Abstract
The epithelium of mammalian tongue hosts most of the taste buds that transduce gustatory stimuli into neural signals. In the field of taste biology, taste bud cells have been described as arising from “local epithelium”, in distinction from many other receptor organs that are derived from neurogenic ectoderm including neural crest (NC). In fact, contribution of NC to both epithelium and mesenchyme in the developing tongue is not fully understood. In the present study we used two independent, well-characterized mouse lines, Wnt1-Cre and P0-Cre that express Cre recombinase in a NC-specific manner, in combination with two Cre reporter mouse lines, R26R and ZEG, and demonstrate a contribution of NC-derived cells to both tongue mesenchyme and epithelium including taste papillae and taste buds. In tongue mesenchyme, distribution of NC-derived cells is in close association with taste papillae. In tongue epithelium, labeled cells are observed in an initial scattered distribution and progress to a clustered pattern between papillae, and within papillae and early taste buds. This provides evidence for a contribution of NC to lingual epithelium. Together with previous reports for the origin of taste bud cells from local epithelium in postnatal mouse, we propose that NC cells migrate into and reside in the epithelium of the tongue primordium at an early embryonic stage, acquire epithelial cell phenotypes, and undergo cell proliferation and differentiation that is involved in the development of taste papillae and taste buds. Our findings lead to a new concept about derivation of taste bud cells that include a NC origin.
Keywords: tongue, taste papillae, taste bud, neural crest, Wnt1-Cre, P0-Cre
Introduction
The vertebrate neural crest (NC) is a multipotent cell population derived from the lateral ridges of the neural plate in the early embryo (Bronner-Fraser, 1993; Barembaum and Bronner-Fraser, 2005). NC cells disperse from the dorsal neural tube and migrate extensively, giving rise to a wide variety of differentiated cell types that include neurons (sensory, autonomic, enteric nervous system), glia, fibroblasts, bone, cartilage, connective tissue and dermis of the head (Bronner-Fraser, 1993). Important contributions of NC cells have been demonstrated in formation of mammalian craniofacial structures including branchial arches (Chai and Maxson, 2006; Cordero et al., 2011), where the tongue forms. However, there is not a detailed study or firm demonstration about the nature of NC-derived cells in the developing tongue or taste organs.
Taste buds in the mammalian tongue reside in specialized gustatory organs, the taste papillae (Mistretta and Hill, 1995). Taste buds transduce gustatory stimuli into neural signals, and each bud includes about 60 specialized cells with both neuronal and epithelial features. Data from anatomical studies (Farbman, 1965; Bradley and Stern, 1967; Farbman and Mbiene, 1991) and transgenic phenotype analyses (Stone et al., 1995; Okubo et al., 2009) have demonstrated that taste bud cells are derived from the surrounding local epithelium in mammals. In a cell lineage analysis in postnatal mouse tongue, an inducible Cre driven by the promoter of K14, a basal epithelial cell marker, labeled a large population of taste bud cells (Okubo et al., 2009). In an earlier cell lineage analysis in a transgenic mouse line using mosaicism in β-galactosidase (β-gal) activity by X-chromosome inactivation (Stone et al., 1995), taste bud cell populations matched that of the surrounding epithelial tissue in terms of β-gal activity; this led to a conclusion that taste bud progenitors arise from local epithelium. Overall, these reports contribute to the widely accepted idea that taste bud cells arise from local epithelium.
However, a question remains: What is the derivation and developmental timing for the “local” epithelial cells that contribute to taste bud cell populations? For example, because X-chromosome inactivation is complete in most ectodermal and mesodermal tissues by 9.5 dpc (Tan et al., 1993; Tam et al., 1994) there is a possibility that NC cells, at an earlier embryonic stage, migrate into and reside in the epithelium of the tongue primordium, and then proliferate and differentiate in taste papilla residences that will include taste buds. In humans, an absence or reduction of taste papillae has been found in several neurologic disorders (Axelrod and Gold-von Simson, 2007; Gardiner et al., 2008; Rubin and Anderson, 2008) that affect the development and maintenance of sensory and autonomic neurons which in general have a neurogenic origin from both ectodermal placode and neural crest. In one of these neuropathies, familial dysautonomia, absolute absence of fungiform and vallate taste papillae (Smith et al., 1965a, b) suggests a causal link with NC defects.
Recently, cell lineage tracing using NC-specific Cre recombinase and lacZ or EGFP reporter mice has facilitated genetic marking of NC. Multiple models have been developed for NC derivation assays, e.g., Wnt1-Cre (Danielian et al., 1998), P0-Cre (Yamauchi et al., 1999), HtPA-Cre (Pietri et al., 2003), Pax3-Cre (Li et al., 2000; Engleka et al., 2005), Sox10-Cre (Ludwig et al., 2004; Stine et al., 2009), in which the transgenes mark a population of pre-migratory and/or post-migratory NC cells. Use of these model systems has yielded new data on NC roles in mice, e.g., demonstration of Merkel cells from epidermal lineage (Morrison et al., 2009); distinct genesis of skin-derived precursors in craniofacial and dorsal skin from NC and mesoderm respectively (Jinno et al., 2010); and, NC and placodal derivation of the otic vesicle (Freyer et al., 2011). Also, a dual origin of sensory organs is recently demonstrated with use of Wnt1-Cre and P0-Cre lines to show that NC-derived cells contribute to the placodally-derived olfactory epithelium (Katoh et al., 2011). Whereas prior thinking attributed a NC cell contribution to structural elements only of the olfactory organ, use of transgenic mouse lines demonstrated a NC contribution to embryonic and postnatal olfactory epithelium and to olfactory ensheathing cells.
However, inconsistencies have been noted in different models presumably because of the variation in labeled NC cell populations (Nakamura et al., 2006; Olaopa et al., 2011; Wang et al., 2011). Indeed, none of the established mouse lines for NC derivation assay labels all NC-derived cells, or labels NC-derived cells exclusively from other cell lineages. Therefore, conclusions must be carefully drawn from a single mouse line and comparative studies are needed for confirmation. In recent comparative studies of NC contributions to specific lineages, Wnt1-Cre and P0-Cre lines are widely used (Nakamura et al., 2006; Yoshida et al., 2006; Nagoshi et al., 2008; Morikawa et al., 2009; Nagoshi et al., 2011; Katoh et al., 2011; Olaopa et al., 2011).
To learn whether NC cells migrate into lingual epithelium at early embryonic stages and then contribute to taste papillae and taste buds, we made a thorough examination of both Wnt1-Cre and P0-Cre with two reporters, R26R and ZEG, across different stages, from E11.5 when tongue swellings just emerge, through postnatal (P) day 10 when taste buds become mature. We find distribution, though infrequently, of Wnt1-Cre labeled NC-derived cells in taste papillae and taste buds, in contrast to the recent report that NC does not supply cells to taste buds using Wnt1-Cre mice (Thirumangalathu et al., 2009). Furthermore, using another well-characterized mouse line, P0-Cre, for confirmation and comparison, we demonstrate that NC-derived cells are abundantly distributed in lingual epithelium including taste papillae and taste buds. Our data necessitate a revised concept for the embryonic origins of tongue epithelium and developing taste organs that includes a NC derivation. P0-Cre labeled cells appear in the epithelium of the tongue primordium as early as E11.5 when tongue swellings just emerge. Together with previously reported data of taste bud cell origin, we propose that NC cells migrate into the epithelium of tongue primordium at an early embryonic stage, acquire epithelial cell phenotypes, and undergo proliferation and differentiation for the formation of taste papillae and taste buds.
Materials and methods
Animals and tissue processing
Animals were maintained and used in compliance with institutional animal care protocols and in accordance with National Institutes of Health Guidelines for care and use of animals in research.
