Abstract
Formation of the vertebrate brain ventricles requires both production of cerebrospinal fluid (CSF), and its retention in the ventricles. The Na,K-ATPase is required for brain ventricle development, and we show here that this protein complex impacts three associated processes. The first requires both the alpha subunit (Atp1a1) and the regulatory subunit, Fxyd1, and leads to formation of a cohesive neuroepithelium, with continuous apical junctions. The second process leads to modulation of neuroepithelial permeability, and requires Atp1a1, which increases permeability with partial loss of function and decreases it with overexpression. In contrast, fxyd1 overexpression does not alter neuroepithelial permeability, suggesting that its activity is limited to neuroepithelium formation. RhoA regulates both neuroepithelium formation and permeability, downstream of the Na,K-ATPase. A third process, likely to be CSF production, is RhoA-independent, requiring Atp1a1, but not Fxyd1. Consistent with a role for Na,K-ATPase pump function, the inhibitor ouabain prevents neuroepithelium formation, while intracellular Na+ increases after Atp1a1 and Fxyd1 loss of function. These data include the first reported role for Fxyd1 in the developing brain, and indicate that the Na,K-ATPase regulates three aspects of brain ventricle development essential for normal function - formation of a cohesive neuroepithelium, restriction of neuroepithelial permeability, and production of CSF.
Keywords: brain ventricles; Na,K-ATPase; Fxyd1; Atp1a1; neuroepithelium; permeability
INTRODUCTION
The vertebrate brain ventricular system comprises an essential set of interconnected cavities, filled with cerebrospinal fluid (CSF). Initially, CSF is produced by the neuroepithelium lining the ventricles (Welss, 1934), and later also by the choroid plexus, a series of vascularized secretory organs (Brown et al., 2004; Speake et al., 2001). Brain ventricle development requires a cohesive neuroepithelium with apical and basal polarity that can retain fluid, a correctly shaped epithelium, production of CSF, and expansion of the epithelium to accommodate the CSF (Ciruna et al., 2006; Gutzman and Sive, 2010; Hong and Brewster, 2006; Lowery and Sive, 2005; Lowery and Sive, 2009; Zhang et al., 2010). We previously demonstrated a requirement for the Na,K-ATPase during zebrafish brain ventricle development. Specifically, the snakehead (snkto273a) mutant, corresponding to a point mutation in the alpha subunit (atp1a1) of the Na,K-ATPase, fails to inflate its brain ventricles (Lowery and Sive, 2005). This led to the hypothesis that snkto273a ventricles fail to inflate because CSF is not produced (Lowery and Sive, 2005; Zhang et al., 2010).
The Na,K-ATPase is a protein complex composed of an alpha, beta, and FXYD subunit. Alpha and beta subunits form a heterodimer that is the minimal functional unit for catalytic activity and ion transport. FXYD subunits differentially regulate the stability of the alpha subunit, maximum catalytic activity, and apparent affinity for Na+, K+, and ATP in a tissue specific manner (Mishra et al., 2011). FXYD1, also called phospholemman, decreases the apparent K+ and Na+ affinity of Na,K-ATPase (Crambert et al., 2002). However, in both adult mouse cardiac myocytes and in Xenopus oocyte systems, phosphorylation of FXYD1 at Ser68 by PKA can increase apparent Na+ affinity and pump current (Bibert et al., 2008; Pavlovic et al., 2007). FXYD and alpha subunit proteins colocalize (Bossuyt et al., 2006; Feschenko et al., 2003; Lansbery et al., 2006), and the crystal structure of Na,K-ATPases further demonstrates that alpha and FXYD subunits physically interact (Morth et al., 2007; Shinoda et al., 2009). In rats, FXYD1 is enriched in the adult brain, specifically in the cerebellum, choroid plexus, and ependymal lining of the ventricles (Feschenko et al., 2003).
Several studies, in tissues outside the nervous system, suggest that one function of the Na,K-ATPase is to direct formation of a normal epithelium. Analysis of Drosophila septate junction formation suggested that regulation of these junctions by the Na,K-ATPase occurs in a pump-independent manner (Paul et al., 2007). Conversely, tissue culture assays in MDCK cells show that levels of intracellular [Na+] ([Na+]i), regulated by the Na,K-ATPase alpha subunit, are correlated with the amount of RhoA-GTP and epithelial junction integrity (Rajasekaran et al., 2001). The Na,K-ATPase has been implicated in regulation of tight junction proteins such as occludins and claudins, and thereby, regulation of paracellular permeability (Rajasekaran et al., 2007; Zhang et al., 2010).
In this study, we clarify the mechanisms by which the Na,K-ATPase alpha subunit, Atp1a1, regulates brain ventricle development and show for the first time a role for Fxyd1 during brain development. The data demonstrate that the Na,K-ATPase acts as a key regulator of brain ventricle formation by impacting three processes: neuroepithelium formation, neuroepithelial permeability and CSF production.
MATERIALS AND METHODS
Fish lines and maintenance
Danio rerio fish were raised and bred according to standard methods (Westerfield et al., 2001). Embryos were kept at 28.5°C and staged accordingly (Kimmel et al., 1995). Lines used: wild type AB, snakehead (snkto273a) (Jiang et al., 1996), and heart and mind (hadm883) (Ellertsdottir et al., 2006).
Antisense morpholino oligonucleotide (MO) injection
Start site or splice-site blocking morpholino (MO) antisense oligonucleotides (Gene Tools, LLC) (Draper et al., 2001; Nasevicius and Ekker, 2000) were injected into one cell stage embryos as previously described (Graeden and Sive, 2009). The translational start site MO targets bases −11 to +14 of atp1a1 (5’-TCTCCTCGTCCATTTTGCTGCTTT-3’) (Yuan and Joseph, 2004) or −7 to −31 of fxyd1 (5’-GTGGATTTAGCCTTTATCAAGCAGA-3’). Splice-site blocking MOs include, atp1a1 (exon5-intron6), 5’- AATATAATATCAATAAGTACCTGGG-3’, and fxyd1 (intron4-exon5) 5’-CTGTGATAATCTAGAGAGAGAGACA-3’. Concentrations used were 0.5 ng or 1 ng atp1a1 start site MO, 2.5 ng or 7.5 ng atp1a1 splice site MO, 7.5 ng of fxyd1 start site MO, and 0.5 ng or 1 ng fxyd1 splice site MO. Standard control MO used is 5′-CCTCTTACCTCAGTTACAATTTATA-3 ′ and p53 morpholino 5′-GCGCCATTGCTTTGCAAGAATTG-3′ (Gene Tools, LLC).
cDNA constructs
Full-length atp1a1 cDNA constructs in pCS2+ were obtained from the Chen lab (Shu et al., 2003). The Quik Change II XL site directed mutagenesis kit (Stratagene) was used to modify wild type atp1a1, to correspond to the mutation associated with snkto273a, which would lead to a G to A substitution at nucleotide 812 of the coding sequence (with the resulting clone called atp1a1GA) (Lowery and Sive, 2005). Primers used for mutagenesis: snkma, 5’-CCGCACAGTCATGGATCGTATGCCACTCTCG-3’ and snkmb, 5’-CGAGAGTGGCAATACGATCCATGACTGTGCGG-3’. For rescue of the atp1a1 start site morphants, the translational start site of atp1a1-pCS2+ was mutated from 5’-AAAAGCAGCAAAATGGGACGAGGAGAA-3’ to 5’-AAGAGTAGTAAGATGGGTCGGGGCGAA-3’ resulting in six mismatches within MO target sequence thus preventing MO binding.
The full-length fxyd1 was obtained from an EST clone (ID 6894873, Open Biosystems) and subcloned into Stu1 site in pCS2+ with a minimal Kozak consensus sequence adjacent to the initiating ATG. This cDNA clone corresponds to a clone previously identified by Sweadner and Rael (Sweadner and Rael, 2000), who suggested the provisional terminology FXYD9dr. However, there is no FXYD9 in any mammalian species, and since this FXYD gene is most closely related to human FXYD1 (Phospholemman), we have more accurately named the zebrafish gene fxyd1.
C-terminal FLAG tagged Fxyd1 were generated using PCR. Briefly, primers were designed to add a linker (S-G-G-G-G-S) followed by the FLAG tag (DYKDDDDK) between the last codon and stop codon using full length fxyd1 in pCS2+ as template. Primers used were:
fxyd1FLAGCtermF, 5’-GTCCTTGTAGTCAGAGCCGCCTCCACCAGAGCATTCACTCGCCCTCGCTGTGTC -3’,
fxyd1FLAGCtermR 5’-GACGATGACAAGTAAAAACCTGCTGACCTGAACCAATCAGAGGAG-3’.
pCS2+RhoAV14 and pCS2+RhoAN19 were kindly provided by R. Winklbauer (University of Toronto) and K. Symes (Boston University).
Capped atp1a1, atp1a1GA, fxyd1, RhoAV14, RhoAN19, mGFP and fxyd1-FLAG were transcribed in vitro using the SP6 mMessage mMachine kit (Ambion) after linearization. Embryos were injected at the one-cell stage with 50 pg RhoAV14, fxyd1-FLAG, or fxyd1, 200 pg of atp1a1 or atp1a1GA, 20 pg RhoAN19, or 10–200 pg of mGFP mRNA.
RT-PCR
RNA was extracted from morphant and control embryos using Trizol reagent (Invitrogen), followed by chloroform extraction and isopropanol precipitation. RNA was pelleted by centrifugation, re-suspended in water and precipitated with LiCl2. cDNA synthesis was performed using Super Script III Reverse Transcriptase (Invitrogen) plus random hexamers. PCR was then performed using primers which amplified the exonic and intronic sequence surrounding the splice MO target. Primers used include: atp1a1 test F 5’-CTCTTTCAAGAATTTGGTTCCC-3’, atp1a1 test R 5’-CTCAATAGAGATGGGGGTGC-3’, fxyd1L 5’-CACAACCACGCATCAAACTT-3’, fxyd1R 5’-CCGTCTCCTCTGATTGGTTC-3’. Primers used for detection of fxyd1 reverse transcript include: fxyd1oppL 5’-CGGGTCGTTTATAAGCATTGA-3’ and fxyd1oppR 5’-TACGGTGGATCCTCCACAAC-3’.
