Abstract
Antibody rejection is often accompanied by non-donor HLA specific antibodies (NDSA) and self-reactive antibodies that develop alongside donor-specific antibodies (DSA). To determine the source of these antibodies, we immortalized 107 B cell clones from a kidney transplant recipient with humoral rejection. Two of these clones reacted to HLA class I or MICA. Both clones were also reactive to self antigens and a lysate of a kidney cell line, hence revealing a pattern of polyreactivity. Monoclonality was verified by the identification of a single rearranged immunoglobulin heavy chain variable region (VH) sequence for each clone. By tracking their unique CDR3 sequence, we found that one such polyreactive clone was highly expanded in the patient blood, representing ~0.2% of circulating B cells. The VH sequence of this clone showed evidence of somatic mutations that were consistent with its memory phenotype and its expansion. Lastly, the reactivity of the expanded polyreactive B cell clone was found in the patient serum at time of rejection.
In conclusion, we provide here proof of principle at the clonal level that human antibodies can cross-react to HLA and self. Our findings strongly suggest that polyreactive antibodies contribute to DSA, NDSA as well as autoantibodies, in transplant recipients.
Keywords: Humoral rejection, polyreactive antibodies, human
Introduction
Antibodies reactive to kidney grafts have been implicated in transplant rejection for more than 40 years, yet much remains to be elucidated regarding their generation, fine specificities and function [1–3]. The main targets of these humoral responses are HLA class I and class II molecules expressed on donor cells that are mismatched with those of the recipients. These antigens are recognized as non-self by the host immune system due to their polymorphic allelic determinants and lead to the generation of donor specific antibodies (DSA). Serological responses to other targets have also been reported following kidney transplantation, including non-donor specific antibodies (NDSA) [4–7], i.e. antibodies reactive to HLA molecules not expressed by the donor cells. How NDSA develop is still uncertain. The most widely accepted explanation is that NDSA and DSA cross-react to public epitopes shared by multiple HLA [8–11]. In other instances, especially in the absence of DSA, the generation of NDSA is thought to result from previous exposure to non-donor HLA, for example via blood transfusion and pregnancies.
An abundant literature also attests to the development of antibodies to self-antigens alongside anti-HLA antibodies in transplant recipients [12–22]. In a recent study, we used a combination of solid phase techniques including protein microarrays, to characterize serological profiles of kidney transplant recipients with chronic humoral rejection (CHR) [23]. Our experiments showed that patients’ sera reacted to numerous autoantigens, at the time of CHR. Moreover, serum reactivity patterns appeared remarkably unique for each individual with only minimal overlap between patients. These complex serological responses, combining allogeneic and autoimmune components are intriguing and difficult to explain.
In the present study, we investigated the antibody response associated with graft rejection at the cellular level by isolating, cloning and characterizing B cells responsible for the production of graft reactive antibodies. We examined the nature of these antibodies and looked for a possible relation between allo- and autoreactivity.
Materials and Methods
Patient characteristics and biological samples
The patient studied in this report is a 43 year old male who underwent explantation of his second kidney transplant consecutive to rejection. His original disease was focal segmental glomerulosclerosis (FSGS). The first kidney failed 12 years after the initial transplant, consecutive to cyclosporine vasculopathy and focal acute cellular rejection. The second kidney graft was transplanted 5 years after rejection of the first one and functioned for about 1.5 years. It was removed because of suspected pyelonephritis. Sections from the nephrectomy specimen showed a dense mononuclear inflammatory cell infiltrate with marked tubulitis and interstitial hemorrhage. The infiltrate contained focal aggregates of neutrophils. C4d staining was negative in peritubular and glomerular capillaries. In summary, the nephrectomy showed acute cellular rejection, type III, superimposed on chronic rejection. The patient had a slight positive CMV serology 4 months post-transplant. The serology became negative after a month of treatment. The patient received 2 blood transfusions 3 year prior to the second transplant. HLA typing of the blood donor was not documented. The HLA typing of the patient and of both donors are as follow: Recipient: A2,26; B8,70; Bw6; Cw2,3; DR11,13; DQ3; Donor 2nd kidney: A1; B8,44; Bw4,6; Cw5,7; DR15,17; DQ1,2; Donor 1st kidney: A2; B7,44; Bw4,6; Cw5,7; DR4,6; DQ1,3. Anti-Class I and Class II antibodies were detected at low level before and after the second transplant. Reactivity to both Class I and class II was significantly higher at time of nephrectomy. The collection of all clinical specimens and medical information used in the study was approved by the Massachusetts General Hospital internal review board.
Isolation and immortalization of B cell clones
Multiple fragments of the explanted kidney graft were dissociated mechanically to obtain a cell suspension. Although the graft specimen was thoroughly rinsed with PBS, blood was still present in the tissue at time of dissociation. Mononuclear cells were isolated from the cell suspension by gradient centrifugation (Ficoll-paque™ PLUS, GE Healthcare, Piscataway, NJ). EBV-transformed B cell clones were generated by incubating immunopurified B cells (CD20 MicroBeads, Miltenyi Biotec, Auburn, CA) with supernatant from EBV–producing B95-8 marmoset cells in presence of the TLR9 agonist CpG 2006 at 2.5μg/ml [24]. Limiting dilution cloning was performed in 96 well plates by plating serial dilutions of cells on top of feeder cells consisting of 50,000 irradiated monocytic THP-1 cells per well in 96 well plates. Once established, clones were maintained in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum, 4 mM glutamine, 1 mM sodium pyruvate, 10 mM Hepes, and antibiotics. Cells growing in plates where less than 30% of wells contained growing cells were presumed clonal. Clonality was confirmed by molecular analysis of Ig heavy chain transcripts as described below.
