Abstract
Precise profiling of polar lipids including gangliosides and sulfatides is a necessary step in understanding the diverse physiological role of these lipids. We have established an efficient method for the profiling of polar lipids using reverse-phase nano-HPLC-Chip Q-TOF/MS. A microfluidic chip design provides improved chromatographic performance, efficient separation, and stable nanospray while the advanced high resolution mass spectrometer allowed for the identification of complex isobaric polar lipids such as NeuAc- and NeuGc-containing gangliosides. Lipid classes were identified based on the characteristic fragmentation product ions generated during data-dependent MS/MS experiments. Each class was monitored by a post processing precursor ion scan. Relatively simple quantitation and identification of intact ions was possible due to the reproducible retention times provided by the nano-HPLC-Chip. The method described in this paper was used to profile polar lipids from mouse brain, which was found to contain 17 gangliosides and 13 sulfatides. Types and linkages of the monosaccharides and their acetyl modifications were identified by low energy collision induced dissociation (CID) (40V), and the type of sphingosine base was identified by higher energy CID (80V). Accurate mass measurements and chromatography unveiled the degree of unsaturation and hydroxylation in the ceramide lipid tails.
Keywords: Ganglioside, Sulfatide, Polar Lipids, Mouse Brain, HPLC Chip, Quadrupole Time of Flight, Mass Spectrometry, Multiple Precursor Ion Scan
Introduction
Lipids are a vital class of biomolecules in living organisms from bacteria to humans that are used as energy stores, as membrane components, and as signaling molecules.1, 2 Lipid functions remain poorly understood in molecular detail largely because they are structurally and functionally diverse, with each class having a range of unique biological roles. Membrane polar lipids are structurally composed of a discrete set of hydrophilic headgroups that alter their biophysical properties in the membrane ensemble and also have the capability to bind to exogenous compounds. These interactions are thought to have a range of consequences from influencing their dynamics in the membrane to conferring various biological properties.3-5 Gangliosides and sulfatides are two of the less understood polar lipids that appear to be involved in diverse biological processes that are mediated not only by the carbohydrate head group structures but also complexities in the ceramide lipid tail.6, 7 The ceramide imbeds in the membrane and operates as a component of the functional architecture and an intracellular regulator, while the saccharides interact with the environment, serving in one sense as an identification tag. As gangliosides undergo changes in a disease-specific as well as age dependent manner, they represent potential biomarkers of risk and development of diseases and their resolution.8-10 It has also been reported that when gangliosides are ingested, they may contribute nutritionally to the prevention of infection and to the immune response.11
Gaining mechanistic and predictive knowledge attainable from gangliosides first requires analytical techniques, however, classical analytical work flow for this complex lipid class is very involved, laborious, and time consuming.12 The structural complexity of these lipids is an impediment to achieving accurate and precise analysis. They have characteristic amine containing lipid backbones called ceramides, in which a fatty acid is linked to a sphingoid base. An anionic carbohydrate headgroup containing sialic acid residues is attached at the primary hydroxyl of the ceramide.13, 14 Considerable structural heterogeneity exists, arising from the variation in the length of the ceramide tail and the composition and structure of the saccharide moieties. The ceramides can have different total numbers of carbon atoms, double bonds, and hydroxylations, as well as different positions for the unsaturation and hydroxylation. While the number of saccharide residues in the polar head is generally small, the types of saccharides, linkages, and the branching provides even more diversity.15 Moreover, conventional lipid extraction methods such as Folch and Bligh Dyer rely on extracting lipids into the organic methanol chloroform layer, whereas gangliosides and likely a part of other very polar lipids are extracted into the aqueous layer, therefore overlooking them as key polar lipids forming a subset of overall membrane lipids in conventional profiling.16
Electrospray ionization mass spectrometry (ESI-MS) is an attractive analytical strategy for the identification and quantitation of complex lipid molecules, offering the advantage of generating profile information with high sensitivity. Mass spectrometry methods that employ direct infusion target the analysis of crude extracts, mainly by ESI-MS and ESI-MS/MS, without prior chromatographic separation. This approach is characterized by its simplicity and high throughput potential.17, 18 However, isobaric species overlap in the spectra, which is more problematic in the case of complex lipids such as gangliosides. Moreover, signal suppression effects may occur and some low level species may not be detected. To overcome these limitations, liquid chromatography (LC) has been coupled directly to MS. Recently, ESI nano-HPLC-Chip MS has been introduced providing high sensitivity and reproducibility, as well as ease of use.19-22 The goal of the nano-HPLC-Chip is to integrate the sample enrichment and separation columns of a nanoflow LC system with the intricate connections and nanospray emitter used in nanospray MS directly on a polymer chip. This design eliminates the traditional fittings, valves, and connections, providing improved chromatographic performance and a stable nanospray. The nanoLC can separate compounds that may overlap in MS only, as well as helping characterize a compound by the reproducible retention time provided by the chip design.
