Abstract
Ebolavirus causes severe hemorrhagic fever in humans and non-human primates. Entry of ebolavirus is mediated by the viral glycoprotein, GP; however, the required host factors have not been fully elucidated. A screen utilizing a recombinant Vesicular Stomatitis Virus (VSV) encoding Zaire ebolavirus GP identified four Chinese Hamster Ovary (CHO) cell lines resistant to GP-mediated viral entry. Susceptibility to vectors carrying SARS coronavirus S or VSV-G glycoproteins suggests that endocytic and processing pathways utilized by other viruses are intact in these cells. A cathepsin-activated form of the ebolaviral glycoprotein did not overcome the entry restriction, nor did expression of several host factors previously described as important for ebolavirus entry. Conversely, expression of the recently described ebolavirus host entry factor Niemann–Pick Type C1 (NPC1) restored infection. Resistant cells encode distinct mutations in the NPC1 gene, resulting in loss of protein expression. These studies reinforce the importance of NPC1 for ebolavirus entry.
Keywords: Ebolavirus, Viral entry, Ebolavirus GP, Niemann–Pick Type C1—NPC1, CHO-K1, Host factor, Viral glycoprotein
Highlights
► Selected CHO cell lines specifically resistant to ebolavirus GP-mediated infection. ► Host factors previously implicated in ebolavirus entry do not confer susceptibility. ► Resistant cells encode distinct mutations that knock out NPC1 protein expression. ► Expression of NPC1 restores infection. ► NPC1 mutations affecting cholesterol regulation but not protein expression were not selected.
Introduction
Ebolavirus is a non-segmented, negative-sense RNA virus of the Filoviridae family that is associated with outbreaks of hemorrhagic fever in humans and non-human primates in sub-Saharan Africa. Ebolavirus infects a wide range of cell types during an infection, including macrophages, dendritic cells, and hepatocytes. Entry into cells is mediated through the viral glycoprotein (GP), but the process is poorly characterized. Recent studies suggest that the virion enters the cell through macropinocytosis (Hunt et al., 2010, Nanbo et al., 2010, Saeed et al., 2010). Although the trigger for this uptake is not known, several host factors have been identified that could be involved in this step (Hunt et al., 2010, Kondratowicz et al., 2011). Once in the endosome, the virion encounters the cysteine proteases Cathepsin B (CatB) and Cathepsin L (CatL). These proteases cleave GP within a disordered loop (Chandran et al., 2005, Kaletsky et al., 2007, Schornberg et al., 2006). Although this cleavage appears to be necessary for entry, it is not sufficient to induce viral fusion with the host membrane. A subsequent step requiring cathepsins is also necessary (Kaletsky et al., 2007, Schornberg et al., 2006). The substrate of this second cathepsin cleavage has not been defined. Recently, Niemann–Pick Type C1 (NPC1) was identified as a host protein important for entry of Ebola virus (Carette et al., 2011, Cote et al., 2011). NPC1 is thought to interact with the processed form of GP (Cote et al., 2011), but a mechanism of action has not been determined.
The lack of knowledge about host factors involved in entry of ebolavirus has hindered the development of therapeutics targeting this process. Several different screening strategies have been used in the past to identify host factors involved in entry, with varying success. Chinese Hamster Ovary K1 (CHO-K1) cells are a functionally haploid cell line that is naturally genetically diverse. For decades, these properties of CHO-K1 cells have been exploited to isolate variants with alterations in specific processes, such as diphtheria intoxication (Moehring and Moehring, 1977). Such variants were subsequently used to elucidate the mechanism of the given process being studied.
Here we describe the isolation of CHO-K1 cell lines with natural mutations rendering the cells resistant to infection mediated by ebolavirus GP. Multiple clones derived from independent selections were obtained, and four were chosen for further characterization. The cell lines were analyzed for defects in established host factors involved in ebolavirus entry, and all four were found to have a defect in expression of NPC1. Exogenous expression of NPC1 restored susceptibility to infection in all four clones. Additionally, overexpression of several other purported ebolavirus entry factors did not overcome a defect in NPC1. Although NPC1 is not expressed in any of these clones, the mutations leading to this loss of expression are unique in each clone. The fact that we isolated multiple clones with distinct, natural mutations in the NPC1 gene lends weight to the recent evidence suggesting that NPC1 is a key host factor necessary for entry of ebolavirus.
Results
Isolation of CHO-K1 clones resistant to GP-mediated entry
A replication competent VSV encoding ebolavirus Zaire (ZEBOV) GP and mCherry in place of VSV G was constructed. Cellular entry of this recombinant virus, VSV EboGP mCherry, is mediated by the ebolaviral glycoprotein, similar to a previously described vaccine virus, VSVΔG/ZEBOV GP (Feldmann et al., 2007, Garbutt et al., 2004, Jones et al., 2005). The ebolavirus GP requirement for entry was confirmed through loss of infection in the presence of lysosomotropic reagents and CatB- and CatL-specific inhibitors (data not shown). Infection of susceptible cells with this recombinant virus results in efficient killing and is the basis for the selection strategy employed below.
The functionally haploid cell line CHO-K1 was used to identify cells resistant to infection with VSV EboGP mCherry using the strategy outlined in Fig. 1a. Infection of unmutagenized CHO cells with VSV EboGP mCherry at an MOI of 8 produced numerous colonies. In contrast, infection with VSV-GFP, a VSV vector expressing its own glycoprotein and a GFP marker, did not produce any viable colonies. This observation suggests that there is a high level of natural resistance to ebolaviral entry in CHO cells. After expansion, colonies were pooled and cloned by limiting dilution. Clonal cell lines were tested for susceptibility to VSV EboGP mCherry and VSV GFP. Four clonal cell lines (R1, R2, R4, and L2) that displayed complete resistance to VSV EboGP mCherry, while maintaining susceptibility to VSV GFP, were chosen for further characterization. These four cell lines were derived from resistant colonies isolated from separate plates challenged with VSV EboGP mCherry. The R2 and L2 cell lines display a similar morphology to wildtype (WT) CHO-K1 cells while R1 cells are much larger than WT cells with a more fibroblast-like appearance, and the R4 cells are larger and rounder than the CHO-K1 parent.
Fig. 1.
Isolation of cell lines resistant to GP-mediated infection. (a) Selection method for isolating CHO-K1 cells resistant to GP-mediated infection. (b) Resistant cell lines were challenged with VSV GFP or VSV EboGP mCherry. Percent cells infected was quantitated by flow cytometry. Bars represent average of means from three experiments done in triplicate; error bars represent standard error means.
