Abstract
The blue crab Callinectes sapidus has been used as an experimental model organism for the study of regulation of cardiac activity and other physiological processes. Moreover, it is an economically and ecologically important crustacean species. However, there was no previous report on the characterization of its neuropeptidome. To fill in this gap, we employed multiple sample preparation methods including direct tissue profiling, crude tissue extraction and tissue extract fractionation by HPLC to obtain a complete description of the neuropeptidome of C. sapidus. Matrix-assisted laser desorption/ionization (MALDI)-Fourier transform mass spectrometry (FTMS) and MALDI-time-of-flight (TOF)/TOF were utilized initially to obtain a quick snapshot of the neuropeptide profile, and subsequently nanoflow liquid chromatography (nanoLC) coupled with electrospray ionization quadrupole time-of-flight (ESI-Q-TOF) tandem MS analysis of neuropeptide extracts was conducted for de novo sequencing. Simultaneously, the pericardial organ (PO) tissue extract was labeled by a novel N, N-dimethylated leucine (DiLeu) reagent, offering enhanced fragmentation efficiency of peptides. In total, 130 peptide sequences belonging to 11 known neuropeptide families including orcomyotropin, pyrokinin, allatostatin A (AST-A), allatostatin B (AST-B), FMRFamide-like peptides (FLPs), and orcokinin were identified. Among these 130 sequences, 44 are novel peptides and 86 are previously identified. Overall, our results lay the groundwork for future physiological studies of neuropeptides in C. sapidus and other crustaceans.
Keywords: Callinectes sapidus, pericardial organ, de novo sequencing, neuropeptidome, neuropeptides, chemical derivatization
1. Introduction
Crustacean neurosecretory systems synthesize and secrete a diverse class of peptide hormones that play important roles in regulating physiological activities such as reproduction, development, molting, growth, aggression, and adaptation [7, 23, 30–32, 39]. Blue crabs, Callinectus sapidus, are a model organism that is frequently used to study the effects of neuropeptides on many physiological processes. For example, previous research using blue crabs as the experimental model system showed that the rhythmic contractions of heart are neurogenic, driven by rhythmic motor patterns generated by the cardiac ganglion (CG) and could be modulated by neurotransmitters and neuropeptides [10–12, 18]. In another study, blue crabs were employed to study the induction of courtship display behavior by multiple neuropeptides and neuromodulators [43, 44]. Comprehensive neuropeptidomic information would greatly facilitate these studies. In addition to being an important experimental model organism, blue crabs have also long served as a major commercial species in fisheries and aquaculture. However, due to overharvesting and environmental contamination, adult populations of blue crabs are decreasing [1, 29]. A better understanding of the basic biology involved in the blue crab lifecycle and behavior will contribute to improving the blue crab aquaculture and replenishing the declining population. Therefore, profiling the neuropeptidome of blue crabs is essential to expanding our knowledge of neuropeptides implicated in blue crab neurobiology.
The crustacean pericardial organ (PO) is a well-defined neuroendocrine site that controls the secretion of various crustacean neuropeptides [4, 9, 15, 35, 36]. Many studies have reported the identification of specific neuropeptide families in this essential endocrine organ, such as FLPs, orcokinin and allatostatins, etc. [6, 7, 13, 14, 22, 24–28, 36, 37]. Previous studies of neuropeptide families in decapod crustacean provide a foundation and methodologies for blue crab PO neuropeptide profiling using multifaceted mass spectrometric strategies [15, 27]. In this study, we employ two dimensional reversed-phase liquid chromatography (2D RP-LC) and dimethylated leucine (DiLeu) chemical derivatization to complement and augment the aforementioned methodologies. 2D RP-LC provides better neuropeptide separation for MS analysis than 1D RP-LC [41, 46], while derivatization using DiLeu facilitates neuropeptide fragmentation upon tandem MS (MS2) which makes de novo sequencing less complicated compared to unlabeled neuropeptides [16, 17]. DiLeu, a new type of 4-plex isobaric tandem mass (MS2) tag, was recently developed in our lab. MS2 spectra of labeled neuropeptides exhibit intense signature reporter ions (m/z 114) that can be used as a check mark of a labeled neuropeptide. DiLeu labeling also improves neuropeptide fragmentation that is beneficial for de novo sequencing [40, 45].
In this paper, blue crab PO neuropeptidome characterization was carried out by a multifaceted mass spectrometric strategy facilitated with DiLeu labeling. PO tissue and tissue extract as well as fractions of reversed-phase HPLC separation of tissue extract were screened using a high throughput matrix-assisted laser desorption/ionization time-of-flight/ time-of-flight mass spectrometer (MALDI-TOF/TOF). Accurate masses of neuropeptides were determined by a high-resolution, high mass accuracy MALDI Fourier transform mass spectrometer (FTMS). Subsequently, tissue extract and HPLC fractions as well as DiLeu labeled tissue extract were analyzed with nanoflow liquid chromatography electrospray ionization quadrupole time-of-flight (nanoLC-ESI-Q-TOF) mass spectrometer to generate tandem mass spectra for de novo sequencing. Using this combined approach, 130 peptides were identified from the blue crab PO including 44 new peptides to this species. Our data greatly expand the catalog of peptide hormones known to be present in C. sapidus and also provide a foundation for future studies of peptide functions in this species.
2. Materials and methods
2.1 Materials
Methanol (Catalog No.: AC61009-0040 HPLC Grade), acetonitrile (ACN, Catalog No.: AC610010040 HPLC Grade for HPLC and A955-4 Optima for UPLC), formic acid (Catalog No.: AC14793-2500 99% for HPLC and A117-50 Optima for UPLC) and acetic acid (Catalog No.: A38S-212) were purchased from Fisher Scientific (Pittsburgh, PA). Gelatin was purchased from BD (Franklin Lakes, NJ) (Catalog No.: 214340). 2, 5-dihydroxybenzoic acid (DHB) was obtained from MP Biomedicals, Inc. (Solon, OH) (Catalog No.: 212011). α-cyano-4-hydroxy-cinnamic acid (CHCA) was purchased from Sigma-Aldrich (St. Louis, MO) (Catalog No.: 28166-41-8). Acidified methanol was prepared using 90% methanol, 9% glacial acetic acid, and 1% water. All water used in this study was doubly distilled on a Millipore filtration system (Bedford, MA). C18 ziptips were purchased from Millipore (Billerica, MA) (Catalog No.: ZTC18S096). Peptide standards were synthesized by the Peptide Synthesis Facility at the Biotechnology Center, University of Wisconsin at Madison (Madison, WI).
2.2 Animals and dissection
C. sapidus (blue crabs) were obtained from commercial food market and maintained without food for seven days in artificial sea water at 10–12°C. Animals are cold-anesthetized by packing on ice for 15–30min before dissection. The animal was then pinned ventral side up in a Sylgard-lined dissection dish to expose the pericardial cavity. The POs were identified visually as an iridescent web of nerves branching over the muscles surrounding the pericardial cavity and dissected free. All dissection was carried out in chilled (approximately 4°C) physiological saline (composition in mM: NaCl, 440; KCl, 11; MgCl2, 26; CaCl2, 13; Trizma base, 11; maleic acid, 5; pH 7.45).
2.3 Direct tissue analysis and tissue extract analysis
Small pieces of POs were dissected, followed by brief rinsing in acidified methanol for peptide extraction and subsequently rinsing in 10 mg/mL of 2, 5-dihydroxybenzoic acid (DHB), to remove the extracellular salts associated with the tissue samples. For direct tissue analysis, small pieces of tissue were transferred onto a MALDI sample plate followed by application of 150mg/mL DHB matrix solution (in 50/50 methanol /water) for MALDI-FTMS and 5mg/mL CHCA (in 50/50 ACN /water) for MALDI-TOF/TOF MS. For tissue extract analysis, acidified methanol was used for homogenization of neural tissues, followed by centrifugation at 16,000xg using Eppendorf 5415D tabletop centrifuge (Eppendorf AG). The pellet was washed with acidified methanol, supernatant were combined for further drying in Savant SC 110 SpeedVac concentrator (Thermo Electron Corporation) and re-suspended with 50 µL of 0.1% formic acid in water.
2.4 Fractionation of tissue extracts using reversed phase (RP)-HPLC
The resuspended extracts were then vortexed and briefly centrifuged. The resulting supernatants were subsequently fractionated via high performance liquid chromatography (HPLC). HPLC separations were performed using a Rainin Dynamax HPLC system equipped with a Dynamax UV-D II absorbance detector (Rainin Instrument Inc., Woburn, MA). The mobile phases included: Solution A (de-ionized water containing 0.1% formic acid) and Solution B (ACN containing 0.1% formic acid). About 50 µL of extract was injected onto a Macrosphere C18 column (2.1 mm i.d. × 250 mm length, 5 µm particle size; Alltech Assoc. Inc., Deerfield, IL). The separations consisted of a 120 minute gradient of 5–95% Solution B. Fractions were automatically collected every two minutes using a Rainin Dynamax FC-4 fraction collector. Fractions were further dried in a Savant SC 110 SpeedVac concentrator (Thermo Electron Corporation) and resuspended with 10 µL of 0.1% formic acid.