Reverse-phase chromatography has been used to separate gangliosides according to their ceramide portions.23-25 The nano-HPLC-Chip LC provides a user friendly nanoLC-MS for sample limited applications requiring high sensitivity and efficient LC separation. Reverse phase chromatography was employed since separation of individual molecular species in each class is not effective by direct infusion and normal phase liquid chromatography (NPLC). Even if each class is separated by NPLC, it is difficult to distinguish the minor components from the major ones due to ion suppression.26 Moreover, the reverse-phase LC was shown to be more effective due to the high reproducibility and the compatibility of the mobile phase with ESI ionization. NanoLC employing the HPLC-chip packed with C18 has been widely used in fields such as proteomics, glycomics, and lipidomics.27-29 However, to our knowledge, the HPLC Chip has not been used for the separation of very polar lipid such as gangliosides.
Moreover, technologically advanced hybrid analyzers, such as quadrupole time of flight (Q-TOF), provide high mass accuracy and resolution along with fast scan speed. In Q-TOF instruments, the ions selected by the Q1 quadrupole go through the collision cell yielding product ions that are then detected by a TOF mass analyzer. The fragment ion spectra generated from individual precursor ions are detected the analyzer. Therefore, post processing precursor ion scans over a single experimental run can be achieved during data analysis.30 By the use of the high resolution multiple precursor ion scanning method, the limitations of triple quadrupole precursor ion scan, such as low duty cycle and poor scan sensitivity, have been overcome. Furthermore, the high mass resolution of the TOF analyzer significantly increases the specificity and removes false positive molecular assignments. This approach has been used for lipidomics studies by several groups.31-33
In this paper, we present a reverse-phase nano-HPLC-Chip Q-TOF MS strategy and apply it to the identification of multiple classes of polar lipids from murine brain. Several mass spectrometry approaches using imaging MALDI MS, triple quadrupole MS (precursor ion scan and multiple reaction monitoring), high resolution MS (mass defect filter based approach), and Chip ESI-MS have been reported for the analysis of polar lipids, including gangliosides.24, 34, 35-39 In addition, the combination of effective separation and high quality spectra will provide a new tool for the profiling of complicated mixtures of polar lipids, especially gangliosides and sulfatides, found in neurological tissues.
Experimental Section
Chemicals and Materials
HPLC grade methanol and isopropanol were purchased from Sigma (St. Louis, MA). Ammonium acetate and acetic acid were of analytical reagent grade quality from Merck (Darmstadt, Germany). Ganglioside standards GM3 and GD3 from bovine buttermilk and GT1b and GQ1b from bovine brain were purchased from Matreya (Pleasant Gap, PA). Bovine milk gangliosides were prepared as described in Lee et al.40 Mouse brain polar lipids were extracted by the process described in S-Experimental Section.
Instrumentation
A nano-HPLC-Chip Q-TOF system using the Agilent 1200 series microwell plate autosampler, capillary pump, nano pump, HPLC Chip interface, and the Agilent 6520 Q-TOF MS (Agilent Technologies, Inc., Santa Clara, CA) was used.
A nano-HPLC-Chip (G4240 62001, Agilent Technologies, Inc., Santa Clara, CA) with a 40 nL enrichment column and 43 × 0.075 mm ID analytical column was used. Both the enrichment and analytical columns were packed with ZORBAX C18 (5 Km pore size) stationary phase. The nano-HPLC-Chip has been previously described.41 The mobile phases used are water (solvent A) and 15% isopropanol in methanol (v/v) (solvent B), with both containing 20 mM ammonium acetate and 0.1% acetic acid. For loading of the sample onto the enrichment column, the capillary pump was operated at 3 KL/min using 70% solvent B. A gradient based chromatographic separation of the polar lipids was performed on the analytical column and was driven by the nanoliter pump running at 300 nL/min. The gradient used for this separation is as follows: 70% of B until 1 minute after sample injection followed by a linear increase to 80% B after 3 minutes. The gradient further increased over 40 min to 100% B in a linear manner and was kept at 100% B for 5 min. After this time, the amount of solvent B was decreased to 70% and was maintained at this level until completion of the run. The working samples were diluted in water/methanol (1:1, v/v) to corresponding concentrations for injection.
The Agilent 6520 Q-TOF MS was operated in the negative ion mode for MS scans and in both positive and negative ion modes for the MS/MS. The drying gas temperature and gas flow were 325°C and 4 L/min, respectively. Capillary voltage was 1850V for the entire LC run. Fragmentor voltage, the voltage applied to the exit end of the capillary, was set to 175V. The source and ion optic parameters were optimized for minimal in source fragmentation and for the best quality of data. Recorded mass ranges were m/z 500 2500 for MS only and m/z 50 1500 for MS/MS. Acquisition rates were 1 spectrum/s for MS and 3 spectra/s for MS/MS. All mass spectra were internally calibrated using the G1969 85000 ESI tuning mix (Agilent Technologies, Inc., Santa Clara, CA), with reference masses at m/z 680.036 and 1279.995 for negative mode, at m/z 622.029 and 1221.991 for positive mode, chosen to insure the highest mass accuracy. Data dependent MS/MS analysis was performed with collision energies set at 40V for the lower collision energy and 80V for the higher collision energy. The optimum conditions for collision induced dissociation (CID) were selected through the preliminary experiments (S-Experimental Section and S-Figure 1). The precursor ions with 1000 counts/s ion intensity or more were automatically selected for MS/MS.
