Abstract
The cranial neural crest (CNC) is a population of cells that arises from the lateral part of the developing brain, migrates ventrally and coordinates the entire craniofacial development of vertebrates. Many molecules are involved in CNC migration including the transmembrane metalloproteases ADAM13 and 19. We have previously shown that these ADAMs cleave a number of extracellular proteins and modify the transcription of a number of genes, and that both of these activities are important for cell migration. Here we show that the knock down of ADAM13 inhibits CNC migration in vivo but not in vitro, indicating that ADAM13 function is required in the 3-dimentional context of the embryo. We further show that the migration of CNC that do not express ADAM13 and ADAM19 can be rescued in vivo by co-grafting wild type CNC. Furthermore, the migration of CNC lacking ADAM13 can be rescued by mechanically separating the CNC from the surrounding ectoderm and mesoderm. Finally, we show that ADAM13 function is autonomous to CNC tissue, as the migration of morphant CNC can only be rescued by ADAM13 expression in the CNC and not the surrounding tissues. Together our results suggest that ADAM13 changes CNC interaction with the extracellular environment and that this change is necessary for their migration in vivo.
Keywords: ADAM proteases, cranial neural crest, cell migration, Xenopus laevis
Introduction
The neural crest is a pluripotent population of cells specific to vertebrates that arises from the dorsal neural tube during or after neurulation. It undergoes extensive cell migration and gives rise to many derivatives depending of their antero-posterior origin and their final resting place. The most anterior neural crest, called the cranial neural crest (CNC), originates from the midbrain and hindbrain and coordinates craniofacial development (Minoux and Rijli, 2010). Their extensive migration is the result of the coordinated function of cell adhesion molecules, contact mediated cell polarity and guidance cues (Alfandari et al., 2010; Kulesa et al., 2010; Theveneau and Mayor, 2011b). In the amphibian Xenopus laevis, the molecules involved in CNC migration include the α5β1 ••••••••, fibronectin, syndecan4 and the Planar Cell Polarity pathway (Alfandari et al., 2003; Carmona-Fontaine et al., 2008; Matthews et al., 2008). In recent years, we discovered that another family of transmembrane proteins is also involved in CNC migration: the ADAM family.
ADAMs are cell surface metalloproteases that contain a disintegrin and cysteine rich domain. Half of these ADAMs have been shown or are predicted to possess the metalloprotease activity (Alfandari et al., 2009). This includes the subgroup of meltrins composed of ADAM9, 12, 13, 19 and 33. ADAMs are key players in many biological processes including cell adhesion and cell signaling. Their role in cell signaling is linked to their ability to cleave signaling molecules that results in either the activation (EGF, TNF-α, Notch), or termination (ephrins) of signaling pathways (Alfandari et al., 2009). Some ADAM proteins have been shown to regulate cell-cell adhesion by cleaving N- and E-cadherin to promote epithelial to mesenchymal transition (EMT) (Maretzky et al., 2005; Reiss et al., 2005).
We have previously shown that ADAM13 is required for CNC cell migration (Alfandari et al., 2001; Cousin et al., 2011; McCusker et al., 2009). While the single knock down of either ADAM13 or 19 is sufficient to significantly inhibit CNC migration, the double knock down induces an almost complete inhibition that can be rescued by the re-expression of either one of these ADAMs, indicating that these two meltrins have partially redundant function (Cousin et al., 2011; McCusker et al., 2009). The ADAM13 metalloprotease domain is capable of cleaving many substrates, including fibronectin (FN) and Cadherin-11 (Alfandari et al., 2001; Gaultier et al., 2002; McCusker et al., 2009). In a related species, Xenopus tropicalis, ADAM13 cleaves ephrinB ligands and contributes to neural crest cell induction (Wei et al., 2010) but this role does not appear to be conserved in Xenopus laevis (Cousin et al., 2011). While we do not yet know the relevance of FN cleavage during CNC migration, we have shown that Cadherin-11 cleavage is critical (McCusker et al., 2009). The disintegrin and cysteine rich domains are involved in substrate recognition. In the case of FN, the cysteine rich domain of ADAM13 binds to the second Heparin-binding domain (hepII) of FN (Gaultier et al., 2002). The cytoplasmic domain of ADAM13 has multiple functions, one of which is to control the metalloprotease activity of ADAM13 by interacting with the cytoplasmic adaptor protein PACSIN2 (Cousin et al., 2000). Recently, we discovered that this cytoplasmic domain is released in the cell cytosol through a series of proteolytic cleavages, translocates to the nucleus and modifies the transcription of hundreds of genes. Some of these genes, like the cytoplasmic protease Calpain-8a, are critical for CNC migration (Cousin et al., 2011).
To date, two types of assays are used to assess xenopus CNC migration in vivo. In the targeted injection assay, the compounds of interest (mRNA, morpholino (MO) and a lineage tracer like GFP) are injected at the 8 to 16-cell stage in the dorso-animal blastomere that will give rise to the CNC cells (McCusker et al., 2009). This assay offers the advantage of perturbing the development of a finite number of tissues and does not usually perturb earlier events like mesoderm induction or gastrulation. However, the injected dorso-animal cell also gives rise to other tissues in addition to the CNC, including some head ectoderm and dorsal mesoderm. Moreover, not all CNC are targeted. Given these restrictions, the analysis has to be done carefully to account for defects that may be due to the knock down (KD) of the protein in a different tissue, as well as defects that are not apparent because some CNC cells that did not receive the MO may still be migrating. The latter is of particular concern if CNC migration is inferred by in situ hybridization using CNC markers but is not as critical if CNC migration is visualized by GFP, which is only present in cells that also receive the MO. The second assay used to assess CNC migration in vivo is the graft assay. Embryos are injected in one blastomere at the two-cell stage. At the early neurula stage (stage 15 to 17), CNC can be excised and grafted back into a host embryo (Alfandari et al., 2001). The injection at the two-cell stage followed by the graft at neurula stage ensures two things. Firstly, all the CNC cells have received the compounds of interest. Secondly, any defects observed using this assay is truly CNC-specific. Unfortunately, the compound injected at the two-cell stage may also impact earlier stages of development like mesoderm induction or gastrulation, which in turn could affect proper CNC development (Wei et al., 2010). Such problems need to be assessed thoroughly before interpreting any results related to CNC migration.
In this article, we show that the knock down of ADAM13 does not yield the same results by graft or targeted injection assays. We investigated the causes of this difference and uncovered a novel function for ADAM13 that was only hypothesized until now: the modification of the extracellular environment to a state more permissive to CNC migration.
Material and Methods
Embryos
Xenopus laevis embryos were obtained by in vitro fertilization and staged according to Nieuwkoop and Faber (Nieuwkoop, 1967).
Morpholinos
Morpholino antisense oligonucleotides (Gene Tools) were designed to complement the 5′ untranslated sequences of xenopus ADAM13 (TGCTCAGCCGACCCTCCGTCCCCAT), ADAM19 (GAGTCCTGTAGCTCCTTCCATCCGA) and Cadherin-11 (AGTCTTTCTTCATTTTTGGTAGTGT). All were tested for efficiency using specific antibodies to visualize the endogenous protein expression (Cousin et al., 2011; Kashef et al., 2009; McCusker et al., 2009; Neuner et al., 2009). Control-morpholino corresponding to a random generated sequence of 25-mer was used.
Constructs
All ADAM13 constructs used were described in (Cousin et al., 2011). The EC1-3 construct was described in (McCusker et al., 2009).
Antibodies
The following antibodies were used; gA13: goat polyclonal antibody to ADAM13 cytoplasmic domain (Cousin et al., 2011), 1B4: monoclonal antibody to Cadherin-11 cytoplasmic tail (McCusker et al., 2009), 8C8: monoclonal antibody to β1 integrin (Gawantka et al., 1992)
Injections
Capped mRNA were synthesized using SP6 RNA polymerase on DNA linearized with NotI as described before (Cousin et al., 2000). For the CNC migration assays, embryos were injected at the 16-cell stage in one dorsal animal blastomere with 1 ng of morpholino, 0.2 ng of RFP mRNA and 0.08 ng of mRNA encoding various constructs. For graft assays, embryos were injected at the two-cell stage with 5 ng of morpholino, 300 pg of GFP mRNA, 400 pg of ADAM13 mRNA or 200 pg of ADAM19 mRNA constructs. In both cases, embryos were raised at 15°C until they reach tail bud stage (stage 24 to 28) at which time the phenotypes were scored. Embryos were scored for the absence of CNC migration. The percentage of inhibition was normalized to embryos injected with RFP or GFP alone. Error bars correspond to standard deviation assuming unequal variances. Photographs of embryos were taken using a Nikon SMZ dissecting scope equipped with a Nikon color digital camera and the Spot image acquisition software.