Assessment of reactivity to HLA molecules
The reactivity of B cell clone supernatants as well as patient’s serum samples to HLA molecules was assessed using beads coated with either mixed HLA molecules (LABScreen Mixed, One Lambda, Los Angeles, CA), or a single HLA molecules (LABScreen Single Antigen HLA Class I and Class II, One Lambda). Antibodies reactive to beads were detected with an anti-IgG (One Lambda) or IgM (Invitrogen, Carlsbad, CA) PE-conjugated secondary antibody on a Luminex 200 apparatus (Luminex, Austin, TX).
ELISA assays
ELISA assays for the detection of antibodies to double stranded DNA (dsDNA), whole protein extract from human embryonic kidney cell line (HEK-293) and insulin were performed as previously described [23]. Antibody binding was revealed with an HRP–conjugated goat anti–human IgG/M/A (Invitrogen), and developed using 3,3′,5,5′-Tetramethylbenzidine (TMB) (Sigma-Aldrich). Optical density was read at 450 nm.
Immunofluorescence Assays (IFAs)
Slides coated with Hep-2 cells (Bion Enterprises Ltd, Des Plaines, IL) were incubated at room temperature with serum sample diluted at 1/50 or undiluted clone supernatants for 30 min, washed in PBS, stained with fluorescein isothiocyanate-conjugated anti-human IgG/IgM/IgA (Invitrogen), washed with PBS, and visualized by fluorescence microscopy.
Molecular analysis of immunoglobulin heavy chain variable regions
Total RNA was extracted from immortalized clones 3E7, 4G4, and 4G10 using TRIzol reagent (Invitrogen). Superscript III reverse transcriptase kit (Invitrogen) was used to generate cDNA. Variable regions of the Ig heavy chain were amplified by PCR using 6 family-specific forward primers (VH1 to VH6) [25] and a consensus JH reverse primer [26] (Figure S1A). The PCR conditions were as follows: 95°C/5′, (95°C/30″; 56°C/30″; 72°C/30″) × 35 cycles; 72°C/10′. PCR products were cloned in a pCR8/GW/TOPO TA vector (Invitrogen) and used to transform TOP10 chemically competent bacteria (Invitrogen). Twenty representative colonies per clone were harvested and sequenced using the corresponding VH primer to confirm the clonality of the B cells. Sequencing was done at the Massachusetts General Hospital DNA core facility.
Flow cytometry
Immortalized B cell clones were stained with PE-conjugated anti-CD27 antibody (BD Biosciences), washed and analyzed using a FACScan flow cytometer (BD biosciences). To assess their origin, immortalized B cell clones and donor- and recipient-derived EBV cell lines were stained with FITC-conjugated anti-HLA-A2 antibody (BioLegend, San Diego, CA), washed and analyzed using an Accuri flow cytometer (BD biosciences).
Molecular assessment of B cell clones frequency in vivo
Total RNA was extracted from the patient’s PBMC collected at time of nephrectomy as well as from the graft tissue using TRIzol reagent (Invitrogen). First-strand cDNA was then synthesized with the superscript III reverse transcriptase kit (Invitrogen). VH regions corresponding to the 3 clones studied (VH2 for clone 4G10, VH3 for clone 3E7 and VH4 for clone 4G4; Figure S1B) were amplified by PCR using cDNA from blood and graft origin following the same method as described above. PCR products were cloned in a pCR8/GW/TOPO TA vector (Invitrogen) and used to transform TOP10 chemically competent bacteria (Invitrogen). For each PCR product, 940 individual colonies were screened by hybridization for the presence of a plasmid containing a VH insert as well as the corresponding clone-specific CDR3 sequence. The strategy is depicted in Figure S2.
The dot-blot technique used a 96 well-plate vacuum manifold system to spot nylon membranes (BrightStar®-Plus, Ambion, Austin, TX) with 95 plasmid DNA samples, each corresponding to a single colony. One positive control dot (plasmid DNA containing the VH segment of the corresponding clone) and one negative control (empty well) were present on each plate. Membranes were sequentially hybridized; first with a 32P-labelled clonotypic probe corresponding to the unique CDR3 region of each clone (Figure S2) and then with a 32P-labelled JH consensual probe to detect the presence of a VH insert. All DNA samples showing a positive hybridization signal with the clonotypic probes were sequenced to confirm the presence of the unique clonal CDR3 sequences. Results are reported as the ratio of confirmed clonal CDR3 specific dots on total number of VH chain positive dots.
Results
Generation of immortalized B cell clones
To characterize cells responsible for humoral responses to kidney allografts, we generated a series of immortalized B cell clones starting from a patient who underwent transplant nephrectomy for acute and chronic antibody mediated rejection. CD19+ B cells were immunopurified from the explanted graft tissue and immortalized with EBV. One hundred and seven clones were generated and propagated in vitro from the initial sample. All clones produced IgG or IgM in culture. We used these antibodies secreted in the culture supernatant to determine the specificity of the corresponding B cells.