Data Analysis
Data analyses were performed with the MassHunter Qualitative Analysis software ver. B.03.01 (Agilent Technologies, Inc., Santa Clara, CA). Molecular Feature Extraction (MFE) was performed through a mode of the “Find by Molecular Feature” function for non targeted approach. The focused post processing precursor ion scan type analysis was performed through a mode of the “Find by Auto MS/MS”. The software was capable of generating a peak list (m/z, retention time and peak area) taking all ions into account exceeding 1000 counts. For the focused analysis, specific fragment ion masses were selected. For example, in the negative ion mode, NeuAc ions (m/z 290.095) and sulfatide ions (m/z 96.960) were the fragment ions used to determine the Q1 masses representing gangliosides and sulfatides, respectively. The putative target lipids were then confirmed and their structures are elucidated from the corresponding MS and MS/MS data acquired in either negative ion or positive ion mode. The sequential collection and parallel alignment of MS and MS/MS chromatograms made it possible to locate molecules with their molecular ions and fragments. This method results in multiple levels of information, which is extremely useful when trying to confirm the identities of compounds.
Results and discussion
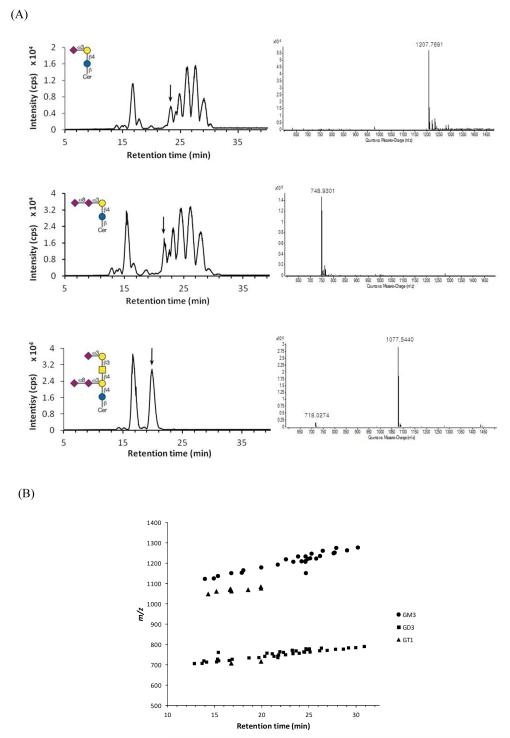
The opposing polarities of head groups and lipid moieties of these lipids required finely balanced proportions of organic solvents to concomitantly meet the requirements for efficient solubilization, chromatographic separation, and ionization. The mixture of 15% isopropanol and methanol (v/v), as the organic mobile phase, provided the best chromatographic performance with gradient. The nano LC chromatograms of gangliosides GM3, GD3 and GT1b, which have one, two and three sialic acids, respectively, are shown in Figure 1(A). The samples were obtained commercially, however the compounds are only available as mixtures with varying lipid chains. The mass chromatograms and mass spectra of the individual peaks indicate that the sample was separated on the basis of the ceramide chain length and the degree of saturation. The detailed assignment of each peak is shown in S-Table 1. In Figure 1(A), the right panel shows one representative spectrum of ganglioside (d38:1) from each species. Accurate identification and quantitation often suffers from uncontrolled sialic acid fragmentation, which is problematic to the identification of their individual species.42 However, we found no sialic acid fragments, indicating little or no in source fragmentation, as shown in Figure 1(A).
Figure 1.
(A) Base peak chromatograms (left panel) and mass spectra (right panel) of ganglioside standards GM3, GD3 and GT1b. A C18 analytical column was used as reverse-phase mode for the separation of ceramide moieties. 20-50 ng samples were injected. Inserts are the sugar structures of each ganglioside. Right panel mass spectra show gangliosides with ceramide d38:1 from LC separation, which is indicated by arrows on the left panel. Ganglioside subspecies and their assignments are shown in S-Table 1. (B) Retention time vs. m/z plot of the ganglioside standards. Ganglioside separation mainly depends on the hydrophobicity of ceramide moieties.
Figure 1(B) shows that the gangliosides with long ceramide chains elute later than those with short chains. For the molecules with different CH2 groups either on the fatty acid chain or on the alkyl chain of the sphingoid base, the retention time was shown to vary linearly with the number of carbon atoms. The length of the ceramide tail mainly influenced the retention behavior with a minor contribution by the oligosaccharide structure. Triplicate experiments were conducted within a single day to study the reproducibility of the method on a run to run basis. The retention time variation was less than 0.3 min in 40 minute runs, and the peak areas differed less than 1% RSD (data not shown).
The sensitivity of the nano-HPLC-Chip Q-TOF MS method for the analysis of gangliosides is shown in S-Figure 2(A). As little as 30 fmol of GT1b yielded a peak from the extracted ion chromatogram supporting the high detection sensitivity that can be obtained through a stable plug-and-play nanospray LC-MS operation. This feature is critical for applications requiring high sensitivity, low sample consumption, and a robust analytical platform. One advantage of the TOF analyzer over other high resolution mass analyzers is its broad linear dynamic range, which is 4-5 orders of magnitude as shown in S-Figure 2(B).