CNC migration in vitro
CNC explants were excised at stage 15 and placed on 96 well plates (Probind, BD Falcon) coated with 10 μg/ml of bovine fibronectin (Sigma) in Danilchik media (53 mM NaCl, 11.7 mM Na2CO3, 4.25 mM potassium gluconate, 2 mM MgSO4, 1 mM CaCl2, 17.5 mM Bicine, 1 mg/ml BSA, pH 8.3), and imaged by time-lapse microscopy using a Zeiss 200M inverted microscope and the OpenLab 4 software (Improvision). Relief substrates were produced on microscope coverslips (22×22 mm) by serial spin coating using 100 nm silicon nitride followed by SU-8 2000.5 at 3000 rpm for 1 min (Brewer Sciences CEE 100CB Spin Coater). Coverslips were baked at 95°C for 2 min, flood exposed 24 mJ/cm2 for 20 sec (Suss MicroTec MA6 Mask aligner) and baked again for 30 min at 170°C. To produce the relief patterns, substrate was spin coated with SU-8 2100 at 2000 rpm for 1 min, and baked for 7 min at 65°C, and 50 min at 95°C prior to exposure with the specific mask (see figure 7) to form the channels (24 mJ/cm2 for 15 s). Following exposure, the coverslips were baked at 65°C for 5 min and 95°C for 15 min before PGMEA development (15 min). Developed coverslips were rinsed with isopropanol and water prior to coating with fibronectin (10 μg/ml) for 2 hr at 20°C or overnight at 4°C. SDF-1 beads were prepared as described in (Theveneau and Mayor, 2011a) using purified mouse SDF-1 protein (ImmunoTools GmbH).
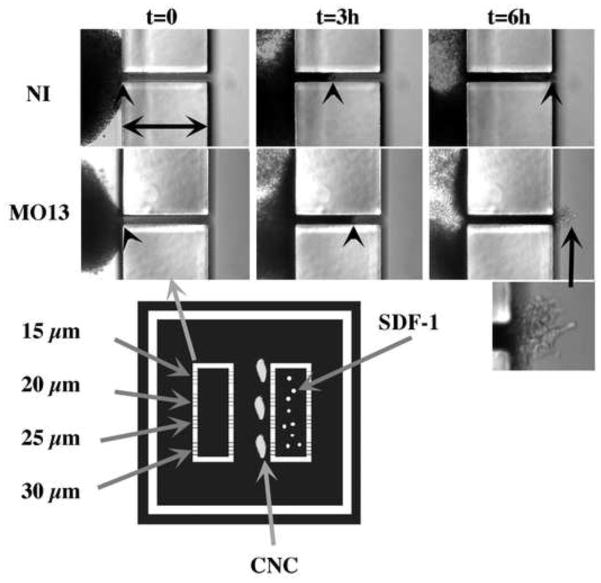
Figure 7. CNC cells can migrate through 20 μm openings.
Migration of CNC through small openings. Photographs of explants (10X) taken from time-lapse movies are represented. The width of the obstacle is 200 μm (horizontal arrows) while the opening is 20 μm. Frames from non-injected control (NI) or morphant (MO13) CNC are presented at 0, 3 and 6 hours of migration. The arrowhead points to the leading edge of cells migrating in the tunnel. The mask used to obtain the substrates is represented below. The white sections represent the walls of the chambers (200 μm high). Each of the two rectangular chambers has four opening for each of their long side ranging from 15 to 30 μm). The center of the rectangles are filled with SDF-1-coated beads and sealed by covering it with a small coverslip fixed with vacuum grease. Explants are placed with their ventral (leading) edge facing each opening.
CNC biotinylation and immunoprecipitation (IP)
CNC were dissected from embryos grown at 14°C for 48 hours post fertilization when they reached stage 15 to 17. Dissected CNC were placed on FN substrate for 2h at 18°C in Danilchik media as previously described (Alfandari et al., 2003). The Danilchik media was thoroughly replaced by 1XMBS with 5 washes and biotinylation was performed at 14°C in 1XMBS containing 1 mg/ml of NHS-lc-Bitotin (Pierce) for 20 min. Excess biotin was removed by 4 washes with 1XMBS 100 mM glycine. Protein extraction was done in 1XMBS, 1% tritonX100, 5 mM EDTA and a protease, phosphatase inhibitor cocktail (HALT protease and phosphatase inhibitor, Fisher). Protein extracts were centrifuged for 30 min at 4°C at 13000 rpm and were pre-cleared using protein-G-agarose beads bound to non-immune Goat and Mouse Ig for 1 h at RT. All IP were performed overnight at 4°C and were sequential. Antibodies used were goat anti-ADAM13 affinity purified on the 37 C-terminal amino acids of the ADAM13 cytoplasmic domain (3 μg/IP), mouse mAb 1B4 to Cad-11 (10 μg/IP) and mouse mAb 8C8 to β1 integrin subunit (5 μg/IP).
Graft procedure
Embryos were injected in one cell at the 2-cell stage and raised at 14°C. Embryos were sorted for optimum lineage tracer expression (GFP or mRFP) at stage 15 (as soon as the anterior neural folds are visible and raised). The vitelline envelopes of selected embryos were removed and embryos were placed in a dish of non-toxic modeling clay (Van Aken, CA) containing 1XMBS 50 μg/ml gentamycin. Embryo-sized cavities were formed in the plasticine using a Pasteur pipet whose tip has been melted into a glass ball. The embryos were placed in the cavities, oriented according to experimenter’s grafting preference and immobilized by pulling back some modeling clay around them. Once immobilized, the ectoderm covering the CNC was peeled off, the CNC were cut out using an eyelash knife and hair loop and grafted into host embryos whose CNC were removed (Figure 2A). Each CNC was grafted according to its proper antero-posterior and dorso-ventral orientation. The grafted embryos were left to heal 30 min in 1X MBS, with an embryo-sized coverslip pressed onto the dissected region of the embryo to maintain the ectoderm flush with the CNC and mesoderm. The embryos were then raised in a tissue culture dish coated with 1% agarose in 0.1XMBS 50 μg/ml gentamycin at 14°C overnight. When double grafts were performed, only half the CNC territory was dissected (in this case the most ventral half to prevent neural tissue contamination).
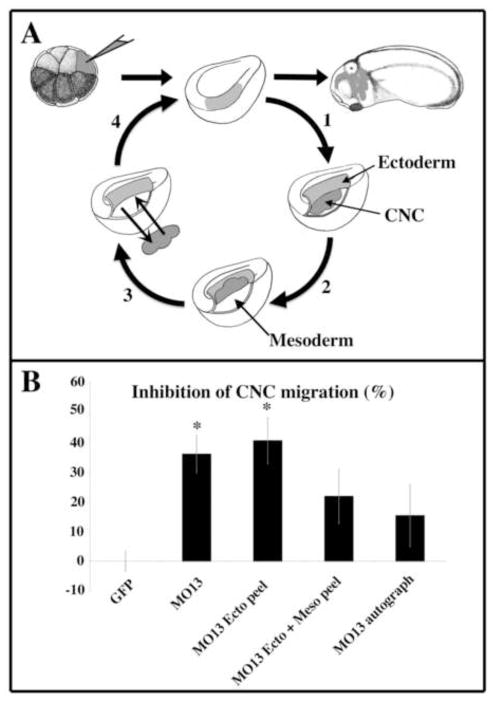
Figure 2. Manually separating CNC from the surrounding tissue partially rescues CNC migration induced by ADAM13 knock down.
(A) Schematics of the experimental procedure. Either GFP mRNA alone, or a mixture of GFP mRNA and ADAM13 morpholino was injected into the dorso-animal cell at the 8-cell stage. At stage 15, embryos were sorted for their expression of GFP and underwent one of the following procedures. 1- the ectoderm was peeled off (ecto peel). 2- the ectoderm was peeled off and the crest lifted away from the underlying mesoderm (ecto + meso peel). 3- the CNC were excised from the embryo and placed back in (autograft). 4- After the procedure, the ectoderm was pulled back on and left to heal. The embryo was left to develop until tailbud stage and scored for their lack of CNC migration. (B) The histogram represents the percentage of embryos with no CNC migration normalized to embryos injected with GFP alone and corresponds to the mean of three independent experiments. As previously reported, ADAM13 KD significantly inhibits CNC migration in the targeted injection assay. The ectoderm peel procedure did not affect this inhibition. However, the ectoderm + mesoderm peel and the autograft procedures rescued CNC migration to levels not statistically different from the GFP control. The number of embryos scored was as follows: GFP; n=159, MO13; n=154, MO13 ecto peel; n= 47; MO13; n=45; MO13 autograft; n=43. The error bar represents standard deviation from the mean. *=Statistically different from GFP (p<0.05).
