, Bastard C. LAZ3, a novel zinc-finger encoding gene, is disrupted by recurring chromosome 3q27 translocations in human lymphomas. Nat Genet. 1993;5:66–70. doi: 10.1038/ng0993-66. [DOI] [PubMed] [Google Scholar]
- Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294:2542–2545. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- Korutla L, Wang P, Jackson TG, Mackler SA. NAC1, a POZ/BTB protein that functions as a corepressor. Neurochem Int. 2009;54:245–252. doi: 10.1016/j.neuint.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Leatherman JL, Dinardo S. Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell. 2008;3:44–54. doi: 10.1016/j.stem.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherman JL, Dinardo S. Germline self-renewal requires cyst stem cells and stat regulates niche adhesion in Drosophila testes. Nat Cell Biol. 2010;12:806–811. doi: 10.1038/ncb2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemercier C, Brocard MP, Puvion-Dutilleul F, Kao HY, Albagli O, Khochbin S. Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor. J Biol Chem. 2002;277:22045–22052. doi: 10.1074/jbc.M201736200. [DOI] [PubMed] [Google Scholar]
- Lin H. The stem-cell niche theory: lessons from flies. Nat Rev Genet. 2002;3:931–940. doi: 10.1038/nrg952. [DOI] [PubMed] [Google Scholar]
- Logarajah S, Hunter P, Kraman M, Steele D, Lakhani S, Bobrow L, Venkitaraman A, Wagner S. BCL-6 is expressed in breast cancer and prevents mammary epithelial differentiation. Oncogene. 2003;22:5572–5578. doi: 10.1038/sj.onc.1206689. [DOI] [PubMed] [Google Scholar]
- Matunis E, Tran J, Gonczy P, Caldwell K, DiNardo S. punt and schnurri regulate a somatically derived signal that restricts proliferation of committed progenitors in the germline. Development. 1997;124:4383–4391. doi: 10.1242/dev.124.21.4383. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE. 2004;2004:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- Mendez LM, Polo JM, Yu JJ, Krupski M, Ding BB, Melnick A, Ye BH. CtBP is an essential corepressor for BCL6 autoregulation. Mol Cell Biol. 2008;28:2175–2186. doi: 10.1128/MCB.01400-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436:871–875. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- Perez-Torrado R, Yamada D, Defossez PA. Born to bind: the BTB protein-protein interaction domain. Bioessays. 2006;28:1194–1202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol. 2005;6:1054–1060. doi: 10.1038/ni1245. [DOI] [PubMed] [Google Scholar]
- Postigo AA, Dean DC. ZEB represses transcription through interaction with the corepressor CtBP. Proc Natl Acad Sci U S A. 1999;96:6683–6688. doi: 10.1073/pnas.96.12.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Parikh N, Hearn SA, Fuller MT, Tazuke SI, Schulz C. Antagonistic roles of Rac and Rho in organizing the germ cell microenvironment. Curr Biol. 2007;17:1253–1258. doi: 10.1016/j.cub.2007.06.048. [DOI] [PubMed] [Google Scholar]
- Schindler C, Darnell JE., Jr. Transcriptional responses to polypeptide ligands: the JAKSTAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- Schulz C, Wood CG, Jones DL, Tazuke SI, Fuller MT. Signaling from germ cells mediated by the rhomboid homolog stet organizes encapsulation by somatic support cells. Development. 2002;129:4523–4534. doi: 10.1242/dev.129.19.4523. [DOI] [PubMed] [Google Scholar]
- Sheng XR, Matunis E. Live imaging of the Drosophila spermatogonial stem cell niche reveals novel mechanisms regulating germline stem cell output. Development. 2011;138:3367–3376. doi: 10.1242/dev.065797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Larson K, Guo D, Lim SJ, Dutta P, Yan SJ, Li WX. Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat Cell Biol. 2008;10:489–496. doi: 10.1038/ncb1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai K, Stark GR, Kerr IM, Darnell JE., Jr. A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science. 1993;261:1744–1746. doi: 10.1126/science.7690989. [DOI] [PubMed] [Google Scholar]