B cell clones reactivity to HLA and MICA molecules
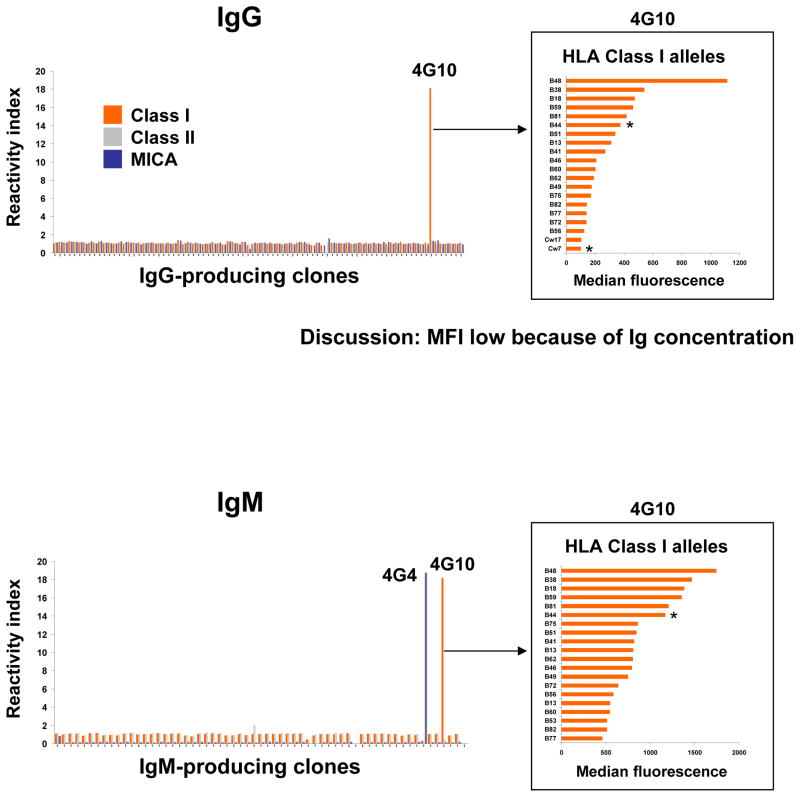
Culture supernatants from the 107 immortalized B cell clones were assessed for HLA reactivity against Class I, Class II, and MICA antigens by Luminex using beads coated with multiple molecules. As shown in Figure 1, one clone (4G4) produced IgM reactive to MICA and one clone (4G10) produced both IgM and IgG reactive to Class I molecules. To further characterize the specificity of both IgM and IgG produced by 4G10 we assessed their reactivity to individual HLA alleles using single antigen beads. Figure 1 (right panels) report the 20 class I antigens toward which 4G10 IgM and IgG were most reactive in descending order. These included HLA-B44, an HLA allele expressed by the donor (noted with an asterisk in Figure 1) as well as several non-donor specific HLA alleles. The highest reactivity was observed towards HLA-B48, a non-donor specific HLA allele. The panel of HLA specificities of 4G10 IgM and IgG were identical, indicating that they originated from the same clone.
Figure 1.
Reactivity of immortalized B cell clones to HLA and MICA molecules. The reactivity to HLA and MICA molecules of the immortalized B cell clones was measured by Luminex with an HLA mixed beads kit using the clones supernatant and anti-IgG or anti-IgM secondary antibodies (upper and lower panel, respectively). Clone 4G10 supernatant, which contains IgG and IgM anti-Class I antibodies was further tested by Luminex using Class I single antigen beads. The 20 Class I alleles towards which 4G10 antibodies reacted the most are depicted (right panels). Results are presented after normalization as fold increase compared to control uncoated bead. HLA Class I alleles expressed by the donor are marked with an asterisk.
B cell clone reactivity to self-antigens
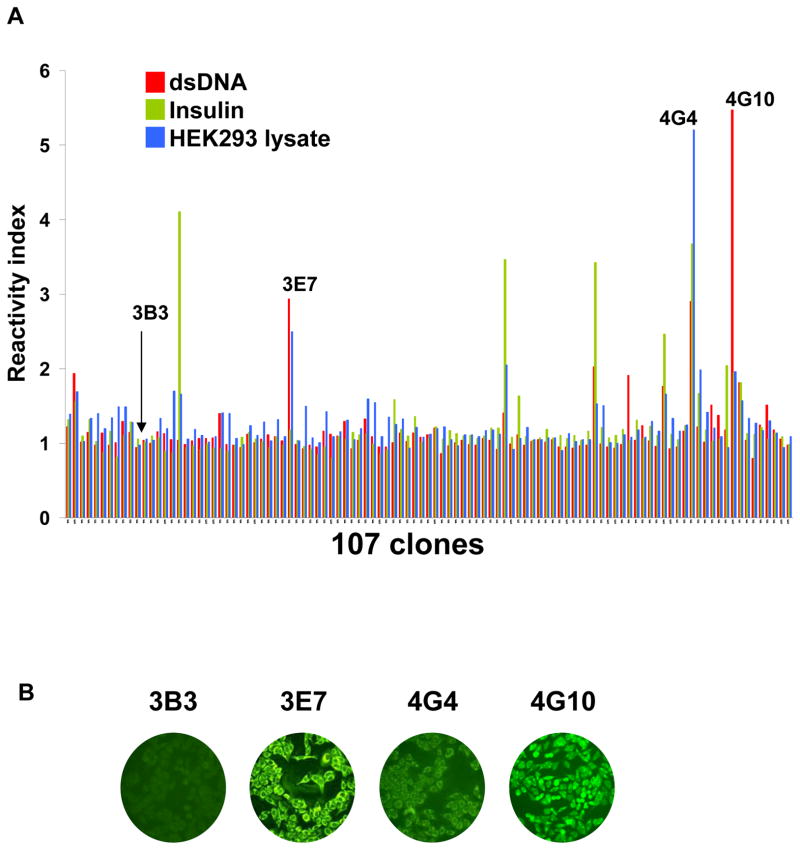
We next assessed the reactivity of the 107 clone’ supernatants to two generic autoantigens, double stranded DNA (dsDNA) and insulin, as well as to a whole protein lysate made from the human kidney cell line HEK293 using ELISA. Seven of the 107 immortalized clones displayed reactivity to one or more of these generic self-antigens (Figure 2A). However, the 2 clones that displayed the highest reactivity to self were clones 4G4 and 4G10 that also reacted to HLA or MICA molecules (Figure 1). Clone 4G4 strongly reacted to HEK293 lysate, dsDNA and insulin whereas clone 4G10 was the most reactive to dsDNA and also showed some reactivity to insulin. These results provided evidence that both clones were polyreactive, i.e. reactive to a broad range of antigens not related to one another. We selected these 2 polyreactive clones, together with another B cell clone showing reactivity to dsDNA and HEK293 lysate but not HLA/MICA molecules (3E7) as well as a non-reactive clone (3B3) for further analysis. As depicted in Figure 2B, supernatants from clones 3E7, 4G4 and 4G10 but not 3B3 reacted to the carcinoma cell line Hep-2 in a comparable manner as self-reactive antibodies developing in patients with autoimmunity.