Transgenic mouse lines
Two tissue-specific Cre mouse lines, Wnt1-Cre (Danielian et al., 1998) and P0-Cre (Yamauchi et al., 1999) that express Cre in a neural crest (NC)-specific manner, were selected. Both Wnt1-Cre and P0-Cre are well characterized and widely used (Nakamura et al., 2006; Yoshida et al., 2006; Nagoshi et al., 2008; Morikawa et al., 2009; Nagoshi et al., 2011; Katoh et al., 2011; Olaopa et al., 2011), with Wnt1-Cre to label pre- and post-migratory NC cells and P0-Cre for post-migratory NC cells. The difference in distribution patterns of Wnt1-Cre and P0-Cre labeled cells is profound in cranial regions, i.e., midbrain and hindbrain regions, that are the main source of NC cells for the formation of cranial-facial structures. Thus, we use these two Cre mouse lines, Wnt1-Cre and P0-Cre, to study NC derivation of developing tongue for confirmation and comparison.
Each Cre mouse line was bred with two reporter lines -- R26R lacZ reporter (Soriano, 1999) and ZEG (lacZ/EGFP) double reporter (Novak et al., 2000). Thus, four bitransgenic mouse strains, Wnt1-Cre/R26R or Wnt1-Cre/ZEG and P0-Cre/R26R or P0-Cre/ZEG were generated, in which Cre dependent excision activates expression of the lacZ or EGFP reporter. The mouse litters were genotyped by PCR, and wild type littermates were used for negative controls of staining. Embryonic (E) mice (E11.5–18.5) and postnatal (P) mice (P1–10) were used. The embryos were staged by vaginal plug detection. E0.5 is noon of the day on which the dam is positive for vaginal plug. P1 is the day when pups are born.
Tissue collection and analysis of labeled cell distributions
Dams and postnatal animals were euthanized with CO2. Embryonic or postnatal mouse heads were removed into cold, sterile phosphate buffered saline (PBS).
E11.5 embryo heads and E12.5-P10 tongues on mandible were fixed in 4% PFA in 0.1 M PBS, pH 7.4, at 4°C for overnight, then transferred to 30% sucrose in PBS at 4°C for 24 hr. Tissues (E11.5 heads, E12.5–E18.5 tongue/mandibles, P1–10 tongues) were frozen in O.C.T. compound (Miles Scientific, Elkhart, IN). Serial sagittal sections were cut at 10 μm, thaw-mounted onto gelatin coated slides and processed for different analyses.
Fig. 1 illustrates the topography (A) of an E15.5 tongue with well-formed taste papillae and orientation (B) of sagittal sections throughout the study. This orientation applies to all images of sections. Analysis of Wnt1-Cre and P0-Cre labeled cell distributions in the developing tongue was performed on serial sagittal sections of tongue or/and mandibles at each stage. Careful examinations were made by reading all sections. Confocal microscopy was performed at different magnifications to illustrate the localization of lacZ gene product β-galactosidase and EGFP labels.
Fig. 1.
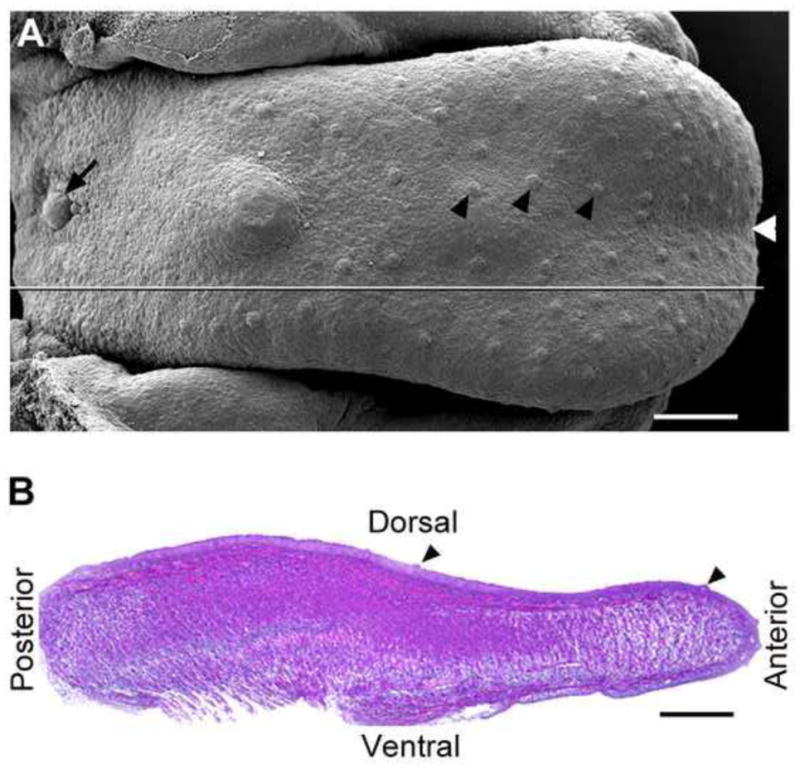
A: Scanning electron microscopy image illustrates the dorsal view of an E15.5 embryonic tongue and papilla types. Black arrowheads point to fungiform papillae on the anterior oral tongue; black arrow points to the single circumvallate papilla in the back. White arrowhead at the tip points to the median furrow. The straight line marks the orientation for sectioning in the sagittal plane. B: H&E stained sagittal section of an E15.5 tongue to illustrate the orientation for all images of tongue sections. Black arrowheads point to fungiform papillae. Scale bars: 250 μm.
X-Gal staining and quantification of labeled taste buds
Tissue sections from R26R lacZ reporter mice were air dried and rehydrated in X-Gal wash buffer (PBS containing 2.0 mM MgCl2), then incubated with freshly prepared X-Gal solution (1 mg/ml X-Gal in 0.1 M PBS with 2.0 mM MgCl2/5 mM potassium ferricyanide/5 mM potassium ferrocyanide), and incubated at 37°C for 1–5 hr. Reacted slides were rinsed in PBS, dehydrated through ethanols, cleared in xylene, and coverslipped for microscopic analysis.
Fungiform taste buds in P0-Cre/R26R mouse tongues were quantified at P1 (n=3) and P10 (n=2). Serial sagittal sections from half of the tongue (from the lateral border to median furrow, about 1 mm in width) were reacted with X-Gal staining and examined with light microscopy. Taste buds were divided into three groups: fully, partially, or not labeled. Taste buds without an obvious X-Gal negative cell are designated as fully labeled. Partially labeled taste buds are those with any X-Gal negative cell (s). The percentage for each group was calculated as taste bud number divided by the total number of examined taste buds, which is about 50 taste buds in each half tongue.
Immunohistochemistry
Antibodies
Primary antibodies and dilutions were: β-glactosidase (ab9361, 1:1000, Abcam, Cambridge, MA); Cre (MAB3120, 1:500, Millipore, Temecula, CA), E-cadherin (AF748, 1:500, R&D Systems, Minneapolis, MN); GFP (A11122, 1:1000, Molecular Probes, Eugene, OR); Keratin 5 (K5) (PRB-160P, 1:5000, Covance, Emeryville, CA); Keratin 8 (K8) (TROMA-I, 1:50, Developmental Studies Hybrydoma Bank, Iowa). Slides treated with no primary antibody or slides with wild type mouse tissue sections treated with the same concentration of primary antibodies were used as controls.
Procedures
For immunohistochemistry on tongue sections, sections were air dried, then rehydrated in PBS. Nonspecific staining was blocked with 10% normal donkey serum in PBS and 0.3% Triton X-100 (Sigma, St. Louis, MO) for 30 min, and then sections were incubated overnight at 4°C in primary antibody at the dilutions described above, in carrier solution (1% normal donkey serum, 0.3% Triton X-100 in PBS). After rinsing in PBS, sections were placed in Alexa fluor® 488, 568/594, or 647 labeled secondary antibody (1:200, Invitrogen, Eugene, OR) for one hour at room temperature. Sections were rinsed with PBS and incubated in DAPI working solution (200 ng/ml in PBS) for 10 min. After rinsing in PBS, slides were air dried and cover slipped with Prolong® Gold antifade mounting medium (Invitrogen, Eugene, OR).
For double labeling with Cre immunoreactions, an extra step for blocking non-specific binding was conducted using the blocking reagent in M.O.M kit (PK-2200, Vector Laboratories, Burlingame, CA), for 1 hr at room temperature before the incubation with primary antibodies.