The susceptibility profiles for clones R1, R2, R4, and L2 were confirmed by examining expression of mCherry or GFP via light microscopy at varying time points post-infection (PI) with VSV EboGP mCherry or VSV GFP, respectively (data not shown). Beginning 6–8 h PI, GFP is visible for R1, R2, R4, L2, and WT CHO-K1 cells infected with VSV GFP. By 24 h PI all cells are infected and most show a cytopathic effect from VSV replication. Infection of WT CHO-K1 cells with VSV EboGP mCherry produces a similar pattern. In contrast, for the R1, R2, R4, and L2 cell lines neither expression of the mCherry marker nor the cytopathic effects of VSV replication is seen with either high or low MOIs of VSV EboGP mCherry at any time point up to 48 h post-infection. Given the rapid kinetics of VSV replication in these cells, even low levels of infection would be apparent by 48 h. A more quantitative analysis using flow cytometry demonstrates that all four cell lines are effectively infected with VSV GFP (Fig. 1b). On the contrary, while an average of 33% of WT CHO-K1 cells express mCherry 12 h after challenge with VSV EboGP mCherry, less than 0.2% of R1, R2, R4, and L2 cells express mCherry (Fig. 1b). This data indicates that the R1, R2, R4, and L2 cells are resistant to infection mediated by Zaire ebolavirus GP, but not VSV G.
CHO-K1 clones are resistant to transduction with HIV pseudotypes bearing ZEBOV GP, but not VSV G
To ensure that the resistance of these clones was specific for GP-mediated entry, we examined whether the clones were susceptible to transduction with HIV pseudotypes encoding GFP and bearing VSV G (HIV/VSV G) or Zaire ebolavirus GP (HIV/ZEBOV GP). Cells were fixed 48 h PI and analyzed by flow cytometry for GFP expression. All four resistant clones had similar levels of transduction with HIV/VSV G as WT CHO-K1 cells, as determined by the percent of cells expressing GFP ( Fig. 2a). In contrast, while on average approximately 26% of WT CHO-K1 cells expressed GFP following transduction with HIV/ZEBOV GP, less than 2% of the resistant cells were transduced. This represents less than ten percent of the level of transduction of WT CHO-K1 cells (Fig. 2a). A similar result was seen when the resistant cells were challenged with HIV pseudotypes bearing Sudan ebolavirus (SEBOV) GP, Cote d'Ivoire ebolavirus (CIEBOV) GP, or Reston ebolavirus (REBOV) GP (Fig. 2b). There was no statistically significant difference in the level of infection for each cell line when comparing across filoviral glycoproteins. Taken together, these data indicate that the resistance to infection observed for R1, R2, R4, and L2 cells is specific for filoviral GP-mediated entry.
Fig. 2.
Resistance is specific to GP-mediated infection. Resistant cell lines were challenged with HIV pseudotypes encoding GFP and bearing VSV G, ZEBOV GP, ZEBOV GP′ (a) SEBOV GP, CIEBOV GP, or REBOV GP (b). Percent cells transformed was quantitated by flow cytometry. Bars represent average of means from three or four experiments done in triplicate; error bars represent standard error means. Asterick indicates statistically significant difference in infection level between GP and GP′ (p=0.04).
Ebolavirus GP is normally processed by endosomal cathepsins during entry into the host cell. This cleavage is believed necessary to activate the fusogenic potential of GP. To test whether the resistance to infection in the isolated CHO cell lines was due to a defect upstream of the ebolavirus GP proteolysis step, we challenged the resistant cells with an HIV pseudotype bearing a mutant form of GP that mimics processed GP (GP′, Fig. 2a). This mutant form, GP′, contains a furin recognition sequence, RRKR, at amino acids 203–206 (Fig. S1a). In the presence of furin, GP′ is cleaved at amino acid 207, producing a GP1 fragment that is six amino acids longer than the CatL processed species (Hood et al., 2010).
Cells were transduced with HIV/ZEBOV GP′ pseudotypes encoding GFP and analyzed by flow cytometry. Although GP′ incorporates into HIV virions at a ratio of approximately 0.5:1 compared to WT GP, pseudotypes bearing GP′ are more infectious (see Fig. 2a, WT cells). The increased susceptibility of WT CHO-K1 cells to HIV/ZEBOV GP′ (average of 45% of cells infected compared to 26% with WT GP) was not seen in the resistant cell lines. The lack of an increase in susceptibility for the resistant cell lines indicates that the resistance to infection seen in these cell lines could not be overcome by prior cleavage of ebolavirus GP (Fig. 2a).
Resistant CHO-K1 cells can bind ZEBOV GP-bearing virus at similar levels to that of WT CHO-K1 cells
To determine whether these cell lines are resistant due to reduced viral binding to the cell surface, we performed a binding assay with HIV/VSV G and HIV/ZEBOV GP. Virus was spun onto cells at 4 °C, followed by washes with cold PBS to remove any unbound virus prior to lysing the cells. Cell lysates were analyzed by quantitative Western blot for HIV p24 levels and compared to a viral input control. The level of binding of HIV/VSV G and HIV/ZEBOV GP pseudotypes to the R1, R2, R4, and L2 cell lines was not statistically significant compared to WT CHO-K1 cells ( Fig. 3), indicating that binding efficiency is not altered in the resistant cells.
Fig. 3.
Pseudotypes bind equally well to resistant cell lines and WT CHO-K1 cells. HIV pseudotypes bearing ZEBOV GP or VSV G were centrifuged onto cells at 4 °C. Unbound virus was washed away and cell lysates were collected. The percent of particles that bound to the cell surface was determined by comparing HIV p24 levels in the cell lysates to the amount of input virus by quantitative fluorescent Western blot. Bars represent average of means from three experiments done in triplicate; error bars represent standard error means.
Cell lines expressing active forms of cathepsin B and cathepsin L remain resistant to ebolavirus entry
As mentioned above, filoviruses require cleavage of GP by CatB and/or CatL to infect cells. Therefore, we tested whether the R1, R2, R4, and L2 cell lines were resistant to GP-mediated infection due to a loss of expression of CatB or CatL. Cells were transfected with a human CatB or human CatL expression plasmid, challenged with VSV EboGP mCherry, and analyzed by flow cytometry. Fig. 4c represents the combined results from three separate experiments. While the percent of cells expressing mCherry was similar to background levels for all conditions with the resistant cell lines, in two of the three experiments there was a small but statistically significant increase in infection for the R4 cells when comparing empty vector to CatL transfected cells (Fig. 4c). However, the percent of cells expressing mCherry was still less than 1%, suggesting that this minor increase is likely not biologically relevant. The lack of infection of R1, R2, R4, and L2 cells in the presence of overexpression of CatB or CatL indicated that a loss of expression of CatB or CatL is not the cause for the resistance to GP-mediated infection. However, this analysis did not rule out the possibility that the cells lacked the ability to activate CatB or CatL.