Scoring of CNC migration in vivo
The presence or absence of fluorescent CNC in the hyoid and branchial arches was assessed at the end of CNC migration, between stage 25 and 28, for each embryo. No qualitative assessment was incorporated into this scoring: either CNC cells were present or absent in these arches, whether they migrated partway or all the way to the ventral side of the embryo. However, partial migrations were seldom observed. The percentage of CNC inhibition was then calculated as follows:
a= number of embryos displaying CNC cells in the pathways in experimental case.
b= number of embryos displaying CNC cells in the pathways in embryos injected with the lineage tracer alone.
n= total number of embryos scored for the experimental (na) or for the control (nb).
%I: Percentage of inhibition.
Results
Targeted Knock down of ADAM13 inhibits CNC migration more robustly than grafting ADAM13 morphant CNC
Using morpholino antisense oligonucleotides (MO), we have previously shown that ADAM13 and 19 double knock down (KD) inhibits CNC migration similarly whether targeted injection (65% inhibition) or CNC graft assay (79% inhibition) are performed (Cousin et al., 2011), (Figure 1A). However, the single KD of ADAM13 inhibits CNC migration significantly better by targeted injection (35%) than by grafting (8%). Since ADAM13 is also expressed in the neural tube (Alfandari et al., 1997), it is possible that the neural expression of ADAM13 is also involved in CNC migration, possibly by facilitating the release of CNC from the neural tube. This neural expression of ADAM13, present in embryos undergoing grafting but not targeted injection, could explain the higher level of CNC migration in grafted embryos. To test this hypothesis, we grafted CNC lacking ADAM13 into either non-injected embryos or embryos knocked down for ADAM13. Results show that the percentage of inhibition of CNC migration between the grafted morphant crest in either wild type (MO13->NI) or morphant (MO13->MO13) embryos is not statistically different (Figure 1B).
Figure 1. ADAM13 knock down in the neural tube does not affect morphant crest migration.
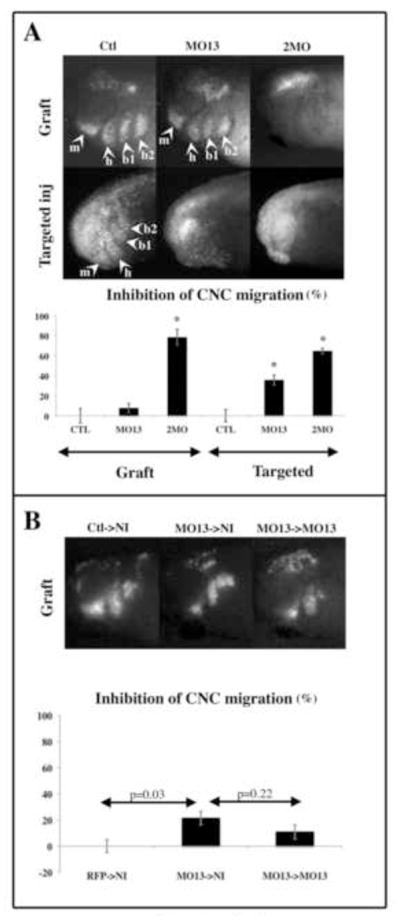
(A) Embryos were injected with mRNA encoding GFP alone (Ctl) or together with antisense morpholino for ADAM13 alone (MO13) or ADAM13 and 19 (2MO). The compounds were injected either in one cell at the 2-cell stage (for graft) or in one dorso-animal cell at the 8–16 cell stage (for targeted injection). The graft procedure was carried out at stage 15 and the embryos were scored for their lack of CNC migration at tailbud stage (26 to 28) for both assays. The pictures show the typical result obtained for each case. The histogram represents the percentage of embryos with no CNC migration normalized to embryos injected with GFP alone and corresponds to the mean of seven (targeted injection) and at least six (graft) independent experiments. The number of embryos scored is as follows. For grafts: Ctl; n=42, MO13; n=25, 2MO; n=61. For targeted injection: Ctl; n=248, MO13; n=258, 2MO; n=281. While 2MO inhibits CNC migration at similar levels for both the grafts and the targeted injection assays, MO13 alone is 5 times more efficient at inhibiting CNC migration in the targeted injection assay than the graft assay. (B) Embryos were injected in one cell at the 2-cell stage with either GFP (Ctl donor), GFP and MO13 (MO13 donor) or RFP and MO13 (MO13 recipient). At stage 15, three types of grafts were performed: 1-GFP expressing CNC were grafted into non-injected embryos (Ctl->NI), 2- GFP+ MO13 injected CNC were grafted into non-injected embryos (MO13->NI), 3- GFP+MO13 injected CNC were grafted into embryos injected with RFP+MO13 (MO13->MO13). The pictures show the typical result obtained for each case. The histogram represents the percentage of embryos with no CNC migration normalized to embryos injected with GFP alone and corresponds to the mean of three independent experiments, representing the scoring of 20 embryos for each case. Results show that morphant CNC display a statistically identical level of migration inhibition whether they are grafted into a wild type embryo or a morphant embryo. The error bar represents standard deviation from the mean. *=Statistically different from controls (p<0.05). Arrowheads point to the segments of the CNC. m: mandibular, h: hyoid, b1 and b2: branchial.
These results show that the difference in the migration of the CNC lacking ADAM13 between the grafting and targeted injection assays is not caused by the presence of ADAM13 in the neural tube or dorsal mesoderm.
Physical disruption of the morphant CNC from the environment rescues CNC migration
We have previously observed that the mandibular crest migration around the optic vesicles is more difficult to inhibit than the migration of the other two segments of the CNC (Alfandari et al., 2001). We hypothesized that this was caused by the optic vesicle pushing the ectoderm away from the mesoderm, hence opening the migration pathway and allowing a better migration of morphant crest (Alfandari et al., 2001). Based on this observation, we hypothesized that the grafting technique introduces a similar bias: mechanically opening the pathways allowing the morphant CNC to migrate.
To test this hypothesis, we performed targeted injection of MO13 as previously described. At stage 15, these embryos were either left alone or subjected to the following grafting procedures: 1- ectoderm was peeled off (Ecto peel), 2- ectoderm peeled off and crest lifted away from the underlying mesoderm (Ecto+Meso peel), 3- crest completely taken out of the embryos and put back into the same embryo (autograft) (Figure 2A). The results show that the inhibition of CNC migration due to the lack of ADAM13 is not rescued when the ectoderm alone is peeled away (Figure 2B). However, the peeling of both ectoderm and mesoderm from the CNC rescues morphant crest migration to the same extent as the autograft (Figure 2B) and the graft into a wild type host embryo (Figure 1). This suggests that while the interaction of CNC with the underlying mesoderm is critical for ADAM13 function, neither the ectoderm nor the neural plate contacts are.
This experiment suggests that the grafting technique is capable of partially compensating for the loss of ADAM13 in the CNC by mechanically altering the extracellular environment of the CNC and making it more amenable to cell migration. However, it should be pointed out that the grafting technique is unable to rescue the CNC migration when both ADAM13 and 19 are knocked down (Figure 1A), indicating that the grafting procedure can compensate for an impaired function of ADAM13 but not a complete loss of function of both proteins. In particular, it is obvious that the grafting procedure may not rescue the expression of genes controlled by ADAM13/19 cytoplasmic domains (Cousin et al., 2011).
ADAM13 and 19 functions are not critical for CNC migration on a two-dimensional surface
The previous results indicate that mechanically altering the environment of the CNC facilitates their migration. This strongly suggests that one role of ADAM13 could be to facilitate CNC migration in a dense three-dimensional ECM pathway. To test this hypothesis, we tested the ability of morphant crests to migrate in vitro on a two-dimensional FN substrate.