In situ hybridization
Standard methods for RNA probe synthesis containing digoxigenin (DIG)-11-UTP, hybridization and single color labeling were used as described (Sagerstrom et al., 1996). After staining, embryos were fixed in 4% paraformaldehyde overnight at 4°C, and washed in PBT. Embryos were either imaged right away or dehydrated in methanol and cleared in a 3:1 benzyl benzoate/benzyl alcohol (BB/BA) solution before mounting and imaging with a Nikon compound microscope or Zeiss dissecting scope.
Brain ventricle injections, dye retention assay, and ventricle size quantification
Brain ventricle injection and imaging have been described previously (Gutzman and Sive, 2009; Lowery and Sive, 2005).
For assaying permeability, 70 kDa MW dextran conjugated to FITC (Invitrogen; 2.5ng/ml in water) was injected into the brain ventricles at 22 hpf and imaged at various time-points as noted in the text. Neuroepithelial permeability was quantified using ImageJ software to measure the distance of the dye front from the forebrain ventricle hinge-point. In Image J, the line tool was used to draw a line from the forebrain hinge-point to dye front at a 10–20° angle from neuroepithelium. This region was chosen because it is the first and most noticeable site of dye leaking out of the wild type neuroepithelium. The net distance the dye front moved over time was calculated by subtracting distance at t=0 from other time points. Statistics were performed with GraphPad InStat software.
Forebrain ventricle area was calculated by measuring pixels per cm2 of forebrain ventricle with ImageJ software. Scanning confocal stacks of the full depth of the forebrain ventricle were taken and analyzed using 3D doctor (Able Software) to reconstruct the forebrain ventricle and calculate volume in µm3.
Immunohistochemistry and Western Blots
Embryos were fixed in 4% PFA or 2% TCA and blocked in 2%NGS/1%Triton-X/1%BSA or 5%NGS/1%Triton-X. 50µm transverse sections were obtained as described previously (Gutzman and Sive, 2010). Na,K-ATPase levels in whole embryos or brains (50 µg of protein) were analyzed via western blot as previously described (Gutzman and Sive, 2010). Antibodies used: Phalloidin-Texas Red/TRITC or Alexa-Fluor 633 (Molecular Probes), aPKC (Santa Cruz), Zo-1 (Invitrogen), FLAG (Sigma), propidium iodide (Invitrogen), and Na,K-ATPase (alpha) (Cell Signaling Technology) and GAPDH (Abcam).
Inhibitor treatments
Dechorionated embryos were incubated in 5 mM ouabain (Sigma) diluted in embryo medium. Not all embryos responded to ouabain soaking at 16 hpf, likely due to difficulty penetrating the embryonic epidermis at this stage. 50 µM ROCK inhibitor (Calbiochem) was injected between the yolk and the brain under the midbrain and hindbrain at 14 hpf or into the brain ventricles at 22 hpf.
Intracellular Na+ Measurement
Dechorionated and deyolked embryos were collected at 24–28 hpf in 300 µl of nuclease free water. Embryos were dounce homogenized, spun at 1000 rpm, and supernatant collected. CoroNa Green (Invitrogen 10 µM) was added to the supernatant and fluorescence readings obtained using a Tecan Safire II microplate reader. A hemocytometer was used to determine number of cells per embryo (Westerfield et al., 2001).
RESULTS
Na,K-ATPase subunits, Atp1a1 and Fxyd1, are required for brain ventricle development
In order to extend our previous findings (Lowery and Sive, 2005) and investigate the mechanisms by which the Na,K-ATPase alpha subunit, Atp1a1, regulates brain ventricle development, we tested the effect of partial loss of atp1a1 using either a second atp1a1 mutant allele, hadm883 (heart and mind) (Ellertsdottir et al., 2006), or a morpholino-modified antisense oligonucleotide (MO) to inhibit zygotic pre-mRNA splicing (splice site MO)(Draper et al., 2001). In all cases, as previously described (Lowery et al., 2009; Lowery and Sive, 2005; Zhang et al., 2010), brain ventricle inflation was reduced at 24 hours post fertilization (hpf) and all images shown are representative of the phenotype observed (Figs. 1A–D; snkto273a - 65% normal ventricles (n=39), hadm883 - 70% normal ventricles (n=55), atp1a1 splice - 20% normal ventricles (n=31)). After more extensive loss of atp1a1, using a previously published MO (Yuan and Joseph, 2004) targeted to the translational start site (start site MO), no brain ventricle lumen was visible (Fig. 1E; 0% normal ventricles (n=60)) (Nasevicius and Ekker, 2000). Consistent with a dose response to Atp1a1 levels, there was a correlation between brain ventricle inflation and the amount of remaining wild type atp1a1 RNA in atp1a1 splice site morphants (Figs. S1C–H).
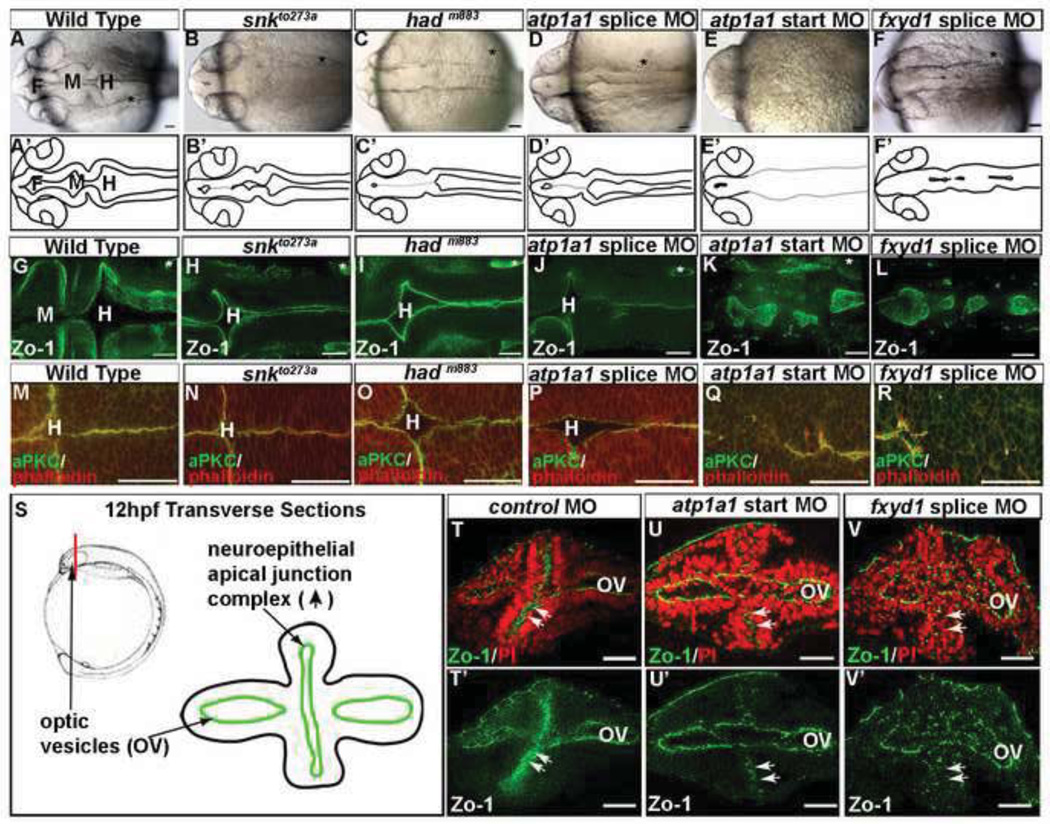
Fig. 1. The Na,K-ATPase is required for brain ventricle development.
(A–F) Loss of function Na,K-ATPase embryos brightfield dorsal view images of wild type (A), partial loss of atp1a1 with mutants snkto273a (B) and hadm883 (C) and splice site MO (D) or severe loss of atp1a1 with start site MO (E) and loss of function fxyd1 splice site MO (F). (G–R) Neuroepithelial formation in Na,K-ATPase morphants/mutants labeled with Zo-1 (G–L) or aPKC (green) and actin (phalloidin; red) (M–R). Images taken at 24 hpf with anterior to the left. (S) Experimental design. Red line indicates region of transverse section in 12 hpf embryo next to corresponding model of transverse section with apically localized junctions (green lines). (T–V) Transverse vibratome sections of 12 hpf embryos stained with Zo 1 (green) and propidium iodide (red) in control (T) atp1a1 start site MO (U), and fxyd1 MO (V). (T’–V’) Zo 1 only. Arrow indicates neuroepithelial apical junction complex. OV = optic vesicle. Asterisk = ear, F = forebrain, M = midbrain, H = hindbrain. Scale bars = 50µm.
The effect of the Atp1a1 splice site MO, as identified by RT-PCR and sequencing of the resulting products, was to generate an early stop codon in the 4th extracellular loop of atp1a1 due to the retention of intron 5 (Fig. S1A). This intron inclusion was predicted to ablate Atp1a1 pumping function. Specificity of the MO phenotypes was shown by phenotypic rescue after co-injection of the MO with 200 pg of the cognate zebrafish mRNA that does not bind the MO sequence (atp1a1 splice rescue- 100% normal ventricles (n=31), atp1a1 start rescue - 100% normal ventricles (n=35), Fig. S1B). The mRNA injected for rescue of the splice site MO had a partial, non-functional target present, while the start site MO mRNA was mutated to prevent MO binding.
