Abstract
We designed a case-control study to determine the plasma homocysteine (Hcy) level and evaluate the potential role of the methylenetetrahydrofolate reductase (MTHFR) C677T gene polymorphism in colorectal cancer (CRC). Total Hcy was quantified using the fluorescence polarization immunoassay (FPIA) on the IMx analyzer. Genomic DNA was analyzed by the real-time polymerase chain reaction (RT-PCR). The plasma levels of Hcy in the CRC group (12.63±3.11 μmol/l) were significantly higher compared with those in the control group (10.87±2.42 μmol/l; P<0.05). The frequency of the MTHFR 677TT genotype in CRC patients was markedly high. The MTHFR 677TT genotype was significantly correlated with an increased risk of CRC (odds ratio, 1.671; 95% confidence interval, 1.094–2.553; P=0.018). This study suggests that the MTHFR C677T polymorphism indicates susceptibility to CRC and is correlated with CRC pathogenesis, suggesting that the homozygous variant MTHFR C677T polymorphism is a candidate risk factor for CRC.
Keywords: methylenetetrahydrofolate reductase, polymorphism, homocysteine, colorectal cancer
Introduction
Colorectal cancer (CRC) is one of the most common malignant diseases worldwide, affecting men and women approximately equally. There are several biological mechanisms by which folate deficiency increases the risk of CRC (1–3). Folate may modulate DNA methylation, which is a significant epigenetic determinant in gene expression, the maintenance of DNA integrity and stability, chromosomal modifications and the development of mutations. A growing body of evidence from in vitro, animal and human studies indicates that folate deficiency is associated with DNA strand breaks, impaired DNA repair, increased mutations and aberrant DNA methylation, and that folate supplementation corrects some of these defects (4).
Findings of previous epidemiological studies suggested that the elevation of plasma homocysteine (Hcy) levels is an independent risk factor for CRC (5,6), although the mechanism remains unknown. A defect in the methylenetetrahydrofolate reductase (MTHFR) gene and a deficiency of folate may lead to the accumulation of Hcy in tissues and blood.
The association between the MTHFR C677T gene polymorphism and genetic susceptibility to CRC has been widely evaluated in recent studies, but with controversial conclusions. Several studies have reported that a homozygous variant genotype of the polymorphism of MTHFR C677T was associated with an increased risk of CRC. Certain studies have reported that individuals with the MTHFR 677TT genotype had a decreased risk of CRC (8), whereas other authors observed no association between the MTHFR C677T genotype and genetic susceptibility to CRC (9–11). Considering the number of studies performed thus far, inconsistent results have been reported on the association of MTHFR C677T gene polymorphisms with genetic susceptibility to CRC. We designed a case-control study to determine the Hcy level and evaluate the potential role of the MTHFR C677T gene polymorphism in CRC.
Materials and methods
Subjects
We conducted a hospital-based case-control study to assess the association between Hcy levels, the MTHFR C677T gene polymorphism and genetic susceptibility to CRC. The cases were 370 patients diagnosed with CRC attending Shandong Provincial Hospital, Shandong University (Jinan, China), between March 2008 and December 2010. These patients had CRC at stages A, B or C, according to Dukes’ classification, and all underwent elective and curative surgery. Of the 370 patients, 204 were male and 166 were female. The mean age was 49.3±8.2 years (range, 35–78). The subsite distribution of the 93 CRCs showed 19 in the right colon, 6 in the transverse colon, 45 in the left colon and 23 in the rectum. Diagnosis of adenocarcinoma was confirmed in all patients by histology and cytological investigations. Histopathological grading of adenocarcinoma was: 5% well-differentiated, 74% moderately differentiated and 21% poorly differentiated. A total of 370 healthy unrelated subjects were recruited into the control group after being interviewed with regard to whether they had been diagnosed with colorectal carcinoma or associated diseases, using age and gender as frequency-matching criteria. The mean age of the normal donors was 47.3±8.1 years (range, 32–74); 196 (53.0%) of the healthy subjects were male and 174 (47.0%) were female. The characteristics of the study population are shown in Table I. Informed consent was obtained from all study subjects following an explanation of the nature of the study. The study was approved by the Shandong University Research Ethics Committee, China.