2.5 MALDI-FTMS and direct tissue analyses
MALDI-FTMS experiments were performed on a Varian/IonSpec ProMALDI Fourier transform mass spectrometer (Lake Forest, CA) equipped with a 7.0 Tesla actively-shielded superconducting magnet. The FTMS instrument contains a high pressure MALDI source where the ions from multiple laser shots can be accumulated in the external hexapole storage trap before the ions are transferred to the ICR cell via a quadrupole ion guide. A 355 nm Nd: YAG laser (Laser Science, Inc., Franklin, MA) was used to create ions in an external source. The ions were excited prior to detection with an rf sweep beginning at 7050 ms with a width of 4 ms and amplitude of 150 V base to peak. The filament and quadrupole trapping plates were initialized to 15 V, and both were ramped to 1V from 6500 to 7000 ms to reduce baseline distortion of peaks. Detection was performed in broadband mode from m/z 108.00 to 2500.00.
Off-line analysis of HPLC fractions was performed by spotting 0.3 µl of HPLC fractions on the MALDI sample plate and adding 0.3 µl of the saturated DHB. The resulting mixture was allowed to crystallize at room temperature. The MALDI-FTMS analysis was then performed as described above.
2.6 MALDI-TOF/TOF
A model 4800 MALDI-TOF/TOF analyzer (Applied Biosystems, Framingham, MA) equipped with a 200 Hz, 355 nm Nd:YAG laser was used for direct peptide profiling of neural tissues and HPLC fraction screening. Acquisitions were performed in positive ion reflectron mode. Instrument parameters were set using the 4000 Series Explorer software (Applied Biosystems). Mass spectra were obtained by averaging 1000 laser shots covering mass range m/z 500–4000. MS/MS was achieved by 1 kV collision induced dissociation (CID) using air as collision gas.
2.7 NanoLC-ESI-Q-TOF
Nanoscale LC-ESI-Q-TOF MS/MS was performed using a Waters nanoAcquity UPLC system coupled to a Q-TOF Micro mass spectrometer (Waters Corp., Milford, MA). 2D-chromatographic separations were performed on a homemade C18 reversed phase capillary column (75 µm i.d. x100 mm length, 3 µm particle size, 100Å) as second dimension separation in addition to the first HPLC separation mentioned above. The mobile phases used were: (A) 0.1% formic acid in deionized water; (B) 0.1% formic acid in ACN. An aliquot of 5.0 µl of a tissue extract or HPLC fraction was injected and loaded onto the trap column (Zorbax 300SB-C18 Nano trapping column, Agilent Technologies, Santa Clara, CA) using mobile phase A at a flow rate of 10 µl/min for 10 minutes. Following this, the stream select module was switched to a position at which the trap column came in line with the analytical capillary column, and a linear gradient of mobile phases A and B was initiated. For neuropeptides, the linear gradient was from 5% buffer B to 45% buffer B over 90 min. The nanoflow ESI source conditions were set as follows: capillary voltage 3200 V, sample cone voltage 35 V, extraction cone voltage 1 V, source temperature 100°C, Data dependent acquisition was employed for the MS survey scan, selection of three precursor ions and subsequent MS/MS of the selected parent ions. The MS scan range was from m/z 400–1800 and the MS/MS scan was from m/z 50–1800.
2.8 DiLeu labeling
Synthesis of DiLeu label is described elsewhere [45]. Tissue extract of five pairs of POs was labeled by 25 µL of DiLeu labeling solution (1mg/ml DiLeu in dry DMF). Sample was labeled at room temperature for 1 h and quenched for 30 min by adding 100 µL of water. Labeled sample was dried under SpeedVac and reconstituted with 40 µL of 0.1% FA for nanoLC-ESI-Q-TOF analysis.
2.9 Peptide sequencing and database searching
De novo sequencing was performed using a combination of MassLynx™ 4.1 PepSeq software (Waters) and manual sequencing. Tandem mass spectra acquired from the ESI-Q-TOF were first deconvoluted using MaxEnt 3 software (Waters) to convert multiply charged ions into their singly charged forms. The resulting spectra were pasted into the PepSeq window for sequencing analysis. The candidate sequences generated by the PepSeq software were compared and evaluated for homology with previously known peptides. The online program blastp (National Center for Biotechnology Information, Bethesda, MD; http://www.ncbi.nlm.nih.gov/BLAST/) was used to search the existing NCBI crustacean protein database, using the candidate peptide sequences as queries. For all searches, the blastp database was set to non-redundant protein sequences (i.e. nr) and restricted to crustacean sequences (i.e. taxid: 6657). Peptides with partial sequence homology were selected for further examination by comparing theoretical MS/MS fragmentation spectra generated by PepSeq with the raw MS/MS spectra. If the fragmentation patterns did not match well, manual sequencing was performed.
2.10 MS imaging
Immediately following dissection, the pericardial organ was rinsed briefly in deionized water to eliminate salt content. Tissues were dehydrated in a desiccator for 1 h prior to matrix application. An airbrush was used to spray coat the tissues with DHB. The airbrush was held perpendicular to the MALDI plate at a distance of 35 cm, and the flow rate of matrix was adjusted so that most of the matrix solvent evaporated before reaching the plate. Five coats of matrix were applied by spraying each sample for 30 s with 1 min dry time between each application.
A model 4800 MALDI-TOF/TOF analyzer was used for all mass spectral analyses. Acquisitions were performed in positive ion reflectron mode. Instrument parameters were set using the 4000 Series Explorer Software (Applied Biosystems). The tissue region to be imaged and the raster step size were controlled using the 4800 imaging application (Novartis, Basel, Switzerland) available through the MALDI MSI web site (www.maldi-msi.org). To generate images, spectra were collected at 100 µm intervals in the x and y dimensions across the surface of the sample. Each mass spectrum was generated by averaging 250 laser shots over the mass range m/z 800–2000. Mass spectra were externally calibrated using peptide standards applied directly to the stainless steel MALDI target. Image files were processed, and extracted ion images were created using the TissueView software package (Applied Biosystems, Framingham, MA).
3. Results
3.1 Enhancing neuropeptidome coverage in C. sapidus using a combination of microscale separation methods and complementary mass spectral techniques
In this study, 87 neuropeptides were identified from the PO without derivatization, including 73 previously known peptides in C. borealis and C. maenas and 14 novel peptides sequenced for the first time. Table 1 showed the complete list of neuropeptides sorted in families. Without derivatization (denoted as U), 87 neuropeptides from 11 neuropeptide families were identified as shown, using three different mass spectrometers by analyzing neural tissue directly, crude tissue extract and HPLC fractions of tissue extract.
Table 1.
Full list of neuropeptides identified from the pericardial organ (PO) of C. sapidus by ESI-Q-TOF (labeled as Q-TOF), MALDI-FTMS (labeled as FT) and MALDI-TOF/TOF (labeled as TOF). A comparison of de novo sequencing of neuropeptides from the PO extract with and without DiLeu labeling reaction is made (labeled as L or U, respectively). Neuropeptides listed in black are previously known peptides; neuropeptides listed in red bold are sequenced for the first time in this study; neuropeptides in grey shade are peptides detected/sequenced by both labeled and unlabeled methods.