Figure 2.
Reactivity of immortalized B cell clones to self-antigens. (A) The reactivity of the 107 B cell clones towards generic self antigens (dsDNA and insulin) as well as towards a whole protein lysate prepared from the kidney cell line HEK293 was measured by ELISA. Results are presented after normalization as fold increase compared to the reference value. (B) Reactivity of the 4 selected immortalized clonal supernatants to permeabilized HEp-2 cells. Staining of HEp-2 cells was revealed using a FITC-conjugated anti-IgG/M secondary antibody. All pictures were acquired using the same settings for consistency purposes.
Analysis of B cell clones’ immunoglobulin heavy chain variable regions
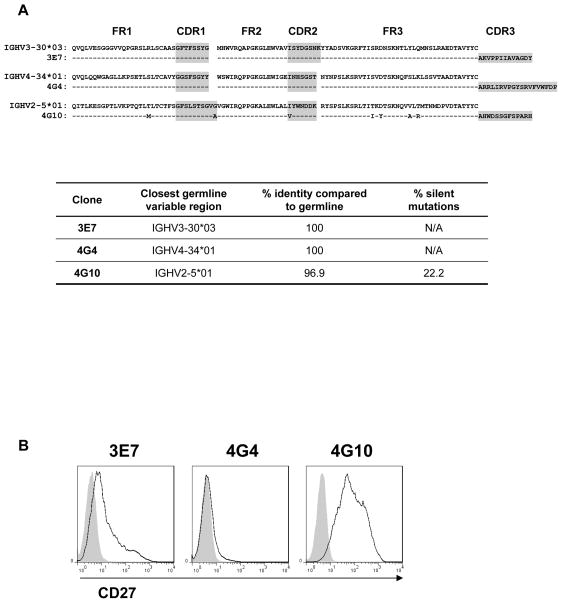
To verify the monoclonality of the selected clones 3E7, 4G4 and 4G10, we analyzed the sequence of their immunoglobulin heavy chain variable region. PCR amplification of the clones cDNA using a series of 6 forward primers corresponding to the 6 immunoglobulin heavy chain variable region (VH) families, generated a unique band for 3E7, 4G4 and 4G10 (Figure S1B). Further analysis of the PCR products revealed a single sequence with a distinct rearranged Complementary Determining Region 3 (CDR3) for each clone. While sequences from clones 3E7 and 4G4 were identical to a germline segment, the sequence of clone 4G10 displayed multiple point mutations upstream of the CDR3 domain (Figure 3A). The mutation frequency for this clone was 3.1% at the nucleic acid level with a percentage of silent mutations of 22.2% (Figure 3A, lower panel). Accumulation of somatic mutations in the variable region of rearranged immunoglobulin genes is a hallmark of differentiated memory B cells. As shown in Figure 3B, mutated clone 4G10 cells highly expressed CD27, consistent with a memory phenotype, whereas clones 3E7 and 4G4 did not (Figure 3B). We also verified that the immortalized clones originated from the recipient. As shown in Figure S3, clones 3B3, 3E7, 4G4 and 4G10 expressed HLA-A2 molecules, a mismatch HLA class I allele between the recipient and the donor of the explanted kidney graft.
Figure 3.
Immunoglobulin heavy chain variable region sequence analysis of selected immortalized clones. (A) The uniquely rearranged Ig VH sequences of the 3 selected polyreactive B cell clones are aligned with their closest germline variable sequence (IMGT/V-Quest [34]). The complementary determining regions 1–3 are highlighted in grey. The table reports the comparison between the clonal sequences and the nearest germline sequence. (B) The expression of the B cell memory marker CD27 at the surface of the 3 selected polyreactive B cell clones was assessed by flow cytometry. Filled gray histograms show the signal generated by staining with the corresponding isotype control.
B cell clone frequency in vivo
The relative frequencies of the 3 polyreactive clones in the patient’s graft and blood were then assessed using a molecular strategy. This approach consisted in screening a large number of PCR amplified VH sequences by hybridization with CDR3 specific probes to detect and quantify sequences corresponding to any of the 3 B cell clones. Approximately 900 sequences of the corresponding VH family generated by PCR using RNA extracted from the graft tissue were first screened for each clone using a dot blot technique (Figure S2). None of the 3 clones could be detected (data not shown). The same technique was then carried out with RNA extracted from the patient’s blood. No positive signals corresponding to the CDR3 sequences of clones 3E7 and 4G4 could be found, indicating that they were either absent from the patient’s blood or represented at very low frequency. In contrast, clone 4G10 CDR3 sequence was detected at a high frequency (4.45%) among VH2 sequences. Since approximately 5% of human B cells use the VH2 segment [27], we estimated the frequency of 4G10 at 0.22% of all blood B cells. This high frequency revealed that clone 4G10 had undergone vigorous clonal expansion in vivo, which is consistent with the memory phenotype and the accumulation of somatic mutations in its rearranged immunoglobulin heavy chain gene. The fact that clone 4G10 was found at high frequency in the blood but not in the graft strongly suggested that it was immortalized from residual blood cells in the explanted kidney tissue.