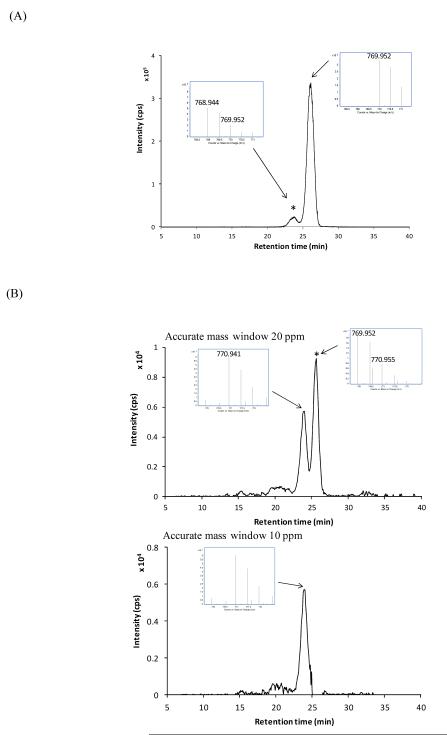
When samples contain lipids with different degrees of unsaturation, the lipid peaks may overlap with the peaks from isotopic species of other lipids. For example, in the case of a lipid with one double bond versus one with two double bonds, the second 13C isotope of that of two double bonds overlap with the mono isotopes of that of one double bond. This is more problematic when the sample contains much greater amounts of lipids with a greater degree of multiple bonds because the overlap interferes with quantitation. To overcome this problem, LC methodology was developed to separate lipids with different degrees of unsaturation. Figure 2(A) shows that the two peaks which correspond to GD3(d41:1) and GD3(d41:2) are baseline resolved. This makes it possible to characterize and quantify the two different compounds accurately. Gangliosides also vary in their carbohydrate moieties as well, which may be composed of different types of monosaccharides. For instance, two kinds of sialic acids, N acetylneuraminic acid (NeuAc) and N glycolylneuraminic acid (NeuGc), exist in nature, with NeuGc found primarily in non human mammals. These structures differ only in that NeuGc has an additional oxygen atom. Here, bovine milk gangliosides that contain both NeuAc and NeuGc were separated and identified (Fig. 2(B)). For example, the calculated m/z for NeuGc-NeuAc GD3 (d40:1) and NeuAc-NeuAc GD3 (d41:1) are 770.941 and 769.952 respectively. Therefore, the third isotope of NeuAc-NeuAc GD3 (d41:1) is not distinguishable from the monoisotope of NeuGc NeuAc GD3 (d40:1) on lower resolution mass spectrometers. An EIC having a mass window of 10 ppm shows a distinct peak, which corresponds to NeuGc NeuAc GD3 (d40:1). The structures of two gangliosides with different sialic acids were further elucidated by tandem MS (S-Figure 3). Fragments at m/z 290.086 for NeuAc and 306.078 for NeuGc were observed. The human body does not have intrinsic NeuGc except possible in the case of cancer cells. It has been proposed that metabolic incorporation of NeuGc from meat and milk from non human sources may contribute to chronic inflammation, perhaps contributing to the increased risks of cardiovascular disease and carcinomas associated with these foods. Therefore, an in depth profiling method provides the necessary analytical tool for understanding the physiological effects of gangliosides in animal products and milk in the human body.43
Figure 2.
Separation of complex gangliosides on the nanoHPLC chip. (A) EIC of bovine milk ganglioside GD3 (d41:1) at m/z 769.952. Separation of the monoisotopic peak of NeuAc-NeuAc GD3 (d41:1) and the third isotopic peak of NeuAc-NeuAc GD3 (d41:2) was achieved. The contribution of 13C was removed by the LC. (B) EIC of NeuGc NeuAc GD3 (d40:1) at m/z 770.941. The separation of NeuAc containing gangliosides and NeuGc containing gangliosides was achieved. The first peak and second peak correspond to the monoisotope of NeuGc NeuAc GD3 (d40:1) and the third isotope of NeuAc-NeuAc GD3 (d41:1), respectively. All the ions are doubly depronated. The isotopic peaks of other compounds are indicated by asterisks (*). Inserts are the spectrum at each retention time.
To apply our method to a biological sample, we used samples from mouse brain, as it is known that neurological tissues are enriched with complex polar lipids.44 The chemically diverse brain membrane lipids are thought to play important roles in various functions. MS was used for untargeted profiling of the polar lipid fraction combined with accurate, mass based quantitation. Positive mode 40V and 80V CID provided the structures of both the headgroup and the ceramide tails. Post processing precursor ion scan of fragment ions allowed detection of major classes of the polar lipids in a single low CID energy experiment in the negative ion mode. The multiple precursor ion scans and reproducible retention time made accurate identification possible.