| Family | [M+H]+ | Sequence | Labeled? | Q-TOF | FT | TOF |
|---|---|---|---|---|---|---|
| AST-A | 569.31 | YAFGLa | L | + | − | − |
| 739.38 | GPYSFGLa | L | + | − | − | |
| 781.39 | DPYAFGLa | L | + | − | − | |
| 796.40 | NPYSFGLa | U/L | + | + | + | |
| 798.41 | NVYSFGLa | L | + | − | − | |
| 810.41 | AGPYSFGLa | U/L | + | + | + | |
| 824.43 | ASPYAFGLa | U | − | − | + | |
| 836.43 | GPPYSFGLa | L | + | − | − | |
| 838.45 | TAPYAFGLa | L | + | − | − | |
| 841.42 | AGQ(GA)YSFGLa | L | + | − | − | |
| 853.42 | NGSYPFGLa | L | + | − | − | |
| 854.40 | DGPYSFGLa | U/L | + | − | + | |
| 870.41 | SGNYNFGLa | L | + | − | − | |
| 883.43 | SNPYSFGLa | U | + | − | − | |
| 893.43 | SGHYNFGLa | U | + | − | − | |
| 897.46 | ARGYDFGLa | U | + | + | − | |
| 909.49 | ARPYSFGLa | U | + | + | + | |
| 911.47 | ARAYDFGLa | U | + | − | − | |
| 918.40 | SDMYSFGLa | L | + | − | − | |
| 923.47 | pERAYSFGLa | U | − | + | − | |
| 928.45 | SSGQYAFGLa | U | + | + | − | |
| 933.48 | TPQANSFGLa | L | + | − | − | |
| 934.40 | SDM(O)YSFGLa | L | + | − | − | |
| 937.52 | PRVYSFGLa | U/L | + | + | + | |
| 939.50 | TRPYSFGLa | U | + | + | + | |
| 962.51 | APQPYAFGLa | U | − | − | + | |
| 986.49 | RNMYSFGLa | L | + | − | − | |
| 998.49 | TSPQYSFGLa | L | + | − | − | |
| 1004.49 | FSGTYNFGLa | U | + | − | + | |
| 1006.55 | APRPYSFGLa | L | + | − | − | |
| 1021.51 | THPTYSFGLa | U/L | + | − | − | |
| 1068.61 | ALTTLYAFGLa | L | + | − | − | |
| 1097.60 | AGLSALYSFGLa | L | + | − | − | |
| 1143.57 | QWLYSMFGLa | L | + | − | − | |
| 1167.58 | AAGLQNYDFGLa | L | + | − | − | |
| 2051.03 |
GSGQYAYGLGKK -AGQYSFGLa |
L | + | − | − | |
|
AST-B |
1031.51 | AWSNLQGAWa | L | + | − | − |
| 1061.52 | AGWSSLKGAWa | L | + | − | − | |
| 1107.52 | AGWSSMRGAWa | U | + | + | + | |
| 1123.52 | AGWSSM(O)RGAWa | U/L | + | − | + | |
| 1165.55 | NWNKFQGSWa | L | + | − | − | |
| 1179.57 | AGWNKFQGSWa | U | − | + | − | |
| 1182.57 | TSWGKFQGSWa | U/L | − | + | + | |
| 1209.58 | TGWNKFQGSWa | L | + | − | − | |
| 1220.58 | SGDWSSLRGAWa | U/L | + | + | + | |
| 1222.58 | GNWNKFQGSWa | U/L | + | + | + | |
| 1252.59 | NNWSKFQGSWa | U/L | + | + | + | |
| 1253.62 | NDWSKFGQSWa | U | + | + | + | |
| 1260.66 | SGKWSNLRGAW | U | − | − | + | |
| 1293.63 | STNWSSLRSAWa | U/L | + | + | + | |
| 1294.63 | STDWSSLRSAWa | U | + | + | − | |
| 1366.63 | NNNWSKFQGSWa | L | + | − | − | |
| 1380.64 | NNNWTKFQGSWa | U/L | + | + | + | |
| 1470.70 | VPNDWAHFRGSWa | U/L | + | + | + | |
| CCAP | 956.38 | PFCNAFTGCa | U/L | + | + | + |
| Myosuppressin | 1125.56 | pQDLDHVFLR | U | + | − | − |
| 817.52 | HVFLRFa | U | + | + | − | |
| 1271.65 | pQDLDHVFLRFa | U/L | + | + | + | |
| 1288.68 | QDLDHVFLRFa | U | − | + | + | |
| Orcomyotropin | 1186.52 | FDAFTTGFGHS | U/L | + | + | + |
| Ork | 1098.52 | EIDRSGFGFA | U | + | + | + |
| 1198.55 | NFDEIDRSGFa | U | + | − | + | |
| 1213.55 | DEIDRSGFGFA | U | + | − | + | |
| 1228.56 | NFDEIDRSSFa | U | + | − | + | |
| 1256.55 | NFDEIDRSGFG | U | + | + | + | |
| 1270.57 | NFDEIDRSGFA | U | + | + | + | |
| 1286.57 | NFDEIDRSSFG | U | + | + | + | |
| 1300.58 | NFDEIDRSSFA | U | + | + | + | |
| 1301.56 | DFDEIDRSSFA | U | + | + | − | |
| 1360.62 | FDEIDRSGFGFA | U | + | − | + | |
| 1403.62 | NFDEIDRSGFGF | U | + | + | + | |
| 1433.63 | NFDEIDRSSFGF | U | + | + | + | |
| 1474.66 | NFDEIDRSGFGFA | U/L | + | + | + | |
| 1502.69 | NFDEIDRSGFGFV | U/L | + | − | + | |
| 1504.67 | NFDEIDRSSFGFA | U | − | − | + | |
| 1532.70 | NFDEIDRSSFGFV | U/L | + | + | + | |
| 1547.68 | NFDEIDRSSFGFN | U/L | − | + | + | |
| 1554.70 | NFDEIDRTGFGFH | U | − | − | + | |
| Others | 844.48 | HLGSLYRa | U | + | + | + |
| 1372.80 | KIFEPLRDKNL | U | − | + | − | |
| Proctolin | 649.37 | RYLPT | U | − | − | + |
| Pyrokinin | 1051.57 | TNFAFSPRLa | U | + | − | − |
| FLPs | 695.40 | NFLRFa | U | − | + | + |
| 735.43 | GPFLRFa | U | − | + | + | |
| 851.50 | RNFLRFa | U/L | + | + | + | |
| 863.45 | pQGNFLRFa | U | − | + | − | |
| 865.52 | RQFLRFa | L | + | − | − | |
| 866.46 | GNNFLRFa | L | + | − | − | |
| 916.52 | THPFLRFa | L | + | − | − | |
| 921.50 | PSM(O)RLRFa | U | − | + | − | |
| 925.49 | DDNFLRFa | L | + | − | − | |
| 926.52 | SKNYLRFa | U | + | − | − | |
| 938.53 | NRSFLRFa | U | + | − | − | |
| 948.54 | PGVNFLRFa | L | + | − | − | |
| 958.48 | FDDFLRFa | L | + | − | − | |
| 965.54 | NRNFLRFa | U/L | + | + | + | |
| 966.53 | DRNFLRFa | U/L | + | + | − | |
| 994.51 | PSDNFLRFa | U | − | − | + | |
| 1005.57 | GRPNFLRFa | L | + | − | + | |
| 1022.56 | GNRNFLRFa | U | + | + | + | |
| 1023.55 | GDRNFLRFa | L | + | − | − | |
| 1031.59 | AHKNFLRFa | U | + | − | − | |
| 1034.55 | NQPNFLRFa | L | + | − | − | |
| 1035.58 | SPRNFLRFa | L | + | − | − | |
| 1048.57 | APQGNFLRFa | U | + | − | − | |
| 1104.61 | GAHKNYLRFa | U/L | + | + | + | |
| 1108.61 | YGAHVFLRFa | L | + | − | − | |
| 1116.55 | YEQDFLRFa | L | + | − | − | |
| 1124.63 | GLSRNYLRFa | U | − | + | + | |
| 1132.60 | NVGSHGFLRFa | L | + | − | − | |
| 1146.61 | GYSKNYLRFa | U | + | + | + | |
| 1147.65 | APQRNFLRFa | U/L | + | + | + | |
| 1158.62 | YGNRSFLRFa | U | + | + | + | |
| 1172.63 | AYNRSFLRFa | U/L | + | − | + | |
| 1181.62 | SENRNFLRFa | L | + | − | − | |
| 1209.61 | DENRNFLRFa | U | − | + | + | |
| 1238.66 | SQPSKNYLRFa | U | + | + | − | |
| 1265.64 | GHDFEVFLRFa | L | + | − | − | |
| RYa | 784.41 | FVGGSRYa | U/L | + | − | + |
| 832.41 | FYANRYa | U | + | + | + | |
| 862.42 | FYSQRYa | L | + | − | + | |
| 943.44 | SGYNANRYa | L | + | − | − | |
| 959.47 | SGFYAPRYa | U | + | − | − | |
| 976.46 | SGFYANRYa | U/L | + | + | + | |
| 988.51 | VGFYANRYa | U | + | − | − | |
| 1027.54 | SRFVGGSRYa | U | + | − | + | |
| 1030.45 | pEGFYSQRYa | U/L | + | + | + | |
| 1114.58 | SSRFVGGSRYa | U/L | + | + | + | |
| SIFa | 1219.62 | DLDSVLDPSIFa | L | + | − | − |
| 1381.73 | GYRKPPFNGSIFa | U | − | − | + |
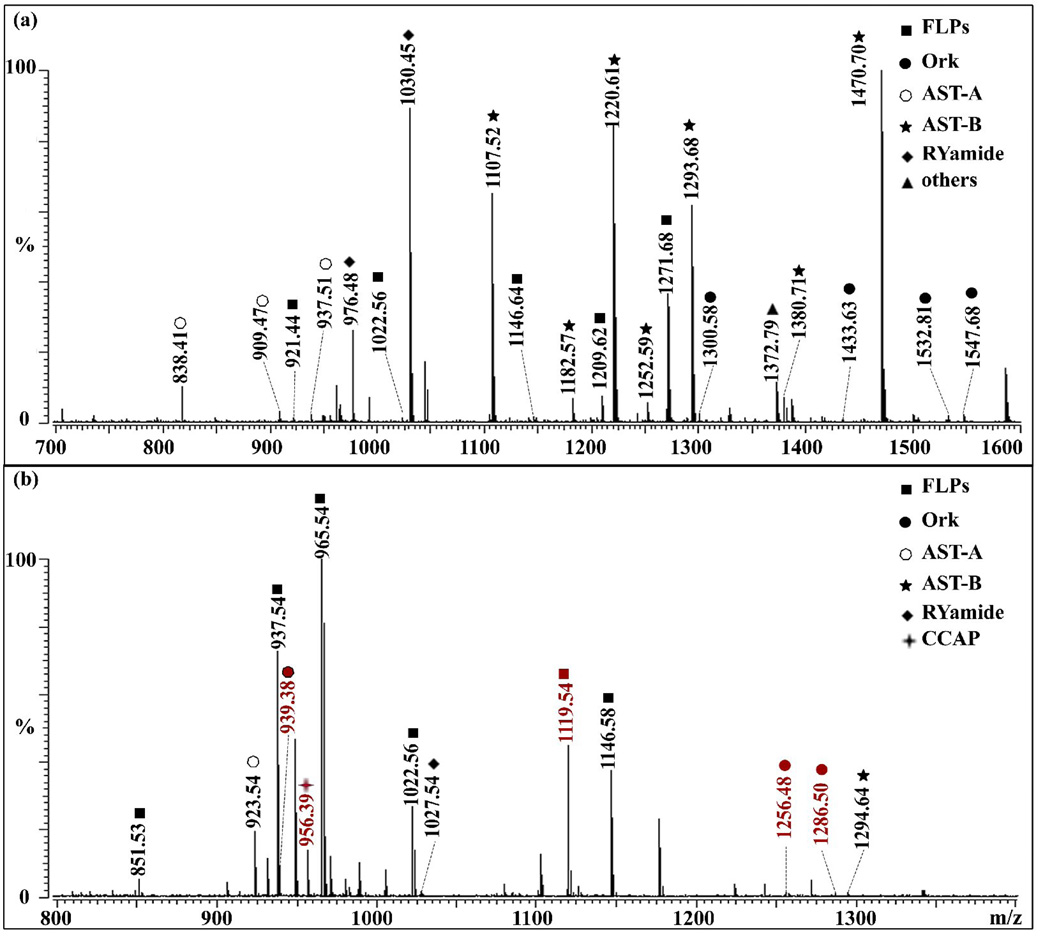
Figure 1 shows representative mass spectra of direct tissue analysis of the PO with MALDI-TOF/TOF. Different areas of tissue give rise to significantly different neuropeptide profiles due to the differential distribution of neuropeptide families and isoforms from the same peptide family [3, 8, 47]. For example, Figure 1(a) shows 38 neuropeptides from 8 families while Figure 1(b) only shows 20 peptides from 6 families from distinct regions of the PO. However, Figure 1(b) shows four unique neuropeptides not detected in Figure 1(a).