The function of zebrafish fxyd1 has not previously been described. fxyd1 is expressed zygotically, that is, mRNA is not detected by RT-PCR at 3 hpf prior to mid-blastula transition (MBT), but is observed at 10 hpf after MBT when zygotic transcription begins. Consistently, we observed, by in situ hybridization, fxyd1 expression within the embryonic brain from 10 hpf to 24hpf (Fig. S1I–L) but not prior to MBT (data not shown). In performing controls for in situ hybridization specificity, an antisense non-protein coding transcript was reproducibly detected, and was confirmed using appropriate primers, by RT-PCR (Fig. S1I–P). This has not previously been reported for any fxyd gene, and the significance is unknown. However, recent data have identified a group of non-coding RNAs which may have profound regulatory function, but further investigation into a putative function of the non-coding transcript will need to be performed in the future (Ulitsky et al., 2011). Further analysis was done on the fxyd1 protein coding transcript, while activity of the non-coding transcript is not known. Loss of function, using a splice site MO targeting the fxyd1 protein coding strand, resulted in absence of brain ventricle inflation in 24 hpf embryos (Fig. 1F, 0/114 wild type), demonstrating a requirement for Fxyd1 during brain ventricle development. Specification of early brain regions in fxyd1 morphants was normal, as judged by in situ hybridization for pax2a, shh and krox20, during somitogenesis (18–22 hpf). By 18 hpf, these embryos showed a shortened antero-posterior axis, suggesting an earlier convergence and extension phenotype. From 24–48 hpf, fxyd1 morphants showed severe heart edema and curved tail (data not shown). As measured by RT-PCR and sequencing of the resulting products, fxyd1 splice site morphants have a deletion of the FXYD and transmembrane domains, due to an excision of exon 5, with no remaining wild type fxyd1 mRNA detectable (Fig. S1A). Specificity was confirmed since the phenotype could be prevented by co-injection of 50pg of the corresponding zebrafish mRNA that does not bind the MO (Fig. S1B; 86% normal ventricles (n=21)). Higher levels of fxyd1 mRNA injected led to a lethal phenotype, indicating that the embryo is sensitive to specific levels of fxyd1 mRNA. A second MO targeting the start site of fxyd1, synergized with low levels of a fxyd1 splice site MO to give a phenotype similar to that seen with high levels of the splice site MO, further confirming specificity of morphant phenotypes (Fig.S1Q–X). The fxyd1 splice site MO was more potent and therefore used for the rest of the study.
The severity of the atp1a1 start site morphants, which lacked brain ventricle inflation and had abnormal neural tube refractivity, led us to examine apical polarity, marked by aPKC, and the apical junction complex using phalloidin (actin) or Zo-1 (Figs. 1G–R). In wild type embryos, junction and polarity proteins form a continuous apical band lining the ventricles (Figs. 1G,M). Partial loss of atp1a1 does not change localization or continuity of the apical junctions (Figs. 1H–J, N–P) likely due to maternal contribution of Atp1a1. However, after greater loss of function elicited by an atp1a1 start site MO, or by fxyd1 splice site MO, apical junction and polarity proteins are discontinuous, with non-polarized cells crossing the midline and multiple small lumens forming (Figs. 1K–L,Q–R).
In order to determine whether apical junctions initially form, localization of Zo-1 was examined in transverse sections of the anterior neural tube at 12 hpf, when apico-basal polarity and junctions are first established (Fig. 1S). In control embryos, Zo-1 is expressed continuously at the apical surface of the neural tube (Fig. 1T, n=8). Conversely, in atp1a1 start site and fxyd1 morphants, Zo-1 expression is patchy and scattered throughout the neuroepithelium (Figs. 1U–V; atp1a1 start MO - n=9, fxyd1 MO – n=10). Therefore, Atp1a1 and Fxyd1 are required during initial neuroepithelial formation.
These data show that Fxyd1 is required for formation of a correctly polarized, continuous neuroepithelium, and are the first demonstration of a function for this gene during brain development. The data also show that partial loss of atp1a1 function leads to failure of ventricle inflation, and that more complete loss of function leads to absence of a polarized and continuous epithelium.
Atp1a1 regulates neuroepithelial permeability
Reduced brain ventricle inflation in atp1a1 partial loss of function embryos may be due to either abnormal CSF production or the inability of the neuroepithelium to retain fluid. We tested whether atp1a1 loss of function increased epithelial permeability, using a dye retention assay. In this assay, FITC-Dextran was injected into the brain ventricles at 22 hpf, and leakage of the dye out of the ventricles was monitored. A 70 kDa FITC-Dextran was chosen as it leaks out of the brain slowly, thereby allowing for identification of conditions that increase or decrease permeability. The distance traveled by the dye over time, was measured from the forebrain hinge-point to the furthest dye front, indicated by the white bar (Figs. 2A–F). This region was chosen because it is the first and most noticeable site of dye leaking out of the wild type brain (Fig. 2B). In snkto273a sibling embryos (wild type and heterozygotes), very little dye movement was observed (Figs. 2A–C,G). However, in snkto273a or hadm883 mutant embryos, or after injection of atp1a1 splice site MO, dye begins to leak out of the neuroepithelium immediately after injection (Figs. 2D–I; snkto273a - n=13, p<.0001; hadm883 - n=15, p<.001; atp1a1 splice MO - n=10, p<.05). Thus, although apical junction proteins properly localize, junctions do not function like wild type. Consistently, the snkto273a brain does not retain a 500 kDa FITC-Dextran whereas the wild type brain does demonstrating that these neuroepithelia are selectively permeable to dyes of different molecular weights (Fig. S2).
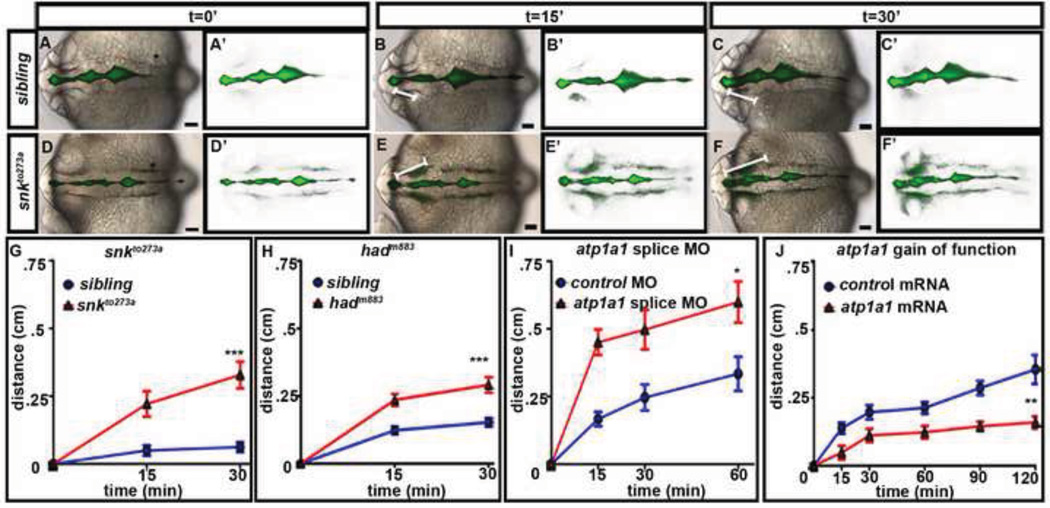
Fig. 2. Atp1a1 regulates neuroepithelial permeability.
(A–F) Dye retention assay in sibling embryos (A–C) vs. snkto273a mutants (D–F). Brightfield dorsal views of embryos ventricle injected with a 70 kDa FITC Dextran (A–F) and corresponding dye only images (A’–F’) over time. The distance the dye front moves measured at each time point indicated by white line. (G–J) Quantification of permeability in snkto273a mutants (G), hadm883 (H), atp1a1 splice site morphants (I), and atp1a1 gain-of-function embryos (J) compared to control injected. Average taken from 3–6 independent experiments and represented by mean +/− SEM. ***= p<0.0001, **= p<0.005, *=p<0.05 compared to control. All images taken at 22–24 hpf with anterior to left. Asterisk = ear. Scale bars = 50µm.
Since permeability is increased in atp1a1 loss-of-function experiments, we hypothesized that the converse would be true, that is, that atp1a1 overexpression in wild type embryos would increase CSF retention relative to controls. We therefore tested neuroepithelial permeability in embryos injected with 200 pg of atp1a1 mRNA and observed a decrease in permeability, consistent with this hypothesis (Fig. 2J, n=10, p<.005). Additionally, forebrain ventricle area and volume increased in embryos overexpressing atp1a1, compared to control injected embryos (Figs. S3A–B, D–E; n=10). We conclude that Atp1a1 regulates neuroepithelial permeability, and correlated with this, controls brain ventricle volume. Thus, part of the snkto273a mutant and atp1a1 splice site morphant phenotypes is likely due to the inability of the neuroepithelium to retain CSF.
Na,K-ATPase pumping is required for brain ventricle development
The Na,K-ATPase is proposed to be a scaffolding complex as well as a pump (Krupinski and Beitel, 2009). Thus, we asked whether pumping is required for brain ventricle development. Treatment with the pump inhibitor, ouabain (Linask and Gui, 1995), at 5 hpf (mid-gastrula stage) resulted in embryos with severely disrupted apical junctions (Figs. 3A–D; 0% normal ventricles (n=50)), similar to the effects of injecting an atp1a1 start site MO. Ouabain treatment at 16 hpf, after neuroepithelium formation, resulted in embryos with reduced ventricle inflation, correctly localized apical junctions (Figs. 3E–F; 25% normal ventricles (n=67)), but increased permeability (Fig. 3G; n=12, p<.05). Consistently, injection of mRNA encoding the putative pump-deficient snkto273a mutant atp1a1 (atp1a1GA) (Lowery and Sive, 2005) did not rescue brain ventricle development after atp1a1 loss of function (Figs. S4A–J) where wild type atp1a1 mRNA rescued both neuroepithelial formation and brain ventricle inflation (Figs. S4K–L). These data show that the pump activity of zebrafish Atp1a1 is essential during brain ventricle development.