Embryos were injected at the two-cell stage with MO13, MO19 or both as previously described. At stage 15, CNC explants were excised and grown in plastic dishes coated with 10μg/ml of FN (Figure 3A). In these conditions, the wild type CNC (Ctl) displayed the typical pattern of migration with an initial phase of sheet migration where all the migrating cells maintain their cell-cell adhesion (t=6h), followed by a second phase where cells have lost their cell-cell adhesions and begin migrating as single cells (t=16h). CNC depleted of ADAM13 protein were capable of migrating with no obvious delays. Interestingly, crest explants depleted of both ADAM13 and 19 proteins migrated as well (Figure 3A). In order to make sure that the lack of defect was not due to an incomplete knock down of ADAM13, the expression of ADAM13, Cadherin-11 and integrin α5β1 in CNC explants was assessed (Figure 3B). After 2 hours of migration on FN, proteins expressed at the cell surface of wild type or ADAM13 morphant CNC were biotinylated and then immunoprecipitated sequentially with antibodies against ADAM13, Cadherin-11 and integrin β1 subunit. Our results show that, as expected, ADAM13 is no longer expressed in morphant CNC while the expression of integrin α5β1 is unaffected. Interestingly, while Cadherin-11 overall expression seems unaffected, its cleavage is inhibited in the morphant CNC. This concurs with our previous findings (McCusker et al., 2009).
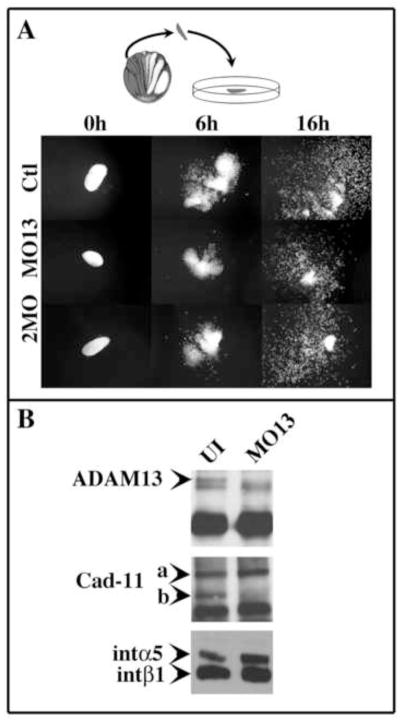
Figure 3. ADAM13 function is dispensable for CNC migration in vitro.
(A) ADAM13 and 19 knock down has no effect on CNC migration in vitro. Embryos were injected in one cell at the two-cell stage with a mixture of GFP mRNA and either control morpholino (Ctl), ADAM13 morpholino (MO13) or ADAM13 and 19 morpholino (2MO). At stage 15, embryos were sorted and CNC explants placed on fibronectin-coated 96-well plates. For each case, pictures of typical explants are shown before migration (t=0), at the end of the sheet migration phase (t=6h) and at the end of the single cell migration phase (t=16h). Morphant CNC cells were capable of migrating in vitro in a pattern indistinguishable form the control CNC. (B) ADAM13 knock down in CNC is efficient and prevents Cadherin-11 cleavage. Twenty CNC were dissected at stage 15 and placed on FN substrate for 2 hours before cell surface proteins were biotinylated. Proteins were extracted and immunoprecipitated sequentially using a goat anti-ADAM13 antibody (gA13), the mouse mAb to Cadherin-11 (1B4) and the mouse mAb to integrin 1 (8C8). Both gA13 and 1B4 recognize the cytodomains of ADAM13 and Cadherin-11, respectively. ADAM13 knock down completely abolishes the expression mature ADAM13 at the cell surface (ADAM13, arrowhead). The full length Cadherin-11 protein is still strongly expressed (Cad-11, arrowhead a) but is no longer cleaved (Cad-11, arrowhead b). Integrin α5β1 cell-surface expression is unaffected by ADAM13 knockdown.
These ex-vivo experiments show that the loss of ADAM13 and 19 does not alter the migration machinery necessary for these cells to migrate, including α5β1 integrin (Alfandari et al., 2003). More importantly, these results demonstrate that ADAM13 function is required in the three-dimensional context of the embryo and that, once taken out of the embryo context, ADAM13 and Cadherin-11 cleavage fragment functions are dispensable. Combined with our previous findings, it is clear that ADAM13 function is not involved in releasing the CNC from the neural tube and is unlikely to be involved in a classical EMT like the one previously described for the trunk neural crest (Shoval et al., 2007). However our observations are compatible with the hypothesis that ADAM13 modifies the interactions of the CNC with its environment, thereby facilitating their migration. Two theories could explain this facilitation. First, ADAM13 could digest partially or modify the ECM proteins like FN to make it more amenable to CNC migration (Machete theory). Second, ADAM13 could loosen the cell-cell interactions within the CNC, making this tissue more flexible and therefore capable to migrate through the dense ECM matrix (Relaxation theory).
ADAM13 and 19 functions are crest autonomous but not cell autonomous
In order to test our two theories (Machete or Relaxation), we performed a series of experiments to study the cell and tissue autonomy of ADAM13 function during CNC migration. One prediction of the Machete model is that wild type CNC should be able to open the pathway for KD cells that are immediately following. This was tested by performing a double graft procedure (Figure 4). Morphant crest (2MO, labeled with GFP) were co grafted with wild type crest (Wt, labeled with RFP) at stage 15 in either one of two configurations. In the first configuration, the wild type CNC were grafted ventrally (Wt leader), while in the second configuration the wild type CNC were grafted dorsally (Wt follower) relative to the morphant CNC. Photographs of each grafted embryo were taken at the end of the grafting procedure to ascertain the successfulness of the graft (Figure 4, stage 15) as well as at the end of the experiment (Figure 4, stage 26). The results show that in both configurations, some of the morphant CNC were capable of migrating following wild type cells (Figure 4, graph). Interestingly, careful analysis of the migration of these explants showed that Wt crest always migrate ahead of morphant crest, regardless of their original position relative to the KD crest (Figure 4, stage 26).
Figure 4. ADAM13 function is not cell autonomous.
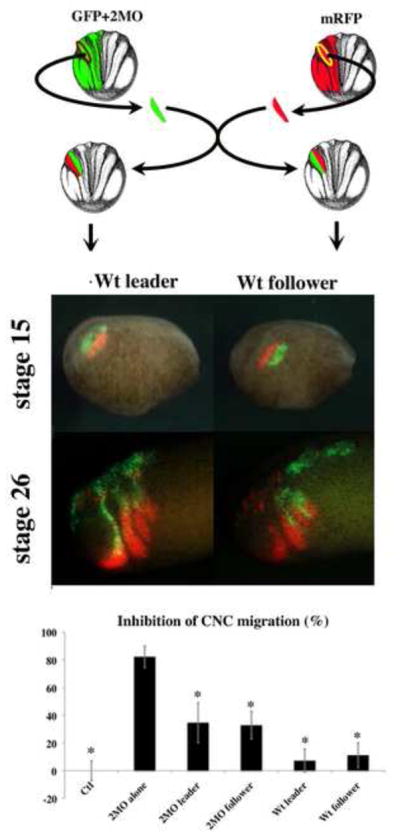
Embryos were injected in one cell of two-cell stage embryos with both GFP mRNA and morpholino against ADAM13 and 19 (GFP+2MO) or with RFP mRNA. At stage 15, CNC expressing the lineage tracer were dissected and grafted into non-injected embryos in either one of the following configurations. Either the RFP expressing explant was grafted ventrally to the morphant crest (Wt leader and 2MO follower) or the RFP expressing crest was grafted dorsally to the morphant crest (Wt follower and 2MO leader). Pictures represent a typical example of each type of grafted embryo before (stage 15) and after migration (stage 26). The histogram represents the percentage of failed migration of each type of CNC (Wt or 2MO) for each grafting configuration. Controls (Ctl) correspond to the graft of GFP expressing CNC alone. Student t-tests between the 2MO CNC grafted alone and all other experimental cases were performed. The experiments show that the co-grafting of wild type CNC significantly rescues morphant CNC migration. The configuration of the graft has no impact on the efficiency of the rescue. The co-grafted morphant CNC had no effect on the ability of wild type CNC to migrate. The histogram represents the average of 7 independent experiments. The number of embryos scored was as follows: Ctl; n=42, 2MO alone; n=40, 2MO leader + Wt follower; n=38; 2MO follower + Wt leader; n=33 n= 47. The error bar represents standard deviation from the mean. * = Statistically different from 2MO alone (p<0.05).