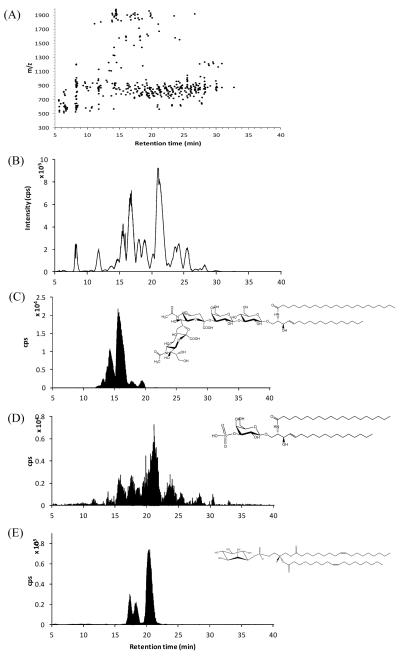
As many as 510 compounds from the mouse brain polar lipid fraction were revealed by the “Find Molecular Feature” algorithm. Figure 3(A) is a two dimensional representation with the mass spectra (m/z) on the y-axis and retention on the x-axis. A vertical line contains the mass spectra obtained at a specific retention time. Figure 3(B) is the base peak chromatogram (BPC) illustrating the approximate profile of the mixture. Due to the high mass accuracy and resolution, it was possible to extract an exact mass ion chromatogram with a narrow mass window of 10 ppm for the diagnostic ions of ganglioside, sulfatide and phosphatidylinositol headgroups. Sialic acid ions (m/z 290.095), bisulfate anions (m/z 96.960) and cyclic inositol phosphate ions (m/z 241.011) were used for ganglioside, sulfatide and phosphatidylinositol scans, respectively, allowing a focused investigation when searching for certain lipid classes. 17 ganglioside ions, 13 sulfatide ions and 15 phosphatidylinositol ions were detected and identified. Gangliosides, which are the most polar lipids in the sample, eluted early in the chromatogram (12 20min). Reproducible retention time allowed both quantitation and structural analysis with consecutive sample injections. Quantitation was performed in the MS mode based on the deprotonated molecular ion, while fragmentation patterns were obtained with MS/MS for the profiling and identification of the polar lipids. Lists of mouse ganglioside, sulfatide, and phosphatidylinositol with the ion intensities are summarized in Table 1 and S-Table 2. Ganglioside profiles showed that GD1 and GT1b are abundant ions in mouse brain. The presence of the O acetyl forms of the GD1, GT1b and GQ1b gangliosides is consistent with the results reported in previous research.34 The major ceramide portions were d18:1/18:0 and d20:1/18:0. GD1 is typically found as two glycan isomers, GD1a and GD1b. We could not separate these two species fully, although tandems MS indicate there are two isomeric glycan species (data not shown). Sulfatide species were also observed by the scan at m/z 96.960. The ceramide portions of sulfatides were unlike those of gangliosides. Both hydroxylated and non hydroxylated sulfatides, which are known to be involved in myelin stability, were observed in the mouse brain.45
Figure 3.
Detection and identification of individual classes of polar lipids in mouse brain extracts. (A) A two dimensional plot of m/z vs. retention time for mouse brain polar lipids. (B) Base peak chromatogram (BPC) from MS analysis, (C) post processing precursor ion scan for m/z 290.095 (gangliosides), (D) for m/z 96.960 (sulfatide), and (E) for m/z 241.011 (phosphatidylinositol). The fragment ions for the individual headgroup being characteristic of their species can be used for identification of their lipids within one class, followed by alignment with the BPC. Gangliosides, which are the most polar lipids, eluted early in the chromatography. EICs with 10 ppm window were used for the scans. Both BPC and EICs were obtained in the negative ion mode. Details are described in the experimental section.
Table 1.
Profiles of mouse brain (A) gangliosides and (B) sulfatides detected.
| (A) | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Retention time (min) |
Measured m/z |
Expected m/z |
Mass error (ppm) |
Charge state |
Structural Assignment | Peak Area |
| 12.8 | 1049.008 | 1049.011 | 2.9 | −2 | GT1b(d18:1/16:0) | 670588 |
| 13.7 | 1062.017 | 1062.019 | 1.9 | −2 | GT1b(d18:2/18:0 or d18:1/18:1) |
2316785 |
| 14.7 | 916.470 | 916.471 | 1.1 | −2 | GD1(d18:1/18:1) | 5262952 |
| 15.1 | 1208.570 | 1208.574 | 3.3 | −2 | GQ1b(d18:1/18:0) | 713035 |
| 15.5 | 1084.029 | 1084.032 | 2.8 | −2 | O-acetyl GT1b(d18:1/18:0) | 925711 |