Results
Analyses of labeled cells were made in tongue tissues from two well-characterized and widely used mouse lines for NC derivation assays, Wnt1-Cre and P0-Cre, each crossed with two reporters (R26R lacZ reporter and ZEG double reporters).
Wnt1-Cre
A thorough examination was conducted in the mouse model, Wnt1-Cre, on the location of NC-derived cells with two reporters (R26R and ZEG) in developing tongue across different stages (E12.5-P1).
Labeled cells in tongue mesenchyme associate with developing taste papillae
In Wnt1-Cre/R26R mice (Fig. 2), lacZ gene product β-galactosidase (β-gal)-positive cells are primarily located in the mesenchyme of embryonic and postnatal tongue. At E12.5–14.5, labeled cells are diffusely distributed throughout the tongue mesenchyme (Fig. 2A, B). However, in E16.5 and P1 tongue, intensely labeled cells are more localized in the mesenchyme just under the lingual epithelium, including the mesenchymal core of all three types of taste papillae, fungiform (Fig. 2C, D, G), foliate (Fig. 2E, H) and circumvallate (Fig. 2F, I).
Fig. 2.
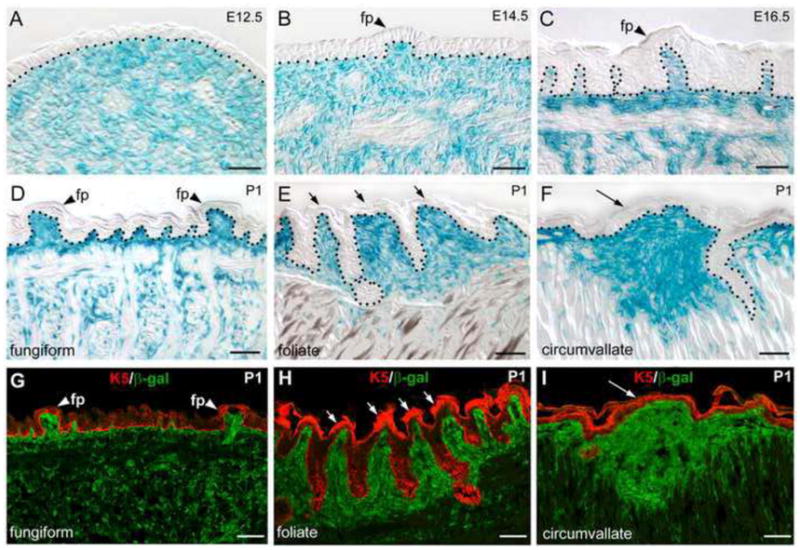
A–F: LacZ expression with X-Gal staining in sagittal sections of Wnt1-Cre/R26R mouse tongue at E12.5, E14.5, E16.5, and P1. Tongue tip is toward to the right. Labeled cells are broadly distributed in tongue mesenchyme at E12.5 (A). Then, progressive restriction of labeled cells are observed in the mesenchyme just under lingual epithelium and the mesenchymal core of fungiform papillae (B–D, fp, arrowheads), foliate (E, short arrows), and circumvallate (F, long arrow). Black dotted lines in A–F demarcate the borders between epithelium and mesenchyme. G–I: Double labeling of β-galactosidase (green) and epithelial cell marker K5 (red) to confirm the mesenchymal location of Wnt1-Cre labeled cells and association with taste papillae. Scale bars: 25μm in A–C; 50 μm in D–I.
Using another reporter (EGFP) mouse line (Wnt1-Cre/ZEG) (Fig. 3), we found that EGFP-positive cells have a similar distribution pattern to that in R26R mouse tongue from E12.5 to P1. In whole tongue images (Fig. 3A, D, G), EGFP labels have a progressive developmental association with taste papillae. In P1 tongue (Fig. 3G), fungiform (arrowheads), foliate (short arrows) and circumvallate (long arrow) are brightly labeled. On sections double labeled with GFP and the epithelial cell marker E-cadherin, distribution of EGFP-positive cells are diffusely located throughout the tongue mesenchyme at E12.5 (Fig. 3B, C). At later stages densely labeled cells are in upper mesenchymal layers under the lingual epithelium (Fig. 3E, H). Brightly labeled cells are especially prominent in the mesenchymal core of taste papillae, fungiform (Fig. 3E, F, H, I, arrowheads), foliate (Fig. 3J) and circumvallate (Fig. 3K).
Fig. 3.
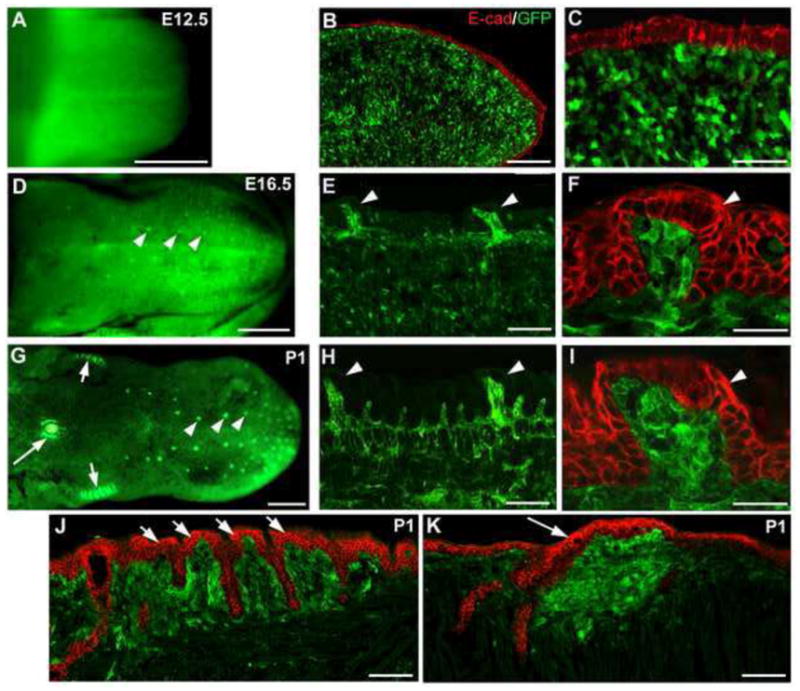
Whole tongue (A, D, G) and single plane confocal images on sections (B, C, E, F, H–K) for EGFP expression to mark NC-derived cells in Wnt1-Cre/ZEG mouse tongue at E12.5, E16.5 and P1. Tongues were sectioned in a sagittal orientation. Epithelial cells are marked with immunoproducts of E-cadherin (E-cad, red). Similar to the R26R reporter, labeled cells are extensively distributed throughout the tongue mesenchyme at E12.5 (A–C). At E16.5 (D–F) and P1 (G–K), EGFP labels are intensely expressed in the mesenchymal cell zone right under lingual epithelium and in the mesenchymal core of taste papillae: see fungiform (white arrowheads), foliate (short arrows) and circumvallate (long arrow) in whole tongue at E16.5 and P1 (G). Scale bars: 500 μm in A, D, G; 100 μm in B, E, H, J, K; 25 μm in C, F, I.
Labeled cells are seen, though rare, in papilla epithelium and taste buds
Whereas NC-derived cells were throughout papilla mesenchyme, and prominent in the core of taste papillae, they were not readily seen in tongue epithelium in Wnt1-Cre mice. However, with thorough microscopic examination of GFP labeling in the epithelium on serial sections of 6 entire tongues at P1 (~200 sections each tongue), GFP labeled cells are seen, though infrequently, in lingual epithelium including taste papillae and early taste buds. As illustrated in Fig. 4, a few labeled cells, among all the examined tongue sections, were observed in the taste bud of fungiform papilla (A–C, white arrows) and in circumvallate epithelium (D–I, white arrows).
Fig. 4.