Figure 1.
Direct tissue analysis of the C. sapidus pericardial organ (PO) by MALDI-TOF/TOF. Panels 1(a) and 1(b) show mass spectra from different regions of the PO tissue. Signals correspond to the protonated molecular ions, [M+H]+, where M is the monoisotopic molecular mass of each peptide. The identified peptides are marked with symbols indicating specific families to which they belong.
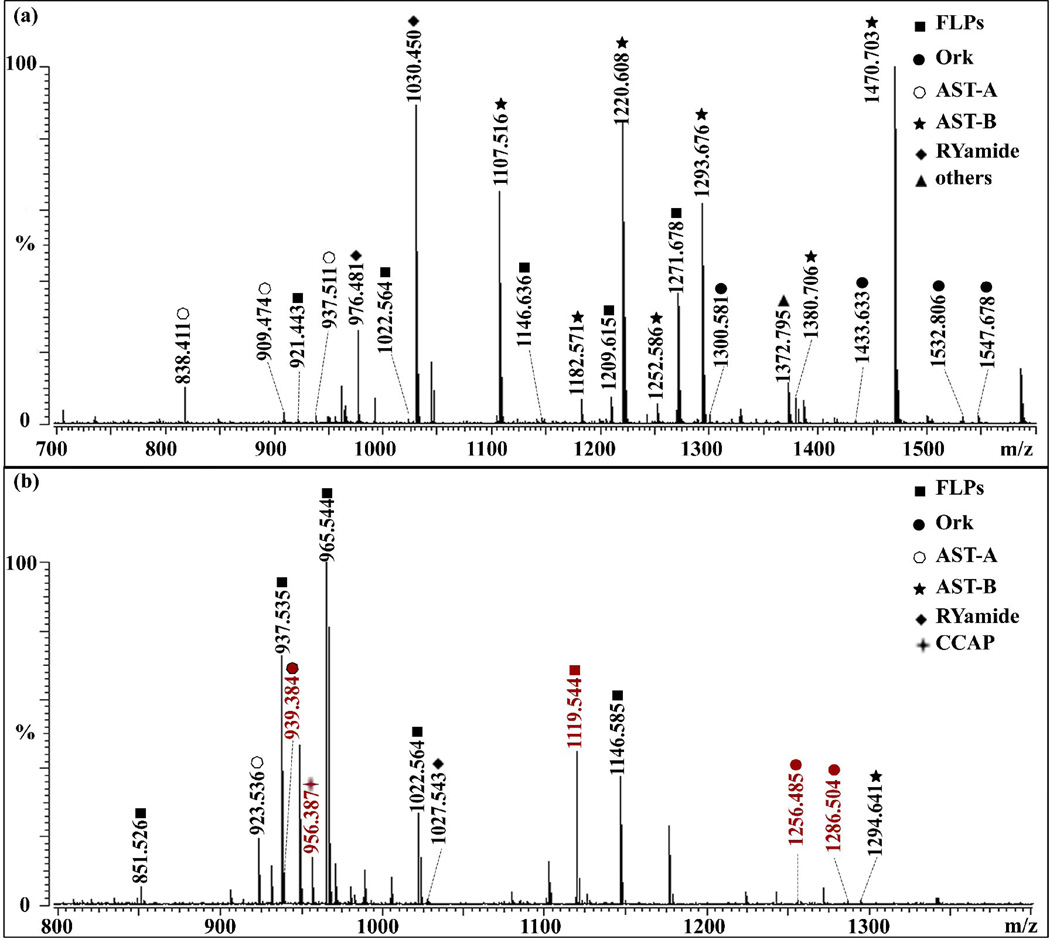
2D RP-LC enabled the detection of many more neuropeptides. Figure 2 shows the mass spectra of the PO crude extract and fractions after off-line HPLC separation. Five additional neuropeptides as shown in red were detected by MALDI-FTMS in a single HPLC fraction #15 compared to crude extract shown in Figure 2(a). Overall, after 2D RP-LC separation, 47 neuropeptides were detected in all 70 HPLC fractions compared to 26 neuropeptides detected in crude extract by MALDI-FTMS. Similarly, 57 neuropeptides were detected in HPLC fractions by ESI-Q-TOF tandem MS compared to 23 neuropeptides identified in crude extract (data not shown).
Figure 2.
Accurate mass measurement of peptide profiles in the tissue extract of (a) C. sapidus PO and (b) HPLC fraction #15 of the extract by MALDI-FTMS. The mass spectral peaks shown in red in (b) represent the neuropeptides that only appear in the HPLC fractions.
3.2 Tandem MS sequencing of peptides in C. sapidus by a multifaceted MS-based strategy
3.2.1 The MS/MS sequencing of orcokinins
Eighteen orcokinins were sequenced from the POs of C. sapidus, among which 15 were previously identified orcokinins from C. borealis or C. maenas, 3 novel orcokinins including DEIDRSGFGFA, DFDEIDRSSFA and NFDEIDRSSFGF were detected in C. sapidus. Most of the sequences identified share the conserved N-terminal sequence of NFDEIDR present in insects and crustaceans. Some of the isoforms such as DFDEIDRSSFA lack the conserved N-terminal motif but exhibit C-terminal sequence homology with other orcokinins. Although most orcokinins contain C-terminal free carboxylic acid, some of them are C-terminally amidated such as NFDEIDRSGFamide and NFDEIDRSSFamide, which occurs via posttranslational modification and may exhibit additional stability of the peptides.
3.2.2 Sequencing of FLPs (FMRFamide-like peptides) in C. sapidus
FLP (FMRFamide-like peptides) is a peptide family with the conserved C-terminal sequence, –XRFamide. Here, 22 FLPs were sequenced with 20 previously identified peptides and two novel peptides LRNLRFamide and APQGNFLRFamide, which were identified in C. sapidus for the first time.
3.2.3 Sequencing of truncated myosuppressin peptides in C. sapidus
Myosuppressin is a subfamily of FLPs (FMRFamide-like peptides). Both QDLDHVFLRFamide and pQDLDHVFLRFamide are known isoforms in crustaceans [38]. Here, both of QDLDHVFLRFamide and pQDLDHVFLRFamide were detected in C. sapidus PO. In addition, two truncated myosuppressin neuropeptides pQDLDHVFLR and HVFLRFa were sequenced via ESI-Q-TOF after 2D-LC separation. In the truncated peptide pQDLDHVFLR, the N-terminal sequence was conserved while the C-terminal motif Famide was missing.
3.2.4 Sequencing of Arg-Tyr-amide (RYamides) in C. sapidus
RYamides is a peptide family possessing the –XRYamide C-termini motif (where X is a variable amino acid). This family was recently discovered to be involved in feeding process [2, 5]. In this study, eight RYamides were identified. Among them, FVGGSRYamide, FYANRYamide, SGFYAPRYamide, SGFYANRYamide, pEGFYSQRYamide and SSRFVGGSRYamide were previously identified in other crustaceans, and two peptides VGFYANRYamide, SRFVGGSRYamide were de novo sequenced for the first time.
3.2.5 Other peptides
In addition to the peptides described above, several previously known peptides such as proctolin (RYLPT m/z 649.37), pyrokinin (TNFAFSPRLamide m/z 1051.57), crustacean cardioactive peptide (CCAP, PFCNAFTGCamide m/z 956.40), Orcomyotropin (FDAFTTGFGHS, m/z 1186.52), were also detected. The recently sequenced peptides HL/IGSL/IYRamide (m/z 844.48) [15] and KIFEPLRDKNL (m/z 1372.79)[26] were also identified (Table 1).