| 15.6 | 708.348 | 708.349 | 1.4 | −3 | GT1b(d18:1/18:0) | 722114 |
| 15.2 | 805.379 | 805.380 | 1.2 | −3 | GQ1b(d18:1/18:0) | 428234 |
| 15.2 | 1229.573 | 1229.575 | 1.6 | −2 | O-acetyl GQ1b(d18:1/18:0) | 266227 |
| 15.2 | 819.383 | 819.384 | 1.2 | −3 | O-acetyl GQ1b(d18:1/18:0) | 241457 |
| 15.6 | 1063.027 | 1063.026 | −0.9 | −2 | GT1b(d18:1/18:0) | 14994553 |
| 15.8 | 734.911 | 734.913 | 2.7 | −2 | GD3(d18:1/18:0) | 709137 |
| 16.5 | 938.483 | 938.484 | 1.1 | −2 | O-acetyl GD1a(d18:1/18:0) | 486074 |
| 16.8 | 917.480 | 917.479 | −1.1 | −2 | GD1(d18:1/18:0) | 36420062 |
| 17.8 | 930.485 | 930.487 | 2.1 | −2 | GD1(d20:1/18:1) | 270179 |
| 18.1 | 1544.869 | 1544.869 | 0.0 | −1 | GM1(d18:1/18:0) | 523277 |
| 18.7 | 1077.039 | 1077.042 | 2.8 | −2 | GT1b(d20:1/18:0) | 1750304 |
| 20.0 | 931.494 | 931.494 | 0.0 | −2 | GD1(d20:1/18:0) | 2495305 |
| (B) | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Retention time (min) |
Measured m/z |
Expected m/z |
Mass error (ppm) |
Charge State |
Structural Assignment | Peak Area |
| 15.3 | 778.512 | 778.514 | 2.6 | −1 | Sulfatide(d18:1/16:0) | 412228 |
| 16.3 | 804.528 | 804.530 | 2.5 | −1 | Sulfatide(d18:1/18:1) | 252356 |
| 17.7 | 822.539 | 822.540 | 1.2 | −1 | Sulfatide(d18:1/h18:0) | 2109640 |
| 18.6 | 806.544 | 806.546 | 2.5 | −1 | Sulfatide(d18:1/18:0) | 7193764 |
| 21.1 | 834.578 | 834.577 | −1.2 | −1 | Sulfatide(d18:1/20:0) | 52812190 |
| 24.4 | 878.601 | 878.603 | 2.3 | −1 | Sulfatide(d18:1/h22:0) | 1119612 |
| 24.7 | 904.616 | 904.618 | 2.2 | −1 | Sulfatide(d18:1/h24:1) | 654469 |
| 25.2 | 862.605 | 862.608 | 3.5 | −1 | Sulfatide(d18:1/22:0) | 828654 |
| 25.5 | 888.623 | 888.624 | 1.1 | −1 | Sulfatide(d18:1/24:1) | 9052460 |
| 26.2 | 892.62 | 892.618 | −2.2 | −1 | Sulfatide(d18:1/h23:0) | 35049 |
| 27.5 | 906.632 | 906.634 | 2.2 | −1 | Sulfatide(d18:1/h24:0) | 990251 |
| 28.1 | 890.638 | 890.640 | 2.2 | −1 | Sulfatide(d18:1/24:0) | 2026942 |
| 28.3 | 930.485 | 920.650 | 1.1 | −1 | Sulfatide(d18:1/h25:0) | 10075 |
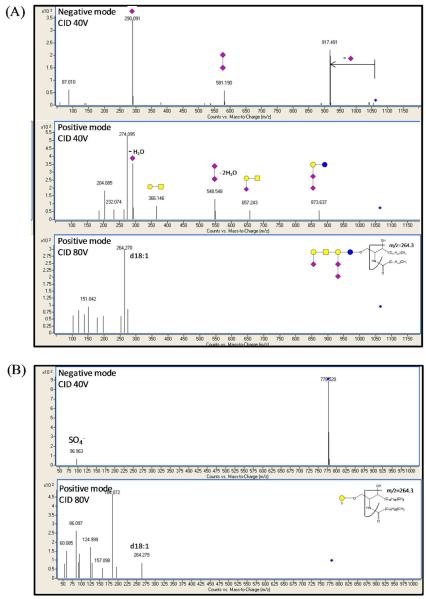
The structures of two peaks elucidated with 40V and 80V CID are shown in Figure 4. In the case of GT1b with sphingosine (d18:1) and palmitic acid (18:0), the glycan and ceramide structures were revealed in the lower and the higher energy CID spectra, respectively. Negative mode MS/MS spectra showed the diagnostic ion at m/z 290.091 had the greatest abundance, which is indicating the presence of sialic acieds. The ion is used for the post processing precursor ion scanning. Tandem MS in the positive mode provides more structural information. Positive mode low energy CID shows the cleavage at the glycosidic linkages, generating B type ions. Positive mode higher energy CID fragmentation was used for the identification of the sphingosine base and fatty acid backbone chains as the positive charge remains on the ceramide moieties. The 80V CID experiments yielded the ion at m/z 264.270, which corresponds to the sphingosine base ion. The product ion analysis revealed that the amide bond and dehydration of the sphingosine base are the most abundant fragment ions. From this product ion data information regarding the number of carbon atoms in the chain, degree of hydroxylation and unsaturation could be determined. With this knowledge about the sphingosine base composition and the original precursor ion m/z, the identity of the fatty acids was determined. For sulfatides, a diagnostic ion was observed at m/z 96.963 in the negative mode low energy CID, while higher energy CID yielded information about the sphingosine base.
Figure 4.
Low (40V) and higher (80V) collision energy MS/MS spectra of (A) ganglioside GT1b (d18:1/18:0) and (B) sulfatide (d18:1/16:0). Low energy spectra and high energy spectra contain various diagnostic ions for the headgroup and ceramide tails, respectively.