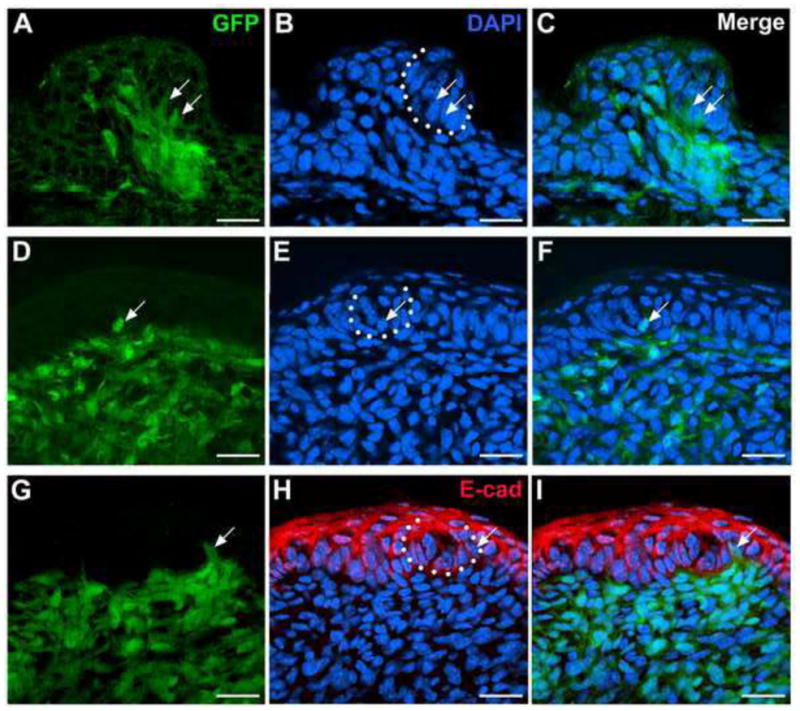
Single plane confocal images of fungiform (A–C) and circumvallate (D–I) papillae in Wnt1-Cre/ZEG mouse tongue at P1. EGFP-positive cells (green) are seen, though infrequently, in the epithelium of taste papillae and within early taste buds (white arrows). In G–I, double labeling of GFP and epithelial cell marker E-cadherin (E-cad, red) was performed to confirm location of the labeled cells in epithelial cells. White dotted lines bracket early taste buds. Scale bars: 20 μm.
In summary, Wnt1-Cre labeled NC-derived cells are primarily distributed in tongue mesenchyme and are in dense association with embryonic and early postnatal taste papillae. Also, a few labeled cells are seen in taste papilla epithelium and early taste buds, in contrast to a previous report (Thirumangalathu et al., 2009).
P0-Cre
Because there are variations in labeling of NC cells across mouse lines with different NC-specific Cre drivers, we also studied another widely-used model for NC derivation, P0-Cre. Analyses were focused on fungiform papillae, which have a distinctive patterned array on the anterior oral tongue. Further, because each mouse fungiform papilla contains a single taste bud, data analysis is straightforward in distinguishing taste bud cells per se from surrounding epithelial cells.
Labeled cells are distributed in tongue mesenchyme, similar to Wnt1-Cre
The R26R reporter mouse line was used to map P0-Cre labeled NC-derived cells in tongues from E11.5 through P10 (Fig. 5). In tongue mesenchyme, the overall distribution pattern of X-Gal staining is similar to that in Wnt1-Cre reporters. Labeled cells are distributed throughout tongue mesenchyme at E11.5–12.5 but have a progressive association with upper mesenchymal cell layers from E13.5–16.5 (Fig. 5, E13.5–16.5). From E18.5-P10, labeled cells are especially dense in the mesenchyme just under the lingual epithelium and within the mesenchymal core of papillae (Fig. 5, E18.5-P10).
Fig. 5.
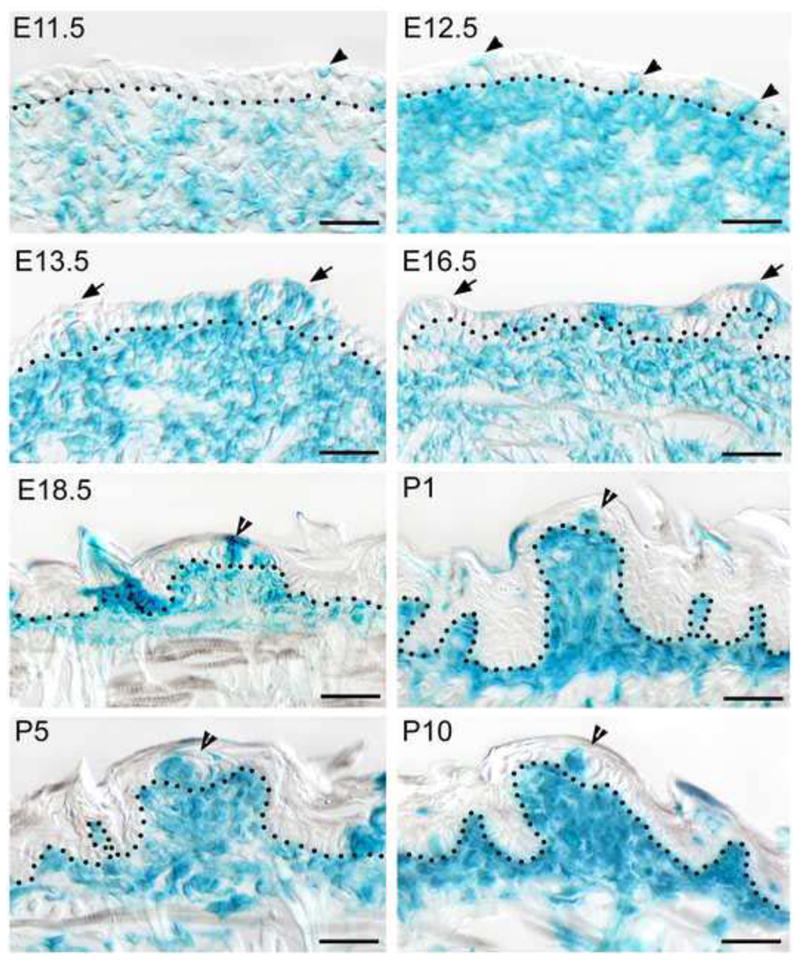
LacZ expression with X-Gal staining in sagittal tongue (or swelling) sections in P0-Cre/R26R mouse from E11.5 through P10. Tongue tip is toward to the right. In the epithelium, labeled cells are singly scattered at E11.5 and E12.5 (arrowheads), clustered within papillae (arrows) and between papillae at E13.5–E16.5. At E18.5-P10, labeled cells are seen in clusters in tongue epithelium and within early taste buds (open arrowheads). In tongue mesenchyme, distribution of labeled cells is overall similar to Wnt1-Cre reporter, i.e., broadly distributed throughout the tongue at E11.5–E13.5, then restricted in the mesenchyme under epithelium, and densely located in the mesenchymal core of taste papillae. Dotted lines demarcate the border between epithelium and mesenchyme. Open arrowheads point to early taste buds with labeled cells. Scale bars: 25 μm.
The dense distribution of Wnt1-Cre and P0-Cre labeled cells just under the papilla epithelium positions these NC-derived cells for potential interaction with the overlying papilla epithelium and suggests the importance of NC-derived mesenchymal cells for the development of taste papillae and taste buds.
Labeled cells are abundant within tongue epithelium including taste papillae and early taste buds
In contrast to data with Wnt1-Cre mice, P0-Cre labeled cells are observed frequently within lingual epithelium, including taste papillae and early taste buds, in R26R reporters (Fig. 5). Single labeled cells are scattered in the epithelium at E11.5 and E12.5 (arrowheads). At E13.5–16.5, numerous labeled cells are observed in clusters, in the epithelium within papillae (see arrows) and between papillae. At E18.5-P10, intensely labeled cells (open arrowheads) are observed in early taste buds.
We quantified labeled taste buds in serial sagittal sections in half tongues at P1 (n=3) and P10 (n=2) (~1 mm thick in about 100 sections from lateral edge to the median furrow) and taste buds were grouped into three types: fully labeled, partially labeled, or without label (Fig. 6). At P1, for about 50 taste buds examined, 3.6% of taste buds are fully labeled, 90% are partially labeled, and 6.4% are without label. At P10, for about 60 taste buds examined in each tongue, taste buds fully labeled, partially labeled or without label are 16.4%, 81.6% and 2.0% respectively. This demonstrates that NC-derived cells contribute to most fungiform taste buds.