3.2.6 The MS/MS sequencing of AST-A and AST-B neuropeptides
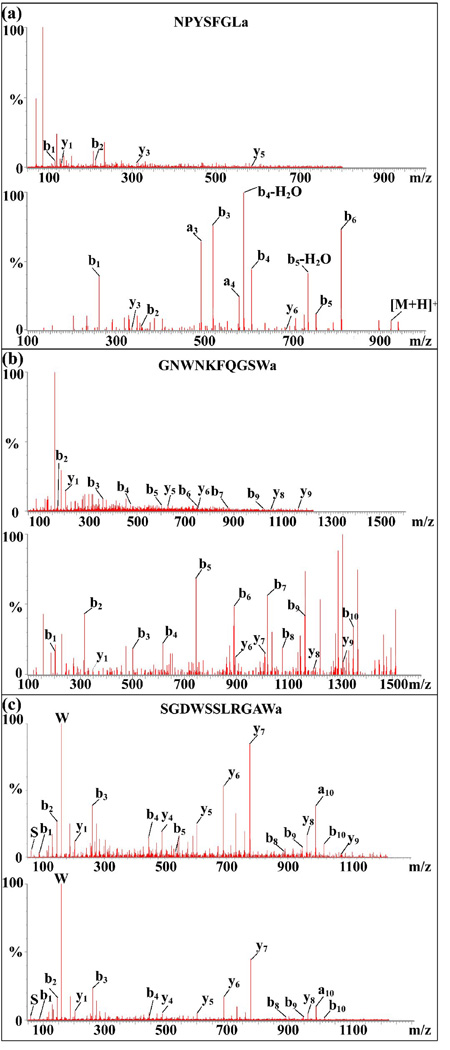
Sixteen AST-A and 13 AST-B neuropeptides were sequenced from the POs of C. sapidus, among which there were 12 previously identified AST-A and 11 AST-B peptides from C. borealis or C. maenas, four novel AST-A and two novel AST-B peptides from C. sapidus. The -YXFGLamide C-termini (where X is a variable amino acid) classify A-type allatostatin while the C-terminal motif -W(X)6Wamide (where X indicates variable amino acids) represents a hallmark of the B-type allatostatin neuropeptides [34]. However, the fragmentation of allatostatin was poor as shown in the top panel of Figure 3(a) and (b), which made the de novo sequencing of these two families particularly difficult and therefore many potential neuropeptides from these families may be missed. In order to obtain more comprehensive neuropeptidome coverage, DiLeu labeling was employed as a derivatization method to facilitate neuropeptide fragmentation upon tandem mass spectrometry (MS2).
Figure 3.
De novo sequencing of allatostatin neuropeptides in C. sapidus PO by ESI-Q-TOF MS/MS. (a) Fragmentation comparison for de novo sequencing of AST-A (NPYSFGLamide m/z 796.40) before (top panel) and after DiLeu labeling (bottom panel) and (b) AST-B (GNWNKFQGSWamide m/z 1222.58) before (top panel) and after DiLeu labeling (bottom panel); (c) confirmation of a novel AST-B peptide sequence elucidation (SGDWSSLRGAWa m/z 1220.58) by comparing peptide fragmentation from the PO tissue extract (top panel) and synthesized peptide standard with proposed sequence (bottom panel).
3.3 DiLeu labeling reaction facilitates de novo sequencing of FLPs and AST family neuropeptides
A total of 73 peptide sequences covering eight known neuropeptide families including AST-A type, AST-B type, FMRFamide-like, RYamide, orcokinin, orcomyotropin, SIFamide and CCAP were identified from the DiLeu labeled PO samples. These sequences are listed in Table 1 (denoted as L). Novel sequences are shown in red while overlapping sequences from both labeled and unlabeled samples are shown in grey shade. Among these 73 sequences, 34 are novel peptides and they belong to five families: AST-A, AST-B, FMRFamide-like, RYamide and SIFamide. As shown in Figure 3(a) and 3(b), the fragmentation is highly improved after DiLeu labeling of these peptides. Figure 3(a) shows tandem MS spectra of AST-A NPYSFGLa before (top panel) and after (bottom panel) labeling. As seen, five fragment ions at very low abundance were observed without labeling. In contrast, all b-ions were detected at very high abundance together with two y-ions upon DiLeu labeling derivatization. The intense signature reporter ion (m/z 114) in MS2 spectrum was used as a marker of labeled peptides. All peptides sequenced after DiLeu labeling contain m/z 114 in their MS2 spectra except the N-terminal pGlu-modified peptides pQDLDHVFLRFa and pEGFYSQRYa which cannot be labeled. One of the novel AST-B type peptides, SGDWSSLRGAWa, was sequenced from both labeled and unlabeled PO samples as shown in Table 1. This peptide was detected at a relatively high level in the PO tissue as shown in Figure 1, as well as in crude tissue extract and HPLC fractions as shown in Figure 2. To further confirm the sequence of this novel peptide, the MS/MS fragmentation patterns of the synthesized standard (Figure 3(c), top panel) and the endogenous peptide from real biological samples (Figure 3(c), bottom panel) are compared. As shown in Figure 3(c), these two spectra exhibit almost identical fragmentation patterns, including y1, y5–y8, b1–b4, b8–b10, which confirms the sequence of this novel peptide. Using DiLeu labeling, 15 novel AST-A and four novel AST-B neuropeptides were sequenced as compared to only four AST-A and two AST-B peptides without labeling. Similarly, 13 novel FLPs were sequenced after DiLeu labeling compared to only two novel ones without labeling.
3.4 MALDI-MS imaging of C. sapidus PO
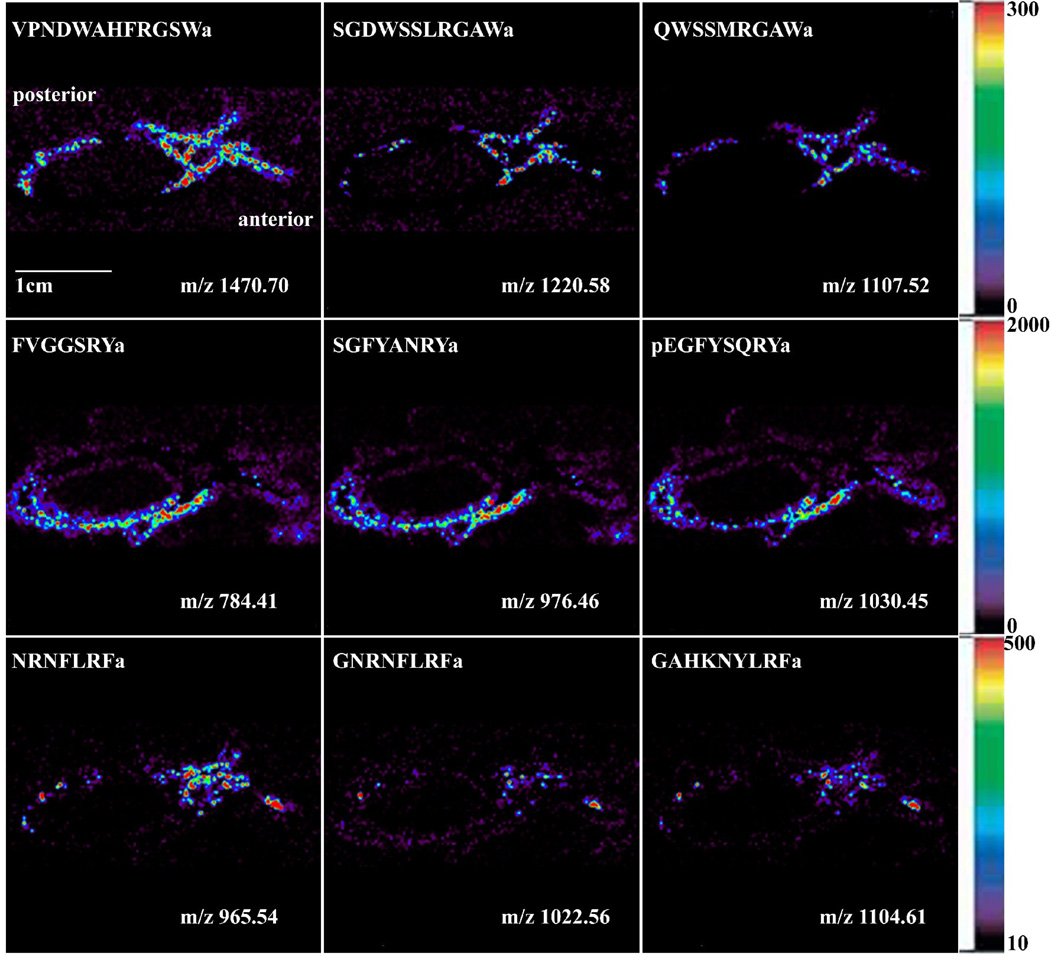
Figure 4 shows the localization of three major neuropeptide families in C. sapidus PO: AST-B, RYamide and FLPs, all of which are shown to be involved in feeding behavior of crustaceans based on previous reports [2, 5, 19, 42]. Several isoforms of the AST-B family display colocalization patterns in the PO, including VPNDWAHFRGSWamide (m/z 1470.70), AGWSSMRGAWamide (m/z 1107.52), and a novel isoform, SGDWSSLRGAWamide (m/z 1220.58), with a high concentration in the posterior bar region, dorsal trunk and anterior bar region. RYamides, including FVGGSRYamide (m/z 784.41), SGFYANRYamide (m/z 976.46) and pEGFYSQRYamide (m/z 1030.45), have been localized in the ventral trunk. The FLPs also show colocalization patterns of several isoforms in the PO, including NRNFLRFamide (m/z 965.54), GNRNFLRFamide (m/z 1022.56) and GAHKNYLRFamide (m/z 1104.61), which are most prevalent in the trunk and posterior bar region of the PO.
