Abstract
Twenty-eight hundred and seventy-two cases of respiratory disease in pigs were analyzed for their etiologic agents. Two or more pathogens were detected from 88.2% of the cases, indicating that porcine reproductive and respiratory syndrome virus (PRRSV) or swine influenza virus (SIV) combined with other bacterial agents was a common cause for porcine respiratory diseases in the mid-western USA.
Porcine respiratory disease (PRD) of nursery and grow-finish pigs is characterized by cough, fever, lethargy, loss of appetite, labored breathing, and possibly death. Mortality can reach up to 15% on some farms, so PRD is considered one of the most economically significant problems in the US swine industry. Combinations of viral and bacterial agents are generally involved in the etiology of porcine respiratory disease. The most common pathogens reported were porcine reproductive and respiratory syndrome virus (PRRSV), Mycoplasma hyopneumoniae, swine influenza virus (SIV), pseudorabies virus (PRV), Actinobacillus pleuropneumoniae, and Haemophilus parasuis (1). Although different bacteria could induce the disease independently, it has commonly been caused by coinfection with bacteria and viruses under field conditions.
Combinations of bacteria and viruses have been reported to work in a synergistic fashion producing more severe respiratory diseases than those induced by each individual agent (2). Coinfections result in pathogenic interaction that have been reported are PRV and Pasteurella multocida (3), PRRSV and M. hyopneumoniae (2), and A. pleuropneumoniae and M. hyopneumoniae (4). In recent years, SIV has shown an increasing role in the development of PRD. Introduction of new SIV subtypes (H3N2 and H1N2) to the US swine population (5,6) has resulted in greater complication in porcine respiratory diseases. Clinical effects of PRRSV have been shown to be exacerbated with concurrent infection with SIV (7).
The purpose of this study was to investigate common etiologic agents associated with porcine respiratory diseases. Diagnostic data from 2872 cases of respiratory disease in pigs received at the Minnesota Veterinary Diagnostic Laboratory (MVDL) between January 2000 and June 2001 were analyzed retrospectively. This period was of particular interest because new subtypes of SIV had been isolated from respiratory samples in pigs (6).
For most cases, lung samples were submitted to the MVDL under chilled condition by an overnight delivery system. There was no consistent information on clinical history and age of the pigs for each sample. The samples were tested for the presence of respiratory pathogens by routine methods: Isolation of PRRSV, SIV, and porcine coronavirus (PRCV) was accomplished by using porcine alveolar macrophages, MARC-145 cells, or both; Madin-Darby canine kidney (MDCK) cells; and swine testicle (ST) cells lines; respectively. Following the observation of cytopathic effects, each isolate was identified by specific tests including immunofluorescent antibody assay with specific antiserum for PRRSV, reverse transcription (RT)-PCR assay for PRCV, and hemagglutination (HA) for SIV. Subtypes of SIV isolates were determined by hemagglutination inhibition (HI) tests, using H1- or H3-monospecific antiserum, 2 multiplex RT-PCR, or both, as previously described (6). Infection with M. hyopneumoniae was detected by means of a nested PCR test (8). Infection with porcine circovirus type 2 (PCV2) was detected by a PCR assay (9). All other bacterial isolations and identifications were carried out at the MVDL by using routine methods.
In the 2872 cases of respiratory disease examined, the percentages of agents diagnosed were PRRSV (35.4%), P. multocida (31.6%), M. hyopneumoniae (27.0%), SIV (22.2%), H. parasuis (22.0%), PCV2 (18.6%), A. pleuropneumoniae (18.2%), Bordetella bronchiseptica (8.0%), and PRCV (1.5%). Of the 2872 cases, a single pathogen was isolated from 338 (11.8%) samples, with PRRSV (3.3%), SIV (3.1%), and M. hyopneumoniae (1.5%) being the most common, while 2 or more pathogens were isolated from 2534 (88.2%) samples.