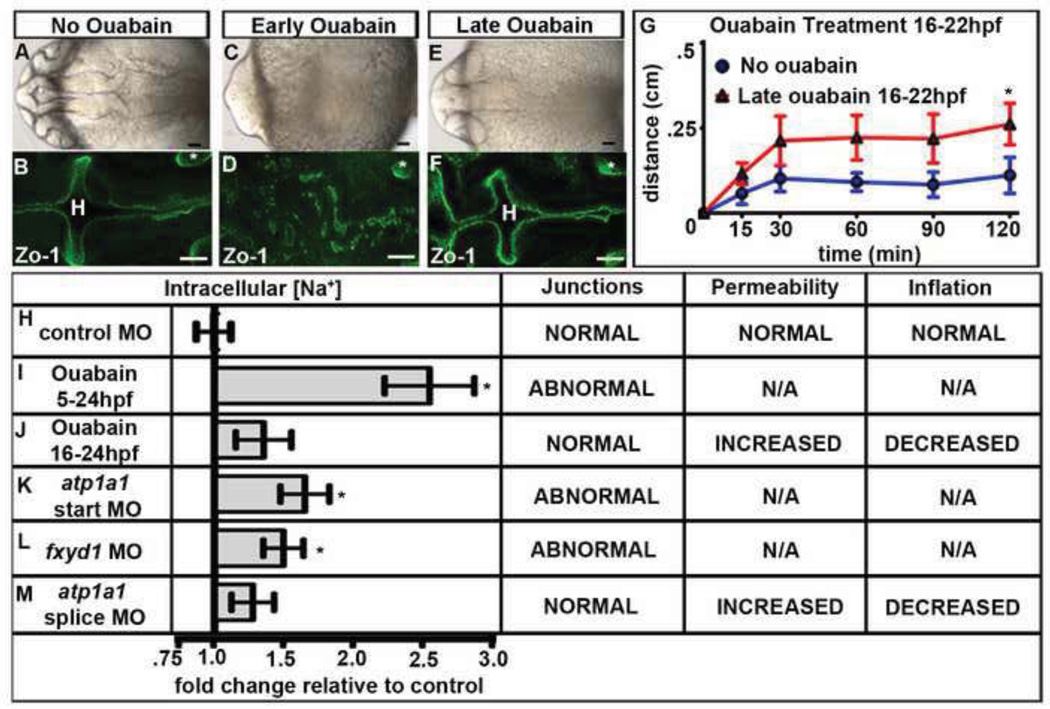
Fig. 3. Na,K-ATPase pumping is required for brain ventricle development.
(A–F) Wild type embryos either untreated (A–B) or ouabain treated from 5–24 hpf (early; C–D) or 16–24 hpf (late; E–F). Brightfield dorsal (A,C,E) or Zo-1 (B,D,F) images. (G) Dye retention assay in untreated (red) vs. late ouabain treated (blue) embryos. Data represented as mean +/− SEM. * = p<.05 compared to control. (H–L) Quantification of fold changes in [Na+]i and corresponding brain ventricle phenotype. Data from 3–8 independent experiments represented as fold change compared to control MO (equal to 1) plotted as a mean +/– SEM. *=p<0.05 compared to control. All embryos at 24 hpf, anterior to left; Asterisk = ear, H = hindbrain. Scale bars = 50µm.
Additionally, we predicted that loss of Atp1a1 function would lead to increased intracellular Na+ concentration ([Na+]i) in the brain, and if Fxyd1 modulates pump function, [Na+]i would also increase in fxyd1 loss of function embryos. Therefore, we measured [Na+]i in whole embryos after loss of function of the Na,K-ATPase. Since inhibiting the Na,K-ATPase using ouabain treatment or morpholinos affects the whole body and atp1a1 and fxyd1 are expressed ubiquitously (Figs. S1O–P) (Canfield et al., 2002; Ellertsdottir et al., 2006), we assume that changes in whole embryo [Na+]i are representative of differences within the brain. Changes in [Na+]i after Na,K-ATPase loss of funtion was quantified relative to [Na+]i levels in control MO injected embryos, whose [Na+]i was set equal to 1. [Na+]i levels in control embryos was constant between several pools of embryos examined (Fig. 3H), indicating that [Na+]i measurements can be compared between experiments. As predicted, we observed a 2.5 fold increase in [Na+]i in embryos treated with ouabain from 5–24 hpf compared to controls (Fig. 3I, p<.005), and a smaller increase in those treated with ouabain from 16–24 hpf (1.3 fold; Fig. 3J). Relative to controls, [Na+]i also increased in embryos injected with high levels of atp1a1 start site MO (1.7 fold; p<.05; Fig. 3K) or fxyd1 MO (1.5 fold; p<.05; Fig. 3L), and slightly after partial loss of atp1a1 via the splice site MO (1.2 fold; Fig. 3M). We found that neuroepithelium abnormality was generally correlated with the highest [Na+]i, (Figs. 3I,K–L).
Interestingly, while both ouabain treatment from 5–24 hpf, and atp1a1 start site MO led to similar phenotypes with regard to development of the neuroepithelium, levels of [Na+]i in ouabain-treated embryos were higher than after injection of the atp1a1 start site MO. This difference is likely due to inhibition of maternal Atp1a1 protein function and that of other alpha subunits by ouabain, whereas the start site MO would not inhibit maternal protein, nor affect other subunits. Consistently, levels of [Na+]i in both snkto273a and hadm883 mutant embryos, were higher than controls (Fig. S4M–N, snkto273a = 2.4 fold increase, p<.05; hadm883 = 2.0 fold increase, p<.001). The atp1a1 missense mutant, snkto273a also showed somewhat higher [Na+]i (Figs. S4M) relative to atp1a1 start site MO (Fig. 3K) (2.4 fold for snkto273a versus 1.7 fold for atp1a1 start site MO relative to controls). We hypothesized that the protein produced in snkto273a, retained some activity affecting [Na+]i but not neuroepithelium formation. Consistently, injection of atp1a1GA mRNA into atp1a1 start site morphants, led to elevated [Na+]i compared to atp1a1 start site MO alone (Fig. S4O, p<.05).
Thus, [Na+]i increases after inhibition of Na,K-ATPase with ouabain or after knockdown of Atp1a1 or Fxyd1 subunits. Moreover, this suggests that Fxyd1 promotes Atp1a1 function, however, we cannot rule out another independent role for Fxyd1.
Atp1a1 and Fxyd1 co-regulate brain ventricle development
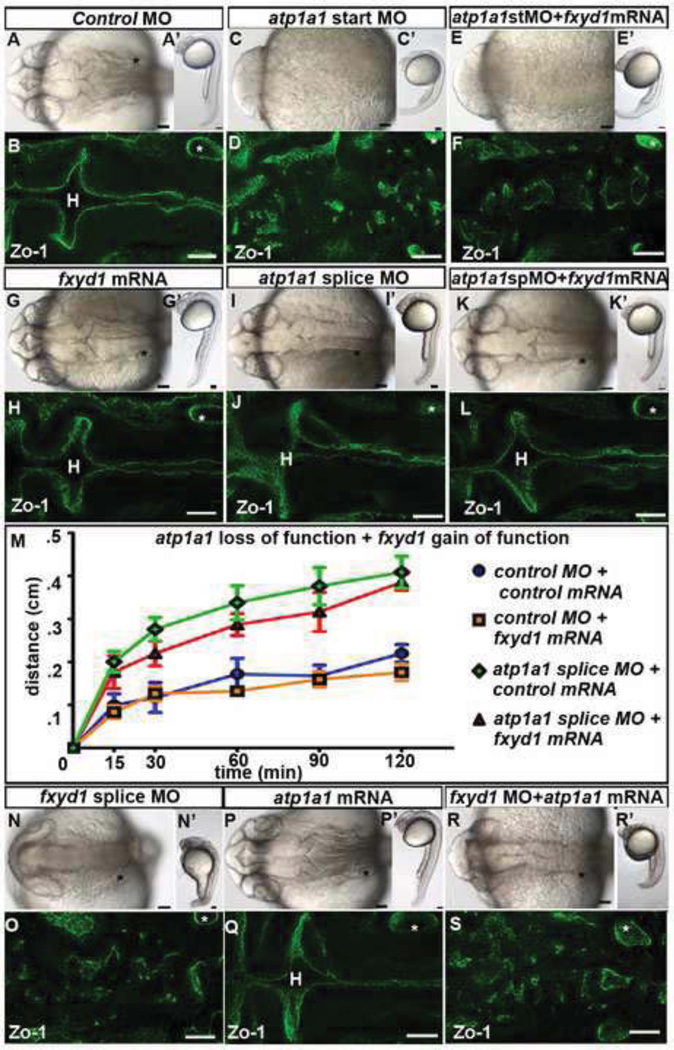
Data from other tissues and in vitro systems, suggest that Atp1a1 and Fxyd1 physically and functionally interact (Bibert et al., 2008; Bossuyt et al., 2006; Crambert et al., 2002; Lansbery et al., 2006; Mishra et al., 2011; Morth et al., 2007; Pavlovic et al., 2007; Shinoda et al., 2009). To determine whether Atp1a1 and Fxyd1 functionally interact, we injected embryos with low, sub-effective concentrations of atp1a1 splice site MO and fxyd1 MO together, which resulted in a non-cohesive neuroepithelium (0% normal ventricles (n=17)), whereas the individual MOs do not give a phenotype (Figs. 4A–D; atp1a1 splice site MO = 100% normal ventricles (n=14); fxyd1 MO = 100% normal ventricles (n=19)) suggesting these subunits functionally interact. Consistently, whereas embryos injected with low atp1a1 or fxyd1 MOs had wild type levels of [Na+]i, the combination of MOs was associated with increased [Na+]i (Figs. 4E–G).