Fig. 4.
Resistant cells do not have defect in CatB or CatL. Overexpression of human CatB (a) or human CatL (b) in resistant cell lines. The procathepsin (Pro), single chain (SC), and double chain (DC) forms are indicated. (c) Resistant cell lines were transiently transfected with an expression plasmid for human CatB or CatL and challenged with VSV EboGP mCherry. Percent cells infected was quantitated by flow cytometry. Bars represent averages from three experiments done in triplicate, error bars represent standard error mean. Asterick indicates statistically significant difference in infection level between CatL and empty vector, p=0.03. (d) Transduction of WT CHO-K1 cells and resistant cell lines with VSV pseudotypes bearing ZEBOV GP or SARS S, as measured by Renilla luciferase activity. Bars represent average of means from three experiments done in triplicate; error bars represent standard error means.
Activation of CatB and CatL requires two sequential processing steps of their procathepsin form to a double chain form. All three forms of the protein, the procathepsin (Pro), the single chain (SC) and the fully active double chain (DC), can be visualized by Western blot analysis using cathepsin antibodies. To determine whether the R1, R2, R4, and L2 cells are capable of processing cathepsins into their active forms, lysates from cells transfected with human CatB or CatL expression plasmids were analyzed by Western blot. Transfected R1, R2, R4, and L2 cells express all three forms of human CatB, indicating that CatB is fully processed in these cells (Fig. 4a). The antibody against CatL recognizes both human and hamster CatL. Therefore, the banding pattern on a Western blot is more complex. However, as can be seen in Fig. 4b, all four resistant clones clearly expressed the active form of endogenous CatL. Together, this data suggests that R1, R2, R4, and L2 cells can process CatB and CatL into their active forms.
Similar to filoviruses, severe acute respiratory syndrome (SARS) coronavirus (CoV) requires cleavage of its glycoprotein, spike (S), by CatL to enter cells (Simmons et al., 2005). Therefore, to directly address whether the cathepsin pathway in R1, R2, R4, and L2 cells can function to activate viral glycoproteins, we challenged the ebolavirus resistant clones with VSV pseudotypes encoding luciferase and bearing SARS-CoV S or ZEBOV GP. Chinese hamster cells lack functional surface receptors for SARS-CoV. Accordingly, the ebolavirus-resistant cell lines were transiently transfected with an expression plasmid for human ACE2, the receptor for SARS-CoV, 48 h prior to transduction with VSV/SARS S or VSV/ZEBOV GP. Untransfected cells were also challenged with VSV/SARS S as a negative control. R1, R2, R4, and L2 cells expressing ACE2 displayed high levels of luciferase activity following challenge with VSV/SARS S (Fig. 4d). Conversely, as before, these cell lines were refractory to transduction mediated by ebolavirus GP, with only background levels of luciferase activity seen following challenge with VSV/ZEBOV GP. These background luciferase values for the mutant cells lines were not statistically different compared to those seen with VSV/SARS S transduction in the absence of human ACE2. The high level of transduction of all four clones with VSV/SARS S indicates that the endosomal uptake and CatL pathways are intact and do not appear to be the mechanism for resistance of these cell lines.
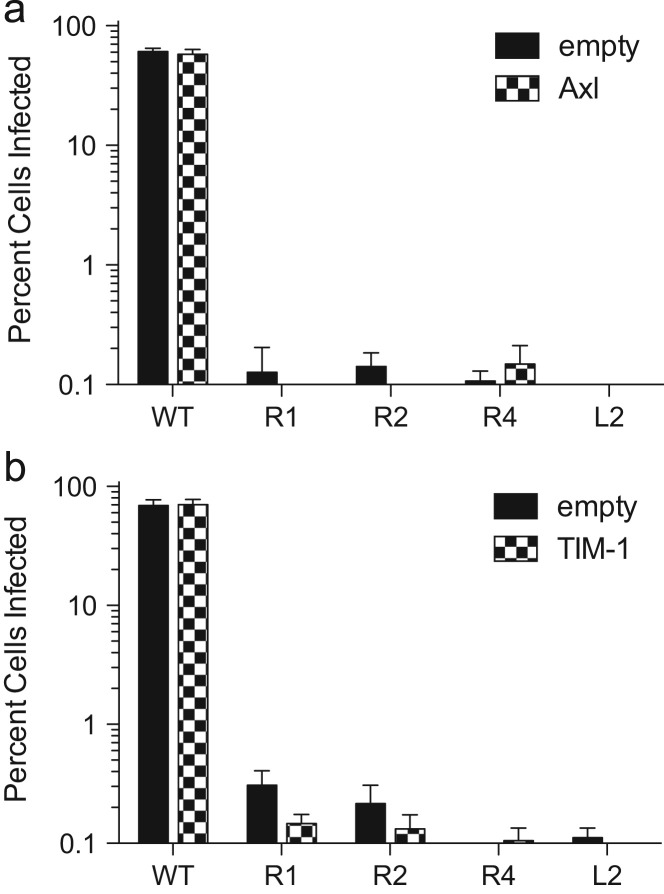
Overexpression of Axl or TIM-1 does not rescue infectivity
In addition to the cathepsins, several other host factors have been identified as potentially important for ebolavirus GP-mediated entry, including Axl and TIM-1 (Brindley et al., 2011, Kondratowicz et al., 2011, Shimojima et al., 2007, Shimojima et al., 2006). To determine if the ebolavirus-resistant cell lines had a defect in either of these proteins, we transfected WT, R1, R2, R4, and L2 cells with expression plasmids for Axl and TIM-1. Expression of Axl and TIM-1 was confirmed, and the cells were subsequently infected with VSV EboGP mCherry and analyzed by flow cytometry. Similar to earlier results seen with overexpression of CatB and CatL, the level of infection for all of the ebolavirus resistant cell lines was not statistically significant when comparing cells expressing Axl or TIM-1 versus empty vector ( Fig. 5). This indicates that a defect in expression of Axl or TIM-1 is likely not the cause of the resistance of the R1, R2, R4, and L2 clones. Moreover, expression of these factors cannot compensate for the deficit in the ebolavirus-resistant cells.