Conclusion
A nano-HPLC-Chip Q-TOF/MS was selected for this study due to the high retention time reproducibility and outstanding abundance reproducibility necessary for successful profiling experiments. While mass accuracy and resolution are important, so are dynamic range, sensitivity, and spectral acquisition speed. The Q-TOF used in this research routinely delivers excellent performance in all of these dimensions simultaneously without the requirement of compromise. To our knowledge, this is the first report to describe the separation of polar lipids through nano-HPLC-Chip in the negative ion mode. Interestingly, in order to reveal the complexity of the mouse brain polar lipids in a relatively accurate and simple manner, we further strengthened the method to include post processing multiple precursor ion scans. This proposed approach allowed for the quantitation and identification of intact polar lipids, including gangliosides, sulfatides, and phosphatidylinositols, opening a promising analytical strategy to uncovering the information hidden within complex lipid signaling.
Supplementary Material
ACKNOWLEDGMENT
This publication was made possible in part by grant support from the University of California Discovery Grant Program, the California Dairy Research Foundation, Dairy Management Inc., NIEHS Superfund P42 ES02710, the Charge study P01 ES11269, and by NIH NICID awards 5R01HD059127 and 1R01HD061923. The Agilent Q-TOF MS instrument was obtained through an NIH grant (S10RR027639).
REFERENCES
- (1).German JB, Gillies LA, Smilowitz JT, Zivkovic AM, Watkins SM. Curr. Opin. Lipidol. 2007;18:66–71. doi: 10.1097/MOL.0b013e328012d911. [DOI] [PubMed] [Google Scholar]
- (2).Wenk MR. Cell. 2010;143:888–895. doi: 10.1016/j.cell.2010.11.033. [DOI] [PubMed] [Google Scholar]
- (3).Rajendran L, Simons KJ. Cell Sci. 2005;118:1099–1102. doi: 10.1242/jcs.01681. [DOI] [PubMed] [Google Scholar]
- (4).Jacobson K, Mouritsen OG, Anderson RG. Nat. Cell. Biol. 2007;9:7–14. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
- (5).Di Paolo G, De Camilli P. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- (6).Schnaar RL. Glycobiology. 1991;1:477–485. doi: 10.1093/glycob/1.5.477. [DOI] [PubMed] [Google Scholar]
- (7).Kannagi R, Nudelman E, Hakomori S. Proc. Natl. Acad. Sci. USA. 1982;79:3470–3474. doi: 10.1073/pnas.79.11.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Hakomori S. Proc. Natl. Acad. Sci. USA. 2002;99:10231–10233. doi: 10.1073/pnas.172380699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Han X, D MH, McKeel DW, Jr., Kelley J, Morris JC. J. Neurochem. 2002;82:809–818. doi: 10.1046/j.1471-4159.2002.00997.x. [DOI] [PubMed] [Google Scholar]
- (10).Kroes RA, He H, Emmett MR, Nilsson CL, Leach FE, 3rd, Amster IJ, Marshall AG, Moskal JR. Proc. Natl. Acad. Sci. USA. 2010;107:12646–12651. doi: 10.1073/pnas.0909862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Rueda R. Br. J. Nutr. 2007;98(Suppl 1):S68–73. doi: 10.1017/S0007114507832946. [DOI] [PubMed] [Google Scholar]
- (12).Morrison IM. Encyclopedia of Analytical Chemistry. John Wiley & Sons, Ltd; 2006. Glycolipid Analysis. [Google Scholar]
- (13).Varki A. Essentials of glycobiology. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2009. [PubMed] [Google Scholar]
- (14).Merrill AH, Jr., Wang MD, Park M, Sullards MC. Trends Biochem. Sci. 2007;32:457–468. doi: 10.1016/j.tibs.2007.09.004. [DOI] [PubMed] [Google Scholar]
- (15).Ledeen RW, Yu RK. Methods Enzymol. 1982;83:139–191. doi: 10.1016/0076-6879(82)83012-7. [DOI] [PubMed] [Google Scholar]
- (16).Sampaio JL, Gerl MJ, Klose C, Ejsing CS, Beug H, Simons K, Shevchenko A. Proc. Natl. Acad. Sci. USA. 2011;108:1903–1907. doi: 10.1073/pnas.1019267108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Han X, Gross RW. Mass Spectrom. Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- (18).Han X, Gross RW. J. Lipid Res. 2003;44:1071–1079. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- (19).Bindila L, Katalinic J. Peter. Mass Spectrom. Rev. 2009;28:223–253. doi: 10.1002/mas.20197. [DOI] [PubMed] [Google Scholar]