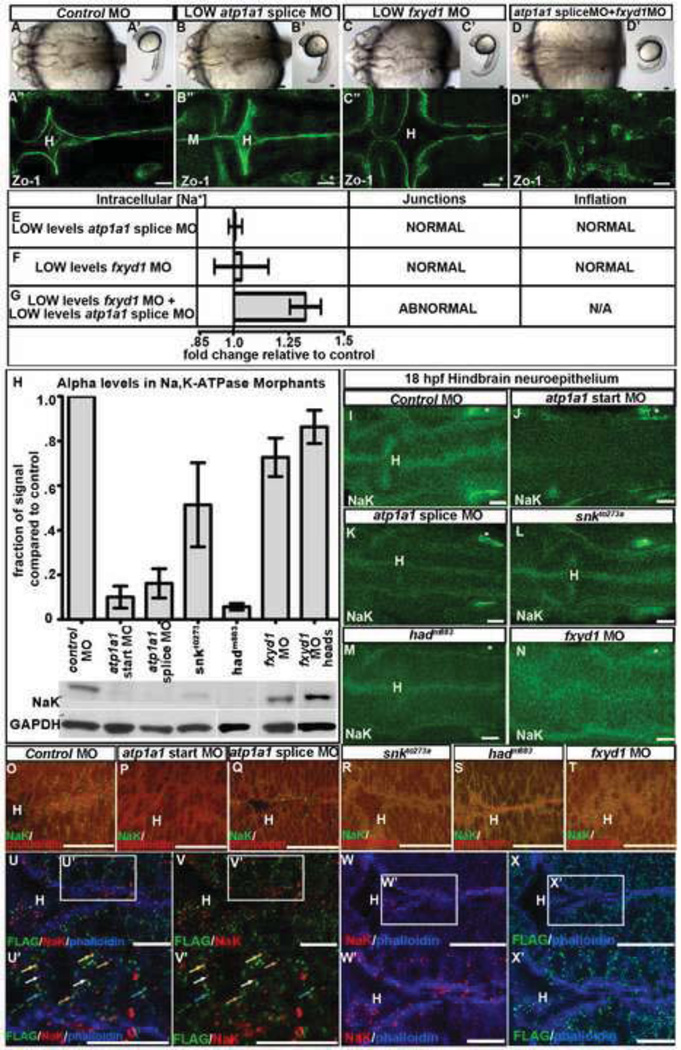
Fig. 4. Atp1a1 and Fxyd1 synergy, interaction and colocalization.
(A–G) Atp1a1 synergizes with Fxyd1. Brightfield dorsal (A–D), lateral (A’–D’) and Zo-1 staining (A’’–D’’; different embryos imaged than A–D) and measurement of [Na+]i with corresponding brain ventricle phenotype (E–G), in control (A), low atp1a1 splice MO (B,E), low fxyd1 MO (C,F) or together (D,G) at 24 hpf. Data representative of 3–6 experiments as fold change relative to control = 1; mean +/− SEM. (H) Western quantification of Atp1a1 (NaK) in 24 hpf whole embryos normalized to GAPDH. Representative of 3–4 independent experiments; mean +/− SEM. (I–T) Atp1a1 (NaK) in 18 hpf control MO (I,O), atp1a1 start site MO (J,P), atp1a1 splice site MO (K,Q), snkto273a (L,R), hadm883 (M,S) and fxyd1 MO (N,T). NaK alone (I–N, green) or with phalloidin (O–T, actin, red). (U–X) Localization of Atp1a1 (NaK; red) and Fxyd1-FLAG (green), phalloidin (blue) in fxyd1 MO embryos rescued with fxyd1-FLAG overexpression and imaged at 24 hpf. (U’–X’) Higher magnification indicated by box in (U–X). Co-localization (yellow arrow), adjacent (orange arrow), Atp1a1 alone (white arrow) and Fxyd1 alone (blue arrow). Anterior to left. Asterisk = ear. H = hindbrain. Scale bars = 10µm (U–X), 50µm (A–T).
Studies in cell culture suggest that FXYD subunits localize, stabilize and prevent degradation of alpha subunit protein (Mishra et al., 2011). In order to ask whether similar functions for Fxyd1 were present in the developing brain, we examined Atp1a1 levels and localization in loss of function embryos by Western blot and immunohistochemisty. As expected, levels of Atp1a1 in atp1a1 loss-of-function embryos were lower than controls and the protein remaining appeared as puncta enriched along the apical surface (Figs. 4H–M,O–S). In fxyd1 morphants, levels of Atp1a1 did not decrease, in either whole embryos or dissected heads (Fig. 4H), indicating that Fxyd1 does not stabilize Atp1a1. However, Atp1a1 expression was dispersed in the fxyd1 morphant neuroepithelium (Figs. 4N,T) demonstrating the necessity for Fxyd1 in correct localization of Atp1a1. Consistently, co-localization of Atp1a1 and Fxyd1 was observed as overlapping or adjacent protein staining of FLAG tagged Fxyd1 and endogenous Atp1a1 (Figs. 4U–V). In addition, Atp1a1 is enriched apically while Fxyd1 is present at higher levels laterally (Figs. 4W–X) suggesting some independent activity.
Consistent with discrete functions for these subunits, overexpression of 200 pg fxyd1 mRNA in atp1a1 start site morphants does not rescue junction formation suggesting that Fxyd1 cannot substitute for Atp1a1 function during neuroepithelial formation (Figs. 5A–H). Overexpression of fxyd1 mRNA alone does not affect neuroepithelial permeability (Figs. 5G,M) or brain ventricle size (Figs. S3C–D). Additionally, injected 50 pg fxyd1 RNA could not restore inflation or alter neuroepithelial permeability when overexpressed after partial loss of Atp1a1 function (Figs. 5G–M). The specific amounts injected were titrated to the maximum that allowed normal embryonic development. Together, these data support the conclusion that Fxyd1 does not normally regulate neuroepithelial permeability or brain ventricle inflation. Further, overexpression of atp1a1 could not substitute for loss of Fxyd1 function in fxyd1 morphants (Figs. 5N–S). The amount of atp1a1 and fxyd1 mRNA used in these assays was the same as was used to rescue the phenotypes caused by atp1a1 or fxyd1 MOs respectively.
Fig. 5. Atp1a1 and Fxyd1 do not substitute for one another.
(A–L) Overexpression of fxyd1 in atp1a1 loss of function embryos. Control (A–B), atp1a1 site start MO (C–D), atp1a1 start site MO + fxyd1 mRNA (E–F), fxyd1 mRNA (G–H), atp1a1 splice site MO (I–J), and fxyd1 mRNA + atp1a1 splice site MO (K–L). (M) Neuroepithelial permeability in control MO (blue), fxyd1 gain of function (orange), atp1a1 splice site MO (red), and fxyd1 mRNA + atp1a1 splice site MO (green). Data represented as mean +/− SEM, p>.05. (N–S) fxyd1 MO (N–O), atp1a1 mRNA (P–Q), and atp1a1 mRNA+ fxyd1 MO (R–S). Brightfield dorsal (A–R) and lateral (A’–R’) views. Zo-1 junction staining (B,D,F,H,J,L,O,Q,S). All images taken at 24 hpf with anterior to left. Asterisk = ear. H = hindbrain. Scale bars = 50µm.
Together, the data demonstrate functional interaction, co-localization and regulation of [Na+]i by zebrafish Fxyd1 and Atp1a1 in the developing neuroepithelium, and show that these proteins have non-redundant functions during ventricle formation.
Constitutively active RhoA rescues neuroepithelial cohesiveness but not brain ventricle inflation
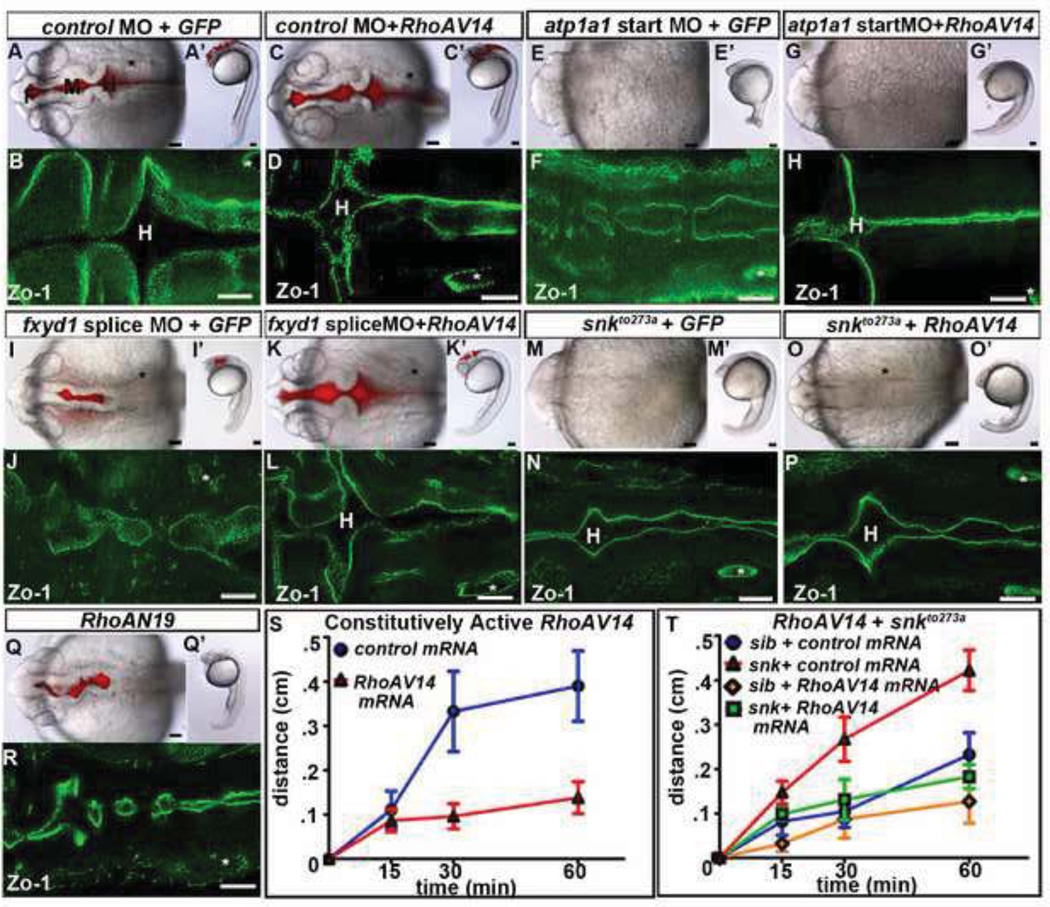
Based on experiments in cell culture (Rajasekaran et al., 2001), we hypothesized that RhoA acts downstream of the Na,K-ATPase during brain ventricle development. Injection of mRNA encoding a constitutively active human RhoA, RhoAV14, resulted in formation of a continuous neuroepithelium in control, atp1a1 start site, and fxyd1 morphants (Figs. 6A–L, S5A–B; atp1a1 start site = 80% normal ventricles (n=12); FXYD1 MO = 80% normal ventricles (n=20)). However, RhoAV14 expression in atp1a1 start site morphants (Figs. 6E–H) or in snkto273a mutants (Figs. 6M–P) did not lead to brain ventricle inflation, suggesting that brain ventricle inflation is a RhoA independent proccess. In contrast, expression of RhoAV14 in fxyd1 morphants not only restored a continuous neuroepithelium, but also led to fully inflated brain ventricles (Figs. 6I–L) further supporting the hypothesis that Fxyd1 is not required for brain ventricle inflation. Expression of a dominant negative RhoA (RhoAN19; Figs. 6Q–R; 0% normal ventricles (n-22)) or incubation in an inhibitor of ROCK (Y27632), a target of active RhoA (Amano et al., 2000) (Figs. S5C–N), resulted in a discontinuous neuroepithelium, supporting a normal requirement for RhoA during brain development. These results suggest that Fxyd1 and Atp1a1 act through RhoA during neuroepithelium formation.