Table I.
Patient characteristics.
| Controls (n=370) n (%) |
Cases (n=370) n (%) |
χ2 | P-value | |
|---|---|---|---|---|
| Age (years) | ||||
| <45 | 67 (18.1) | 51 (13.8) | 4.523 | 0.104 |
| 45–60 | 167 (45.1) | 158 (42.7) | ||
| >60 | 136 (36.8) | 161 (43.5) | ||
| Gender | ||||
| Male | 196 (53.0) | 204 (55.1) | 0.348 | 0.555 |
| Female | 174 (47.0) | 166 (44.9) | ||
| Location | ||||
| Colon | 237 (64.1) | |||
| Rectum | 133 (35.9) | |||
| Dukes’ stage | ||||
| A | 33 (8.9) | |||
| B | 131 (35.4) | |||
| C | 206 (55.7) | |||
Colorectal carcinomas covering Dukes’ stages A, B and C, with more specimens in Dukes’ stage C, representing invasive stage, compared with the less invasive stages corresponding to Dukes’ stages A and B.
Hcy determination and genotyping
Peripheral venous blood was obtained from each subject and genomic DNA was extracted using a DNA extraction kit (Qiagen, Crawley, UK) according to the manufacturer’s instructions. The specimens were stored at −70°C until use. Total Hcy was quantified using the fluorescence polarization immunoassay (FPIA) on the IMx analyzer from Abbott Laboratories (Abbott Park, IL, USA). The IMx Hcy assay is based on the reduction of the plasma samples with dithiothreitol and subsequent conversion of free Hcy to S-adenosyl homocysteine by hydrolase in the presence of added adenosine. The sample and the tracer compete in binding to the monoclonal antibody. This reaction is followed by the detection of S-adenosyl homocysteine by an FPIA. The concentration of Hcy in plasma is inversely correlated with the intensity of the polarized light.
Genomic DNA was detected by real-time polymerase chain reaction (RT-PCR) amplification followed by digestion with the restriction enzyme HinFI, as described by Frosst et al (12). The primers were designed as previously reported, with the following sequences for sense, 5′-GCCCAGCCACTCACTGTTTTA-3′, and antisense, 5′-AGGACGGTGCGGTGAGAGTG-3′, that were used in a 25-μl mixture for amplification. The cycling conditions for PCR were an initial denaturation of 5 min at 94°C, followed by 30 cycles of 40 sec at 94°C, annealing at 56°C for 4 sec and extension at 72°C for 12 sec. A final extension step for 7 min at 72°C was also carried out. The products were stored at 4°C. Amplified products were later mixed and buffered with restriction endonuclease HinFI and sustained in the water bath kettle overnight for digestion. The products were observed following polyacrylamide gel electrophoresis.
Statistical analysis
Statistical analyses were performed using SPSS 13.0 statistical software (SPSS Inc., Chicago, IL, USA) and data were shown as the mean ± standard deviation. Comparisons between the two groups were performed by the independent t-test; χ2 analysis was applied to determine the difference in the genotype and gene frequency. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated from unconditional logistic regression models. P<0.05 was considered to indicate a statistically significant result.