In order to test the tissue autonomy of ADAM13 function, we grafted CNC depleted of both ADAM13 and 19 in either wild type embryos (2MO->Wt) or in embryos mis-expressing the metalloprotease activity of ADAM 13 in the surrounding ectoderm and mesoderm (2MO-> ΔCyto). We chose to express a version of ADAM13 lacking a cytoplasmic domain for two reasons. Firstly, the ΔCyto mRNA produces eight times more active protein than its full-length counterpart (Cousin et al., 2000) ensuring an optimal level of metalloprotease activity present in the pathways. Secondly, the absence of cytoplasmic domain minimizes the risk of changing the transcriptome of the mesoderm and ectoderm, which could result in the mis-expression of multiple genes that could interfere with the interpretation of the data. The results show that CNC migration was not rescued by the expression of ΔCyto-ADAM13 in the pathway (Figure 5). To investigate whether this lack of rescue was due to a lack of transcriptional modulation by ADAM13 cytodomain in the CNC, morphant crest expressing the cytoplasmic domain of ADAM13 (C13) were grafted into embryos expressing ΔCyto in the ectoderm and mesoderm (2MO+C13-> ΔCyto). While these conditions marginally increased CNC migration, the rescue was not statistically significant when compared to the 2MO alone. A similar trend was observed when these CNC were grated into a non-injected embryo (2MO+C13->NI). Interestingly, we previously showed that expression of both C13 and ΔCyto in morphant CNC rescue their migration as efficiently as wild type ADAM13 (Cousin et al., 2011). Therefore, the absence of rescue in these experiments is not due to the dissociation of the proteolytic activity from the cytoplasmic domain function. Lastly, the grafting of morphant CNC expressing C13 in embryos expressing the soluble fragment of Cadherin-11 in the pathways (2MO+C13-> EC1-3) rescued their migration to levels indistinguishable from the positive controls (RFP->NI). Together these experiments allowed us to make three observations. First, wild type CNC are capable of partially rescuing morphant CNC, indicating that part of ADAM13 function is non-cell autonomous supporting the Machete hypothesis. Second, ADAM13 metalloprotease activity needs to be carried out by the CNC in order to migrate properly. In other words, the metalloprotease activity of ADAM13 and 19 is crest autonomous. Third, the lack of ADAM13 metalloprotease activity can be compensated fully by the expression of the cleaved fragment of Cadherin-11 (supporting the Relaxation hypothesis). This indicates that, regardless of any other proteins ADAM13 and 19 may be shedding, the generation of Cadherin-11 cleavage fragment EC1-3 is necessary and sufficient to restore complete CNC migration, as long as the cytoplasmic domain of ADAM13 is also expressed in the CNC.
Figure 5. ADAM 13 metalloprotease function is required crest autonomously.
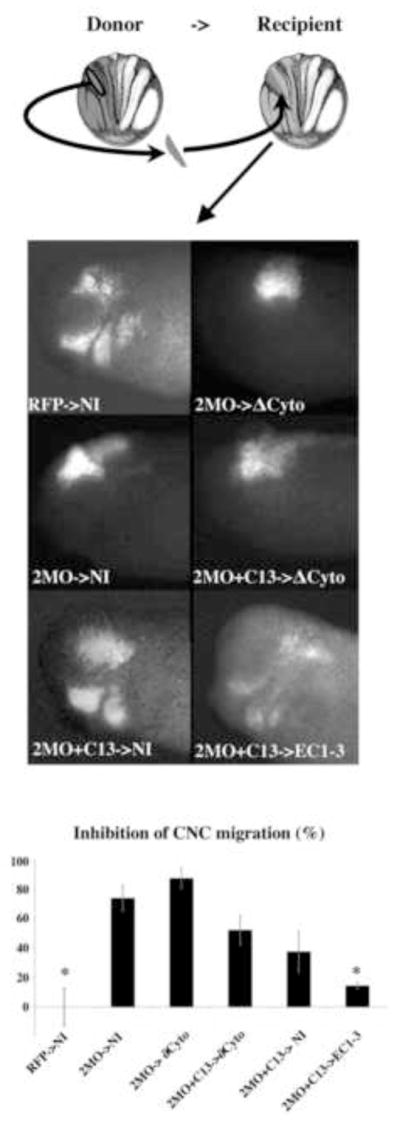
Embryos were injected in one cell of 2-cell stage embryos with the following mixture. Donor embryos were injected with RFP as a lineage tracer and either the morpholinos against ADAM13 and 19 alone (2MO) or the two morpholinos plus the mRNA encoding the cytoplasmic domain of ADAM13 (2MO+C13). The recipient embryos were injected with a mixture of GFP and either a form of ADAM13 lacking the cytoplasmic domain (ΔCyto) or the cleaved form of Cadherin-11 (EC1-3). At stage 15, embryos were sorted for their expression of lineage tracer, CNC explants taken out from donor embryos (RFP, 2MO or 2MO+C13) and grafted into various recipient embryos (NI, ΔCyto or EC1-3). The embryo was left to develop until tailbud stage and scored for their lack of CNC migration. The histogram represents the percentage of embryos displaying no CNC migration normalized to embryos injected with RFP alone and corresponds to the mean of three independent experiments. Results show that morphant crest migration is not rescued if the metalloprotease activity of ADAM13 is not expressed by the CNC themselves (2MO-> ΔCyto). The expression of the cytodomain of ADAM13 in the morphant crest partially rescues CNC migration (2MO+C13->NI). This partial rescue is not enhanced when these CNC are grafted in embryos expressing ΔCyto (2MO+C13-> ΔCyto) confirming that ADAM13 can exert its metalloprotease function only when expressed by the CNC. The rescue is complete when the 2MO+C13 CNC are grafted in embryos expressing EC1-3 (2MO+C13->EC1-3). The number of embryos scored was as follows: RFP->NI; n=15, 2MO->NI; n=14, 2MO-> ΔCyto; n=16, 2MO+C13-> ΔCyto; n=15. The error bar represents standard deviation from the mean. * = Statistically different from 2MO ->NI (p<0.05).
We previously observed that the loss of ADAM13 results in an increase in full length Cadherin-11 (McCusker et al., 2009). To test if the inhibition of CNC migration observed in the morphant embryos was due to the overexpression of Cadherin-11, we used various doses of a morpholino to Cadherin-11 in combination with MO13 and measured both Cadherin-11 protein level and CNC cell migration (Figure 6). The results show that while 1 ng of the Cad11-MO was sufficient to decrease the level of Cad-11 protein in MO13 embryos to that of the control, none of the concentrations tested rescued CNC migration. This confirms that the main role of ADAM13 is to produce the extracellular fragment and not simply to reduce the overall level of Cadherin-11.
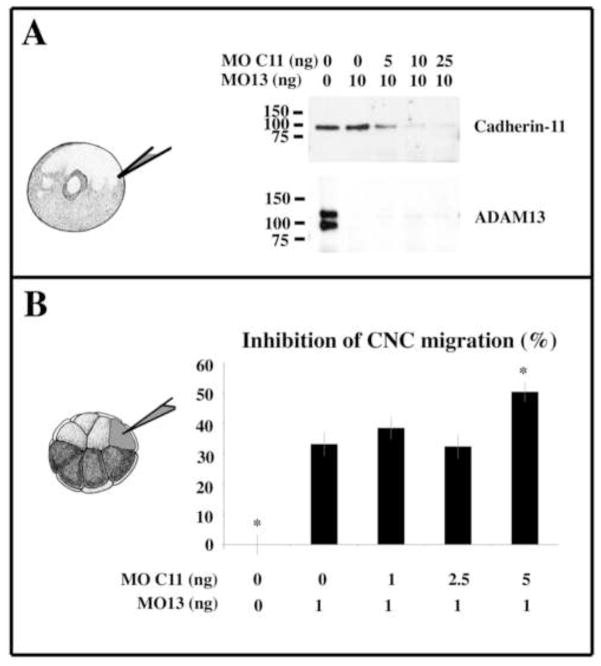
Figure 6. The partial knock down of Cadherin-11 does not rescue CNC migration of ADAM13 morphants.
Embryos were injected at the 1 cell stage with either ADAM13 morpholino (MO13 1ng) alone or together with various doses of morpholino against Cadherin-11 (MOC11) ranging from 5 ng to 25 ng. At stage 20, the proteins of 10 embryo equivalents were extracted, immunoprecipitated with either the cadherin-11 mAb 4B12 or ADAM13 rabbit pAb 6615F and blotted with the cadherin rabbit pAb C11 and the ADAM13 goat pAb 877 respectively. The knock down of ADAM13 alone leads to an increase of the full-length form of Cadherin-11 that can be decreased by a partial cadherin-11 knock down using 5ng of MOC11. (B) Embryos were injected at the 8-cell stage with either ADAM13 morpholino (MO13 1ng) alone or together with various doses of morpholino against Cadherin-11 (MOC11) ranging from 1 ng to 5 ng. The mRNA encoding GFP was either co-injected or injected alone as lineage tracer. Embryos were raised until stage 28 and scored for their lack of CNC migration. The histogram represents the percentage of embryos displaying no CNC migration normalized to embryos injected with RFP alone and corresponds to the mean of four independent experiments. The partial knock down of Cadherin-11 with either 1 or 2.5 ng of MOC11 did not significantly rescue the inhibition of CNC migration induced by ADAM13 knock down. The co-injection of 5 ng of MOC11 produced a significantly more severe inhibition than ADAM13 knock down alone. The error bars represent standard deviation from the mean. The number of embryos scored for each case ranged between 207 and 218. * = Statistically different from MO13 (p<0.05).