Of the 1017 PRRSV-positive samples, PRRSV only was in 96 (3.3%) samples, while coinfections were common with P. multocida in 298 (10.4%) samples, M. hyopneumoniae in 201 (7.0%) samples, A. pleuropneumoniae in 179 (6.2%) samples, H. parasuis in 176 (6.1%) samples, and PCV2 in 67 (2.3%) samples. Of the 636 SIV-positive cases, SIV only was found in 89 (3.1%) samples, while coinfections were recorded with P. multocida in 148 (5.2%) samples, M. hyopneumoniae in 122 (4.3%) samples, PRRSV in 109 (3.8%) samples, and PCV2 in 54 (1.9%) samples. Of 636 SIV isolates, 321 (50.1%) were subtyped as H3N2, 304 (47.8%) as H1N1, and 11 (1.7%) as H1N2.
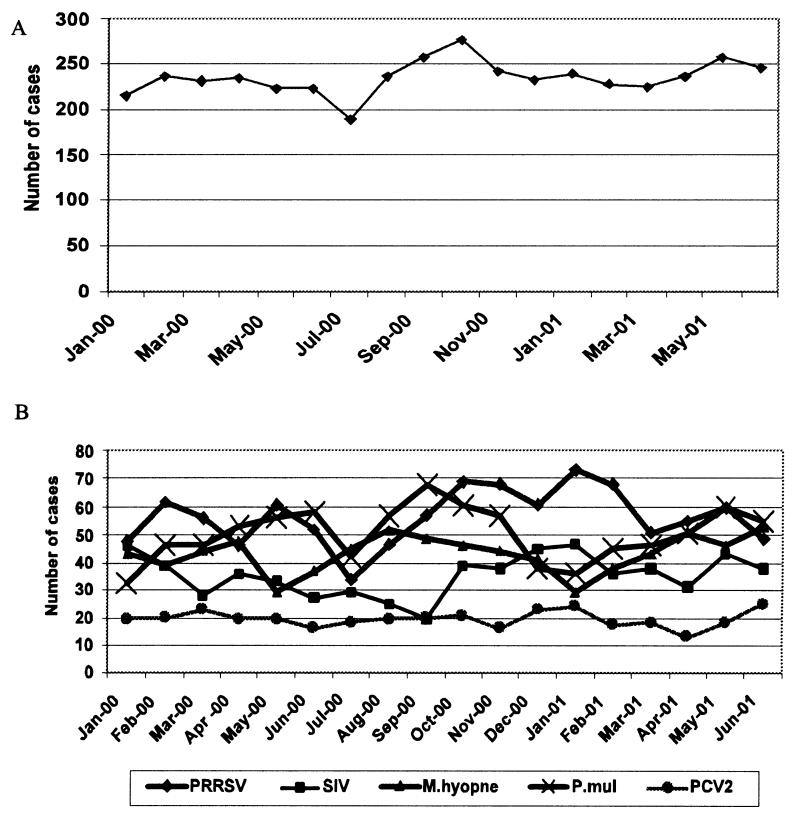
Monthly distribution of the respiratory disease cases submitted is shown in Figure 1. The highest and lowest numbers of cases were submitted between September and October, and between June and July, respectively. Porcine reproductive and respiratory syndrome virus and SIV infections showed the highest incidence during winter months between October and February. Mycoplasma hyopneumoniae and P. multocida infections were the highest in October to November and February to May. However, no seasonal variation was seen for PCV2 infections.
Figure 1. Temporal distribution of 2872 cases of porcine respiratory disease from which samples were submitted to the Minnesota Veterinary Diagnostic Laboratory between January 2000 and June 2001 (A) and distribution of individual respiratory pathogens by month (B).
PRRSV — porcine reproductive and respiratory syndrome virus
SIV — swine influenza virus
M. hyopne — Mycoplasma hyopneumoniae
P. mul — Pasteurella multocida
PCV2 — porcine circovirus 2
The present results show that PRRSV was the most common pathogen involved in over 35% of respiratory disease cases in the mid-western swine farms. Following experimental infection, PRRSV alone had little or no clinical significance in feeder pigs (7), while infection with other viral or bacterial agents resulted in severe clinical disease (2,7). Therefore, clinical problems associated with PRRSV infection under field conditions appeared to be the effects of combined infection with 2 or more respiratory pathogens.