Results
Fig. 1 shows polyacrylamide gel electrophoresis with restriction endonuclease HinFI. There are 3 genotypes, CC, CT and TT, in the MTHFR gene at position 677 in the two groups. Table II shows that the distribution of the MTHFR C677T gene polymorphisms in the controls was in Hardy-Weinberg equilibrium. Table III shows the MTHFR C677T genotype frequencies and allele frequencies between the two groups (χ2=4.690; P=0.030). The frequencies of the C and T allele were 56.1 and 43.9% in the CRC group, and 61.6 and 38.4% in the controls, respectively. The MTHFR C677T frequencies of the CC, CT and TT genotypes were 33.5, 45.1 and 21.4% among the CRC patients and 37.6, 48.1 and 14.3% in the controls, respectively (χ2=6.327; P=0.042). Table IV shows that, compared with the CC genotype, the TT genotype was significantly correlated with an increased risk of CRC (OR, 1.671; 95% CI, 1.094–2.553; P=0.018).
Figure 1.
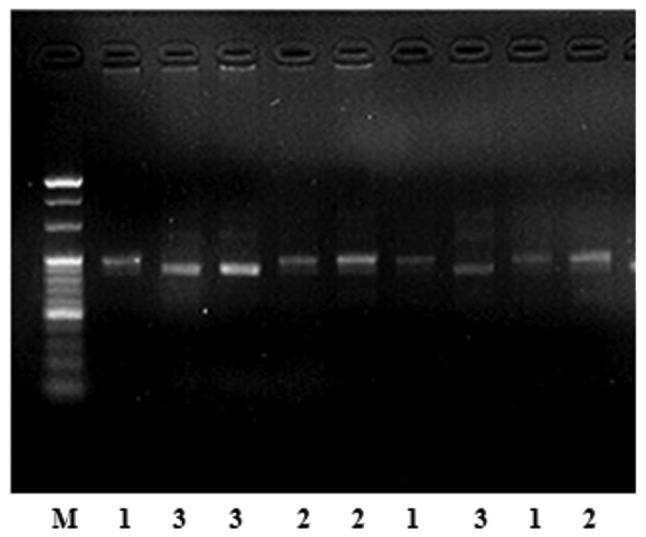
MTHFR C677T gene enzyme-cut electrophoresis with restriction endonuclease HinFI. M, DNA molecular weight marker; lane 1, CC genotype; lane 2, CT genotype; lane 3, TT genotype. MTHFR, methylenetetrahydrofolate reductase.
Table II.
MTHFR C677T genotype distribution in Hardy-Weinberg equilibrium.
| Gene | Genotype | Predicted value | Observed value | χ2 | P-value |
|---|---|---|---|---|---|
| MTHFR C677T | CC | 140 (37.9) | 139 (37.6) | 0.066 | 0.967 |
| CT | 175 (47.4) | 178 (48.1) | |||
| TT | 55 (14.7) | 53 (14.3) |
In the control group, the three genotypes have a distribution in Hardy-Weinberg equilibrium, χ2=0.066, P=0.967. MTHFR, methylenetetrahydrofolate reductase.
Table III.
MTHFR C677T genotype frequency and allele frequency between the two groups.
| Genotypes and alleles | Controls (n=370) n (%) |
Cases (n=370) n (%) |
χ2 | P-value |
|---|---|---|---|---|
| C | 456 (61.6) | 415 (56.1) | 4.690 | 0.030 |
| T | 284 (38.4) | 325 (43.9) | ||
| CC | 139 (37.6) | 124 (33.5) | 6.327 | 0.042 |
| CT | 178 (48.1) | 167 (45.1) | ||
| TT | 53 (14.3) | 79 (21.4) |
TT genotype and T allele were significantly higher in frequency in the CRC group than in the control group. MTHFR, methylenetetrahydrofolate reductase; CRC, colorectal cancer.
Table IV.
MTHFR C677T genotype frequencies and the CRC risk.
| Genotypes | Controls (n=370) n (%) |
Cases (n=370) n (%) |
OR (95% CI) | P-value |
|---|---|---|---|---|
| CC | 139 (37.6) | 124 (33.5) | 1 | |
| CT | 178 (48.1) | 167 (45.1) | 1.052 (0.763–1.450) | 0.758 |
| TT | 53 (14.3) | 79 (21.4) | 1.671 (1.094–2.553) | 0.018 |
| CT+TT | 231 (62.4) | 246 (66.5) | 1.194 (0.883–1.614) | 0.249 |
OR, odds ratio; 95% CI, 95% confidence interval. MTHFR, methylenetetrahydrofolate reductase; CRC, colorectal cancer.