Testing the Relaxation hypothesis
In order to test if the CNC from morphant embryos have increased cell adhesion that would prevent the cells from changing their relative positions to fit through small openings (Relaxation hypothesis), we created a new device that challenges the migration of cells forcing them to migrate through channels of variable sizes (Figure 7, the corresponding time-lapse movies are available in the supplemental materials). CNC explants, which are about 500 μm wide, were placed in front of small tunnels (20 μm) and were attracted to the other side using the chemoattractant SDF-1 (Theveneau et al., 2010). We found that both control and morphant CNC were able to migrate through these openings and reached the opposite side of the tunnel (200 μm) at the same time. This assay was performed during the first phase of migration while cell contacts are maintained, showing that in the absence of ADAM13, cells within the explant are able to modulate their adhesion to fit through the opening.
Discussion
We previously showed that ADAM13 performs at least two functions during CNC migration. First, it binds and cleaves Cadherin-11 to release an extracellular fragment that stimulates CNC migration (McCusker et al., 2009). Second, the cytoplasmic domain of ADAM13 is cleaved and translocates into the nucleus, where it regulates gene expression to promote cell migration (Cousin et al., 2011). In this report, we show that the requirement for ADAM13 function depends on the CNC environment. In vitro on a flat FN substrate, ADAM13 is not required for migration, while it is essential in vivo. Furthermore, physically separating the CNC from the underlying mesoderm in vivo is sufficient to rescue migration of CNC lacking ADAM13.
The comparison of morphant CNC migration in vivo and in vitro shows that ADAM13/19 does not affect the function of other molecules critical for CNC migration. For example, cells lacking ADAM13 and ADAM19 are still capable of migrating on FN, indicating that the proteins involved in CNC cell migration, including integrin 5 1, are still expressed and functional (Alfandari et al., 2003). This indicates that ADAM13/19 acts differently from other ADAMs involved in cell adhesion and migration or invasion (McGinn et al., 2011; Toquet et al., 2010). Another example is that morphant CNC cultured in vitro also segregates into 3 segments, indicating that the mechanisms responsible for the cell sorting of CNC into these segments are still functional (Smith et al., 1997).
ADAM13 function during CNC migration: Machete or Relaxation?
Taken together, our results suggest that the role of ADAM13 is to modulate the interaction of CNC with their environment. Given the known substrates of ADAM13, we propose two mutually non-exclusive functions for ADAM13 in vivo. The first one, named the Machete model, would involve cleaving FN and/or other proteins of the ECM to open or widen existing openings in the dense ECM that surrounds the CNC during their migration. This model is supported by the ability of ADAM13 to bind, cleave and remodel a FN substrate (Alfandari et al., 2001; Gaultier et al., 2002), as well as the ability of wild type CNC to allow morphant CNC to migrate immediately behind them (Figure 4). It is also consistent with the lack of defect observed in ADAM13 KD in vitro or after grafting when the ECM has been manually opened (Figures 3 and 1, respectively). The fact that the ADAM13 cytoplasmic domain also increases the expression of the metalloprotease MMP13 offers yet another way to remodel the ECM (Cousin et al., 2011). The ECM modification by metalloproteinases has been extensively documented for the Matrix Metalloproteinases (MMP). MMPs promote cell migration and metastasis by degrading a number of ECM components. This degradation in turn could reveal cryptic sites, create microtracks or free growth factors necessary for cell migration and invasion (Fowlkes et al., 1995; Giannelli et al., 1997; Ilina et al., 2011; Stamenkovic, 2003; Whitelock et al., 1996; Xu et al., 2001). Some evidence suggests that ADAMs could do the same (Alfandari et al., 2001; Mazzocca et al., 2005). However, since the expression of ADAM13 metalloprotease domain in the migrating pathways cannot rescue morphant crest migration, it is unlikely that ADAM13 plays a role in freeing growth factors crucial for CNC migration such as Sdf-1 (Theveneau et al., 2010; Theveneau and Mayor, 2011b). To further test this hypothesis we measured CNC invasion in Rat tail collagen gels containing fibronectin and SDF-1 beads, but in this assay CNC cells invaded poorly with or without ADAM13 suggesting that the composition of this artificial ECM was not conducive for CNC migration (data not shown). Since the exact composition of the CNC migration pathway is not known, additional tests are required to define a compatible in vitro environment. While multiple invasion studies use Matrigel, this represents a laminin-rich ECM similar to a basement membrane and xenopus CNC do not survive in it for the length of the assay (Alfandari, unpublished).
Our finding that ADAM13 metalloprotease activity is required specifically in the CNC offers a unique twist on the Machete model. If ADAM13’s role in CNC migration was to simply digest the ECM surrounding the CNC, then ADAM13 expression in the tissue surrounding the CNC should rescue their migration, which it doesn’t. However, morphant CNC migration can be rescued if the pathways have been primed by migrating CNC. This suggests that migrating cells reorganize the ECM in a way that is conducive to the migration of morphant cells in vivo. Similar polarization of the ECM has been observed in numerous migrating tissues during development and cancer metastasis. This includes, the developing drosophila follicles, the zebrafish and frog gastrula, as well as during primitive streak formation in avian embryos (Calvo and Sahai, 2011; Davidson et al., 2006; Friedl and Alexander, 2011; Haigo and Bilder; Latimer and Jessen; Zamir et al., 2008). The polarization of the ECM is thought to play multiple roles during morphogenesis. One hypothesis is that migrating cells reorient the ECM to provide directional cues that further facilitate the migration of following cells. This has been elegantly shown in vitro in breast cancer invasion assays on collagen matrices (Ilina et al., 2011). Another possibility is that ECM remodeling constrains tissue shape, which generates polarized biomechanical forces that facilitate directional cell movement. We propose that ADAM13 activity in the leading edge of the CNC is required to both open the pathway through a dense ECM, like a Machete, and to rearrange and reorient the ECM to promote the migration of following cells.
On the other hand, the ability of the EC1-3 domain of Cadherin-11 in combination with the ADAM13 cytoplasmic domain to rescue CNC migration in embryos lacking both ADAM13 and 19 cannot easily be explained by the Machete theory. In a second model named the Relaxation model, the cleaved extracellular fragment of Cadherin-11 would increase the relative fluidity of the CNC tissue so that it may deform to migrate through openings within the ECM. As mentioned before, this model does not eliminate the possibility that ADAM13 also contributes to the opening of these pathways. The control of tissue fluidity could be achieved in two ways. First, ADAM13 would reduce the total amount of Cadherin-11 present at the cell surface and therefore decrease cell-cell adhesion. As seen in Figure 6, simply reducing Cadherin-11 protein level using morpholino does not rescue migration, suggesting that this is not the main role of ADAM13. Second, the released EC1-3 may interact with either Cadherin-11 or other proteins at the CNC surface to favor cell shuffling. In the latter model we envision the EC1-3 as a lubricant that facilitates the movement of cells within a population. In an attempt to test this hypothesis, we challenged CNC explant to migrate during the first phase through opening as small as 20 μm forcing them to rearrange. Surprisingly we did not observe any difference between control and morphant CNC capacity to migrate through such openings. It should be noted that the walls of our tunnels (200 μm long), which are 200 μm high, were also coated with FN and thus it is possible that cells were using the entire surface available to migrate, thereby decreasing their constraint. Additional work will be needed to create an in vitro environment in which ADAM13 function is required.
Alternatively, EC1-3 could directly or indirectly activate a signaling pathway that could change either the shape of the cells or their protrusive activity, ultimately making them more amenable for migrating in a three-dimentional ECM. Our data suggest that the EC1-3, while not a chemoattractant per se, enhances CNC migration in vitro (McCusker, 2010). Soluble N-cadherin has been shown to promote neurite outgrowth by promoting FGF signaling (Boscher and Mege, 2008; Paradies and Grunwald, 1993; Utton et al., 2001). Similarly, the soluble E-cadherin ectodomain shed by MMP-7 promotes invasive behavior of cancer cells (Noe et al., 2001). The extracellular cleavage fragment of E-cadherin was also shown to interact with ErbB receptors and initiate downstream signaling that resulted in cell migration and cell proliferation in cultured cells (Najy et al., 2008). The cleavage of the CAM L1 by ADAM10 and/or 8 releases a soluble fragment capable of promoting neurite outgrowth possibly by binding and activating integrins (Mechtersheimer et al., 2001; Naus et al., 2004). Thus, while our observations with EC1-3 fit with the Relaxation model to promote CNC migration, there are a number of alternative mechanisms possible.