Fig. 5.
Overexpression of Axl or TIM-1 does not restore susceptibility. Resistant cell lines were transiently transfected with an expression plasmid for human Axl (a) or human TIM-1 (b) challenged with VSV EboGP mCherry, and analyzed by flow cytometry for expression of mCherry. Bars represent means from two experiments done in triplicate; error bars represent standard error means.
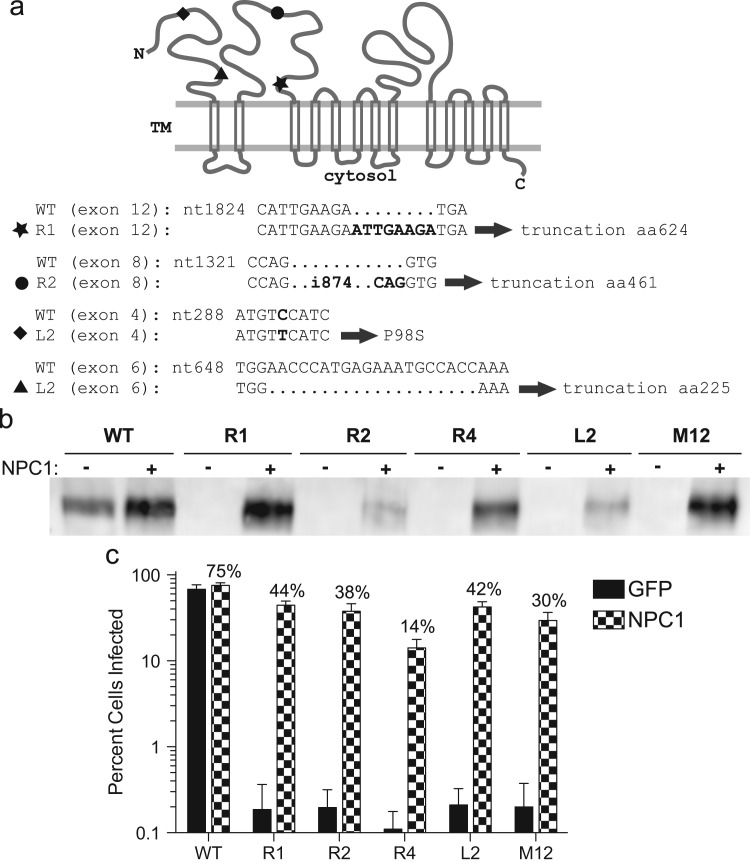
All resistant clones have mutations in the NPC1 gene
Recently the endosomal-resident protein NPC1 was shown to be important for Ebola infection (Carette et al., 2011, Cote et al., 2011). Therefore, we analyzed the resistant CHO-K1 cell lines for defects in NPC1 expression. Expression of full-length NPC1 is readily evident by Western blot analysis of WT CHO-K1 cells ( Fig. 6b) whereas no expression is seen in the four ebolavirus-resistant cell lines. To determine the basis for the loss of expression of NPC1 in the resistant cells we performed RT-PCR on total cellular RNA. Sequence analysis of the cDNA for three of the clones, R1, R2, and L2, reveals that they contain unique frameshift mutations in the NPC1 gene. The R1 cell line encodes an 8-nucleotide (nt) duplication in exon 12, starting at nt 1832 and resulting in a truncated protein predicted to be 624 amino acids long. The R2 cell line contains an 874-nt insertion in exon 8 flanked by a 3-nt repeat (see Fig. 6a). This insert results in a predicted 466 aa product. L2 contains a substitution (P98S) in exon 4 and a 20-nt deletion in exon 6. The deletion results in a truncation of the protein after NPC1 residue number 225. RT-PCR analysis using several sets of primers failed to detect an intact transcript encoding NPC1 in the R4 cell line, suggesting there is a large deletion or major rearrangement of the locus in these cells.
Fig. 6.
Resistant cell lines have mutations in NPC1. (a) Schematic of NPC1 protein. Location of insertions in R1 (star) and R2 (circle), and deletion (triangle) and substitution (diamond) in L2 indicated in diagram at top and in sequence at bottom. (b) Expression of NPC in WT, M12, and resistant cells transduced with pLenti CMV NPC1 Hygro or pLenti CMV GFP Hygro. (c) Resistant cells stably expressing NPC1 or GFP were challenged with VSV EboGP mCherry and infection was quantitated by flow cytometry. Bars represent average of means from three separate experiments done in triplicate; errors bars represent standard error means.
To establish whether the loss of expression of NPC1 in the R1, R2, R4, and L2 cell lines is the cause for their ebolavirus-resistant phenotype, we generated cells that stably express human NPC1. A lentiviral vector encoding the Hygromycin B resistance gene and NPC1 or GFP was used to transduce parental WT CHO-K1 and the four resistant cell lines. Hygromycin B selected cells were used for protein analysis by Western blot or challenged with VSV EboGP mCherry and analyzed by flow cytometry. Varying, but readily detectable, levels of NPC1 were seen in all of the Hygromycin selected cell lines (Fig. 6b). In all cases, stable expression of NPC1 was able to at least partially restore susceptibility to ebolavirus GP-mediated infection (Fig. 6c). The transduced R1, R2, and L2 cell lines were infected by VSV EboGP mCherry at roughly 49–60% that of WT cells (Fig. 6c). While the restoration of susceptibility was less efficient in the R4 line, with an average infection level of about 19% of WT, this level was still approximately100-fold above that seen without NPC1 expression. A similar trend with the R4 clone showing the lowest degree of restoration was seen when the cells transiently expressed NPC1 (data not shown). By comparison, stable expression of NPC1 in M12 cells, a CHO cell line carrying a deletion of the NPC1 locus (Millard et al., 2000), resulted in ∼40% level of infection in WT cells. While the basis for the varying degrees of susceptibility in the transduced cells is unclear, these results demonstrate that restoration of NPC1 expression in these cell lines confers susceptibility to ebolavirus infection.
Discussion
Multiple screening strategies, including genetic complementation and comparative gene expression analysis, have been used in the past to identify host factors important for entry of ebolavirus. Often the results were ambiguous, with only a modest increase in infection seen in the presence of the host factor. In some instances the predicted host factor was not expressed in all susceptible cell types (Chan et al., 2001, Kondratowicz et al., 2011).