Fig. 6. Constitutively active RhoA rescues neuroepithelial formation.
(A–H) Constitutively active RhoAV14 (C,D,G,H,K,L,O,P), or control mRNA (GFP; A,B,E,F,I,J,M,N) in control MO (A–D), atp1a1 start site MO (E–H), fxyd1 MO (I–L), and snkto273a (M–P). (Q–R) Overexpression of dominant negative RhoAN19. (S–T) Dye retention assay in wild type embryos expressing control mRNA (blue), or RhoAV14 (red) (S) and sibling (sib) + control mRNA (red), snkto273a + control mRNA (blue), sib + RhoAV14 (orange), and snkto273a + RhoAV14 (green) (T). Brightfield dorsal (A–Q) and lateral (A’–Q’) views. Ventricle injection of 2000 kDa Rhodamine Dextran dye (A,C,I,K,Q). Zo −1 (B,D,F,H,J,L,N,P,R). All images taken at 24 hpf with anterior to left. F = forebrain, M = midbrain, H = hindbrain. Asterisk = ear. Scale bars = 50µm.
One explanation for the inability of RhoAV14 to restore brain ventricle inflation to snkto273a mutants, despite a continuous neuroepithelium, is that the neuroepithelium was leaky and could not retain CSF. In wild type embryos, expression of RhoAV14 increased dye retention, and therefore decreased permeability (Fig. 6S; p<.05). Significantly, RhoAV14 expression also decreased permeability of snkto273a neuroepithelium, into the wild type range (Fig. 6T; p<.05), indicating that a RhoA is required for regulation of neuroepithelial permeability. Further, although RhoA overexpression in snkto273a embryos rescued neuroepithelial permeability, brain ventricle inflation remained reduced suggesting that a RhoA-insensitive process is necessary for brain ventricle inflation.
These results indicate that RhoA signaling acts downstream of Atp1a1 and Fxyd1 to regulate neuroepithelium formation and permeability. The data also delineate a RhoA-insensitive step in brain ventricle inflation, likely CSF production, which is dependent on Atp1a1, but not on Fxyd1 function.
DISCUSSION
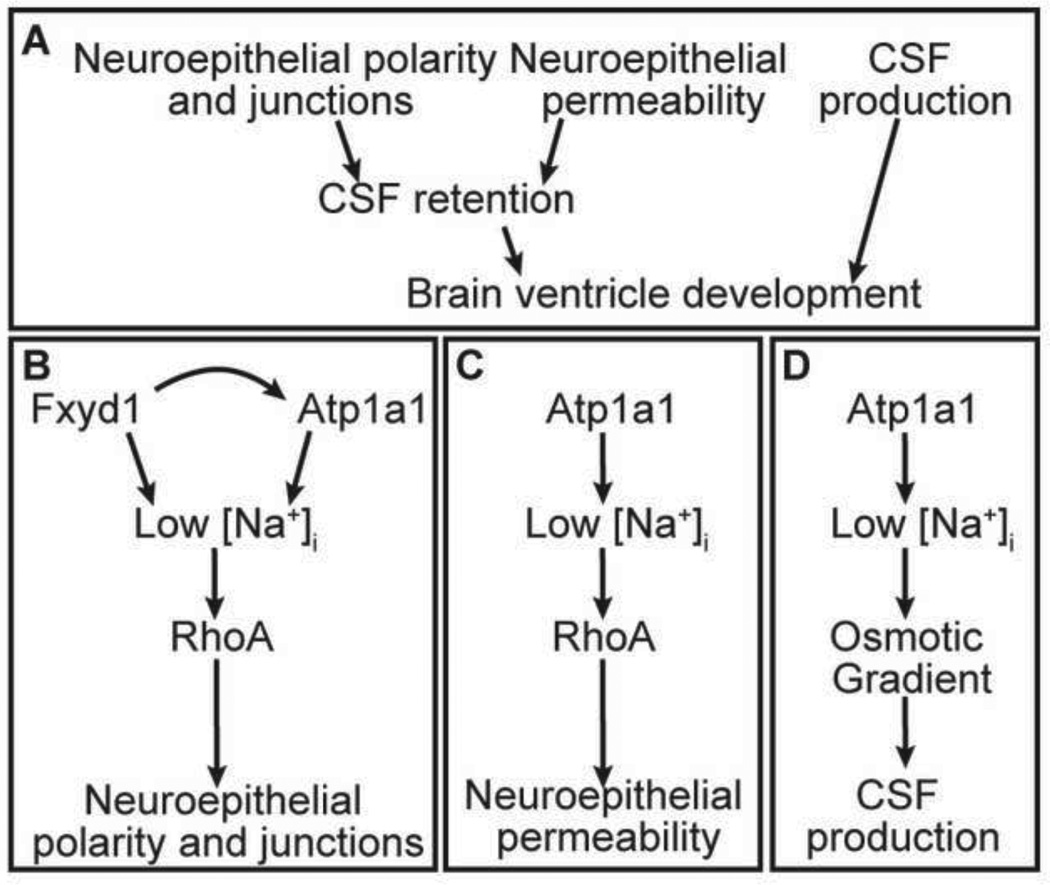
The Na,K-ATPase has a well-known role as a modulator of membrane potential in neurons and is essential for generating an action potential. However, this protein complex has additional activities, and this study uncovers three processes in the developing brain that are regulated by Na,K-ATPase function to culminate in the formation of the brain ventricular system: neuroepithelial polarity and junction formation, neuroepithelial permeability and CSF production (Fig. 7A). The ability of a single protein complex to regulate multiple aspects of brain ventricle formation and inflation suggests that the Na,K-ATPase is a pivotal regulator of ventricle volume.
Fig. 7. Model for requirement of Na,K-ATPase during brain ventricle development.
(A) Brain ventricle development is a three step process that requires establishment of neuroepithelial polarity and junctions and regulation of neuroepithelial permeability allowing for CSF retention, in addition to CSF production. Brain ventricle inflation occurs via CSF retention and CSF production. (B) Neuroepithelial formation requires RhoA which acts downstream of Fxyd1 and Atp1a1. (C) Neuroepithelial permeability requires RhoA which acts downstream of only Atp1a1. (D) CSF production is RhoA - insensitive and requires Atp1a1.
Dissecting the activities of the Na,K-ATPase during brain ventricle formation
Several experimental approaches allowed for division of the Na,K-ATPase function into activities relevant for brain ventricle development, occurring sequentially and coordinately after neural tube closure. Formation of continuous apical neuroepithelial polarity and junctions requires RhoA activity downstream of both Atp1a1 and Fxyd1 (Fig. 7B). Na,K-ATPase pumping appears to be important in establishing rather than maintaining junctions since adding the pump inhibitor, ouabain, after initial junctions had formed, did not disrupt the neuroepithelium.
By further exploring the interface with RhoA signaling, we showed that Atp1a1 had two additional activities. RhoA could substitute for one of these functions, modulation of neuroepithelial permeability (Fig. 7C), however, the other was a RhoA-independent role uncovered by failure of RhoA to substitute for Atp1a1 during ventricle inflation, despite promoting wild type neuroepithelial permeability. This activity is likely to be CSF production (Fig. 7D). Once the neuroepithelium has formed, equilibrium between drainage from the ventricles and CSF production would maintain a normal ventricular volume. The amount of active Atp1a1 may regulate the equilibrium, since different levels of this protein can increase or decrease permeability and ventricle size. Thus, the strong phenotypes observed in Atp1a1 mutants, are likely a composite of both impaired CSF production and retention.
The role of Fxyd1 during brain ventricle development
This is the first report of a requirement for Fxyd1 function in the developing brain. FXYD1 knockout mice show no apparent brain phenotype, suggesting functional redundancy not present in zebrafish. FXYD1 adult null mice had increased cardiac mass, larger cardiac myocytes and, consistent with results in zebrafish, show 50% reduced Na,K-ATPase activity in mutant hearts relative to wild type (Jia et al., 2005). Fxyd1 appears to have a distinct role during brain ventricle formation, since it is required for neuroepithelium formation but, unlike Atp1a1, not for subsequent steps that lead to ventricle inflation. Consistently, overexpression of Atp1a1 does not substitute for Fxyd1, and vice versa. While synergy between atp1a1 and fxyd1 loss of function supports a major role for Fxyd1 as a regulator of Atp1a1 pumping, Fxyd1 may also have additional activity, that could modulate [Na+]i, for example, by inhibiting the Na+/Ca2+ exchanger (Zhang et al., 2003).