- (20).Ninonuevo M, An H, Yin H, Killeen K, Grimm R, Ward R, German B, Lebrilla C. Electrophoresis. 2005;26:3641–3649. doi: 10.1002/elps.200500246. [DOI] [PubMed] [Google Scholar]
- (21).Chu CS, Ninonuevo MR, Clowers BH, Perkins PD, An HJ, Yin H, Killeen K, Miyamoto S, Grimm R, Lebrilla CB. Proteomics. 2009;9:1939–1951. doi: 10.1002/pmic.200800249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Froehlich JW, Barboza M, Chu C, Lerno LA, Jr., Clowers BH, Zivkovic AM, German JB, Lebrilla CB. Anal. Chem. 2011;83:5541–5547. doi: 10.1021/ac2003888. [DOI] [PubMed] [Google Scholar]
- (23).Gazzotti G, Sonnino S, Ghidoni R, Kirschner G, Tettamanti G. J. Neurosci. Res. 1984;12:179–192. doi: 10.1002/jnr.490120206. [DOI] [PubMed] [Google Scholar]
- (24).Ikeda K, Shimizu T, Taguchi R. J. Lipid Res. 2008;49:2678–2689. doi: 10.1194/jlr.D800038-JLR200. [DOI] [PubMed] [Google Scholar]
- (25).Sorensen LK. Rapid Commun. Mass Spectrom. 2006;20:3625–3633. doi: 10.1002/rcm.2775. [DOI] [PubMed] [Google Scholar]
- (26).Houjou T, Yamatani K, Imagawa M, Shimizu T, Taguchi R. Rapid Commun. Mass Spectrom. 2005;19:654–666. doi: 10.1002/rcm.1836. [DOI] [PubMed] [Google Scholar]
- (27).Yin H, Killeen K, Brennen R, Sobek D, Werlich M, van de Goor T. Anal. Chem. 2005;77:527–533. doi: 10.1021/ac049068d. [DOI] [PubMed] [Google Scholar]
- (28).Hardouin J, Duchateau M, Caron R. Joubert, Caron M. Rapid Commun. Mass Spectrom. 2006;20:3236–3244. doi: 10.1002/rcm.2725. [DOI] [PubMed] [Google Scholar]
- (29).Alley WR, Jr., Madera M, Mechref Y, Novotny MV. Anal. Chem. 2010;82:5095–5106. doi: 10.1021/ac100131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Chernushevich IV, Loboda AV, Thomson BA. J. Mass Spectrom. 2001;36:849–865. doi: 10.1002/jms.207. [DOI] [PubMed] [Google Scholar]
- (31).Ejsing CS, Duchoslav E, Sampaio J, Simons K, Bonner R, Thiele C, Ekroos K, Shevchenko A. Anal. Chem. 2006;78:6202–6214. doi: 10.1021/ac060545x. [DOI] [PubMed] [Google Scholar]
- (32).Ekroos K, Chernushevich IV, Simons K, Shevchenko A. Anal. Chem. 2002;74:941–949. doi: 10.1021/ac015655c. [DOI] [PubMed] [Google Scholar]
- (33).Schwudke D, Oegema J, Burton L, Entchev E, Hannich JT, Ejsing CS, Kurzchalia T, Shevchenko A. Anal. Chem. 2006;78:585–595. doi: 10.1021/ac051605m. [DOI] [PubMed] [Google Scholar]
- (34).Colsch B, Woods AS. Glycobiology. 2010;20:661–667. doi: 10.1093/glycob/cwq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Sugiura Y, Shimma S, Konishi Y, Yamada MK, Setou M. PLoS One. 2008;3:e3232. doi: 10.1371/journal.pone.0003232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).He H, Conrad CA, Nilsson CL, Ji Y, Schaub TM, Marshall AG, Emmett MR. Anal. Chem. 2007;79:8423–8430. doi: 10.1021/ac071413m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Zamfir A, Vukelic Z, Bindila L, Katalinic J. Peter, Almeida R, Sterling A, Allen M. J. Am. Soc. Mass Spectrom. 2004;15:1649–1657. doi: 10.1016/j.jasms.2004.08.002. [DOI] [PubMed] [Google Scholar]
- (38).Almeida R, Mosoarca C, Chirita M, Udrescu V, Dinca N, Vukelic Z, Allen M, Zamfir AD. Anal. Biochem. 2008;378:43–52. doi: 10.1016/j.ab.2008.03.039. [DOI] [PubMed] [Google Scholar]
- (39).Schiopu C, Flangea C, Capita F, Serb A, Vukelic Z, Bognar S. Kalanj, Sisu E, Prybylski M, Zamfir AD. Anal. Bioanal. Chem. 2009;395:2465–2477. doi: 10.1007/s00216-009-3188-8. [DOI] [PubMed] [Google Scholar]
- (40).Lee H, An HJ, Lerno LA, Jr., German JB, Lebrilla CB. Int. J. Mass Spectrom. 2011;305:138–150. doi: 10.1016/j.ijms.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Yin H, Killeen K. J. Sep. Sci. 2007;30:1427–1434. doi: 10.1002/jssc.200600454. [DOI] [PubMed] [Google Scholar]
- (42).Penn SG, Cancilla MT, Green MK, Lebrilla CB. Eur. Mass Spectrom. 1997;3:67–79. [Google Scholar]
- (43).Varki A. Proc. Natl. Acad. Sci. USA. 2010;107(Suppl 2):8939–8946. doi: 10.1073/pnas.0914634107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Piomelli D, Astarita G, Rapaka R. Nat. Rev. Neurosci. 2007;8:743–754. doi: 10.1038/nrn2233. [DOI] [PubMed] [Google Scholar]
- (45).Fewou SN, Bussow H, Wiemers N. Schaeren, Vanier MT, Macklin WB, Gieselmann V, Eckhardt M. J. Neurochem. 2005;94:469–481. doi: 10.1111/j.1471-4159.2005.03221.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.