Fig. 6.
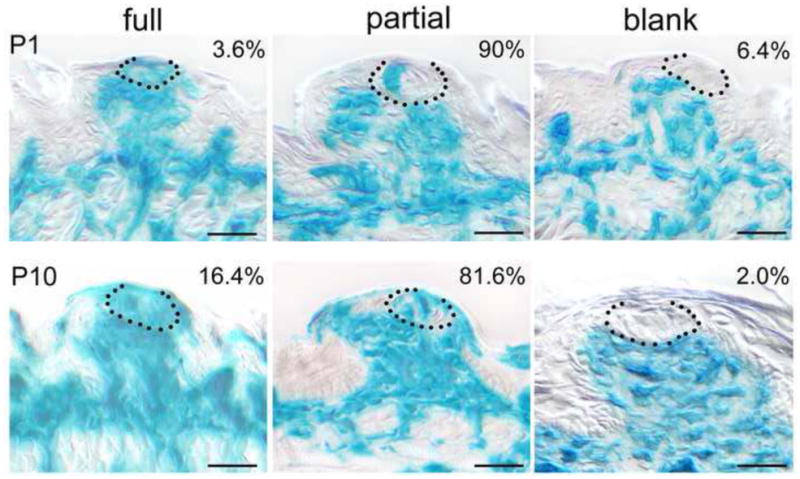
P0-Cre labeled fungiform taste buds in sagittal tongue sections of R26R reporter mice at P1 and P10. Representative images of early taste buds (black dotted outlines) in fungiform papillae for fully (left column), partially (middle) labeled, and without labels (right). The number on the top right corner in each image is the average percentage of the labeled taste buds divided by the total number of taste buds. Quantification was done in serial sections (10 μm thick, ~100 sections); n=3 tongues for P1 and n=2 for P10 data. Scale bars: 25 μm.
Labeled cells are positive for taste cell marker, K8, in early taste buds
In neonatal (P1–5) P0-Cre/R26R and P0-Cre/ZEG mouse tongue, double labeling of β-gal or GFP and the pan taste cell marker, keratin 8 (K8), was used to determine co-localization of reporter label and taste cell marker in epithelial cells (Fig. 7). Distribution of ZEG reporter (Fig. 7D–F) labeled cells was consistent with that in the R26R lacZ reporter (Fig. 7A–C) mouse line, i.e., GFP labeled cells are clustered in tongue epithelium, within taste papillae. Labeled cells are observed in early taste buds (Fig. 7A, D) and co-localize with the taste cell marker, K8 (Fig. 7C, F). These results indicate that NC-derived cells within taste buds are taste cells.
Fig. 7.
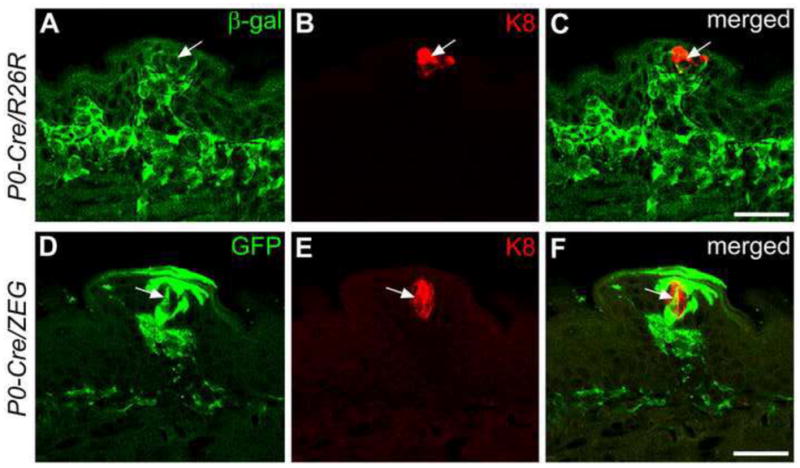
Single plane confocal images of fungiform papillae and taste buds with double labeling of β-gal (A) or GFP (D) (green) and K8 (B, E, red) immunoreactions at P1–5 in P0-Cre driven R26R (A–C) or ZEG (D–F) mouse tongue. β-gal and GFP positive cells are observed within taste buds and some of the cells (white arrows) are also labeled with K8. Scale bars: 25 μm.
No Cre immunoreactivity in the labeled tongue epithelial and mesenchymal cells
The abundant labeled cells in tongue epithelium in P0-Cre/R26R mouse lead to a new concept about derivation of tongue epithelial cells and taste bud cells. The faithful expression of a given promoter for Cre recombinase is a basis of accurate lineage tracing experiments and thus we confirmed the specificity of P0-Cre activity in two aspects. This is critical because specificity of a given promoter may be altered depending on the chromosome integration site. Firstly, we have demonstrated previously that Cre immunoreactivity was highly co-localized with NC cell markers in the migrating NC cells at E9.0 in the P0-Cre/R26R mouse line (Wang et al., 2011). Secondly, to exclude the possibility that tongue epithelium and mesenchyme are labeled by ectopic Cre activity after migration of NC, we double labeled for Cre and β-gal immunoreactivity in E11.5 head and P1 tongue sections (Fig. 8).
Fig. 8.
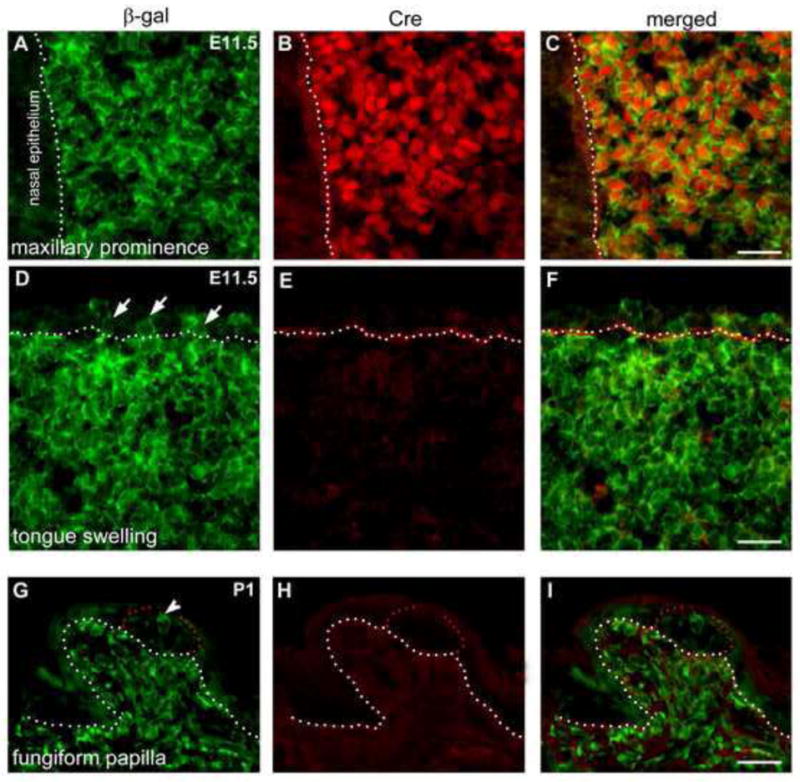
Maxillary prominence (A–C) and tongue swelling (D–F) at E11.5 and fungiform papillae at P1 (G–I) in P0-Cre/R26R mouse line. Sections were double labeled with antibodies against β-gal (green) and Cre (red). Anterior tip of the upper jaw and tongue are toward to the right. White dotted lines demarcate the border between epithelium and mesenchyme. Red dots outline early taste buds in fungiform papilla at P1 (G–I). Short arrows point to β-gal labeled epithelial cells (D) and arrowhead (G) points to labeled cells in early taste buds. Scale bars: 25 μm.