The plasma levels of Hcy in the group of patients with CRC (12.63±3.11 μmol/l) was significantly higher than that in the control group (10.87±2.42 μmol/l; P<0.05). The levels of Hcy among the different genotypes are shown in Table V. In the CRC group (F=37.346; P<0.001), the level of Hcy in subjects with the CT or TT genotypes (14.47±3.92 μmol/l) was significantly higher than that in subjects with the CC genotype (11.67±2.03 μmol/l; P<0.05). In the control group (F=46.241; P<0.001), the level of Hcy in subjects with the CT or TT genotypes (12.08±3.53 μmol/l) was significantly higher than that in subjects with the CC genotype (9.54±1.72 μmol/l; P<0.05). In each genotype, the level of Hcy was higher in the CRC group than in the control group (t=22.179 for CC, P<0.001; t=10.966 for CT, P<0.001; t=2.449 for TT, P=0.016; t=7.654 for CT+TT, P<0.001).
Table V.
Plasma Hcy levels among the different genotypes.
| Genotypes | Cases (μmol/l) | Controls (μmol/l) | t | P-value |
|---|---|---|---|---|
| CC | 11.67±2.03 | 9.54±1.72 | 22.179 | <0.001 |
| CT | 12.94±2.68a | 9.98±2.33 | 10.966 | <0.001 |
| TT | 15.36±3.44a,b | 13.79±3.85a,b | 2.449 | 0.016 |
| CT+TT | 14.47±3.29a,b,c | 12.08±3.53a,b,c | 7.654 | <0.001 |
| F | 37.346 | 46.241 | - | - |
| P | <0.001 | <0.001 | - | - |
P<0.05 vs. CC;
P<0.05 vs. CT;
P<0.05 vs. TT.
Hcy, homocysteine.
Discussion
In the general population, hyperhomocysteinemia has been found to be associated with the occurrence of adenomas and CRC (6). However, in the absence of low serum folate, the presence of a mutation of the MTHFR gene which is responsible for hyperhomocysteinemia has not been identified as an oncogenic risk factor. Hcy is formed from methionine and is either catabolized in the vitamin B6-dependent transsulfuration pathway or remethylated into methionine (13). This latter reaction is catalyzed by methionine synthase, which requires 5-methyltetrahydrofolate as a substrate and vitamin B12 as a co-factor; 5-methyltetrahydrofolate is formed by the reduction of 5,10-methylenetetrahydrofolate by MTHFR, which is a regulating enzyme in Hcy metabolism. A 677C-T mutation was detected in the MTHFR gene and homozygosity for this genotype was found to be associated with a decreased specific enzyme activity and elevated Hcy.
MTHFR is an essential enzyme in the metabolism of folic acid and catalyzes the irreversible reduction of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate (14). A change of C to T at nucleotide 677 in MTHFR C677T results in an amino acid sequence change of an alanine to valine, and this protein is associated with reduced enzyme activity that leads to reduced plasma folate levels. The low enzyme activity of MTHFR C677T variant genotypes is associated with DNA hypomethylation, which may induce genomic instability and thereby affect the expression of oncogenes or tumor suppressor genes.
It has been shown that Hcy levels are positively associated with the proliferation rates of cells in a variety of tumors (15,16), as well as with oxidative damage to cells. Hcy has been considered to possess antioxidant properties through its rate-limiting role in the biosynthesis of glutathione, the intracellular antioxidant and detoxifying agent (17,18). However, evidence from in vivo and in vitro studies suggests that Hcy acts as a pro-oxidant agent that causes DNA oxidative damage as a result of the overproduction of free radicals and hydrogen peroxide, leading to gene mutation and subsequent cancer development. Elevated levels of Hcy are also associated with several metabolic disorders, including high body mass index, high plasma triglyceride levels, hypertension and the abnormal oxidation of low-density lipoproteins (19), which may lead to the development of several types of cancer, including CRC.