Collective cell migration: CNC and cancer
Xenopus CNC migration is biphasic (Alfandari et al., 2003). A collective migration phase of 5 hours is followed by a single cell migration. This behavior appear uncommon among vertebrates since the CNC of most species, including the Mexican Axolotl, appear to migrate as single cell from the start (Epperlein et al., 2000). On the other hand, in species with monophasic CNC migration like zebrafish, contact between migrating CNC has been shown to be critical to maintain cell identity suggesting that while cell cohesion is not as obvious as in xenopus, it is a conserved and essential characteristic of these pluripotent cells (Schilling et al., 2001). Moreover, such behavior has been observed in some cancer undergoing metastasis, in particular mesenchymal tumors and melanoma (Alexander et al., 2008; Hegerfeldt et al., 2002). This has been correlated with the ability of the cells to perform partial EMT, allowing them to migrate and invade without losing the contact with their neighbor (Friedl and Alexander, 2011). This collective migration gives the migrating cells a better ability to invade the surrounding tissue. Using the collective force of cell migration, they are capable of remodeling the surrounding tissue either mechanically or biochemically to facilitate the migration of the group. To our knowledge, no role of ADAM metalloproteases or the cleavage of a type II cadherin like Cadherin-11 has been associated with these types of tumor. Since the cleavage of Cadherin-11 and the release of its EC1-3 fragment into the extracellular environment is critical for CNC migration, it would be interesting to see if such cleavage also occur in the aforementioned cancer.
In conclusion, our findings support a role for ADAM13 modulating the direct or indirect (via the ECM) interaction with the underlying mesoderm. This role involves the cleavage and release of the extracellular domain of Cadherin-11. The receptor of the Cadherin-11 fragment remains to be identified, as does the exact mechanism by which this promotes cell migration. More generally, the comparison of data obtained in vitro versus in vivo shows that ADAM13 function is only required in the three-dimensional context of the embryo and cannot be studied using solely tissue culture assays. At a more fundamental level, this finding emphasizes the fact that while two-dimensional assays yield interesting and fundamental results, three-dimensional assays need to be developed in order to fully understand the function of proteins during cell migration.
Supplementary Material
Highlights.
ADAM13 function is essential in vivo but not in vitro.
ADAM13 acts both cell autonomously and non-cell autonomously.
CNC cells lacking ADAM13 and 19 can follow wild type CNC.
Disruption of CNC/mesoderm attachment rescues the migration of morphant CNC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Alexander S, Koehl GE, Hirschberg M, Geissler EK, Friedl P. Dynamic imaging of cancer growth and invasion: a modified skin-fold chamber model. Histochemistry and cell biology. 2008;130:1147–1154. doi: 10.1007/s00418-008-0529-1. [DOI] [PubMed] [Google Scholar]
- Alfandari D, Cousin H, Gaultier A, Hoffstrom BG, DeSimone DW. Integrin alpha5beta1 supports the migration of Xenopus cranial neural crest on fibronectin. Dev Biol. 2003;260:449–464. doi: 10.1016/s0012-1606(03)00277-x. [DOI] [PubMed] [Google Scholar]
- Alfandari D, Cousin H, Gaultier A, Smith K, White JM, Darribere T, DeSimone DW. Xenopus ADAM 13 is a metalloprotease required for cranial neural crest-cell migration. Curr Biol. 2001;11:918–930. doi: 10.1016/s0960-9822(01)00263-9. [DOI] [PubMed] [Google Scholar]
- Alfandari D, Cousin H, Marsden M. Mechanism of Xenopus cranial neural crest cell migration. Cell Adh Migr. 2010;4:553–560. doi: 10.4161/cam.4.4.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfandari D, McCusker C, Cousin H. ADAM function in embryogenesis. Semin Cell Dev Biol. 2009;20:153–163. doi: 10.1016/j.semcdb.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfandari D, Wolfsberg TG, White JM, DeSimone DW. ADAM 13: a novel ADAM expressed in somitic mesoderm and neural crest cells during Xenopus laevis development. Dev Biol. 1997;182:314–330. doi: 10.1006/dbio.1996.8458. [DOI] [PubMed] [Google Scholar]
- Boscher C, Mege RM. Cadherin-11 interacts with the FGF receptor and induces neurite outgrowth through associated downstream signalling. Cell Signal. 2008;20:1061–1072. doi: 10.1016/j.cellsig.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Calvo F, Sahai E. Cell communication networks in cancer invasion. Curr Opin Cell Biol. 2011;23:621–629. doi: 10.1016/j.ceb.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Carmona-Fontaine C, Matthews HK, Kuriyama S, Moreno M, Dunn GA, Parsons M, Stern CD, Mayor R. Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature. 2008;456:957–961. doi: 10.1038/nature07441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin H, Abbruzzese G, Kerdavid E, Gaultier A, Alfandari D. Translocation of the cytoplasmic domain of ADAM13 to the nucleus is essential for Calpain8-a expression and cranial neural crest cell migration. Dev Cell. 2011;20:256–263. doi: 10.1016/j.devcel.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin H, Gaultier A, Bleux C, Darribere T, Alfandari D. PACSIN2 is a regulator of the metalloprotease/disintegrin ADAM13. Dev Biol. 2000;227:197–210. doi: 10.1006/dbio.2000.9871. [DOI] [PubMed] [Google Scholar]
- Davidson LA, Marsden M, Keller R, Desimone DW. Integrin alpha5beta1 and fibronectin regulate polarized cell protrusions required for Xenopus convergence and extension. Curr Biol. 2006;16:833–844. doi: 10.1016/j.cub.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Epperlein H, Meulemans D, Bronner-Fraser M, Steinbeisser H, Selleck MA. Analysis of cranial neural crest migratory pathways in axolotl using cell markers and transplantation. Development. 2000;127:2751–2761. doi: 10.1242/dev.127.12.2751. [DOI] [PubMed] [Google Scholar]
- Fowlkes JL, Thrailkill KM, Serra DM, Suzuki K, Nagase H. Matrix metalloproteinases as insulin-like growth factor binding protein-degrading proteinases. Prog Growth Factor Res. 1995;6:255–263. doi: 10.1016/0955-2235(95)00017-8. [DOI] [PubMed] [Google Scholar]
- Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- Gaultier A, Cousin H, Darribere T, Alfandari D. ADAM13 disintegrin and cysteine-rich domains bind to the second heparin-binding domain of fibronectin. J Biol Chem. 2002;277:23336–23344. doi: 10.1074/jbc.M201792200. [DOI] [PubMed] [Google Scholar]
- Gawantka V, Ellinger-Ziegelbauer H, Hausen P. Beta 1-integrin is a maternal protein that is inserted into all newly formed plasma membranes during early Xenopus embryogenesis. Development. 1992;115:595–605. doi: 10.1242/dev.115.2.595. [DOI] [PubMed] [Google Scholar]
- Giannelli G, Falk-Marzillier J, Schiraldi O, Stetler-Stevenson WG, Quaranta V. Induction of cell migration by matrix metalloprotease-2 cleavage of laminin-5. Science. 1997;277:225–228. doi: 10.1126/science.277.5323.225. [DOI] [PubMed] [Google Scholar]
- Haigo SL, Bilder D. Global tissue revolutions in a morphogenetic movement controlling elongation. Science. 331:1071–1074. doi: 10.1126/science.1199424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegerfeldt Y, Tusch M, Brocker EB, Friedl P. Collective cell movement in primary melanoma explants: plasticity of cell-cell interaction, beta1-integrin function, and migration strategies. Cancer Res. 2002;62:2125–2130. [PubMed] [Google Scholar]
- Ilina O, Bakker GJ, Vasaturo A, Hofmann RM, Friedl P. Two-photon laser-generated microtracks in 3D collagen lattices: principles of MMP-dependent and -independent collective cancer cell invasion. Physical biology. 2011;8:015010. doi: 10.1088/1478-3975/8/1/015010. [DOI] [PubMed] [Google Scholar]