Figure 4.
Neuropeptide localization in C. sapidus PO via MALDI-mass spectrometric imaging (MSI) images of selected peptides: AST-B peptide family includes VPNDWAHFRGSWamide (m/z 1470.70), AGWSSMRGAWamide (m/z 1107.52) and SGDWSSLRGAWamide (m/z 1220.58) at the top panels, RYamide family includes FVGGSRYamide (m/z 784.41), SGFYANRYamide (m/z 976.46) and pEGFYSQRYamide (m/z 1030.45) at the middle panels and FLPs includes NRNFLRFamide (m/z 965.54), GNRNFLRFamide (m/z 1022.56) and GAHKNYLRFamide (m/z 1104.61) at the bottom panel.
4. Discussion
4.1 Enhancing neuropeptidome coverage in C. sapidus using a combination of microscale separation methods and complementary mass spectral techniques
The combined mass spectrometric approach involving MALDI-FTMS, MALDI-TOF/TOF and nanoLC-ESI-Q-TOF was employed in this study for comprehensive characterization of neuropeptides expressed in the PO of C. sapidus. With the high resolution and mass accuracy of MALDI-FTMS, high sensitivity of MALDI-TOF/TOF and de novo sequencing capability of nanoLC-ESI-Q-TOF, 87 neuropeptides were identified from the PO without derivatization, including 73 previously known peptides in C. borealis and C. maenas and 14 novel peptides sequenced for the first time as listed in Table 1. The use of multiple mass spectrometers offers complementary detection of neuropeptides. NanoLC-ESI-Q-TOF identified many neuropeptides that were not detected by MALDI-FTMS and MALDI-TOF/TOF, thanks to the employment of the 2D RP-LC separation, where the additional LC separation further reduces the sample complexity. Moreover, the ionization mode of ESI gives rise to multiply-charged ions offering more efficient fragmentation, which makes it a better ionization method than MALDI for de novo sequencing. On the other hand, MALDI-FTMS and MALDI-TOF/TOF enabled the detection of some neuropeptides that were missed in ESI-Q-TOF which may be due to greater sample loss caused by an additional LC separation, especially for low abundance neuropeptides. Overall, 66 neuropeptides were identified in ESI-Q-TOF, 54 were identified in MALDI-FTMS and 66 were detected in MALDI-TOF/TOF.
In order to obtain more complete neuropeptide profiles, multiple sample preparation methods including direct tissue profiling, tissue extract analysis, and fractions from off-line HPLC separation were used in this study. As an initial screening method, direct tissue analysis by MALDI-TOF/TOF and MALDI-FTMS provides a quick snapshot of the peptide profiles and accurate mass measurements to identify numerous previously known neuropeptides from multiple peptide families. However, its coverage is limited due to the heterogeneity of tissues. Tissue extract analysis, on the other hand, can provide a more complete profile of peptides present in the whole tissue, especially for larger size tissues with heterogeneity such as the PO or the brain. The indispensability of tissue extract analysis can be better illustrated in Figure 1 which shows that different neuropeptides are present in different areas of the PO as revealed by direct tissue analysis. In tissue extract analysis, tissues were homogenized and extracted with acidified methanol, which usually requires more tissue samples than the direct tissue analysis. Nevertheless, tissue extract analysis reduces the individual variability of a single tissue. More importantly, it is compatible with further separation and its coupling to MS detection such as nanoLC-ESI-Q-TOF analysis.
For complex neural tissues such as the PO, the spectral quality of direct tissue analysis and crude tissue extract analysis is often compromised by salt and lipid interference. Further separation is necessary to reduce the interference and generate better neuropeptide profiles. In this study, 2D RP-LC including an off-line reversed phase HPLC and an on-line nanoLC coupled to ESI-Q-TOF was employed prior to MS analysis to reduce chemical complexity and offer an expanded dynamic range in the MS detection. Employing the additional off-line HPLC besides the online nanoLC results in the detection of many more peptides in ensuing fractions compared to the crude tissue extract, due to the extra separation step and further elimination of lipid interference and chemical complexity. As shown in Figure 2 more neuropeptides were detected in HPLC fractions compared to crude tissue extract by MALDI-FTMS. This result highlights the advantage of fractionation of complex tissue extracts to improve peptide coverage. With 2D RP-LC separation, 57 neuropeptides were identified in HPLC fractions via ESI-Q-TOF tandem MS. As a comparison, 23 neuropeptides were detected in crude tissue extract, separated solely by nanoLC coupled to ESI-Q-TOF, suggesting better separation power of 2D RP-LC. On the other hand, many neuropeptides were only detected in crude tissue extract, which largely due to sample loss during HPLC fractionation and the relatively low concentration after HPLC fractionation. This significantly impacts the detection of certain neuropeptides, especially those with low abundances.
The combination of microscale separation methods and complementary mass spectral techniques provides enhanced neuropeptidome coverage. However, for certain neuropeptides, such as neuropeptides from AST-A and AST-B peptide families, the identification and de novo sequencing suffer from their poor fragmentation as shown in top panel of Figure 3(a) and (b). Previous work in our lab showed that N, N-dimethylation facilitates neuropeptide fragmentation upon tandem MS (MS2), which makes de novo sequencing less complicated compared to unlabeled neuropeptides [16, 17]. Here, we employed dimethylated leucine (DiLeu) chemical modification, a novel labeling technique developed in our lab recently [45], to provide complementary sequence information for these peptide families.
4.2 DiLeu labeling reaction enhances fragmentation efficiency and facilitates de novo sequencing of FLPs and AST neuropeptides in C. sapidus
Our previous study showed that DiLeu labeling offers improved fragmentation efficiency and thus facilitates neuropeptide de novo sequencing [45] which is essential for neuropeptide discovery of C. sapidus due to the limited genomic information for this species. Here, DiLeu labeling was used as a parallel approach to label free method presented in previous section, providing complementary information for de novo sequencing to enhance the confidence of peptide fragment assignments and discover novel neuropeptide family members.
Thirty sequences in 7 families were found to overlap by the direct comparison of labeled and unlabeled samples (grey shaded in Table 1). Two sequences in AST-A and AST-B type neuropeptide families were selected to demonstrate the MS2 fragmentation improvement (Figure 3(a) and (b)). The substantial enhancement of N-terminal fragment ion signals and simplified fragmentation patterns are likely due to a combination of stabilized b/a-type fragment ions by dimethyl groups and reduced gas-phase fragment ion cyclization and subsequent sequence scrambling with blocked N-terminus upon DiLeu labeling [20]. Our results indicated that DiLeu labeling significantly enhanced the fragmentation of the AST-A, AST-B types neuropeptides and FLPs, leading to more confident peptide fragment assignments. One novel AST-B type sequence (SGDWSSLRGAWa m/z 1220.58) was identified among these overlapping peptides. This novel sequence was also confirmed by reductive dimethylation of blue crab PO extract (data not shown). Because two chemical modification approaches and the label free method yielded the same peptide sequence, the confidence of the correctly assigned novel sequence is very high. Further confirmation of this sequence was performed by comparing MS2 pattern of synthesized standard and putative peptide from real sample, in this case the HPLC fraction of the PO extract. The results in Figure 3(c) show almost identical fragmentation patterns and confirm the assigned sequence.
Thirty-eight novel sequences were elucidated from DiLeu labeled PO samples covering five neuropeptide families. The greater number of identified peptide sequences is due to a combination of higher ionization efficiency and better fragmentation of peptides after DiLeu labeling. In general, N, N-dimethylation of amino acids makes their N-terminal nitrogen atoms more electron rich than non-derivatized amino acids. Coupling dimethylated amino acids to the N-terminus of a peptide increases its basicity, which improves ionization efficiency compared to non-derivatized peptides. Increased ionization efficiency enabled peptides present in lower abundances to be detected, thus resulting in more peptides being observed after DiLeu labeling. Greater peptide precursor ion signals also helped improve fragmentation and enabled better sequence elucidation. Overall, DiLeu labeling showed great utility in neuropeptide discovery and de novo sequencing.
4.3 MALDI-MS imaging of neuropeptides in C. sapidus PO
Previous immunocytochemical [21, 33] and mass spectral [15, 24, 26, 27] studies showed that a rich source of neurohormones exists in the PO of crustaceans and they are sorted as families based on their sequence homology. Typically, isoforms of the same peptide family are co-localized since they are encoded by the same neuropeptide precursor gene. However, differential localization of members from the same peptide family has been reported [3, 8, 47], which may be caused by tissue-specific processing of neuropeptide prohormones and may suggest different function of isoforms despite their sequence similarity. MALDI mass spectral imaging (MSI) can distinguish specific isoforms of the same peptide family in contrast to immunocytochemistry. Furthermore, it offers higher throughput due to its capability of simultaneously detecting multiple peptide families in a single experiment. In our imaging study, neuropeptide localization was found to be highly correlated to peptide family with isoforms from the same peptide family sharing similar distribution patterns (Figure 4). While most neuropeptide localization patterns in C. sapidus were similar to those observed in other crustaceans, the trend in C. sapidus was not always the same as in C. borealis. For example, RYamide, which was found to be concentrated in ventral trunk in C. sapidus PO, was most concentrated in the anterior bar region in C. borealis, which suggested the potential differences in functions of the RYamide peptides in C. sapidus from those in C. borealis. Further physiological investigations would be needed to elucidate these potential functional differences.