Swine influenza virus was the second most frequently identified virus in this study, and the problems associated with SIV infection appear to be increasing under field conditions. The H3N2 subtype was the most dominant, whereas H1N2 was newly identified in association with porcine respiratory diseases in this study. Although a high seasonal prevalence was shown, SIV was isolated from samples collected throughout the year (Figure 1). This suggests that SIV has now become an endemic respiratory pathogen. Because SIV causes damage to the respiratory epithelium lining the airways, the virus could easily interact with other pathogens. An interaction between SIV and M. hyopneumoniae in the induction of pneumonia in susceptible swine was reported recently (2).
Pasteurella multocida and M. hyopneumoniae were also identified in high association with respiratory diseases in pigs. Pasteurella multocida was the second most frequently identified bacterium in this study, although P. multocida is generally considered an opportunistic pathogen in the respiratory tract. Mycoplasma hyopneumoniae still appears to be a significant bacterium despite the wide use of commercial vaccines on swine farms.
Porcine circovirus 2 infection has recently been reported in association with porcine respiratory diseases, with the virus being identified as the third most common pathogen among the diagnostic cases of porcine respiratory diseases (10). In contrast to the other pathogens, single infection and seasonal high distribution of PCV2 were not observed in this study. At this time, the pathogenic role of PCV2 alone is not clear, although synergism between PCV2 and other pathogens is possible in the respiratory system.
Both PRRSV and SIV appear to be the major primary viral pathogens involved in porcine respiratory diseases, and they interact with different bacterial pathogens. Any management procedure that will break the opportunity for combined infections would be beneficial, since over 88% of the cases of respiratory disease were caused by coinfection. Other factors associated with respiratory diseases in pigs, such as ventilation and animal density, should also be considered in the control of respiratory diseases in pigs.
Footnotes
Acknowledgments
The authors thank Dr. Jim Collins for helpful discussions. Technical assistance of Sigrun Haugerud, Wendy Wiese, and Karen Olsen is gratefully acknowledged.CVJ
Address all correspondence and reprint requests to Dr. H.S. Joo; E-mail address: jooxx001@umn.edu
References
- 1.Halbur PG. Porcine respiratory diseases. Proc 15th Congr Int Pig Vet Soc 1998:1–10.
- 2.Thacker EL, Halbur PG, Ross RF, Thanawongnuwech R, Thacker BJ. Mycoplasma hyopneumoniae potentiation of porcine reproductive and respiratory syndrome virus-induced pneumonia. J Clin Microbiol 1999;37:620–627. [DOI] [PMC free article] [PubMed]
- 3.Fuentes MC, Pijoan C. Pneumonia in pigs induced by intranasal challenge exposure with pseudorabies virus and Pasteurella multocida. Am J Vet Res 1987;48:1446–1448. [PubMed]
- 4.Yagihashi T, Nunoya T, Mitui T, Tajima M. Effect of Mycoplasma hyopneumoniae infection on the development of Haemophilus pleuropneumoniae pneumonia in pigs. Nippon Juigaku Zasshi 1984;46:705–713. [DOI] [PubMed]
- 5.Zhou NN, Senne DA, Landgraf JS, et al. Emergence of H3N2 reassortant influenza A viruses in North American pigs. Vet Microbiol 2000;74:47–58. [DOI] [PubMed]
- 6.Choi YK, Goyal SM, Joo HS. Prevalence of swine influenza virus subtypes on swine farms in the United States. Arch Virol 2002;147:1209–1220. [DOI] [PubMed]
- 7.Van Reeth K, Nauwynck H, Pensaert M. Dual infections of feeder pigs with porcine reproductive and respiratory syndrome virus followed by porcine respiratory coronavirus or swine influenza virus: a clinical and virological study. Vet Microbiol 1996;48:325–335. [DOI] [PMC free article] [PubMed]
- 8.Calsamiglia M, Pijoan C, Trigo A. Application of a nested polymerase chain reaction assay to detect Mycoplasma hyopneumoniae from nasal swabs. J Vet Diagn Invest 1999;11:246–251. [DOI] [PubMed]
- 9.Johnson CS, Joo HS, Direksin K, Yoon KJ, Choi YK. Experimental in utero inoculation of late-term swine fetuses with porcine cirocovirus type 2. J Vet Diagn Invest 2002;14:507–512. [DOI] [PubMed]
- 10.Harms PA, Halbur PG, Sorden SD. Three cases of porcine respiratory disease complex associated with porcine circovirus type 2 infection. J Swine Health Prod 2002;10:27–30.