A number of studies concerning the molecular mechanisms of hyperhomocysteinemia in oncogenesis have been performed. It is known that Hcy levels are raised in colic tissue in inflammatory bowel disease (20). The increase in homocysteinemia may present an oncogenic risk in two ways. Firstly, it increases cellular oxidative stress by stimulating and increasing the secretion of proinflammatory cytokines, including TNF and IL-12 (21). Chronic inflammation is now known to increase carcinogenic risk. Secondly, hyperhomocysteinemia is responsible for a fault in cellular methylation (22). Hypomethylated DNA is unstable and subject to breakage of DNA strands. In the case of the hypomethylation of DNA, there is an increase in the number of non-methylated cytosines, which are deaminated in uracyls and therefore are incorporated more easily. Uracyl DNA glycosylase repairs these errors by excising the abnormally present uracyls in the DNA, thus creating temporary breaks (23). The likelihood of the two DNA strands breaking is increased, thus increasing carcinogenic risk.
The results of our study have shown that the frequency of MTHFR TT homozygotes was 21.4% in our CRC group, which was higher than in a UK study (14.0%) (24). Compared with wild-type individuals, carriers of the MTHFR 677TT genotype are more likely to develop CRC. The percentage of subjects with the T allele has been found to be 43.9%. The results of MTHFR genotyping in different populations showed an overall T allele prevalence of 32%. In North America (25), this value was 35%. It is thought that the TT genotype rarely occurs in African-Americans (26), whereas among healthy Japanese (27) its prevalence is 14.7%. Our study showed that, in controls, the prevalence of this polymorphism was 14.3% and that of the T allele was 38.4%, markedly higher than in a Polish study (28), with 4.4 and 21.5%, respectively. The observed trend (overall TT versus CC: OR, 1.671; 95% CI, 1.094–2.553; P=0.018) indicates a pathogenic effect. The plasma level of Hcy in the CRC group (12.63±3.11 μmol/l) was significantly higher than that in the control group (10.87±2.42 μmol/l; P<0.05). In the CRC group, the level of Hcy in subjects with the CT or TT genotypes was significantly higher than in subjects with the CC genotype. In the control group, the level of Hcy in subjects with the CT or TT genotypes was significantly higher than in subjects with the CC genotype. The plasma Hcy level was associated with the polymorphism of MTHFR C677T. In our study, the frequency of the MTHFR 677TT genotype in the CRC group was significantly higher than that in the control group, suggesting an increased risk for CRC in patients with the MTHFR 677TT genotype. The T allele frequency in the CRC group was increased when compared with healthy control subjects, which was consistent with the results of other studies.
In conclusion, the results of this study suggest that the MTHFR C677T polymorphism indicates susceptibility to CRC and is correlated with CRC pathogenesis. The plasma Hcy level in CRC patients was higher than that in healthy control subjects and the frequency of the MTHFR 677TT genotype in CRC patients was markedly increased when compared with healthy subjects. These findings suggest that the MTHFR C677T polymorphism is a candidate genetic risk factor for CRC.