In this study we employed an alternative approach by selecting for natural mutations that rendered cells normally susceptible to infection resistant. Because CHO-K1 cells are functionally haploid at most alleles, a mutation in a single allele of a gene is likely to result in a phenotypic effect on the affected protein. By not mutagenizing the cells prior to selection, we limited our screen to naturally occurring mutations that disrupted a function vital for viral entry. In doing so, we also increased the likelihood of selecting for genetic variants with mutations in a single gene. Four independent cell lines that demonstrated complete resistance to infection with VSV EboGP mCherry were identified and selected for further characterization.
Previously, another CHO-K1 clone, CHO B2, was also shown to be resistant to ebolavirus GP-mediated entry (Schornberg et al., 2009). This resistance was attributed to a defect in processing of CatB and CatL caused by a lack of expression of α5β1 integrin on the cell surface. Unlike CHO B2 cells, our cell lines express the active, DC form of CatB and CatL. Furthermore, the R1, R2, R4, and L2 cell lines are susceptible to infection by pseudotypes bearing the SARS CoV spike protein, confirming that both endosomal internalization and CatL are functional in these cells. Although there was a slight, albeit statistically significant, increase in susceptibility of the R4 cell line to VSV EboGP mCherry when human CatL was overexpressed, the level of infection was still less than one percent, suggesting that the increase is not biologically relevant.
Several host factors, in addition to CatL and CatB, have been identified previously as being important for GP-mediated entry of Ebola, including Axl and TIM-1 (Brindley et al., 2011, Kondratowicz et al., 2011, Shimojima et al., 2007, Shimojima et al., 2006). Overexpression of these proteins in our resistant cell lines did not restore susceptibility, suggesting that the R1, R2, R4, and L2 cells do not have a defect in either of these genes. Recent evidence demonstrates that Axl can facilitate entry of some viruses by a Gas6/phosphatidylserine mediated mechanism resembling apoptotic body uptake, and this process is most evident when a primary entry pathway is disrupted or impaired (Morizono et al., 2011). Axl can also provide an alternative entry pathway for Lassa virus in cells deficient for the primary receptor α-dystroglycan by a process that does not require Gas6 (Shimojima et al., 2012). Our results indicate that Axl overexpression does not provide an alternative means of entry into NPC1 deficient, ebolavirus-resistant mutant CHO-K1 cell lines.
During entry into target cells, ebolavirus likely uses multiple host factors to accomplish the steps of attachment, uptake, trafficking, and fusion. Defects in host proteins involved in any of these steps could potentially confer cellular resistance to infection. Consequently, the relatively unbiased selection method employed here could result in isolation of mutants with defects in several different genes, as was seen with a similar screen looking at diphtheria toxin resistance (Moehring and Moehring, 1977). However, all four of the independently isolated ebolavirus GP-resistant CHO cell lines contained mutations in the same gene, NPC1. Although all of the resistant cell lines characterized had mutations in NPC1, the mutations were all unique, indicating that they arose from discrete events. Furthermore, exogenous expression of NPC1 restored susceptibility of the resistant cells, indicating that the loss of expression of NPC1 was sufficient to render these cells resistant. It is possible that these resistant cell lines could have an additional defect in another gene. However, given the high level of restoration seen with exogenous expression of NPC1, any such mutation is likely to only play a minor role, if any, in susceptibility to entry.
Recently, two groups independently identified NPC1 as a key component in ebolavirus GP-mediated infection (Carette et al., 2011, Cote et al., 2011). Carette et al. used an insertional mutagenesis approach to identify cells resistant to infection by ebolavirus GP. Although they utilized a recombinant VSV-EboGP vector similar to the one employed here, the insertional mutagenesis strategy employed could have biased the results toward NPC1 because of the large target size (>55 kilobases) of the NPC1 gene. Similarly, the studies by Cote and colleagues used a compound library screen to identify NPC1, and thus were biased toward “druggable” targets. Our findings, using a selection strategy with a bias for naturally occurring mutations, also identified NPC1 in multiple ebolavirus-resistant cell lines. Despite performing a selection screen with VSV-EboGP using four independent pools of CHO-K1 cells, and in contrast to the selection screen of Carette et al. (2011), the current studies did not identify any other targets of ebolavirus entry aside from NPC1. The fact that our screen did not identify the HOPS (hemolytic fusion and protein sorting) endosomal sorting complex proteins found by Carette et al. (2011) may suggest that CHO cells with defects in these genes are rare or that the screen was not saturating. Taken as a whole, three strategies, each with a different selection bias, identified the same host factor, confirming the critical role of NPC1 in Ebola entry.
NPC1, an important player in cholesterol homeostasis, is a 1278-amino acid protein that has 13 predicted membrane-spanning domains and is normally localized within late endosomes. Mutational analysis of NPC1 in human patients has revealed over 100 different alterations, widely distributed throughout the gene, which affect cholesterol regulatory function (Carstea et al., 1997, Garver and Heidenreich, 2002). The majority are missense mutations, several of which result in essentially normal amounts of NPC1 protein but impair cholesterol control (Millat et al., 2001, Ribeiro et al., 2001). Missense and deletion mutations that affect NPC1 localization to the late endosome have also been identified (Blom et al., 2003). However, in our studies all of the CHO mutants selected for resistance to ebolavirus GP abrogated NPC1 protein expression. This suggests that simple mutations that impair the cholesterol regulatory function of NPC1 do not cripple the protein's role in ebolaviral entry.
Conclusions
We demonstrate that CHO cells selected for resistance to infection mediated by the ebolavirus glycoprotein harbor defects in NPC1. In four independent cell lines examined, the mutation within the NPC1 gene aborgates expression of the NPC1 protein and restoration of protein expression restores sensitivity to ebolavirus GP mediated infection. These results underscore the importance of NPC1 in ebolaviral entry.