4.4 Conservation and variation of neuropetidome between multiple crustacean species
Previous study in our lab characterized the neuropeptidomes of several crustacean species such as Cancer borealis and Carcinus maenas [26–28]. The neuropeptidome of C. sapidus is comparable to those characterized in C. borealis and C. maenas, with a large portion of neuropeptides being highly conserved. However, many novel neuropeptide isoforms are discovered and unique to C. sapidus as those highlighted in Table 1. As expected, FLPs, AST-A, AST-B and orcokinins are the major peptide families found in the POs for all three crustacean species examined here. Nonetheless, subsets of different peptide isoforms are uniquely present in each species. In order to further understand the degree of similarities of these neuropeptidomes, we compared four major neuropeptide family members. Among them, over 80% of orcokinins are conserved between these three species; approximately 60% FLPs, 50% AST-A and 70% AST-B detected in C. sapidus can also be detected in Cancer borealis and Carcinus maenas. The conservation for majority of the neuropeptidome elucidates the high degree of sequence homology in signaling peptides and close phylogenetic relationship of these decapod crustacean species, yet the variation of their neuropeptidomes may contribute to species diversity of decapod crustaceans.
5. Conclusions
In this study we combined multiple sample preparation methods, multifaceted mass spectral techniques and dimethylated leucine (DiLeu) labeling strategies to comprehensively characterize the neuropeptides present in the pericardial organ of the blue crab C. sapidus. In total, 130 peptides from twelve families were identified and 44 were new to this species. This study reported for the first time the neuropeptidome of blue crabs and greatly expanded the number of known peptides in this important crustacean species. The research presented here provides a strong foundation for future studies on the physiological roles played by these signaling molecules in a well-defined neural network.
Highlights.
The first comprehensive report on discovery of neuropeptides in the pericardial organ of blue crab Callinectes sapidus
130 peptides from 11 families including 44 novel ones were discovered and sequenced
A combination of multifaceted mass spectrometry (MS) approach and chemical derivatization was employed for peptidomic analysis
Our results lay the groundwork for future neuropeptide physiology studies on C. sapidus and other crustaceans
Acknowledgements
We thank Tyler Greer in the Li Laboratory for critical reading of the manuscript. The authors wish to thank the University of Wisconsin-Biotechnology Center Mass Spectrometry Facility for access to the MALDI TOF/TOF instrument. We also want to thank the University of Wisconsin School of Pharmacy Analytical Instrumentation Center for access to the MALDI FTICR MS instrument. This work was supported in part by National Science Foundation (CHE- 0957784), National Institutes of Health through grants 1R01DK071801 and R56 DK071801. L.L. acknowledges an H.I. Romnes Faculty Research Fellowship.
Abbreviations
- 2D RP-LC
Two dimensional reversed-phase liquid chromatography
- ACN
acetonitrile
- AST-A
Allatostatin A
- AST-B
Allatostatin B
- CCAP
crustacean cardioactive peptide
- CG
cardiac ganglion
- CHCA
α-cyano-4-hydroxy-cinnamic acid
- DHB
2, 5-dihydroxybenzoic acid
- DiLeu
dimethylated leucine
- FLPs
FMRFamide-like peptides
- HPLC
high performance liquid chromatography
- MALDI-TOF/TOF
matrix-assisted laser desorption/ionization time-of-flight/ time-of-flight mass spectrometer
- MALDI-FTMS
matrix-assisted laser desorption/ionization Fourier transform mass spectrometer
- MS
mass spectrometry
- MS2
Tandem mass spectrometry
- nanoLC-ESI-Q-TOF
nanoflow liquid chromatography electrospray ionization quadrupole time-of-flight
- PO
pericardial organ
- RPCH
red pigment concentrating hormone
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abbe GR, Stagg C. Trends in blue crab (Callinectes sapidus Rathbun) catches near Calvert Cliffs, Maryland, from 1968 to 1995 and their relationship to the Maryland commercial fishery. J. Shellfish Res. 1996;15:751–758. [Google Scholar]
- 2.Chen RB, Hui LM, Cape SS, Wang JH, Li LJ. Comparative neuropeptidomic analysis of food intake via a multifaceted mass spectrometric approach. ACS Chem. Neurosci. 2010;1:204–214. doi: 10.1021/cn900028s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen RB, Hui LM, Sturm RM, Li LJ. Three dimensional mapping of neuropeptides and lipids in crustacean brain by mass spectral imaging. J. Am. Soc. Mass Spectrom. 2009;20:1068–1077. doi: 10.1016/j.jasms.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen RB, Jiang XY, Conaway MCP, Mohtashemi I, Hui LM, Viner R, et al. Mass spectral analysis of neuropeptide expression and distribution in the nervous system of the lobster Homarus americanus. J. Proteome Res. 2010;9:818–832. doi: 10.1021/pr900736t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen RB, Ma MM, Hui LM, Zhang J, Li LJ. Measurement of neuropeptides in crustacean hemolymph via MALDI mass spectrometry. J. Am. Soc. Mass Spectrom. 2009;20:708–718. doi: 10.1016/j.jasms.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christie AE, Lundquist CT, Nassel DR, Nusbaum MP. Two novel tachykinin-related peptides from the nervous system of the crab Cancer borealis. J. Exp. Biol. 1997;200:2279–2294. doi: 10.1242/jeb.200.17.2279. [DOI] [PubMed] [Google Scholar]
- 7.Christie AE, Stemmler EA, Dickinson PS. Crustacean neuropeptides. Cell. Mol. Life Sci. 2010;67:4135–4169. doi: 10.1007/s00018-010-0482-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeKeyser SS, Kutz-Naber KK, Schmidt JJ, Barrett-Wilt GA, Li LJ. Imaging mass spectrometry of neuropeptides in decapod crustacean neuronal tissues. J. Proteome Res. 2007;6:1782–1791. doi: 10.1021/pr060603v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dircksen H, Bocking D, Heyn U, Mandel C, Chung JS, Baggerman G, et al. Crustacean hyperglycaemic hormone (CHH)-like peptides and CHH-precursor-related peptides from pericardial organ neurosecretory cells in the shore crab Carcinus maenas, are putatively spliced and modified products of multiple genes. Biochem. J. 2001;356:159–170. doi: 10.1042/0264-6021:3560159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fort TJ, Brezina V, Miller MW. Modulation of an integrated central pattern generator-effector system: Dopaminergic regulation of cardiac activity in the blue crab Callinectes sapidus. J. Neurophysiol. 2004;92:3455–3470. doi: 10.1152/jn.00550.2004. [DOI] [PubMed] [Google Scholar]
- 11.Fort TJ, Brezina V, Miller MW. Regulation of the crab heartbeat by FMRFamide-like peptides: Multiple interacting effects on center and periphery. J. Neurophysiol. 2007;98:2887–2902. doi: 10.1152/jn.00558.2007. [DOI] [PubMed] [Google Scholar]
- 12.Fort TJ, Garcia-Crescioni K, Agricola HJ, Brezina V, Miller MW. Regulation of the crab heartbeat by crustacean cardioactive peptide (CCAP): Central and peripheral actions. J. Neurophysiol. 2007;97:3407–3420. doi: 10.1152/jn.00939.2006. [DOI] [PubMed] [Google Scholar]
- 13.Fu Q, Christie AE, Li LJ. Mass spectrometric characterization of crustacean hyperglycemic hormone precursor-related peptides (CPRPs) from the sinus gland of the crab Cancer productus. Peptides. 2005;26:2137–2150. doi: 10.1016/j.peptides.2005.03.040. [DOI] [PubMed] [Google Scholar]
- 14.Fu Q, Goy MF, Li LJ. Identification of neuropeptides from the decapod crustacean sinus glands using nanoscale liquid chromatography tandem mass spectrometry. Biochem. Biophys. Res. Commun. 2005;337:765–778. doi: 10.1016/j.bbrc.2005.09.111. [DOI] [PubMed] [Google Scholar]
- 15.Fu Q, Kutz KK, Schmidt JJ, Hsu YWA, Messinger DI, Cain SD, et al. Hormone complement of the Cancer productus sinus gland and pericardial organ: An anatomical and mass spectrometric investigation. J. Comp. Neurol. 2005;493:607–626. doi: 10.1002/cne.20773. [DOI] [PubMed] [Google Scholar]