Connection between Na,K-ATPase function, RhoA signaling, neuroepithelium formation and permeability
How does the Na,K-ATPase modulate RhoA function, which impacts both neuroepithelial polarity and permeability? One possibility is that depolarization of the cell by the Na,K-ATPase leads to RhoA activation. In cell culture, depolarization of epithelial cells activates the Ras/MEK/ERK pathway (Waheed et al., 2010), by promoting GEF activity and increasing RhoA-GTP levels. In kidney tubular cells (LLC-PK1) and MDCK cells, depolarization activates RhoA and ROCK leading to Myosin Light Chain phosphorylation (Szaszi et al., 2005). However, in no case is the mechanism connecting depolarization and RhoA activation understood.
Unlike the case in Drosophila trachea formation (Paul et al., 2007), our data indicate a requirement for Na,K-ATPase pump function in epithelium formation. Multiple differences between fly and vertebrate junctions (Knust and Bossinger, 2002) likely explain these different requirements for the Na,K-ATPase. Consistent with our study, Na,K-ATPase pumping is required for formation of continuous junctions and lumens in the zebrafish heart and gut (Bagnat et al., 2007; Cibrian-Uhalte et al., 2007).
We propose that RhoA regulates paracellular permeability in the zebrafish neuroepithelium, based on the size selectivity of our dye retention assay. However, we cannot rule out some contribution of vesicular transcellular pathways. Consistently, Claudin5a, a barrier claudin, and component of the tight junction complex responsible for paracellular ion transport and selectivity, is required for brain ventricle inflation and permeability (Terry et al., 2010; Zhang et al., 2010). In cell culture, RhoA activation promotes claudin phosphorylation and regulates permeability by modulating claudin-claudin interactions or recycling tight junction components (Yamamoto et al., 2008). Thus, there are plausible connections between the Na,K-ATPase, RhoA and claudins.
Modulation of CSF production by the Na,K-ATPase
Ventricle “inflation” is a scorable phenotype, but not a discrete process, as it includes formation of a non-leaky epithelium as well as CSF production, which appears to be a RhoA-insensitive process. This is likely to be mediated by the osmotic gradient formed as a result of a membrane potential difference, under control of the Na,K-ATPase and other pumps and channels (Fig. 7D) (Brown et al., 2004; Pollay et al., 1985). Production of CSF requires both movement of water into the ventricular lumen, as well as secretion of proteins and other factors. The role of the Na,K-ATPase in water movement may occur either via Aquaporins in the plasma membrane or through the paracellular pathway. Although Aquaporin 1 is strongly expressed in the choroid plexus and knockout mice have abnormal CSF production (Oshio et al., 2003), analysis of Aquaporin activity during initial ventricle inflation has not been explored. There is little data to suggest whether Na,K-ATPase regulates protein secretion into the ventricular lumen, although one report implicates Na,K-ATPase during FGF2 secretion in primate cells (Dahl et al., 2000). Analysis of the role of Na,K-ATPase during protein secretion and water movement that results in brain ventricle inflation will be a future direction of this study.
Significance of Na,K-ATPase activity during brain ventricle volume control
Either an increase or decrease in CSF volume can be pathological throughout life (Lowery and Sive, 2009), due to either changes in pressure and CSF composition (Desmond et al., 2005; Gato et al., 2005). Since the amount of functional Na,K-ATPase can regulate brain ventricle size in a graded manner, this pump may play a “volume sensor” and homeostatic role. The molecular basis for non-obstructive hydrocephalus is not clear, and our data suggest possible input from Na,K-ATPase function. In addition to the regulation described in this study, Na,K-ATPase activity can be fine-tuned by differential expression of beta and Fxyd subunits which modulate pump activity, expression level and correct cellular localization (Geering, 2001; Geering, 2006; Wilson et al., 2000). Therefore a slight disruption of Na,K-ATPase subunits could lead to drastic changes in activity and abnormal CSF levels.
Supplementary Material
Na,K-ATPase function coordinates three aspects of brain ventricle development.
The FXYD1 subunit is required for neuroepithelium formation.
Atp1a1 regulates neuroepthelium formation, permeability, and CSF production.
RhoA acts downstream of Na,K-ATPase in neuroepithelium formation and permeability.
CSF production is a RhoA-insensitive process.
ACKNOWLEDGEMENTS
This work was supported by the National Institute for Mental Health, and National Science Foundation. Special thanks to Dr. Jen Gutzman, Dr. Iain Cheeseman and Sive lab members for many useful discussions and constructive criticism, and to Olivier Paugois for expert fish husbandry.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Amano M, Fukata Y, Kaibuchi K. Regulation and functions of Rho associated kinase. Exp Cell Res. 2000;261:44–51. doi: 10.1006/excr.2000.5046. [DOI] [PubMed] [Google Scholar]
- Bagnat M, Cheung ID, Mostov KE, Stainier DY. Genetic control of single lumen formation in the zebrafish gut. Nat Cell Biol. 2007;9:954–960. doi: 10.1038/ncb1621. [DOI] [PubMed] [Google Scholar]
- Bibert S, Roy S, Schaer D, Horisberger JD, Geering K. Phosphorylation of phospholemman (FXYD1) by protein kinases A and C modulates distinct Na,K-ATPase isozymes. J Biol Chem. 2008;283:476–486. doi: 10.1074/jbc.M705830200. [DOI] [PubMed] [Google Scholar]
- Bossuyt J, Despa S, Martin JL, Bers DM. Phospholemman phosphorylation alters its fluorescence resonance energy transfer with the Na/K-ATPase pump. J Biol Chem. 2006;281:32765–32773. doi: 10.1074/jbc.M606254200. [DOI] [PubMed] [Google Scholar]
- Brown PD, Davies SL, Speake T, Millar ID. Molecular mechanisms of cerebrospinal fluid production. Neuroscience. 2004;129:957–970. doi: 10.1016/j.neuroscience.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield VA, Loppin B, Thisse B, Thisse C, Postlethwait JH, Mohideen MA, Rajarao SJ, Levenson R. Na,K-ATPase alpha and beta subunit genes exhibit unique expression patterns during zebrafish embryogenesis. Mech Dev. 2002;116:51–59. doi: 10.1016/s0925-4773(02)00135-1. [DOI] [PubMed] [Google Scholar]
- Cibrian-Uhalte E, Langenbacher A, Shu X, Chen JN, Abdelilah-Seyfried S. Involvement of zebrafish Na+,K+ ATPase in myocardial cell junction maintenance. J Cell Biol. 2007;176:223–230. doi: 10.1083/jcb.200606116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruna B, Jenny A, Lee D, Mlodzik M, Schier AF. Planar cell polarity signalling couples cell division and morphogenesis during neurulation. Nature. 2006;439:220–224. doi: 10.1038/nature04375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crambert G, Fuzesi M, Garty H, Karlish S, Geering K. Phospholemman (FXYD1) associates with Na,K-ATPase and regulates its transport properties. Proc Natl Acad Sci U S A. 2002;99:11476–11481. doi: 10.1073/pnas.182267299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JP, Binda A, Canfield VA, Levenson R. Participation of Na,K-ATPase in FGF-2 secretion: rescue of ouabain-inhibitable FGF-2 secretion by ouabain-resistant Na,K-ATPase alpha subunits. Biochemistry. 2000;39:14877–14883. doi: 10.1021/bi001073y. [DOI] [PubMed] [Google Scholar]
- Desmond ME, Levitan ML, Haas AR. Internal luminal pressure during early chick embryonic brain growth: descriptive and empirical observations. Anat Rec A Discov Mol Cell Evol Biol. 2005;285:737–747. doi: 10.1002/ar.a.20211. [DOI] [PubMed] [Google Scholar]
- Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30:154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- Ellertsdottir E, Ganz J, Durr K, Loges N, Biemar F, Seifert F, Ettl AK, Kramer-Zucker AK, Nitschke R, Driever W. A mutation in the zebrafish Na,K-ATPase subunit atp1a1a.1 provides genetic evidence that the sodium potassium pump contributes to left-right asymmetry downstream or in parallel to nodal flow. Dev Dyn. 2006;235:1794–1808. doi: 10.1002/dvdy.20800. [DOI] [PubMed] [Google Scholar]
- Feschenko MS, Donnet C, Wetzel RK, Asinovski NK, Jones LR, Sweadner KJ. Phospholemman, a single-span membrane protein, is an accessory protein of Na,K-ATPase in cerebellum and choroid plexus. J Neurosci. 2003;23:2161–2169. doi: 10.1523/JNEUROSCI.23-06-02161.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gato A, Moro JA, Alonso MI, Bueno D, De La Mano A, Martin C. Embryonic cerebrospinal fluid regulates neuroepithelial survival, proliferation, and neurogenesis in chick embryos. Anat Rec A Discov Mol Cell Evol Biol. 2005;284:475–484. doi: 10.1002/ar.a.20185. [DOI] [PubMed] [Google Scholar]
- Geering K. The functional role of beta subunits in oligomeric P-type ATPases. J Bioenerg Biomembr. 2001;33:425–438. doi: 10.1023/a:1010623724749. [DOI] [PubMed] [Google Scholar]
- Geering K. FXYD proteins: new regulators of Na-K-ATPase. Am J Physiol Renal Physiol. 2006;290:F241–F250. doi: 10.1152/ajprenal.00126.2005. [DOI] [PubMed] [Google Scholar]