As a positive control, Cre immunoreactivity was intense in the maxillary prominence at E11.5 in the mass of mesenchymal cells anterior to the nasal epithelium (Fig. 8A–C). In contrast, in the E11.5 tongue swellings from the same head section as in A–C, there was no Cre immunoreactivity observed in the β-gal immunoreactive cells (Fig. 8D–F); neither the mesenchymal nor epithelial cells are positive for Cre immunoreactivity. Also, in the newborn mouse tongue no Cre immunoreactivity was observed in these Cre-driven reporter labeled cells in either epithelium or mesenchyme. At P1 (Fig. 8G–I), β-gal immunoreactive cells are densely distributed in the mesenchymal core of taste papillae, and in mesenchyme under lingual epithelium. In tongue epithelium, β-gal positive cells are clustered within and between papillae, and within early taste buds (Fig. 8G, arrowhead). The labeled cells are negative for Cre immunoreactivity (Fig. 8H).
Specific P0-Cre activity in migrating NC cells at early stages and negative Cre immunoreactivity in tongue tissues at later stages demonstrate the faithfulness of the P0-Cre mouse line for cell lineage tracing. Therefore, the label is not driven by ectopic Cre activity and thus, these cells are most likely derived from P0-expressing NC cells.
Discussion
The mammalian tongue hosts taste sensory end organs, taste buds, which reside in three types of papillae: fungiform, circumvallate and foliate. The taste buds are collections of diverse cell types and taste bud cells have been described as arising from the surrounding local epithelium (Stone et al., 1995; Okubo et al., 2009). However, descriptions of the embryonic origin of the lingual epithelium have been controversial, and there is no detailed study to investigate the distribution of neural crest (NC) derived cells in developing tongue epithelium or mesenchyme.
In the present study, we found that NC-derived cells are distributed within lingual epithelium and in mesenchyme in close association with taste papillae in both Wnt1-Cre and P0-Cre mice, two well-characterized and widely used transgenic lines for NC derivation assays (Nakamura et al., 2006; Yoshida et al., 2006; Nagoshi et al., 2008; Morikawa et al., 2009; Nagoshi et al., 2011; Katoh et al., 2011; Olaopa et al., 2011). Identification of NC-derived cells within tongue epithelium and early taste buds is novel to the field and the findings lead to a new concept about the embryonic origin of taste bud cells that includes a NC derivation. Furthermore, the large proportion (about 95%) of labeled early taste buds in P0-Cre/R26R mouse tongue at P1–10 suggests a significant contribution of NC-derived cells to early taste buds.
The specificity of P0-Cre activity was confirmed in P0-Cre/R26R mice. At early embryonic stages (E9.0 and E9.5), Cre expression and NC cell markers are highly co-localized in the migratory NC cells in the hindbrain region, indicating specific expression of P0-Cre transgene in migrating NC cells (Wang et al., 2011). However, negative Cre reactivity of the labeled cells in the E11.5 and P1 tongue, as we showed, excludes the possibility that ectopic Cre activity in the cells drives reporter gene expression for the label. Thus, P0-Cre labeled cells in tongue epithelium, including taste bud cells, are most likely derived from ancestor NC cells.
NC-derived cell contribution to tongue epithelium including taste papillae and taste buds
Descriptions about the embryonic origin of tongue epithelium have been confusing. It has been proposed that the epithelium of the anterior two thirds or oral tongue is ectodermally derived like other regions of oral epithelium (Mistretta and Hill, 1995). However, it is also stated that the entire lingual epithelium is of endodermal origin (Luo et al., 2009). Based on the fact that the oral tongue forms on branchial arches I and II, it is reasonable to speculate that the oral tongue epithelium has the same embryonic origin as these arches which have an ectodermal derivation.
In the present study, our findings demonstrate the NC derivation of tongue epithelial cells, of ectodermal origin. Both Wnt1-Cre and P0-Cre labeled NC-derived cells are observed in lingual epithelium and within early taste buds in the developing tongue, although in different proportions. P0-Cre labeled cells are seen in most taste buds. Double labeling with a taste cell marker demonstrated that labeled cells are taste cells. Our results strongly suggest a NC derivation of a large population of lingual epithelial cells and fungiform taste bud cells.
P0-Cre labeled cells are distributed in tongue epithelium in a pattern of single, scattered elements at E11.5 when the tongue swellings just emerge. This suggests that a population of NC cells might migrate into the epithelium of tongue primordium at an early embryonic stage, potentially even before the tongue emerges. The migration of cranial NC cells into the epithelium of tongue primordium is supported by an early study of NC contribution to tooth formation, with focal DiI labeling in rat embryos at the midbrain and anterior hindbrain crests followed by embryo cultures for 30 or 60 hr. Labeled crest cells from posterior midbrain and anterior hindbrain migrated by the end of the 4-somite stage to the epithelium and mesenchyme of branchial arch I, where tooth buds and tongue normally develop (Imai et al., 1996).
We speculate that after migrating into the epithelium of the tongue primordium, NC-derived single, scattered cells in the epithelium acquire epithelial phenotypes and serve as precursors. The distribution pattern of P0-Cre labeled cells in tongue epithelium from single to a later, clustered pattern in taste papillae and between papillae suggests a process of cell proliferation and differentiation of the scattered single precursors. These phenotypic, epithelial NC-derived cells become taste bud cells later at postnatal stages.
This new finding of NC origin of taste bud cells does not contradict previously reported conclusions that taste bud cells arise from local epithelium; rather, the data can be readily combined with reports from phenotype analyses with X-chromosome inactivation (Stone et al., 1995) and cell lineage mapping with the epithelial cell marker K14 (Okubo et al., 2009) in postnatal mice. As described earlier, X-chromosome inactivation is complete by 9.5 dpc (Tan et al., 1993; Tam et al., 1994) and there is a possibility that NC cells, at an earlier embryonic stage, migrate into and reside in the epithelium of the tongue primordium, and then proliferate and differentiate in taste papilla residences that will include taste buds. In the present study, the single, scattered distribution pattern of P0-Cre labeled NC derived cells in the epithelium of tongue swellings at E11.5 suggests an early migration of NC cells into the epithelium of tongue primordium. The NC-derived cells in the epithelium of tongue primordium acquire epithelial phenotypes and undergo cell proliferation and differentiation to become taste papilla and taste bud cells, which can readily explain the data from cell lineage mapping with the epithelial cell marker K14 in postnatal mice (Okubo et al., 2009). Taken together, our data provide a better understanding for the origin of “local tongue epithelium”, which is in part from NC at an early embryonic stage before the tongue emerges.
In summary, we propose that development of oral tongue epithelium, taste papillae and taste buds needs to incorporate contributions from embryonic ectoderm and NC to the tongue and contributions of NC-derived cells to epithelium including taste papillae and taste buds.
Differences between Wnt1-Cre and P0-Cre labeled cells in tongue epithelium
The difference is profound in distribution patterns of NC-derived cells in tongue epithelium between Wnt1-Cre and P0-Cre lines, i.e., abundant epithelial P0-Cre labels versus few cells in Wnt1-Cre mouse. Differences in distributions between Wnt1-Cre and P0-Cre labeled cells are also found in other systems, e.g., P0-Cre labeled cells are seen in the epithelium of tooth bud, whereas Wnt1-Cre labels are not observed (Wang et al., 2011); in the developing cardiovascular system, the proportions of Wnt1-Cre and P0-Cre labeled cells are different in different regions (Nakamura et al., 2006). In the otic vesicle, few labeled cells are seen in Wnt1-Cre mice compared to Pax3-Cre line (Freyer et al., 2011). Also, there are no phenotypes in the developing heart with conditional deletion of Pax3 driven by Wnt1-Cre but P0-Cre driven Pax3 deletion leads to significant heart defects (Olaopa et al., 2011). These data suggest that Wnt1 marks a more restricted subpopulation of NC cells than other promoters including P0. In the developing tongue, Wnt1-Cre labels few tongue epithelial cells and these cells might be easily missed if a study is not detailed and fully focused on NC derivation analyses. This could help to explain why investigators previously concluded that NC does not contribute to taste bud cells (Thirumangalathu et al., 2009).
