Abstract
The molecular mechanisms underlying the reduced penetrance seen in the nonsense-mediated decay–positive (NMD+) BMPR2 mutation–associated hereditary pulmonary arterial hypertension (HPAH) remain unknown. We reasoned that the cellular and genetic mechanisms behind this phenomenon could be uncovered by combining expression profiling with Connectivity Map (cMap) analysis. Cultured lymphocytes from 10 patients with HPAH and 10 matched familial control subjects, all with NMD+ BMPR2 mutations, were subjected to expression analysis. For each group, the expression data were combined before analysis. This generated a signature of 23 up-regulated and 12 down-regulated genes in patients with HPAH compared with control subjects (the “PAH penetrance signature”). Although gene set enrichment analysis of this signature was not uniquely informative, cMap analysis identified drugs with expression signatures similar to the PAH penetrance signature. Several of these drugs were predicted to influence reactive oxygen species (ROS) formation. This hypothesis was tested and confirmed in the same cells initially subjected to the expression analysis using quantitative biochemical detection of ROS concentration. We conclude that expression of the PAH penetrance signature represents an increased risk of developing clinical HPAH and that ROS formation may play a role in pathogenesis of HPAH. These results provide the first molecular insights into NMD+ BMPR2 related HPAH penetrance and highlight the potential utility of cMap analyses in pulmonary research.
Keywords: hereditary pulmonary arterial hypertention, ROS, connectivity map, expression signature, BMPR2
Clinical Relevance
Even though BMPR2 mutations are known to cause hereditary pulmonary arterial hypertension (HPAH), only 20% of mutation carriers get disease. We present the first expression analysis of patients with HPAH and carriers (with NMD+ BMPR2 mutations). Our study clearly demonstrates that a layered bioinformatics approach using cMap analysis can generate meaningful data and testable hypotheses. Our data suggest that ROS formation may be a determinant of HPAH penetrance.
Pulmonary arterial hypertension (PAH) is a progressive, fatal disease, and most patients with PAH have a poor prognosis despite standard-of-care therapies (1). PAH is characterized by vascular remodeling of the distal pulmonary arteries (100–200 μM in size) via smooth muscle hypertrophy and intimal endothelial cell proliferation, effectively decreasing the surface area of the pulmonary vasculature (2, 3). The resulting increase in pulmonary vascular resistance leads to the failure of a progressively overloaded right ventricle and, eventually, death. The heritable form of PAH (HPAH) is usually (>80% of the time) due to germline mutations in the Bone Morphogenetic Protein Receptor 2 (BMPR2) (4–7), whereas 5 to 25% of patients diagnosed as having idiopathic (I)PAH have a detectable germline mutation in BMPR2 as well (5, 12–15).
In HPAH, BMPR2 mutations can produce stable transcripts or premature termination codons, resulting in the mutated transcript being rapidly degraded through the nonsense-mediated decay (NMD) pathway (8). NMD is an mRNA surveillance system that degrades transcripts containing premature termination codons to prevent translation of unnecessary or harmful transcripts (9). Thus, patients with PAH with NMD-positive (NMD+) BMPR2 mutations have disease due to haploinsufficiency, whereas patients whose mutations are NMD negative (NMD−) may have disease due to a dominant negative mechanism.
BMPR2 mutations constitute the largest known risk for developing PAH; however, relatives within HPAH kindreds who are BMPR2 mutation carriers have only a 20% chance of developing the disease. We have previously shown that the expression of nonmutated wild-type BMPR2 allele transcript may be one molecular mechanism of this observed reduced penetrance; however, it is likely that there are unknown additional factors and pathways that influence disease risk (10, 11). Identification of such pathways that differ between affected mutation carriers and unaffected mutation carriers represents a strategy for gaining additional molecular insights into HPAH penetrance and possible discovery of new treatment options. Previous approaches to identify molecular pathways important in HPAH penetrance have relied on tissues collected in end-stage disease (typically during autopsy or transplant), all profoundly compromised by drug and end-stage disease effects. While providing significant contributions to our understanding of end stage HPAH, these approaches are open to the legitimate criticism that end-stage lung changes may not reflect initiating disease mechanisms (12).
The Broad Institute's Connectivity Map (cMap) is a public database (www.broad.mit.edu/cmap/) that contains approximately 7,000 gene expression profiles from four cell lines—MCF7 (breast cancer epithelial cell line), PC3 (prostate cancer epithelial cell line), HL60 (myeloid cell line), and SLMEL5 (melanoma cell line)—treated with over 1,300 FDA-approved small-molecule drugs. The cMap is based on the hypothesis that a connection can be made between a disease process, a disease-modifying gene, and a drug that influences the expression of that gene (13–15). Of particular relevance to this study is that the pathways affected were not particularly sensitive to cell type (14). By knowing the molecular targets/effects of the drug, the shared gene expression profile between the drug and the disease can thus point to potentially useful biochemical and cellular pathways to investigate further as disease modifiers. Thus, the cMap database is a hypothesis generation tool that cannot provide clues toward possible treatment options and uncover new potential pathways of importance in the disease of interest.