References
- 1.Choi SW, Mason JB. Folate and carcinogenesis: an integrated scheme. J Nutr. 2000;130:129–132. doi: 10.1093/jn/130.2.129. [DOI] [PubMed] [Google Scholar]
- 2.Duthie SJ. Folic acid deficiency and cancer: mechanisms of DNA instability. Br Med Bull. 1999;55:578–592. doi: 10.1258/0007142991902646. [DOI] [PubMed] [Google Scholar]
- 3.Mason JB, Choi SW. The mechanisms by which folate depletion enhances colorectal carcinogenesis: a unified scheme. Nestle Nutr Workshop Ser Clin Perform Programme. 2000;4:87–101. doi: 10.1159/000061830. [DOI] [PubMed] [Google Scholar]
- 4.Whiteside MA, Heimburger DC, Johanning GL. Micronutrients and cancer therapy. Nutr Rev. 2004;62:142–147. doi: 10.1111/j.1753-4887.2004.tb00036.x. [DOI] [PubMed] [Google Scholar]
- 5.Peyrin-Biroulet L, Barraud H, Ancel D, Petit-Laurent F, Bigard MA, Gueant JL, Bronowicki JP. Folate metabolism and colorectal carcinogenesis. Gastroenterol Clin Biol. 2004;28:582–592. doi: 10.1016/s0399-8320(04)95015-4. [DOI] [PubMed] [Google Scholar]
- 6.Kato I, Dnistrian AM, Schwartz M, et al. Serum folate, homocysteine and colorectal cancer risk in women: a nested case-control study. Br J Cancer. 1999;79:1917–1922. doi: 10.1038/sj.bjc.6690305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen K, Song L, Jin MJ, Fan CH, Jiang QT, Yu WP. Association between genetic polymorphisms in folate metabolic enzyme genes and colorectal cancer: a nested case-control study. Zhonghua Zhong Liu Za Zhi. 2006;28:429–432. (In Chinese) [PubMed] [Google Scholar]
- 8.Le Marchand L, Wilkens LR, Kolonel LN, Henderson BE. The MTHFR C677T polymorphism and colorectal cancer: the multiethnic cohort study. Cancer Epidemiol Biomarkers Prev. 2005;14:1198–1203. doi: 10.1158/1055-9965.EPI-04-0840. [DOI] [PubMed] [Google Scholar]
- 9.Kim JK, Kim S, Han JH, et al. Polymorphisms of 5,10-methylenetetrahydrofolate reductase and risk of stomach cancer in a Korean population. Anticancer Res. 2005;25:2249–2252. [PubMed] [Google Scholar]
- 10.Zeybek U, Yaylim I, Yilmaz H, et al. Methylenetetrahydrofolate reductase C677T polymorphism in patients with gastric and colorectal cancer. Cell Biochem Funct. 2007;25:419–422. doi: 10.1002/cbf.1317. [DOI] [PubMed] [Google Scholar]
- 11.Vollset SE, Igland J, Jenab M, et al. The association of gastric cancer risk with plasma folate, cobalamin, and methylenetetrahydrofolate reductase polymorphisms in the European Prospective Investigation into Cancer and Nutrition. Cancer Epidemiol Biomarkers Prev. 2007;16:2416–2424. doi: 10.1158/1055-9965.EPI-07-0256. [DOI] [PubMed] [Google Scholar]
- 12.Frosst P, Blom HJ, Milos R, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 13.Finkelstein JD. The metabolism of homocysteine: pathways and regulation. Eur J Pediatr. 1998;157:40–44. doi: 10.1007/pl00014300. [DOI] [PubMed] [Google Scholar]
- 14.Gudnason V, Stansbie D, Scott J, Bowron A, Nicaud V, Humphries S. C677T (thermolabile alanine/valine) polymorphism in methylenetetrahydrofolate reductase (MTHFR): its frequency and impact on plasma homocysteine concentration in different European populations. Atherosclerosis. 1998;136:347–354. doi: 10.1016/s0021-9150(97)00237-2. [DOI] [PubMed] [Google Scholar]
- 15.Hultdin J, Van Guelpen B, Bergh A, Hallmans G, Stattin P. Plasma folate, vitamin B12, and homocysteine and prostate cancer risk: a prospective study. Int J Cancer. 2005;113:819–824. doi: 10.1002/ijc.20646. [DOI] [PubMed] [Google Scholar]