Patient’s serum reactivity to HLA and self-antigens
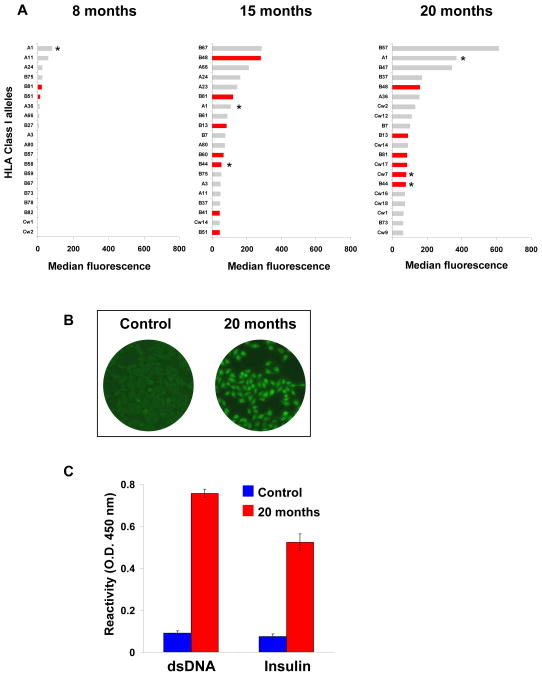
We then examined the contribution of polyreactive antibodies secreted by high frequency clone 4G10 to the reactivity of the patient’s serum to HLA class I molecules and self antigens. As depicted in Figure 5A, multiple HLA class I alleles were recognized by the patient serum at 15 months and 20 months post transplantation, including donor specific alleles (A1, B44). The serum also reacted to several non-donor specific HLA class I molecules including those recognized by clone 4G10 (Figure 5A). Seven out of the 20 class I alleles towards which the serum reacted the most at 15 and 20 months post-transplant, were also recognized by 4G10 monoclonal antibody (Figure 1 and red histograms in Figure 5A). Moreover, the serum was also reactive to Hep-2 cells, dsDNA and insulin as antibodies produced by clone 4G10 did (Figures 5B and 5C). The correspondence between the serum reactivity pattern and that of clone 4G10, together with the observation that this clone was highly expanded in the blood, strongly suggest that antibodies produced by 4G10 are present in the serum and contribute to its overall reactivity.
Figure 5.
Patient’s serum reactivity. (A) The reactivity of the patient’s serum IgG to HLA class I molecules was tested by Luminex. The 20 alleles towards which the serum was most reactive are depicted for 3 time points post transplantation. Class I alleles towards which clone 4G10 was most reactive are depicted as red bars. Class I alleles expressed by the donor are labeled with an asterisk. (B) The patient’s serum IgG reactivity to Hep-2 cell was determined using a sample collected at time of transplant nephrectomy (20 months post-transplant). The serum of a representative non-CHR patient was used as control. Staining was revealed using a FITC-conjugated anti-IgG secondary. (C) Serum reactivity to self antigens. The patient’s serum collected 20 months post-transplantation was assessed for reactivity to dsDNA and insulin by ELISA. A serum sample collected from a non CHR patient was used as control. The reactivity was revealed using an HRP-conjugated anti-IgG secondary antibody.
Discussion
Antibodies are key elements in the host response to solid organ grafts and are involved in both acute and chronic forms of rejection. It is commonly accepted that these antibodies are either alloreactive, i.e. reactive to polymorphic antigens disparate between donor and recipient, such as donor HLA molecules, or autoreactive, i.e. recognizing self-determinants such as vimentin [16, 21] or myosin [20]. The distinction between allo- and autoreactivity is well entrenched and seldom questioned as it relies on the assumption that each antibody is specific to a single antigen, be it self or non-self. Here, we describe another sort of antibodies detected amidst kidney graft rejection that are neither alloreactive nor autoreactive but rather polyreactive in that they react to a broad range of antigens as dissimilar as nucleic acids and HLA molecules.
Antibodies cross-reactive to multiple HLA alleles have already been reported in transplant recipients. It has been proposed that they recognize “public” epitopes shared among polymorphic HLA alleles [8–11]. Likewise, the monoclonal antibody secreted by immortalized B cell clone 4G10 recognizes several HLA class I molecules in Luminex based assays. However, its reactivity pattern exceeds the sole recognition of public HLA epitopes as it also reacts to DNA, Hep-2 cell cytoplasmic structures and HEK 293 protein lysate. The fact that the patient serum reacted to the same non-donor specific HLA molecules than that recognized by clone 4G10, together with the demonstration that this clone was highly expanded in the blood, suggested that polyreactive 4G10 antibodies contributed to the serum reactivity. Remarkably, reactivity to HLA-B44, a donor allele recognized by polyreactive 4G10 antibodies, represents only a small component of the overall serum reactivity. It is plausible that additional high frequency polyreactive clones produced antibodies targeting other HLA molecules, including HLA-A1, the donor specific allele towards which the serum was the most reactive. At this point however, we cannot discriminate which components of the serum result from monospecific or polyreactive antibodies.