- Graeden E, Sive H. Live imaging of the zebrafish embryonic brain by confocal microscopy. J Vis Exp. 2009 doi: 10.3791/1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzman JH, Sive H. Zebrafish brain ventricle injection. J Vis Exp. 2009 doi: 10.3791/1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzman JH, Sive H. Epithelial relaxation mediated by the myosin phosphatase regulator Mypt1 is required for brain ventricle lumen expansion and hindbrain morphogenesis. Development. 2010;137:795–804. doi: 10.1242/dev.042705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E, Brewster R. N-cadherin is required for the polarized cell behaviors that drive neurulation in the zebrafish. Development. 2006;133:3895–3905. doi: 10.1242/dev.02560. [DOI] [PubMed] [Google Scholar]
- Jia LG, Donnet C, Bogaev RC, Blatt RJ, McKinney CE, Day KH, Berr SS, Jones LR, Moorman JR, Sweadner KJ, Tucker AL. Hypertrophy, increased ejection fraction, and reduced Na-K-ATPase activity in phospholemman-deficient mice. Am J Physiol Heart Circ Physiol. 2005;288:H1982–H1988. doi: 10.1152/ajpheart.00142.2004. [DOI] [PubMed] [Google Scholar]
- Jiang YJ, Brand M, Heisenberg CP, Beuchle D, Furutani-Seiki M, Kelsh RN, Warga RM, Granato M, Haffter P, Hammerschmidt M, Kane DA, Mullins MC, Odenthal J, van Eeden FJ, Nusslein-Volhard C. Mutations affecting neurogenesis and brain morphology in the zebrafish, Danio rerio. Development. 1996;123:205–216. doi: 10.1242/dev.123.1.205. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Knust E, Bossinger O. Composition and formation of intercellular junctions in epithelial cells. Science. 2002;298:1955–1959. doi: 10.1126/science.1072161. [DOI] [PubMed] [Google Scholar]
- Krupinski T, Beitel GJ. Unexpected roles of the Na-K-ATPase and other ion transporters in cell junctions and tubulogenesis. Physiology (Bethesda) 2009;24:192–201. doi: 10.1152/physiol.00008.2009. [DOI] [PubMed] [Google Scholar]
- Lansbery KL, Burcea LC, Mendenhall ML, Mercer RW. Cytoplasmic targeting signals mediate delivery of phospholemman to the plasma membrane. Am J Physiol Cell Physiol. 2006;290:C1275–C1286. doi: 10.1152/ajpcell.00110.2005. [DOI] [PubMed] [Google Scholar]
- Linask KK, Gui YH. Inhibitory effects of ouabain on early heart development and cardiomyogenesis in the chick embryo. Dev Dyn. 1995;203:93–105. doi: 10.1002/aja.1002030110. [DOI] [PubMed] [Google Scholar]
- Lowery LA, De Rienzo G, Gutzman JH, Sive H. Characterization and classification of zebrafish brain morphology mutants. Anat Rec (Hoboken) 2009;292:94–106. doi: 10.1002/ar.20768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery LA, Sive H. Initial formation of zebrafish brain ventricles occurs independently of circulation and requires the nagie oko and snakehead/atp1a1a.1 gene products. Development. 2005;132:2057–2067. doi: 10.1242/dev.01791. [DOI] [PubMed] [Google Scholar]
- Lowery LA, Sive H. Totally tubular: the mystery behind function and origin of the brain ventricular system. Bioessays. 2009;31:446–458. doi: 10.1002/bies.200800207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra NK, Peleg Y, Cirri E, Belogus T, Lifshitz Y, Voelker DR, Apell HJ, Garty H, Karlish SJ. FXYD proteins stabilize Na,K-ATPase: amplification of specific phosphatidylserine-protein interactions. J Biol Chem. 2011;286:9699–9712. doi: 10.1074/jbc.M110.184234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morth JP, Pedersen BP, Toustrup-Jensen MS, Sorensen TL, Petersen J, Andersen JP, Vilsen B, Nissen P. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene 'knockdown' in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Oshio K, Song Y, Verkman AS, Manley GT. Aquaporin-1 deletion reduces osmotic water permeability and cerebrospinal fluid production. Acta Neurochir Suppl. 2003;86:525–528. doi: 10.1007/978-3-7091-0651-8_107. [DOI] [PubMed] [Google Scholar]
- Paul SM, Palladino MJ, Beitel GJ. A pump-independent function of the Na,K-ATPase is required for epithelial junction function and tracheal tube size control. Development. 2007;134:147–155. doi: 10.1242/dev.02710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovic D, Fuller W, Shattock MJ. The intracellular region of FXYD1 is sufficient to regulate cardiac Na/K ATPase. FASEB J. 2007;21:1539–1546. doi: 10.1096/fj.06-7269com. [DOI] [PubMed] [Google Scholar]
- Pollay M, Hisey B, Reynolds E, Tomkins P, Stevens FA, Smith R. Choroid plexus Na+/K+-activated adenosine triphosphatase and cerebrospinal fluid formation. Neurosurgery. 1985;17:768–772. doi: 10.1227/00006123-198511000-00007. [DOI] [PubMed] [Google Scholar]
- Rajasekaran SA, Barwe SP, Gopal J, Ryazantsev S, Schneeberger EE, Rajasekaran AK. Na-K-ATPase regulates tight junction permeability through occludin phosphorylation in pancreatic epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2007;292:G124–G133. doi: 10.1152/ajpgi.00297.2006. [DOI] [PubMed] [Google Scholar]
- Rajasekaran SA, Palmer LG, Moon SY, Peralta Soler A, Apodaca GL, Harper JF, Zheng Y, Rajasekaran AK. Na,K-ATPase activity is required for formation of tight junctions, desmosomes, and induction of polarity in epithelial cells. Mol Biol Cell. 2001;12:3717–3732. doi: 10.1091/mbc.12.12.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagerstrom CG, Grinbalt Y, Sive H. Anteroposterior patterning in the zebrafish, Danio rerio: an explant assay reveals inductive and suppressive cell interactions. Development. 1996;122:1873–1883. doi: 10.1242/dev.122.6.1873. [DOI] [PubMed] [Google Scholar]
- Shinoda T, Ogawa H, Cornelius F, Toyoshima C. Crystal structure of the sodium-potassium pump at 2.4 A resolution. Nature. 2009;459:446–450. doi: 10.1038/nature07939. [DOI] [PubMed] [Google Scholar]
- Shu X, Cheng K, Patel N, Chen F, Joseph E, Tsai HJ, Chen JN. Na,K-ATPase is essential for embryonic heart development in the zebrafish. Development. 2003;130:6165–6173. doi: 10.1242/dev.00844. [DOI] [PubMed] [Google Scholar]
- Speake T, Whitwell C, Kajita H, Majid A, Brown PD. Mechanisms of CSF secretion by the choroid plexus. Microsc Res Tech. 2001;52:49–59. doi: 10.1002/1097-0029(20010101)52:1<49::AID-JEMT7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Sweadner KJ, Rael E. The FXYD gene family of small ion transport regulators or channels: cDNA sequence, protein signature sequence, and expression. Genomics. 2000;68:41–56. doi: 10.1006/geno.2000.6274. [DOI] [PubMed] [Google Scholar]
- Szaszi K, Sirokmany G, Di Ciano-Oliveira C, Rotstein OD, Kapus A. Depolarization induces Rho-Rho kinase-mediated myosin light chain phosphorylation in kidney tubular cells. Am J Physiol Cell Physiol. 2005;289:C673–C685. doi: 10.1152/ajpcell.00481.2004. [DOI] [PubMed] [Google Scholar]
- Terry S, Nie M, Matter K, Balda MS. Rho signaling and tight junction functions. Physiology (Bethesda) 2010;25:16–26. doi: 10.1152/physiol.00034.2009. [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waheed F, Speight P, Kawai G, Dan Q, Kapus A, Szaszi K. Extracellular signal-regulated kinase and GEF-H1 mediate depolarization-induced Rho activation and paracellular permeability increase. Am J Physiol Cell Physiol. 2010;298:C1376–C1387. doi: 10.1152/ajpcell.00408.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welss P. Secretory activity of the inner layer of the embryonic mid-brain of the chick, as revealed by tissue culture. The Anatomical Record. 1934;58:299–302. [Google Scholar]
- Westerfield M, Sprague J, Doerry E, Douglas S, Grp Z. The Zebrafish Information Network (ZFIN): a resource for genetic, genomic and developmental research. Nucleic Acids Research. 2001;29:87–90. doi: 10.1093/nar/29.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PD, Devuyst O, Li X, Gatti L, Falkenstein D, Robinson S, Fambrough D, Burrow CR. Apical plasma membrane mispolarization of NaK-ATPase in polycystic kidney disease epithelia is associated with aberrant expression of the beta2 isoform. Am J Pathol. 2000;156:253–268. doi: 10.1016/s0002-9440(10)64726-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Ramirez SH, Sato S, Kiyota T, Cerny RL, Kaibuchi K, Persidsky Y, Ikezu T. Phosphorylation of claudin-5 and occludin by rho kinase in brain endothelial cells. Am J Pathol. 2008;172:521–533. doi: 10.2353/ajpath.2008.070076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Joseph EM. The small heart mutation reveals novel roles of Na+/K+-ATPase in maintaining ventricular cardiomyocyte morphology and viability in zebrafish. Circ Res. 2004;95:595–603. doi: 10.1161/01.RES.0000141529.48143.6e. [DOI] [PubMed] [Google Scholar]
- Zhang J, Piontek J, Wolburg H, Piehl C, Liss M, Otten C, Christ A, Willnow TE, Blasig IE, Abdelilah-Seyfried S. Establishment of a neuroepithelial barrier by Claudin5a is essential for zebrafish brain ventricular lumen expansion. Proc Natl Acad Sci U S A. 2010;107:1425–1430. doi: 10.1073/pnas.0911996107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XQ, Qureshi A, Song J, Carl LL, Tian Q, Stahl RC, Carey DJ, Rothblum LI, Cheung JY. Phospholemman modulates Na+/Ca2+ exchange in adult rat cardiac myocytes. Am J Physiol Heart Circ Physiol. 2003;284:H225–H233. doi: 10.1152/ajpheart.00698.2002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.