- 16.Martínez-Poveda B, Chavarría T, Sánchez-Jiménez F, Quesada AR, Medina MA. An in vitro evaluation of the effects of homocysteine thiolactone on key steps of angiogenesis and tumor invasion. Biochem Biophys Res Commun. 2003;311:649–653. doi: 10.1016/j.bbrc.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 17.Chavarría T, Rodríguez-Nieto S, Sánchez-Jiménez F, Quesada AR, Medina MA. Homocysteine is a potent inhibitor of human tumor cell gelatinases. Biochem Biophys Res Commun. 2003;303:572–575. doi: 10.1016/s0006-291x(03)00382-6. [DOI] [PubMed] [Google Scholar]
- 18.Schroecksnadel K, Frick B, Fiegl M, Winkler C, Denz HA, Fuchs D. Hyperhomocysteinaemia and immune activation in patients with cancer. Clin Chem Lab Med. 2007;45:47–53. doi: 10.1515/CCLM.2007.012. [DOI] [PubMed] [Google Scholar]
- 19.Lever M, George PM, Dellow WJ, Scott RS, Chambers ST. Homocysteine, glycine betaine, and N,N-dimethylglycine in patients attending a lipid clinic. Metabolism. 2005;54:1–14. doi: 10.1016/j.metabol.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Morgenstern I, Raijmakers MT, Peters WH, et al. Homocysteine, cysteine, and glutathione in human colonic mucosa: elevated levels of homocysteine in patients with inflammatory bowel disease. Dig Dis Sci. 2003;48:2083–2090. doi: 10.1023/a:1026338812708. [DOI] [PubMed] [Google Scholar]
- 21.Poddar R, Sivasubramanian N, DiBello PM, et al. Homocysteine induces expression and secretion of monocyte chemoattractant protein-1 and interleukin-8 in human aortic endothelial cells: implications for vascular disease. Circulation. 2001;103:2717–2723. doi: 10.1161/01.cir.103.22.2717. [DOI] [PubMed] [Google Scholar]
- 22.Kim YI, Pogribny IP, Basnakian AG, et al. Folate deficiency in rats induces DNA strand breaks and hypomethylation within the p53 tumor suppressor gene. Am J Clin Nutr. 1997;65:46–52. doi: 10.1093/ajcn/65.1.46. [DOI] [PubMed] [Google Scholar]
- 23.Blount BC, Mack MM, Wehr CM, et al. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci USA. 1997;94:3290–3295. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams M, Smith PD, Martin D, Thompson JR, Lodwick D, Samani NJ. Genetic analysis of thermolabile methylenetetrahydrofolate reductase as a risk factor for myocardial infarction. QJM. 1996;89:437–444. doi: 10.1093/qjmed/89.6.437. [DOI] [PubMed] [Google Scholar]
- 25.Zuntar I, Topić E, Vukosavić D, et al. Croatian population data for the C677T polymorphism in methylenetetrahydrofolate reductase: frequencies in healthy and atherosclerotic study groups. Clin Chim Acta. 2003;335:95–100. doi: 10.1016/s0009-8981(03)00283-3. [DOI] [PubMed] [Google Scholar]
- 26.Motulsky AG. Nutritional ecogenetics: homocysteine-related arteriosclerotic vascular disease, neural tube defects, and folic acid. Am J Hum Genet. 1996;58:17–20. [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Miyaki K, Araki J, Nakayama T, Muramatsu M. The relation between nicotinamide N-methyltransferase gene polymorphism and plasma homocysteine concentration in healthy Japanese men. Thromb Res. 2007;121:55–58. doi: 10.1016/j.thromres.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 28.Goracy I, Cyrylowski L, Kaczmarczyk M, et al. C677T polymorphism of the methylenetetrahydrofolate reductase gene and the risk of ischemic stroke in Polish subjects. J Appl Genet. 2009;50:63–67. doi: 10.1007/BF03195654. [DOI] [PubMed] [Google Scholar]