We have recently reported on the increase of serum reactivity to a wide variety of self-antigens in patients undergoing CHR [23]. In light of our present study, this observation can be rationally explained by the presence of polyreactive antibodies such as 4G10. Additional studies are now warranted to extend our findings to other transplant recipients patients and determine the exact contribution of polyreactive B cells and the antibodies they produce to humoral responses post-transplant. In particular, circulating polyreactive antibodies could explain the detection of NDSA or even high Panel Reactive Antibody (PRA) measurements often observed in transplant recipients with graft rejection.
The reason why polyreactive IgG develop during CHR is still unclear. Polyreactive IgM, known as “natural IgM”, are commonly observed in healthy individuals and are thought to be produced by B1 B cells (for review: [28]). Our experiments demonstrated the unexpectedly high frequency of clone 4G10 in the blood of a patient with ongoing rejection. This frequency implied a considerable expansion in vivo that was consistent with the accumulation of somatic mutations found in the immunoglobulin heavy chain region as well as the expression of the CD27 memory marker by the corresponding immortalized B cell clone. We hypothesize that this clone is not unique but rather, is representative of a discrete subset of memory, somatically mutated polyreactive B cells, producing “natural IgM” under physiological conditions. Upon activation in the context of inflammation associated with graft rejection or as a result of common infection related to immunosuppression, these polyreactive B cells would expand, undergo class switch recombination and produce IgG. Remarkably, clone 4G10 underwent class switch recombination in vitro and produced both IgM and IgG in the culture supernatant. Although this was likely the result of EBV immortalization and TLR9 stimulation by CpG [29, 30], it revealed the capacity of 4G10 cells to expand, undergo class switch recombination and secrete antibodies upon stimulation.
An important question is whether our immortalization and culture conditions would skew B cell populations. For example, the differentiation stage of isolated B cells could affect their ability to be transformed. To address this critical issue, we carried out a series of immortalization tests on transitional, naïve and memory B cells sorted from healthy donor samples. The immortalization rate was comparable for all subsets, showing that no significant bias would be introduced through the immortalization process. Moreover, the percentage of polyreactive B cells among the 107 clones is consistent with the frequency of polyreactive B cells described by the group of Michel Nussenzweig showing, using a molecular approach, that the percentage of polyreactive B cells was contained between 4 and 7% of all B cells (for review: [31]). Polyreactive antibodies have been described for many years, especially as a first line of defense against bacterial infection [28, 32, 33]. Their implication in humoral responses to solid organ grafts, however, has not been examined and will warrant additional mechanistic studies to determine whether they play and active role in tissue destruction or whether production of these antibodies is more a consequence of graft dysfunction. In this case, it is also possible that they play a beneficial role in participating in mechanisms of wound healing.
Overall, we provide here the proof of principle for the existence of polyreactive human monoclonal immunoglobulin reacting to multiple HLA alleles as well as DNA and other self structures. We also found that such polyreactive antibodies can be produced by memory, somatically mutated B cell clones highly expanded in the blood of kidney transplant recipients experiencing graft rejection. Further investigation will advance our knowledge of these neglected elements of B cell immunity following solid organ transplantation and their precise contribution to the pathophysiology of graft rejection.
Supplementary Material
Figure S1. (A) Forward primers used for the PCR amplification of the 6 immunoglobulin VH families as well as the consensus JH reverse primer. (B) PCR amplification of the 6 immunglobulin heavy chain variable regions using cDNA prepared from purified healthy human CD19+ or from the 3 selected polyreactive B cell clones.
Figure S2. (A) Strategy developed to determine the frequency of the 3 polyreactive B cell clones in the patient’s blood. (B) Sequence of the 3 clonotypic probes used to determine the frequency of the 3 selected polyreactive B cell clones.
Figure S3. Assessment of the immortalized B cell clones’ origin. EBV-B cell lines established from donor recipient blood specimens as well as the 4 B cells clones under study were stained for HLA-A2. This HLA allele is expressed by the recipient but not by the donor of the explanted graft.
Figure 4.
B cell clones frequencies in the blood. Frequency of the 3 polyreactive clones was assessed in the blood by hybridization of plasmid cloned PCR amplified VH segments using dot blot assays as described in Figure S3. Bacterial colonies grown in 96 well microplates were transferred onto membranes and screened with the corresponding clonotypic probes (Figure S2). Ten membranes were screened for each clone. The upper left corner dot in each plate contained a control plasmid. Each positive dot was further analyzed to verify the presence of the unique clonal CDR3 sequence. All weakly positive dots observed for 3E7 were false positive. Each membrane was also screened for the presence of a VH sequence using a consensual JH probe. The ratio of positive clonotypic sequence on total VH sequence is reported for each clone.
Acknowledgments
This work was supported by the Fahd and Nadia Alireza’s Research Fund, the Roche Organ Transplantation Research Foundation (ROTRF) and the National Institute of Health, National Institute of Diabetes, Digestive and Kidney Diseases Grant DK083352.