Comparing the regions of NC cell origins between Wnt1-Cre (Danielian et al., 1998) and P0-Cre (Yamauchi et al., 1999) lines, the difference is particularly obvious in the regions of midbrain and hindbrain which are known as the main NC cell source for the formation of branchial arch I where the oral tongue forms. At embryonic stage E8.5, a time when cranial crest cells are emerging, Wnt1-Cre labels NC cells extensively in midbrain but many fewer cells in hindbrain (Echelard et al., 1994; Danielian et al., 1998). In contrast, P0-Cre abundantly labels migratory NC cells in hindbrain, but sparsely labels midbrain crest (Yamauchi et al., 1999). In the present study, although both Wnt1-Cre and P0-Cre labeled cells are seen in tongue epithelium, P0-Cre labeled cells are far more abundant than Wnt1-Cre. Our findings implicate the hindbrain as the primary region of NC cells that contribute to lingual epithelium including taste papillae and taste buds.
NC-derived cell contribution to tongue mesenchyme
The mesenchymal components of the oral tongue are known to form primarily from the first branchial arch (Paulson et al., 1985). Although it has been demonstrated that NC cells contribute to formation of mammalian craniofacial structures including branchial arches (Chai et al., 2000; Cordero et al., 2011), there is not clarity about distribution patterns of NC-derived cells in the mesenchyme of developing tongue. Our studies demonstrate NC-derived cell distributions in tongue mesenchyme with similar patterns across different embryonic and postnatal stages in both Wnt1-Cre and P0-Cre reporter mouse lines. The distribution of NC-derived cells in tongue mesenchyme is diffuse and then progressively restricted to connective tissue just subjacent to the lingual epithelium, and is particularly dense in the mesenchymal core of taste papillae. This distribution pattern positions NC-derived mesenchymal cells to interact with overlying epithelium for regulating cell proliferation and differentiation. Further studies are needed to elucidate the roles of these lingual mesenchymal cells.
Dermal papilla cells of hair and whisker follicle in the face have a demonstrated neural crest derivation (Jinno et al., 2010). Our data on NC-derived cells in the mesenchymal core of taste papillae add another craniofacial derivative to the list of cell types with NC origins.
Summary
Our results strongly suggest a NC contribution to tongue epithelium and mesenchyme with two widely used, NC-specific promoter driven reporter mouse lines, Wnt1-Cre and P0-Cre. The distribution of NC-derived cells in tongue epithelium, taste papillae and taste buds implicates a NC derivation of taste organs which is a new concept that includes a NC contribution to taste bud cells. Figure 9 summarizes the basic elements of this proposed distribution and contribution. We propose that NC cells from around the neural tube (A) migrate into the epithelium and mesenchyme of the tongue primordium at an early embryonic stage before the tongue emerges. The migrated NC cells (B) are in the mesenchyme and scattered singly in the epithelium of tongue primordium and serve as precursors. They acquire epithelial phenotypes and undergo cell proliferation and differentiation to achieve a later, clustered pattern in taste papillae and between papillae (C). A subpopulation of these phenotypic, epithelial NC-derived cells become taste bud cells later at postnatal stages (D). In tongue mesenchyme, NC-derived cells are progressively associated with taste papillae suggesting an interactive role of these NC-derived mesenchymal cells with tongue epithelium for papilla and taste bud development and maintenance.
Fig. 9.
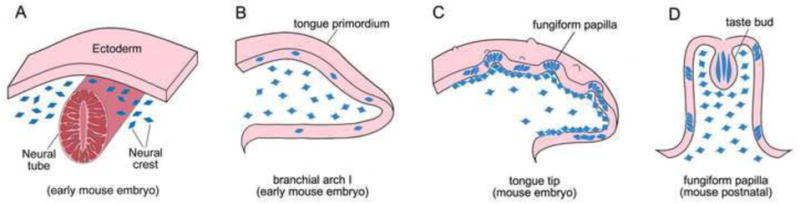
Diagram to illustrate distribution of NC cells and NC-derived cells in developing tongue. A: Cross section of an early stage embryo at a cranial level. The delaminated, migratory NC cells (blue) are located at both sides lateral to neural tube (red), under the ectodermal sheet (pink). B: Sagittal section of branchial arch I (tongue primordium) before tongue emerges. The anterior tip is toward to the right and dorsal surface up (also applies to C and D). Migrated NC cells (blue) are scattered in the epithelium and broadly distributed in the mesenchyme. C: Sagittal section of anterior region of oral tongue with developing fungiform papillae on the dorsal surface. NC derived cells (blue) are clustered in tongue epithelium within and between papillae. In tongue mesenchyme, NC derived cells are more restricted under the epithelium. D: A fungiform papilla with single taste bud at the apex. NC derived cells are within taste buds and in the surrounding papilla epithelium. Also, NC derived cells are densely distributed in the mesenchymal core of papillae. We propose that NC cells (A, blue cells) migrate into the epithelium and mesenchyme of tongue primordium (B) at early embryonic stage. The NC cells in tongue epithelium acquire epithelial phenotype and undergo cell proliferation and differentiation to become clusters within and between papillae (C). A population of NC-derived cells is within early taste buds (D). In tongue mesenchyme, NC-derived cells are progressively restricted to connective tissues under tongue epithelium and densely distributed in the mesenchymal core of taste papillae (C, D).
Highlights.
Wnt1-Cre labeled cells: rarely seen in taste papilla epithelium and taste buds.
P0-Cre labeled cells: abundant in tongue epithelium, taste papillae and taste buds.
P0-Cre labeled cells in epithelium progress from single to clusters of cells.
P0-Cre labeled cells: in most early postnatal taste buds and co-localize with K8.
Labeled mesenchymal cells: associated with lingual epithelium and papillae.
Acknowledgments
The research was supported by NIDCD, NIH Grant R03DC009055 to HXL; NIDCR NIH Grant K99DE021054 to YK; NIDCR, NIH Grant R01DE020843 to YM; NIDCD, NIH Grant R01DC000456-21 to CMM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Axelrod FB, Gold-von Simson G. Hereditary sensory and autonomic neuropathies: types II, III, and IV. Orphanet J Rare Dis. 2007;2:1–12. doi: 10.1186/1750-1172-2-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barembaum M, Bronner-Fraser M. Early steps in neural crest specification. Semin Cell Dev Biol. 2005;16:642–646. doi: 10.1016/j.semcdb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Bradley RM, Stern IB. The development of the human taste bud during the foetal period. J Anat. 1967;101:743–752. [PMC free article] [PubMed] [Google Scholar]
- Bronner-Fraser M. Neural crest cell migration in the developing embryo. Trends Cell Biol. 1993;3:392–397. doi: 10.1016/0962-8924(93)90089-j. [DOI] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas PJr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Chai Y, Maxson RE., Jr Recent advances in craniofacial morphogenesis. Dev Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- Cordero DR, Brugmann S, Chu Y, Bajpai R, Jame M, Helms JA. Cranial neural crest cells on the move: their roles in craniofacial development. Am J Med Genet A. 2011;155A:270–279. doi: 10.1002/ajmg.a.33702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Echelard Y, Vassileva G, McMahon AP. Cis-acting regulatory sequences governing Wnt-1 expression in the developing mouse CNS. Development. 1994;120:2213–2224. doi: 10.1242/dev.120.8.2213. [DOI] [PubMed] [Google Scholar]
- Engleka KA, Gitler AD, Zhang M, Zhou DD, High FA, Epstein JA. Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Dev Biol. 2005;280:396–406. doi: 10.1016/j.ydbio.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Farbman AI. Fine structure of the taste bud. J Ultrastruct Res. 1965;12:328–350. doi: 10.1016/s0022-5320(65)80103-4. [DOI] [PubMed] [Google Scholar]