- Kashef J, Kohler A, Kuriyama S, Alfandari D, Mayor R, Wedlich D. Cadherin-11 regulates protrusive activity in Xenopus cranial neural crest cells upstream of Trio and the small GTPases. Genes Dev. 2009;23:1393–1398. doi: 10.1101/gad.519409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesa PM, Bailey CM, Kasemeier-Kulesa JC, McLennan R. Cranial neural crest migration: new rules for an old road. Dev Biol. 2010;344:543–554. doi: 10.1016/j.ydbio.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latimer A, Jessen JR. Extracellular matrix assembly and organization during zebrafish gastrulation. Matrix Biol. 29:89–96. doi: 10.1016/j.matbio.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Maretzky T, Reiss K, Ludwig A, Buchholz J, Scholz F, Proksch E, de Strooper B, Hartmann D, Saftig P. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and beta-catenin translocation. Proc Natl Acad Sci U S A. 2005;102:9182–9187. doi: 10.1073/pnas.0500918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews HK, Marchant L, Carmona-Fontaine C, Kuriyama S, Larrain J, Holt MR, Parsons M, Mayor R. Directional migration of neural crest cells in vivo is regulated by Syndecan-4/Rac1 and non-canonical Wnt signaling/RhoA. Development. 2008;135:1771–1780. doi: 10.1242/dev.017350. [DOI] [PubMed] [Google Scholar]
- Mazzocca A, Coppari R, De Franco R, Cho JY, Libermann TA, Pinzani M, Toker A. A secreted form of ADAM9 promotes carcinoma invasion through tumor-stromal interactions. Cancer Res. 2005;65:4728–4738. doi: 10.1158/0008-5472.CAN-04-4449. [DOI] [PubMed] [Google Scholar]
- McCusker C, Cousin H, Neuner R, Alfandari D. Extracellular cleavage of cadherin-11 by ADAM metalloproteases is essential for Xenopus cranial neural crest cell migration. Mol Biol Cell. 2009;20:78–89. doi: 10.1091/mbc.E08-05-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker CD. Molecular and Cellular Biology Program. University of Massachusetts; Amherst: 2010. Beyond Cell Adhesion: Exploring the role of cadherin-11 extracellular processing by ADAM metalloproteases in cranial neural rest migration; p. 212. [Google Scholar]
- McGinn O, English W, Roberts S, Ager A, Newham P, Murphy G. Modulation of integrin alpha4beta1 by ADAM28 promotes lymphocyte adhesion and transendothelial migration. Cell Biol Int. 2011 doi: 10.1042/CBI20100885. [DOI] [PubMed] [Google Scholar]
- Mechtersheimer S, Gutwein P, Agmon-Levin N, Stoeck A, Oleszewski M, Riedle S, Postina R, Fahrenholz F, Fogel M, Lemmon V, Altevogt P. Ectodomain shedding of L1 adhesion molecule promotes cell migration by autocrine binding to integrins. J Cell Biol. 2001;155:661–673. doi: 10.1083/jcb.200101099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoux M, Rijli FM. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development. 2010;137:2605–2621. doi: 10.1242/dev.040048. [DOI] [PubMed] [Google Scholar]
- Najy AJ, Day KC, Day ML. The ectodomain shedding of E-cadherin by ADAM15 supports ErbB receptor activation. J Biol Chem. 2008;283:18393–18401. doi: 10.1074/jbc.M801329200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naus S, Richter M, Wildeboer D, Moss M, Schachner M, Bartsch JW. Ectodomain shedding of the neural recognition molecule CHL1 by the metalloprotease-disintegrin ADAM8 promotes neurite outgrowth and suppresses neuronal cell death. J Biol Chem. 2004;279:16083–16090. doi: 10.1074/jbc.M400560200. [DOI] [PubMed] [Google Scholar]
- Neuner R, Cousin H, McCusker C, Coyne M, Alfandari D. Xenopus ADAM19 is involved in neural, neural crest and muscle development. Mech Dev. 2009;126:240–255. doi: 10.1016/j.mod.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus Laevis (Daudin) 2. Amsterdam: North-Holland; 1967. [Google Scholar]
- Noe V, Fingleton B, Jacobs K, Crawford HC, Vermeulen S, Steelant W, Bruyneel E, Matrisian LM, Mareel M. Release of an invasion promoter E-cadherin fragment by matrilysin and stromelysin-1. J Cell Sci. 2001;114:111–118. doi: 10.1242/jcs.114.1.111. [DOI] [PubMed] [Google Scholar]
- Paradies NE, Grunwald GB. Purification and characterization of NCAD90, a soluble endogenous form of N-cadherin, which is generated by proteolysis during retinal development and retains adhesive and neurite-promoting function. J Neurosci Res. 1993;36:33–45. doi: 10.1002/jnr.490360105. [DOI] [PubMed] [Google Scholar]
- Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D, Saftig P. ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and beta-catenin nuclear signalling. EMBO J. 2005;24:742–752. doi: 10.1038/sj.emboj.7600548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling TF, Prince V, Ingham PW. Plasticity in zebrafish hox expression in the hindbrain and cranial neural crest. Dev Biol. 2001;231:201–216. doi: 10.1006/dbio.2000.9997. [DOI] [PubMed] [Google Scholar]
- Shoval I, Ludwig A, Kalcheim C. Antagonistic roles of full-length N-cadherin and its soluble BMP cleavage product in neural crest delamination. Development. 2007;134:491–501. doi: 10.1242/dev.02742. [DOI] [PubMed] [Google Scholar]
- Smith A, Robinson V, Patel K, Wilkinson DG. The EphA4 and EphB1 receptor tyrosine kinases and ephrin-B2 ligand regulate targeted migration of branchial neural crest cells. Curr Biol. 1997;7:561–570. doi: 10.1016/s0960-9822(06)00255-7. [DOI] [PubMed] [Google Scholar]
- Stamenkovic I. Extracellular matrix remodelling: the role of matrix metalloproteinases. J Pathol. 2003;200:448–464. doi: 10.1002/path.1400. [DOI] [PubMed] [Google Scholar]
- Theveneau E, Marchant L, Kuriyama S, Gull M, Moepps B, Parsons M, Mayor R. Collective chemotaxis requires contact-dependent cell polarity. Dev Cell. 2010;19:39–53. doi: 10.1016/j.devcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E, Mayor R. Beads on the run: beads as alternative tools for chemotaxis assays. Methods Mol Biol. 2011a;769:449–460. doi: 10.1007/978-1-61779-207-6_30. [DOI] [PubMed] [Google Scholar]
- Theveneau E, Mayor R. Collective cell migration of the cephalic neural crest: The art of integrating information. Genesis. 2011b;49:164–176. doi: 10.1002/dvg.20700. [DOI] [PubMed] [Google Scholar]
- Toquet C, Colson A, Jarry A, Bezieau S, Volteau C, Boisseau P, Merlin D, Laboisse CL, Mosnier JF. ADAM15 to alpha5beta1 integrin switch in colon carcinoma cells: A late event in cancer progression associated with tumor dedifferentiation and poor prognosis. Int J Cancer. 2010 doi: 10.1002/ijc.25891. [DOI] [PubMed] [Google Scholar]
- Utton MA, Eickholt B, Howell FV, Wallis J, Doherty P. Soluble N-cadherin stimulates fibroblast growth factor receptor dependent neurite outgrowth and N-cadherin and the fibroblast growth factor receptor co-cluster in cells. J Neurochem. 2001;76:1421–1430. doi: 10.1046/j.1471-4159.2001.00140.x. [DOI] [PubMed] [Google Scholar]
- Wei S, Xu G, Bridges LC, Williams P, White JM, DeSimone DW. ADAM13 induces cranial neural crest by cleaving class B Ephrins and regulating Wnt signaling. Dev Cell. 2010;19:345–352. doi: 10.1016/j.devcel.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelock JM, Murdoch AD, Iozzo RV, Underwood PA. The degradation of human endothelial cell-derived perlecan and release of bound basic fibroblast growth factor by stromelysin, collagenase, plasmin, and heparanases. J Biol Chem. 1996;271:10079–10086. doi: 10.1074/jbc.271.17.10079. [DOI] [PubMed] [Google Scholar]
- Xu J, Rodriguez D, Petitclerc E, Kim JJ, Hangai M, Moon YS, Davis GE, Brooks PC. Proteolytic exposure of a cryptic site within collagen type IV is required for angiogenesis and tumor growth in vivo. J Cell Biol. 2001;154:1069–1079. doi: 10.1083/jcb.200103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir EA, Rongish BJ, Little CD. The ECM moves during primitive streak formation--computation of ECM versus cellular motion. PLoS Biol. 2008;6:e247. doi: 10.1371/journal.pbio.0060247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.