Abbreviations
- CDR3
Complementary Determining Region 3
- CHR
Chronic Humoral Rejection
- DSA
Donor-Specific Antibodies
- HEK
Human Embryonic Kidney
- Mab
monoclonal antibody
- MICA
MHC Class I-related Chain A
- NDSA
Non-Donor-Specific Antibodies
- PRA
Panel Reactive Antibody
- VH
immunoglobulin heavy chain variable region
Footnotes
Disclosure
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
References
- 1.Akalin E, Dinavahi R, Dikman S, de Boccardo G, Friedlander R, Schroppel B, et al. Transplant glomerulopathy may occur in the absence of donor-specific antibody and C4d staining. Clin J Am Soc Nephrol. 2007 Nov;2(6):1261–7. doi: 10.2215/CJN.02420607. [DOI] [PubMed] [Google Scholar]
- 2.Terasaki P, Mizutani K. Antibody mediated rejection: update 2006. Clin J Am Soc Nephrol. 2006 May;1(3):400–3. doi: 10.2215/CJN.02311205. [DOI] [PubMed] [Google Scholar]
- 3.Terasaki PI. Humoral theory of transplantation. Am J Transplant. 2003 Jun;3(6):665–73. doi: 10.1034/j.1600-6143.2003.00135.x. [DOI] [PubMed] [Google Scholar]
- 4.Briggs D, Zehnder D, Higgins RM. Development of non-donor-specific HLA antibodies after kidney transplantation: frequency and clinical implications. Contributions to nephrology. 2009;162:107–16. doi: 10.1159/000170843. [DOI] [PubMed] [Google Scholar]
- 5.Hourmant M, Cesbron-Gautier A, Terasaki PI, Mizutani K, Moreau A, Meurette A, et al. Frequency and clinical implications of development of donor-specific and non-donor-specific HLA antibodies after kidney transplantation. J Am Soc Nephrol. 2005 Sep;16(9):2804–12. doi: 10.1681/ASN.2004121130. [DOI] [PubMed] [Google Scholar]
- 6.Mao Q, Terasaki PI, Cai J, Briley K, Catrou P, Haisch C, et al. Extremely high association between appearance of HLA antibodies and failure of kidney grafts in a five-year longitudinal study. Am J Transplant. 2007 Apr;7(4):864–71. doi: 10.1111/j.1600-6143.2006.01711.x. [DOI] [PubMed] [Google Scholar]
- 7.Opelz G. Non-HLA transplantation immunity revealed by lymphocytotoxic antibodies. Lancet. 2005 Apr-May;365(9470):1570–6. doi: 10.1016/S0140-6736(05)66458-6. [DOI] [PubMed] [Google Scholar]
- 8.Cai J, Terasaki PI, Mao Q, Pham T, El-Awar N, Lee JH, et al. Development of nondonor-specific HLA-DR antibodies in allograft recipients is associated with shared epitopes with mismatched donor DR antigens. Am J Transplant. 2006 Dec;6(12):2947–54. doi: 10.1111/j.1600-6143.2006.01560.x. [DOI] [PubMed] [Google Scholar]
- 9.El-Awar N, Terasaki PI, Nguyen A, Sasaki N, Morales-Buenrostro LE, Saji H, et al. Epitopes of human leukocyte antigen class I antibodies found in sera of normal healthy males and cord blood. Human immunology. 2009 Oct;70(10):844–53. doi: 10.1016/j.humimm.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 10.Mao Q, Terasaki PI, Cai J, El-Awar N, Rebellato L. Analysis of HLA class I specific antibodies in patients with failed allografts. Transplantation. 2007 Jan 15;83(1):54–61. doi: 10.1097/01.tp.0000250492.55775.83. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki N, Idica A, Terasaki PI. Mimetic human leukocyte antigen epitopes: shown by monoclonal antibodies and extra antibodies produced on transplantation. Transplantation. 2008 Oct 15;86(7):912–8. doi: 10.1097/TP.0b013e318183783b. [DOI] [PubMed] [Google Scholar]
- 12.Dragun D, Muller DN, Brasen JH, Fritsche L, Nieminen-Kelha M, Dechend R, et al. Angiotensin II type 1-receptor activating antibodies in renal-allograft rejection. The New England journal of medicine. 2005 Feb 10;352(6):558–69. doi: 10.1056/NEJMoa035717. [DOI] [PubMed] [Google Scholar]
- 13.Forman JP, Lin J, Pascual M, Denton MD, Tolkoff-Rubin N. Significance of anticardiolipin antibodies on short and long term allograft survival and function following kidney transplantation. Am J Transplant. 2004 Nov;4(11):1786–91. doi: 10.1046/j.1600-6143.2004.00602.x. [DOI] [PubMed] [Google Scholar]
- 14.Glotz D, Lucchiari N, Pegaz-Fiornet B, Suberbielle-Boissel C. Endothelial cells as targets of allograft rejection. Transplantation. 2006 Jul 15;82(1 Suppl):S19–21. doi: 10.1097/01.tp.0000231348.55262.5a. [DOI] [PubMed] [Google Scholar]
- 15.Joosten SA, Sijpkens YW, van Ham V, Trouw LA, van der Vlag J, van den Heuvel B, et al. Antibody response against the glomerular basement membrane protein agrin in patients with transplant glomerulopathy. Am J Transplant. 2005 Feb;5(2):383–93. doi: 10.1111/j.1600-6143.2005.00690.x. [DOI] [PubMed] [Google Scholar]
- 16.Jurcevic S, Ainsworth ME, Pomerance A, Smith JD, Robinson DR, Dunn MJ, et al. Antivimentin antibodies are an independent predictor of transplant-associated coronary artery disease after cardiac transplantation. Transplantation. 2001 Apr 15;71(7):886–92. doi: 10.1097/00007890-200104150-00011. [DOI] [PubMed] [Google Scholar]
- 17.Le Bas-Bernardet S, Hourmant M, Coupel S, Bignon JD, Soulillou JP, Charreau B. Non-HLA-type endothelial cell reactive alloantibodies in pre-transplant sera of kidney recipients trigger apoptosis. Am J Transplant. 2003 Feb;3(2):167–77. doi: 10.1034/j.1600-6143.2003.00021.x. [DOI] [PubMed] [Google Scholar]