- Farbman AI, Mbiene JP. Early development and innervation of taste bud-bearing papillae on the rat tongue. J Comp Neurol. 1991;304:172–186. doi: 10.1002/cne.903040203. [DOI] [PubMed] [Google Scholar]
- Freyer L, Aggarwal V, Morrow BE. Dual embryonic origin of the mammalian otic vesicle forming the inner ear. Development. 2011;138:5403–5414. doi: 10.1242/dev.069849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner J, Barton D, Vanslambrouck JM, Braet F, Hall D, Marc J, Overall R. Defects in tongue papillae and taste sensation indicate a problem with neurotrophic support in various neurological diseases. Neuroscientist. 2008;14:240–250. doi: 10.1177/1073858407312382. [DOI] [PubMed] [Google Scholar]
- Imai H, Osumi-Yamashita N, Ninomiya Y, Eto K. Contribution of early-emigrating midbrain crest cells to the dental mesenchyme of mandibular molar teeth in rat embryos. Dev Biol. 1996;176:151–165. doi: 10.1006/dbio.1996.9985. [DOI] [PubMed] [Google Scholar]
- Jinno H, Morozova O, Jones KL, Biernaskie JA, Paris M, Hosokawa R, Rudnicki MA, Chai Y, Rossi F, Marra MA, Miller FD. Convergent genesis of an adult neural crest-like dermal stem cell from distinct developmental origins. Stem Cells. 2010;28:2027–2040. doi: 10.1002/stem.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh H, Shibata S, Fukuda K, Sato M, Satoh E, Nagoshi N, Minematsu T, Matsuzaki Y, Akazawa C, Toyama Y, Nakamura M, Okano H. The dual origin of the peripheral olfactory system: placode and neural crest. Mol Brain. 2011;4:34. doi: 10.1186/1756-6606-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen F, Epstein JA. Neural crest expression of Cre recombinase directed by the proximal Pax3 promoter in transgenic mice. Genesis. 2000;26:162–164. doi: 10.1002/(sici)1526-968x(200002)26:2<162::aid-gene21>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Ludwig A, Schlierf B, Schardt A, Nave KA, Wegner M. Sox10-rtTA mouse line for tetracycline-inducible expression of transgenes in neural crest cells and oligodendrocytes. Genesis. 2004;40:171–175. doi: 10.1002/gene.20083. [DOI] [PubMed] [Google Scholar]
- Luo X, Okubo T, Randell S, Hogan BL. Culture of endodermal stem/progenitor cells of the mouse tongue. In Vitro Cell Dev Biol Anim. 2009;45:44–54. doi: 10.1007/s11626-008-9149-2. [DOI] [PubMed] [Google Scholar]
- Mistretta CM, Hill DL. Development of the taste system. Basic neurology. In: Doty R, editor. Handbook of Olfaction and Gustation. Marcel Dekker Press; New York: 1995. pp. 635–668. [Google Scholar]
- Morikawa S, Mabuchi Y, Niibe K, Suzuki S, Nagoshi N, Sunabori T, Shimmura S, Nagai Y, Nakagawa T, Okano H, Matsuzaki Y. Development of mesenchymal stem cells partially originate from the neural crest. Biochem Biophys Res Commun. 2009;379:1114–1119. doi: 10.1016/j.bbrc.2009.01.031. [DOI] [PubMed] [Google Scholar]
- Morrison KM, Miesegaes GR, Lumpkin EA, Maricich SM. Mammalian Merkel cells are descended from the epidermal lineage. Dev Biol. 2009;336:76–83. doi: 10.1016/j.ydbio.2009.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi N, Shibata S, Hamanoue M, Mabuchi Y, Matsuzaki Y, Toyama Y, Nakamura M, Okano H. Schwann cell plasticity after spinal cord injury shown by neural crest lineage tracing. Glia. 2011;59:771–784. doi: 10.1002/glia.21150. [DOI] [PubMed] [Google Scholar]
- Nagoshi N, Shibata S, Kubota Y, Nakamura M, Nagai Y, Satoh E, Morikawa S, Okada Y, Mabuchi Y, Katoh H, Okada S, Fukuda K, Suda T, Matsuzaki Y, Toyama Y, Okano H. Ontogeny and multipotency of neural crest-derived stem cells in mouse bone marrow, dorsal root ganglia, and whisker pad. Cell Stem Cell. 2008;2:392–403. doi: 10.1016/j.stem.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Colbert MC, Robbins J. Neural crest cells retain multipotential characteristics in the developing valves and label the cardiac conduction system. Circ Res. 2006;98:1547–1554. doi: 10.1161/01.RES.0000227505.19472.69. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Okubo T, Clark C, Hogan BL. Cell lineage mapping of taste bud cells and keratinocytes in the mouse tongue and soft palate. Stem Cells. 2009;27:442–450. doi: 10.1634/stemcells.2008-0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaopa M, Zhou HM, Snider P, Wang J, Schwartz RJ, Moon AM, Conway SJ. Pax3 is essential for normal cardiac neural crest morphogenesis but is not required during migration nor outflow tract septation. Dev Biol. 2011;356:308–321. doi: 10.1016/j.ydbio.2011.05.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietri T, Eder O, Blanche M, Thiery JP, Dufour S. The human tissue plasminogen activator-Cre mouse: a new tool for targeting specifically neural crest cells and their derivatives in vivo. Dev Biol. 2003;259:176–187. doi: 10.1016/s0012-1606(03)00175-1. [DOI] [PubMed] [Google Scholar]
- Rubin BY, Anderson SL. The molecular basis of familial dysautonomia: overview, new discoveries and implications for directed therapies. Neuromolecular Med. 2008;10:148–156. doi: 10.1007/s12017-007-8019-5. [DOI] [PubMed] [Google Scholar]
- Smith A, Farbman A, Dancis J. Absence of taste-bud papillae in familial dysautonomia. Science. 1965a;147:1040–1041. doi: 10.1126/science.147.3661.1040. [DOI] [PubMed] [Google Scholar]
- Smith A, Farbman A, Dancis J. Tongue in familial dysautonomia, a diagnostic sign. Am J Dis Child. 1965b;110:152–154. doi: 10.1001/archpedi.1965.02090030162010. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. 1999;21:70–71. doi: 10.1038/5007. Erratum in: Nat Genet 1993 Jul;4(3):320. [DOI] [PubMed] [Google Scholar]
- Stine ZE, Huynh JL, Loftus SK, Gorkin DU, Salmasi AH, Novak T, Purves T, Miller RA, Antonellis A, Gearhart JP, Pavan WJ, McCallion AS. Oligodendroglial and pan-neural crest expression of Cre recombinase directed by Sox10 enhancer. Genesis. 2009;47:765–770. doi: 10.1002/dvg.20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone LM, Finger TE, Tam PP, Tan SS. Taste receptor cells arise from local epithelium, not neurogenic ectoderm. Proc Natl Acad Sci U S A. 1995;92:1916–1920. doi: 10.1073/pnas.92.6.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PP, Williams EA, Tan SS. Expression of an X-linked HMG-lacZ transgene in mouse embryos: implication of chromosomal imprinting and lineage-specific X-chromosome activity. Dev Genet. 1994;15:491–502. doi: 10.1002/dvg.1020150608. [DOI] [PubMed] [Google Scholar]
- Tan SS, Williams EA, Tam PP. X-chromosome inactivation occurs at different times in different tissues of the post-implantation mouse embryo. 1993;3:170–174. doi: 10.1038/ng0293-170. Erratum in: Nat Genet 1993 Jul;4(3):320. [DOI] [PubMed] [Google Scholar]
- Thirumangalathu S, Harlow DE, Driskell AL, Krimm RF, Barlow LA. Fate mapping of mammalian embryonic taste bud progenitors. Development. 2009;136:1519–1528. doi: 10.1242/dev.029090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SK, Komatsu Y, Mishina Y. Potential contribution of neural crest cells to dental enamel formation. Biochem Biophys Res Commun. 2011;415:114–119. doi: 10.1016/j.bbrc.2011.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Abe K, Mantani A, Hitoshi Y, Suzuki M, Osuzu F, Kuratani S, Yamamura K. A novel transgenic technique that allows specific marking of the neural crest cell lineage in mice. Dev Biol. 1999;212:191–203. doi: 10.1006/dbio.1999.9323. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Shimmura S, Nagoshi N, Fukuda K, Matsuzaki Y, Okano H, Tsubota K. Isolation of multipotent neural crest-derived stem cells from the adult mouse cornea. Stem Cells. 2006;24:2714–2722. doi: 10.1634/stemcells.2006-0156. [DOI] [PubMed] [Google Scholar]