Materials and methods
Cell culture and plasmids
CHO-K1 cells were maintained in Kaighn's modified F-12K nutrient mixture with 10% heat-inactivated fetal bovine serum (FBS), L-glutamine, penicillin, and streptomycin. M12 cells were kindly provided by Daniel Ory (Millard et al., 2000). BHK cells were maintained in Eagle's Minimum Essential Medium (MEM) with 10% FBS, penicillin, and streptomycin. Vero and 293T cells were maintained in high glucose Dulbecco's modified Eagle's medium (DMEM) with 10% FBS, penicillin, and streptomycin. All cultures were grown in 5% CO2.
pCAGGS-EboGP-V5 (strain Zaire 1976 Mayinga), pCB6-VSV G, pCB6-EboZ GP, pCAGGS SARS S and pSPAX have been previously described (Simmons et al., 2002, Simmons et al., 2004). pCAGGS-EboGPrrkr-V5 was generated by site directed mutagenesis of pCAGGS-EboGP-V5. pLenti CMV GFP Hygro, hCathepsinL, and hCathepsinB were obtained from Addgene (Cambridge, MA). The cDNA for TIM-1 was obtained from Open Biosystems and was cloned into pLKO by PCR. VSV-XN2 rLuc, VSV-L, VSV-N, and VSV-P expression plasmids, all under the control of the T7 promoter, were kindly provided by Robert Doms. VSV-XN2 EboGP mCherry was made by overlapping extension PCR. The flanking primer at the 5′ end contained an MluI restriction site and was used to amplify ebolavirus Zaire GP. The flanking primer at the 3′ end contained an NheI restriction site and was used to amplify mCherry. The internal primers were designed such that an additional junction sequence, flanked by KpnI and XhoI, was inserted between GP and mCherry. The rLuc gene was removed from VSV-XN2 rLuc by cleavage with MluI and NheI and replaced with the GP-KpnI-junction-XhoI-mCherry sequence. pLenti CMV NPC1 Hygro was constructed by removing the GFP gene from pLenti CMV GFP Hygro and inserting into its place the NPC1 cDNA (Origene). pSport6 Axl (clone ID 5205825) was from Open Biosystems. pcDNA3.1 furin and pcDNA3.1-ACE2 were previously described (Simmons et al., 2005, Wool-Lewis and Bates, 1999).
Generation of VSV EboGP mCherry
VSV EboGP mCherry was generated using a protocol similar to that described in Lawson et al. with the following changes (Lawson et al., 1995). Briefly, early passage BHK cells were infected with Modified Vaccinia Ankara T7 (MVA-T7), a gift from Bernard Moss. After 30 min cells were transfected with VSV-XN2 EboGP mCherry, pVSV-N, pVSV-L, and pVSV-P using Lipofectamine 2000 (Invitrogen). Three days post-transfection cells were scraped from the dish. Cells and supernatant were subjected to three rounds of freeze-thawing. The resulting mixture was spun at 1250g to remove cellular debris and then passed through a 0.2 um filter before being added to naïve BHK cells. Clarified and filtered supernatant was passaged onto BHK cells two additional times. After three rounds of infection on BHK cells, supernatant was transferred to naïve Vero cells along with neutralizing antibodies (VMC-2 and VMC-25, kindly provided by Gary Cohen and Roz Eisneberg) against Vaccinia. Antibodies were included in two additional passages on Vero cells to ensure removal of MVA.
Working stocks of VSV EboGP mCherry and VSV GFP were expanded on Vero cells grown in 850 cm2 roller bottles. Supernatant was clarified by centrifugation at 1250g for 5 min followed by ultracentrifugation at 6000 rpm in an SW28 rotor for 10 min at 4 °C. Supernatant was then transferred to new tubes and concentrated through a 20% sucrose cushion by ultracentrifugation at 28,000 rpm for two hours in an SW28 rotor at 4 °C. Viral Pellets were resuspended in hepes-buffered saline (HBS) at 4 °C overnight and stored at −80 °C. Concentrations for both viral stocks were determined by plaque assay on Vero cells. VSV GFP was kindly provided by Sara Cherry.
Isolation of ebolavirus GP-resistant CHO-K1 cells
CHO-K1 cells were plated in 10 cm plates. At ∼85% confluency, cells were infected with VSV EboGP mCherry at an MOI of 8. Plates were maintained for three weeks until resistant colonies contained between 100 and 500 cells. Colonies were transferred to 96-well plates using sterile paper discs soaked in trypsin. Cells were allowed to grow to confluence before being re-plated at approximately one cell per well of a 96-well plate. Clones were expanded and challenged separately with VSV EboGP mCherry and VSV GFP. Four cell lines, R1, R2, R4, and L2, originating from resistant colonies isolated from separate 10 cm plates, were chosen for further analysis.
Pseudovirion preparation
To make human immunodeficiency virus (HIV) pseudotypes, 293T cells were transfected using a standard calcium phosphate method with 8 μg of the HIV vector pSPAX, 8 μg of pLenti CMV GFP Hygro, and either 6 μg of pCB6-VSV G or 10 μg of pCAGGS-Ebola GP-V5. For HIV/ZEBOV GPrrkr, 8 μg of pSPAX, 8 μg of pLenti CMV GFP Hygro, 20 μg of pCAGGS-Ebola GPrrkr-V5, and 4 μg of pcDNA3.1 furin were used. Viral supernatants were collected 48 h post-transfection and concentrated through a 20% sucrose cushion by ultracentrifugation at 28,000 rpm in an SW28 rotor for 2 h at 4 °C. Pellets were resuspended in HBS at 4 °C overnight. Incorporation of ebolavirus glycoproteins was determined by quantitative western blotting with either a polyclonal anti-V5 rabbit antibody (Bethyl) or a polyclonal anti-GP1 rabbit antiserum. VSV pseudotypes were made by Graham Simmons as previously described (Ray et al., 2010). Briefly, 293T cells were transfected with pCAGGS SARS S or pCB6-EboZ GP 30 h prior to infection with VSVΔG virions derived from VSV-XN2 rLuc. Supernatant was harvested 48 h post-infection, filtered through a 0.45 μm filter, and stored at −80 °C.
Determination of specificity of resistance
All infections were done in triplicate. Cells were plated in 24-well plates at equivalent densities. For VSV EboGP mCherry and VSV GFP, 2.15×106 or 2.35×105 plaque forming units (pfu), respectively, were added to each well. Cells were fixed 12 h later and analyzed by flow cytometry for mCherry or GFP expression. For HIV pseudotypes, cells were transduced with equivalent amounts of virus based on quantitative western blotting with antibody specific for HIV-1 p24 (NIH AIDS Research and Reference Program, Cat#6458). Cells were fixed 48 h post-challenge and analyzed by flow cytometry for GFP expression.
Flow cytometry
CHO-K1 cells were detached from the plate with phosphate-buffered saline lacking Ca++ and Mg++ (PBS−/−) but containing 0.5 mM EDTA. Cells were pelleted at 4 °C for 2 min at 1100 rpm then resuspended in flow wash buffer (PBS−/− with 1% bovine calf serum and 0.05% sodium azide) with 2% paraformaldehyde. For each sample, 10,000 events were collected using a Becton Dickinson LSRII and analyzed using FlowJo software (Tree Star, Inc.).