- 18.Linke AT, Marchant B, Marsh P, Frampton G, Murphy J, Rose ML. Screening of a HUVEC cDNA library with transplant-associated coronary artery disease sera identifies RPL7 as a candidate autoantigen associated with this disease. Clinical and experimental immunology. 2001 Oct;126(1):173–9. doi: 10.1046/j.1365-2249.2001.01654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lucchiari N, Panajotopoulos N, Xu C, Rodrigues H, Ianhez LE, Kalil J, et al. Antibodies eluted from acutely rejected renal allografts bind to and activate human endothelial cells. Human immunology. 2000 May;61(5):518–27. doi: 10.1016/s0198-8859(00)00109-9. [DOI] [PubMed] [Google Scholar]
- 20.Warraich RS, Pomerance A, Stanley A, Banner NR, Dunn MJ, Yacoub MH. Cardiac myosin autoantibodies and acute rejection after heart transplantation in patients with dilated cardiomyopathy. Transplantation. 2000 Apr 27;69(8):1609–17. doi: 10.1097/00007890-200004270-00015. [DOI] [PubMed] [Google Scholar]
- 21.Wheeler CH, Collins A, Dunn MJ, Crisp SJ, Yacoub MH, Rose ML. Characterization of endothelial antigens associated with transplant-associated coronary artery disease. J Heart Lung Transplant. 1995 Nov-Dec;14(6 Pt 2):S188–97. [PubMed] [Google Scholar]
- 22.Zhang Q, Reed EF. Non-MHC antigenic targets of the humoral immune response in transplantation. Current opinion in immunology. Oct;22(5):682–8. doi: 10.1016/j.coi.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porcheray F, DeVito J, Yeap BY, Xue L, Dargon I, Paine R, et al. Chronic humoral rejection of human kidney allografts associates with broad autoantibody responses. Transplantation. 2010 May 27;89(10):1239–46. doi: 10.1097/TP.0b013e3181d72091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Traggiai E, Becker S, Subbarao K, Kolesnikova L, Uematsu Y, Gismondo MR, et al. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nature medicine. 2004 Aug;10(8):871–5. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell MJ, Zelenetz AD, Levy S, Levy R. Use of family specific leader region primers for PCR amplification of the human heavy chain variable region gene repertoire. Molecular immunology. 1992 Feb;29(2):193–203. doi: 10.1016/0161-5890(92)90100-c. [DOI] [PubMed] [Google Scholar]
- 26.van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003 Dec;17(12):2257–317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 27.Zheng J, Huang J, Mao Y, Liu S, Sun X, Zhu X, et al. Immunoglobulin gene transcripts have distinct VHDJH recombination characteristics in human epithelial cancer cells. The Journal of biological chemistry. 2009 May 15;284(20):13610–9. doi: 10.1074/jbc.M809524200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou ZH, Tzioufas AG, Notkins AL. Properties and function of polyreactive antibodies and polyreactive antigen-binding B cells. Journal of autoimmunity. 2007 Dec;29(4):219–28. doi: 10.1016/j.jaut.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He B, Qiao X, Cerutti A. CpG DNA induces IgG class switch DNA recombination by activating human B cells through an innate pathway that requires TLR9 and cooperates with IL-10. J Immunol. 2004 Oct 1;173(7):4479–91. doi: 10.4049/jimmunol.173.7.4479. [DOI] [PubMed] [Google Scholar]
- 30.Lin L, Gerth AJ, Peng SL. CpG DNA redirects class-switching towards “Th1-like” Ig isotype production via TLR9 and MyD88. European journal of immunology. 2004 May;34(5):1483–7. doi: 10.1002/eji.200324736. [DOI] [PubMed] [Google Scholar]
- 31.Yurasov S, Hammersen J, Tiller T, Tsuiji M, Wardemann H. B-cell tolerance checkpoints in healthy humans and patients with systemic lupus erythematosus. Annals of the New York Academy of Sciences. 2005 Dec;1062:165–74. doi: 10.1196/annals.1358.019. [DOI] [PubMed] [Google Scholar]
- 32.Michael JG, Whitby JL, Landy M. Studies on natural antibodies to gram-negative bacteria. The Journal of experimental medicine. 1962 Jan 1;115:131–46. doi: 10.1084/jem.115.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou ZH, Zhang Y, Hu YF, Wahl LM, Cisar JO, Notkins AL. The broad antibacterial activity of the natural antibody repertoire is due to polyreactive antibodies. Cell host & microbe. 2007 Mar 15;1(1):51–61. doi: 10.1016/j.chom.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic acids research. 2008 Jul 1;36(Web Server issue):W503–8. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. (A) Forward primers used for the PCR amplification of the 6 immunoglobulin VH families as well as the consensus JH reverse primer. (B) PCR amplification of the 6 immunglobulin heavy chain variable regions using cDNA prepared from purified healthy human CD19+ or from the 3 selected polyreactive B cell clones.
Figure S2. (A) Strategy developed to determine the frequency of the 3 polyreactive B cell clones in the patient’s blood. (B) Sequence of the 3 clonotypic probes used to determine the frequency of the 3 selected polyreactive B cell clones.
Figure S3. Assessment of the immortalized B cell clones’ origin. EBV-B cell lines established from donor recipient blood specimens as well as the 4 B cells clones under study were stained for HLA-A2. This HLA allele is expressed by the recipient but not by the donor of the explanted graft.