- 16.Fu Q, Li LJ. Fragmentation of peptides with N-terminal dimethylation and imine/methylol adduction at the tryptophan side-chain. J. Am. Soc. Mass Spectrom. 2006;17:859–866. doi: 10.1016/j.jasms.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Fu Q, Li LJ. Investigation of several unique tandem mass spectrometric fragmentation patterns of NFDEIDR, an orcokinin analog, and its N-terminal dimethylated form. Rapid Commun. Mass Spectrom. 2006;20:553–562. doi: 10.1002/rcm.2337. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Crescioni K, Fort TJ, Stern E, Brezina V, Miller MW. Feedback from peripheral musculature to central pattern generator in the neurogenic heart of the crab Callinectes sapidus: role of mechanosensitive dendrites. J. Neurophysiol. 2010;103:83–96. doi: 10.1152/jn.00561.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garside CS, Nachman RJ, Tobe SS. Injection of Dip-allatostatin or Dip-allatostatin pseudopeptides into mated female Diploptera punctata inhibits endogenous rates of JH biosynthesis and basal oocyte growth. Insect Biochem. Mol. Biol. 2000;30:703–710. doi: 10.1016/s0965-1748(00)00041-2. [DOI] [PubMed] [Google Scholar]
- 20.Harrison AG, Young AB, Bleiholder C, Suhai S, Paizs B. Scrambling of sequence information in collision-induced dissociation of peptides. J. Am. Chem. Soc. 2006;128:10364–10365. doi: 10.1021/ja062440h. [DOI] [PubMed] [Google Scholar]
- 21.Hsu YWA, Messinger DI, Chung JS, Webster SG, de la Iglesia HO, Christie AE. Members of the crustacean hyperglycemic hormone (CHH) peptide family are differentially distributed both between and within the neuroendocrine organs of Cancer crabs: implications for differential release and pleiotropic function. J. Exp. Biol. 2006;209:3241–3256. doi: 10.1242/jeb.02372. [DOI] [PubMed] [Google Scholar]
- 22.Jensen ON. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr. Opin. Chem. Biol. 2004;8:33–41. doi: 10.1016/j.cbpa.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Keller R. Crustacean neuropeptides - structures, functions and comparative aspects. Experientia. 1992;48:439–448. doi: 10.1007/BF01928162. [DOI] [PubMed] [Google Scholar]
- 24.Li LJ, Kelley WP, Billimoria CP, Christie AE, Pulver SR, Sweedler JV, et al. Mass spectrometric investigation of the neuropeptide complement and release in the pericardial organs of the crab Cancer borealis. J. Neurochem. 2003;87:642–656. doi: 10.1046/j.1471-4159.2003.02031.x. [DOI] [PubMed] [Google Scholar]
- 25.Li LJ, Pulver SR, Kelley WP, Thirumalai V, Sweedler JV, Marder E. Orcokinin peptides in developing and adult crustacean stomatogastric nervous systems and pericardial organs. J. Comp. Neurol. 2002;444:227–244. doi: 10.1002/cne.10139. [DOI] [PubMed] [Google Scholar]
- 26.Ma MM, Bors EK, Dickinson ES, Kwiatkowski MA, Sousa GL, Henry RP, et al. Characterization of the Carcinus maenas neuropeptidome by mass spectrometry and functional genomics. Gen. Comp. Endocrinol. 2009;161:320–334. doi: 10.1016/j.ygcen.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma MM, Chen RB, Sousa GL, Bors EK, Kwiatkowski MA, Goiney CC, et al. Mass spectral characterization of peptide transmitters/hormones in the nervous system and neuroendocrine organs of the American lobster Homarus americanus. Gen. Comp. Endocrinol. 2008;156:395–409. doi: 10.1016/j.ygcen.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma MM, Wang JH, Chen RB, Li LJ. Expanding the Crustacean neuropeptidome using a multifaceted mass spectrometric approach. J Proteome Res. 2009;8:2426–2237. doi: 10.1021/pr801047v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendonca JT, Verani JR, Nordi N. Evaluation and management of blue crab Callinectes sapidus (Rathbun, 1896) (Decapoda - Portunidae) fishery in the Estuary of Cananeia, Iguape and Ilha Comprida, Sao Paulo, Brazil. Braz. J. Biol. 2010;70:37–45. doi: 10.1590/s1519-69842010000100007. [DOI] [PubMed] [Google Scholar]
- 30.Mercier J, Doucet D, Retnakaran A. Molecular physiology of crustacean and insect neuropeptides. J. Pestic. Sci. 2007;32:345–359. [Google Scholar]
- 31.Mykles DL, Adams ME, Gade G, Lange AB, Marco HG, Orchard I. Neuropeptide action in onsects and crustaceans. Physiol. Biochem. Zool. 2010;83:836–846. doi: 10.1086/648470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz MW, Woods SC, Porte D, Seeley RJ, Baskin DG. Central nervous system control of food intake. Nature. 2000;404:661–671. doi: 10.1038/35007534. [DOI] [PubMed] [Google Scholar]
- 33.Skiebe P. Allatostatin-like immunoreactivity in the stomatogastric nervous system and the pericardial organs of the crab Cancer pagurus, the lobster Homarus americanus, and the crayfish Cherax destructor and Procambarus clarkii. J. Comp. Neurol. 1999;403:85–105. [PubMed] [Google Scholar]
- 34.Stay B, Tobe SS. The role of allatostatins in juvenile hormone synthesis in insects and crustaceans. Annu. Rev. Entomol. 2007;52:277–299. doi: 10.1146/annurev.ento.51.110104.151050. [DOI] [PubMed] [Google Scholar]
- 35.Stemmler EA, Bruns EA, Gardner NP, Dickinson PS, Christie AE. Mass spectrometric identification of pEGFYSQRYamide: A crustacean peptide hormone possessing a vertebrate neuropeptide Y (NPY)-like carboxy-terminus. Gen. Comp. Endocrinol. 2007;152:1–7. doi: 10.1016/j.ygcen.2007.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stemmler EA, Cashman CR, Messinger DI, Gardner NP, Dickinson PS, Christie AE. High-mass-resolution direct-tissue MALDI-FTMS reveals broad conservation of three neuropeptides (APSGFLGMRamide, GYRKPPFNGSIFamide and pQDLDHVFLRFamide) across members of seven decapod crustaean infraorders. Peptides. 2007;28:2104–2115. doi: 10.1016/j.peptides.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 37.Stemmler EA, Peguero B, Bruns EA, Dickinson PS, Christie AE. Identification, physiological actions, and distribution of TPSGFLGMRamide: a novel tachykinin-related peptide from the midgut and stomatogastric nervous system of Cancer crabs. J. Neurochem. 2007;101:1351–1366. doi: 10.1111/j.1471-4159.2007.04520.x. [DOI] [PubMed] [Google Scholar]
- 38.Stevens JS, Cashman CR, Smith CM, Beale KM, Towle DW, Christie AE, et al. The peptide hormone pQDLDHVFLRFamide (crustacean myosuppressin) modulates the Homarus americanus cardiac neuromuscular system at multiple sites. J. Exp. Biol. 2009;212:3961–3976. doi: 10.1242/jeb.035741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sweedler JV, Li L, Rubakhin SS, Alexeeva V, Dembrow NC, Dowling O, et al. Identification and characterization of the feeding circuit-activating peptides, a novel neuropeptide family of Aplysia. J. Neurosci. 2002;22:7797–7808. doi: 10.1523/JNEUROSCI.22-17-07797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang JH, Zhang YZ, Xiang F, Zhang ZC, Li LJ. Combining capillary electrophoresis matrix-assisted laser desorption/ionization mass spectrometry and stable isotopic labeling techniques for comparative crustacean peptidomics. J. Chromatogr. A. 2010;1217:4463–4470. doi: 10.1016/j.chroma.2010.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang N, Xie CH, Young JB, Li L. Off-line two-dimensional liquid chromatography with maximized sample loading to reversed-phase liquid chromatography-electrospray ionization tandem mass spectrometry for shotgun proteome analysis. Anal. Chem. 2009;81:1049–1060. doi: 10.1021/ac802106z. [DOI] [PubMed] [Google Scholar]
- 42.Wilson CH, Christie AE. Distribution of C-type allatostatin (C-AST)-like immunoreactivity in the central nervous system of the copepod Calanus finmarchicus. Gen. Comp. Endocrinol. 2010;167:252–260. doi: 10.1016/j.ygcen.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wood DE, Derby CD. Coordination and neuromuscular control of rhythmic behaviors in the blue crab Callinectes sapidus. J. Comp. Physiol. A. 1995;177:307–319. doi: 10.1007/BF00192420. [DOI] [PubMed] [Google Scholar]
- 44.Wood DE, Derby CD. Distribution of dopamine-like immunoreactivity suggests a role for dopamine in the courtship display behavior of the blue crab Callinectes sapidus. Cell Tissue Res. 1996;285:321–330. doi: 10.1007/s004410050649. [DOI] [PubMed] [Google Scholar]
- 45.Xiang F, Ye H, Chen RB, Fu Q, Li LJ. N,N-Dimethyl Leucines as novel isobaric tandem mass tags for quantitative proteomics and peptidomics. Anal. Chem. 2010;82:2817–2825. doi: 10.1021/ac902778d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Gao MX, Tang J, Yang PY, Liu YK, Zhang XM. Improvements in protein identification confidence and proteome coverage for human liver proteome study by coupling a parallel mass spectrometry/mass spectrometry analysis with multi-dimensional chromatography separation. Anal. Chim. Acta. 2006;566:147–156. [Google Scholar]
- 47.Zimmerman TA, Rubakhin SS, Romanova EV, Tucker KR, Sweedler JV. MALDI mass spectrometric imaging using the stretched sample method to reveal neuropeptide distributions in Aplysia nervous tissue. Anal. Chem. 2009;81:9402–9409. doi: 10.1021/ac901820v. [DOI] [PMC free article] [PubMed] [Google Scholar]