Virus binding assay
CHO cells grown to confluence in 24-well plates were cooled to 4 °C. Equivalent amounts of particles (based on HIV p24 levels) were spun onto cells at 1250g for 30 min at 4 °C. Cells were then washed twice with cold PBS−/− and lysed in 30 μl of lysis buffer (1% Triton X-100, 50 mM Tris pH 8, 5 mM EDTA, 150 mM NaCl) with complete protease inhibitor cocktail (Roche). Lysates were cleared by centrifugation at 13,000 rpm for 2 min at 4 °C. Entire lysates were mixed with reducing SDS buffer, boiled for 5 min, and separated on a 4–15% Criterion PAGE gel (Bio-Rad). Proteins were transferred to a PVDF membrane (Millipore). Membranes were probed with a monoclonal antibody against HIV-1 p24. Protein was detected with goat anti-mouse IRdye800CW (Rockland) and viewed on a LI-COR Odyssey scanner. To determine the percent of input virus that bound to cells, p24 levels from lysates were compared to the p24 level in a lane containing the same amount of virus as was initially added to each well of cells.
Overexpression assays for CatL, CatB, Axl and TIM-1
WT and resistant CHO-K1 cells in 6-well plates were transfected with 1–2 μg of hCathepsinL, hCathepsinB, pSport6 Axl, pLKO TIM-1, or pcDNA3.1(+) using Lipofectamine LTX with Plus reagent. Cells were re-plated into 24-well plates 24 h post-transfection. At 48 h post-transfection cells were infected with VSV EboGP mCherry. Cells were fixed 12 h post-infection and analyzed by flow cytometry for mCherry expression. Protein expression was confirmed by Western blotting using antibodies specific for CatL, CatB (Athens Research and Technology), Axl, or TIM-1 (R&D Systems).
Generation of stable cell populations expressing human NPC1
WT CHO-K1, M12, and resistant CHO clones were challenged with HIV pseudotypes bearing VSV G and containing a plenti CMV NPC1 Hygro vector. At 48 h post-infection transduced cells were selected with 500 μg/ml Hygromycin B. Stable Hygromycin-resistant cells were subsequently infected with VSV EboGP mCherry and analyzed by flow cytometry as described above.
Luciferase assay
WT and resistant CHO-K1 cells in 6-well plates were transfected with 1 μg of pcDNA3.1-ACE2 using Lipofectamine LTX and Plus reagent. Transfected cells were re-plated into 96-well plates 24 h later and infected with equal amounts of VSV/ZEBOV GP or VSV/SARS S supernatant in triplicate 48 h post-transfection. At 12 h post-infection, media was removed from the wells and replaced with 20 ul Renilla Luciferase Assay Lysis Buffer (Promega). Plates were rocked for 30 min before lysates were transferred to new wells containing 100 μl of Renilla Luciferase Assay Reagent. Luminescence was measured on a Luminoskan Ascent (Thermo Electron Corporation) with Ascent 2.5 software.
Statistical analysis
For the luciferase and binding assays, data was analyzed by repeated measures, two-way ANOVA with Bonferroni post-tests using GraphPad PRISM software. All other data was analyzed by permutation test with a significance cut off of p=0.05. Number of permutations calculated ranged from 67 to 162.
Acknowledgments
The authors would like to thank Sudil Mahendra for assistance with the virus binding assays, Kenneth Briley, Jr., for assistance in generating the NPC1 lentiviral vector, Shane Jensen for assistance with the statistical analysis, and members of the Bates lab for comments on the manuscript. Thanks also to Sara Cherry, Daniel Ory, Bob Doms, Graham Simmons, Roz Eisenberg and Gary Cohen who provided reagents. This work was funded by Public Health Service Grants T32AI007324 (KH and NHVB), T32AI055400 (JRF and RLK), and R01AI081913 and U54AI57168 (PB).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2012.05.018.
Appendix A. Supplementary materials
References
- Blom T.S., Linder M.D., Snow K., Pihko H., Hess M.W., Jokitalo E., Veckman V., Syvanen A.C., Ikonen E. Defective endocytic trafficking of NPC1 and NPC2 underlying infantile Niemann–Pick type C disease. Hum. Mol. Genet. 2003;12:257–272. doi: 10.1093/hmg/ddg025. [DOI] [PubMed] [Google Scholar]
- Brindley M.A., Hunt C.L., Kondratowicz A.S., Bowman J., Sinn P.L., McCray P.B., Jr., Quinn K., Weller M.L., Chiorini J.A., Maury W. Tyrosine kinase receptor Axl enhances entry of Zaire ebolavirus without direct interactions with the viral glycoprotein. Virology. 2011;415:83–94. doi: 10.1016/j.virol.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J.E., Raaben M., Wong A.C., Herbert A.S., Obernosterer G., Mulherkar N., Kuehne A.I., Kranzusch P.J., Griffin A.M., Ruthel G., Dal Cin P., Dye J.M., Whelan S.P., Chandran K., Brummelkamp T.R. Ebola virus entry requires the cholesterol transporter Niemann–Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstea E.D., Morris J.A., Coleman K.G., Loftus S.K., Zhang D., Cummings C., Gu J., Rosenfeld M.A., Pavan W.J., Krizman D.B., Nagle J., Polymeropoulos M.H., Sturley S.L., Ioannou Y.A., Higgins M.E., Comly M., Cooney A., Brown A., Kaneski C.R., Blanchette-Mackie E.J., Dwyer N.K., Neufeld E.B., Chang T.Y., Liscum L., Strauss J.F., 3rd, Ohno K., Zeigler M., Carmi R., Sokol J., Markie D., O'Neill R.R., van Diggelen O.P., Elleder M., Patterson M.C., Brady R.O., Vanier M.T., Pentchev P.G., Tagle D.A. Niemann–Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science. 1997;277:228–231. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- Chan S.Y., Empig C.J., Welte F.J., Speck R.F., Schmaljohn A., Kreisberg J.F., Goldsmith M.A. Folate receptor-alpha is a cofactor for cellular entry by Marburg and Ebola viruses. Cell. 2001;106:117–126. doi: 10.1016/s0092-8674(01)00418-4. [DOI] [PubMed] [Google Scholar]
- Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote M., Misasi J., Ren T., Bruchez A., Lee K., Filone C.M., Hensley L., Li Q., Ory D., Chandran K., Cunningham J. Small molecule inhibitors reveal Niemann–Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann H., Jones S.M., Daddario-DiCaprio K.M., Geisbert J.B., Stroher U., Grolla A., Bray M., Fritz E.A., Fernando L., Feldmann F., Hensley L.E., Geisbert T.W. Effective post-exposure treatment of Ebola infection. PLoS Pathog. 2007;3:e2. doi: 10.1371/journal.ppat.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbutt M., Liebscher R., Wahl-Jensen V., Jones S., Moller P., Wagner R., Volchkov V., Klenk H.D., Feldmann H., Stroher U. Properties of replication-competent vesicular stomatitis virus vectors expressing glycoproteins of filoviruses and arenaviruses. J. Virol. 2004;78:5458–5465. doi: 10.1128/JVI.78.10.5458-5465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garver W.S., Heidenreich R.A. The Niemann–Pick C proteins and trafficking of cholesterol through the late endosomal/lysosomal system. Curr. Mol. Med. 2002;2:485–505. doi: 10.2174/1566524023362375. [DOI] [PubMed] [Google Scholar]
- Hood C.L., Abraham J., Boyington J.C., Leung K., Kwong P.D., Nabel G.J. Biochemical and structural characterization of cathepsin L-processed Ebola virus glycoprotein: implications for viral entry and immunogenicity. J. Virol. 2010;84:2972–2982. doi: 10.1128/JVI.02151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt C.L., Kolokoltsov A.A., Davey R.A., Maury W. The Tyro3 receptor kinase Axl enhances macropinocytosis of Zaire ebolavirus. J. Virol. 2010;85:334–347. doi: 10.1128/JVI.01278-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S.M., Feldmann H., Stroher U., Geisbert J.B., Fernando L., Grolla A., Klenk H.D., Sullivan N.J., Volchkov V.E., Fritz E.A., Daddario K.M., Hensley L.E., Jahrling P.B., Geisbert T.W. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat. Med. 2005;11:786–790. doi: 10.1038/nm1258. [DOI] [PubMed] [Google Scholar]
- Kaletsky R.L., Simmons G., Bates P. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J. Virol. 2007;81:13378–13384. doi: 10.1128/JVI.01170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratowicz A.S., Lennemann N.J., Sinn P.L., Davey R.A., Hunt C.L., Moller-Tank S., Meyerholz D.K., Rennert P., Mullins R.F., Brindley M., Sandersfeld L.M., Quinn K., Weller M., McCray P.B., Jr., Chiorini J., Maury W. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc. Natl. Acad. Sci. USA. 2011;108:8426–8431. doi: 10.1073/pnas.1019030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson N.D., Stillman E.A., Whitt M.A., Rose J.K. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. USA. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard E.E., Srivastava K., Traub L.M., Schaffer J.E., Ory D.S. Niemann–Pick type C1 (NPC1) overexpression alters cellular cholesterol homeostasis. J. Biol. Chem. 2000;275:38445–38451. doi: 10.1074/jbc.M003180200. [DOI] [PubMed] [Google Scholar]
- Millat G., Marcais C., Tomasetto C., Chikh K., Fensom A.H., Harzer K., Wenger D.A., Ohno K., Vanier M.T. Niemann–Pick C1 disease: correlations between NPC1 mutations, levels of NPC1 protein, and phenotypes emphasize the functional significance of the putative sterol-sensing domain and of the cysteine-rich luminal loop. Am. J. Hum. Genet. 2001;68:1373–1385. doi: 10.1086/320606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehring T.J., Moehring J.M. Selection and characterization of cells resistant to diphtheria toxin and pseudomonas exotoxin A: presumptive translational mutants. Cell. 1977;11:447–454. doi: 10.1016/0092-8674(77)90063-0. [DOI] [PubMed] [Google Scholar]
- Morizono K., Xie Y., Olafsen T., Lee B., Dasgupta A., Wu A.M., Chen I.S. The soluble serum protein Gas6 bridges virion envelope phosphatidylserine to the TAM receptor tyrosine kinase Axl to mediate viral entry. Cell Host Microbe. 2011;9:286–298. doi: 10.1016/j.chom.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbo A., Imai M., Watanabe S., Noda T., Takahashi K., Neumann G., Halfmann P., Kawaoka Y. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010:6. doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray N., Whidby J., Stewart S., Hooper J.W., Bertolotti-Ciarlet A. Study of Andes virus entry and neutralization using a pseudovirion system. J. Virol. Methods. 2010;163:416–423. doi: 10.1016/j.jviromet.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Ribeiro I., Marcao A., Amaral O., Sa Miranda M.C., Vanier M.T., Millat G. Niemann–Pick type C disease: NPC1 mutations associated with severe and mild cellular cholesterol trafficking alterations. Hum. Genet. 2001;109:24–32. doi: 10.1007/s004390100531. [DOI] [PubMed] [Google Scholar]
- Saeed M.F., Kolokoltsov A.A., Albrecht T., Davey R.A. Cellular entry of Ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 2010:6. doi: 10.1371/journal.ppat.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornberg K., Matsuyama S., Kabsch K., Delos S., Bouton A., White J. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornberg K.L., Shoemaker C.J., Dube D., Abshire M.Y., Delos S.E., Bouton A.H., White J.M. α5β1-Integrin controls ebolavirus entry by regulating endosomal cathepsins. Proc. Natl. Acad. Sci. 2009;106:8003–8008. doi: 10.1073/pnas.0807578106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima M., Ikeda Y., Kawaoka Y. The mechanism of Axl-mediated Ebola virus infection. J. Infect. Dis. 2007;196(Suppl. 2):S259–S263. doi: 10.1086/520594. [DOI] [PubMed] [Google Scholar]
- Shimojima M., Stroher U., Ebihara H., Feldmann H., Kawaoka Y. Identification of cell surface molecules involved in dystroglycan-independent lassa virus cell entry. J. Virol. 2012;86:2067–2078. doi: 10.1128/JVI.06451-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima M., Takada A., Ebihara H., Neumann G., Fujioka K., Irimura T., Jones S., Feldmann H., Kawaoka Y. Tyro3 family-mediated cell entry of Ebola and Marburg viruses. J. Virol. 2006;80:10109–10116. doi: 10.1128/JVI.01157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Wool-Lewis R.J., Baribaud F., Netter R.C., Bates P. Ebola virus glycoproteins induce global surface protein down-modulation and loss of cell adherence. J. Virol. 2002;76:2518–2528. doi: 10.1128/jvi.76.5.2518-2528.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wool-Lewis R.J., Bates P. Endoproteolytic processing of the Ebola virus envelope glycoprotein: cleavage is not required for function. J. Virol. 1999;73:1419–1426. doi: 10.1128/jvi.73.2.1419-1426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]