- Singh SR, Zheng Z, Wang H, Oh SW, Chen X, Hou SX. Competitiveness for the niche and mutual dependence of the germline and somatic stem cells in the Drosophila testis are regulated by the JAK/STAT signaling. J Cell Physiol. 2010;223:500–510. doi: 10.1002/jcp.22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry NA, Tulina N, Matunis E, DiNardo S. Novel regulators revealed by profiling Drosophila testis stem cells within their niche. Dev Biol. 2006;294:246–257. doi: 10.1016/j.ydbio.2006.02.048. [DOI] [PubMed] [Google Scholar]
- Tran J, Brenner TJ, DiNardo S. Somatic control over the germline stem cell lineage during Drosophila spermatogenesis. Nature. 2000;407:754–757. doi: 10.1038/35037613. [DOI] [PubMed] [Google Scholar]
- Tran TH, Utama FE, Lin J, Yang N, Sjolund AB, Ryder A, Johnson KJ, Neilson LM, Liu C, Brill KL, Rosenberg AL, Witkiewicz AK, Rui H. Prolactin inhibits BCL6 expression in breast cancer through a Stat5a-dependent mechanism. Cancer Res. 2010;70:1711–1721. doi: 10.1158/0008-5472.CAN-09-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 2001;294:2546–2549. doi: 10.1126/science.1066700. [DOI] [PubMed] [Google Scholar]
- Walker SR, Nelson EA, Frank DA. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene. 2007;26:224–233. doi: 10.1038/sj.onc.1209775. [DOI] [PubMed] [Google Scholar]
- Wong CW, Privalsky ML. Components of the SMRT corepressor complex exhibit distinctive interactions with the POZ domain oncoproteins PLZF, PLZF-RARalpha, and BCL-6. J Biol Chem. 1998;273:27695–27702. doi: 10.1074/jbc.273.42.27695. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Jones DL, Fuller MT. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 2003;301:1547–1550. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
- Ye BH, Cattoretti G, Shen Q, Zhang J, Hawe N, de Waard R, Leung C, Nouri-Shirazi M, Orazi A, Chaganti RS, Rothman P, Stall AM, Pandolfi PP, Dalla-Favera R. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- Ye BH, Rao PH, Chaganti RS, Dalla-Favera R. Cloning of bcl-6, the locus involved in chromosome translocations affecting band 3q27 in B-cell lymphoma. Cancer Res. 1993;53:2732–2735. [PubMed] [Google Scholar]
- Zhang Q, Wang HY, Marzec M, Raghunath PN, Nagasawa T, Wasik MA. STAT3- and DNA methyltransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. Proc Natl Acad Sci U S A. 2005;102:6948–6953. doi: 10.1073/pnas.0501959102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Western blot analysis of Ken. Western blot probed with anti-Ken antibody of protein lysates prepared from hs-ken adults that were not heat-shocked (Lane 1) or heat-shocked for 45 minutes at 37°C and then allowed to recover for the time indicated (Lanes 2-8). Ken is transiently detected as a ~67 kDa band. Note the presence of a smaller non-specific band in all lanes (serves as a loading control).
Supplemental Figure 2. Ken overexpression does not cause ectopic Upd expression. (A, B) In situ hybridization with upd antisense probe on whole testes. (A) In wild-type, upd mRNA is expressed specifically in the hub. (B) In heat-shocked hs-ken testes, upd mRNA is present only in the hub, its normal expression domain. Scale bar, 10 m.
Supplemental Figure 3. Ken overexpression partially rescues CySC loss induced by Stat92E- or Zfh1-RNAi. (A-D) Testes immunostained with anti-Vasa (red), anti-ZFH1 (green), and DAPI (blue). Hubs are outlined. Scale bar, 10 m. All testes are from flies that were raised at the indicated temperature for 1 week. (A) Control c587>stat92E-RNAi testes raised at the restrictive temperature (18°C) are wild-type in appearance. (B) Control testes from c587>stat92E-RNAi flies that also contain a UAS-Ken construct appear wild-type. (C, C’) When shifted to the permissive temperature (31°C), c587>stat92E-RNAi induces loss of CySCs and their daughters which also leads to subsequent loss of their associated germ cells. (D, D’) Ken overexpression in the CySC lineage partially rescues CySC (and corresponding germ cell) loss induced by c587>stat92E-RNAi. (E) Graph depicting CySC (and early germ cell) loss caused by stat92E- or zfh1-RNAi. Simultaneous Ken overexpression partially rescues the RNAi-mediated phenotypes.