To identify molecular pathways that might contribute to disease penetrance, we compared expression profiles of cultured lymphocytes (CLs) derived from patients with HPAH (positive for NMD+ BMPR2 mutation) to familial-matched individuals without HPAH (positive for NMD+ BMPR2 mutation), thereby normalizing any potential effects of the BMPR2 mutation itself. Not only is the comparison of expression signatures of affected versus unaffected NMD+ BMPR2 mutation carriers completely novel, but our subsequent analysis, using the cMap database in PAH (specifically) and in pulmonary disease (generally), is novel as well. Our data show that the cMap can be used to discover cellular pathways important in a disease, which would not be discoverable using conventional methodology.
Materials and Methods
PAH Patient Samples
CLs from 20 subjects with NMD+ BMPR2 mutations (10 patients with HPAH and 10 unaffected carriers) were used in this study (5, 8, 16). Cells were established as previously described (17, 18). BMPR2 mutation and NMD status were experimentally determined and validated as previously described (5, 8).
Microarray and Gene Set Enrichment Analysis
RNA was extracted and used to probe Affymetrix human exon 1.0 ST arrays. Raw data were then analyzed by the Partek package at the Vanderbilt Functional Genomics Shared Resource to produce expression intensities for each probe set. To study pathway level differences between groups, gene set enrichment analysis (GSEA) was conducted on the microarray gene expression data (19). We used the Gene Ontology Biological Process (c5 from the MSigDB database at http://www.broadinstitute.org/ gsea/msigdb/index.jsp) as our gene set database.
In Silico Functional Analysis of PAH Penetrance Signature Using Ingenuity Pathway Analysis
We used the Ingenuity Pathway Analysis (IPA) in the Ingenuity System (www.ingenuity.com) to explore the functional features of our gene signature. Functional networks and enriched pathways were generated as previously described (20).
Defining Gene Signature for cMap Query and Analysis
To define a gene signature that represents the differences between patients and carriers, we adopted a two-step procedure. First, for each gene, we calculated the correlations of its expression values with the phenotypic categories. The correlation coefficients ± 0.5 were used to define a raw positive signature and a raw negative signature. These two signatures were combined, and genes were ordered by the absolute magnitudes of the correlations. This combined gene list was subjected to an optimization process as previously described (21). In brief, subsets of the pool signature were tested for their prediction power in differentiating the patient and carrier samples. We initially selected the top five genes from the preordered pool signature, and then five genes were added to the previous subset to form a new subset. Each subset was tested for its performance in predicting a sample's category. The prediction was evaluated using the false-positive and false-negative rates. A false-positive prediction is defined as the sample that is a carrier but is predicted to be a patient; a false-negative prediction is defined as the one that is a patient but is predicted to be a carrier. For each subset signature, all samples were iterated to count the false-positive and false-negative predictions. The subset (35 genes; see Figure E1 in the online supplement) that yielded the lowest false-positive and false-negative rates was selected as the final representative gene signature. We then converted our signature to the cMap-required format using the Affymetrix web tool NetAffx (http://www.affymetrix.com/). This signature was submitted to cMap query engine for connected profiles. The top 20 related small molecules were manually examined.
Reactive Oxygen Species Measurement
Cellular reactive oxygen species (ROS) generation was measured by two methods as described previously: (1) oxidation of dihydroethidium and analysis by flow cytometer (22) and (2) analysis of 2′,7′-dichlorofluorescein diacetate (Invitrogen, Grand Island, NY) at an excitation/emission wavelength of 490/530 nm using a microplate reader (Spectramax M5; Molecular Devices, Sunnyvale, CA) (23).
Results
Comparison of Expression Profiles Identifies a PAH Penetrance Signature
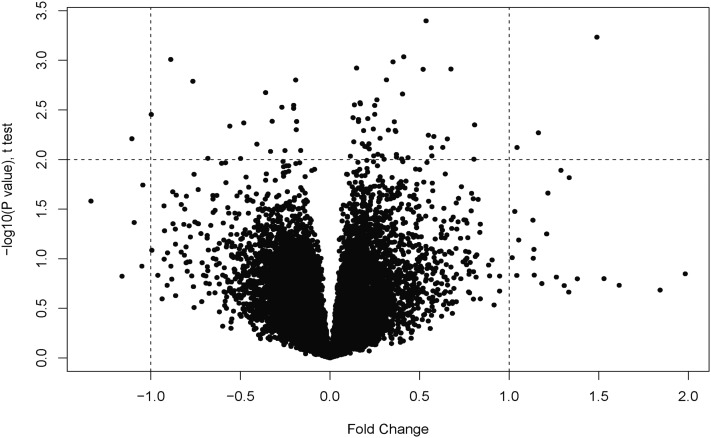
To gain insights into pathways that play a role in HPAH penetrance CLs from 10 patients with HPAH and 10 corresponding familial control subjects (“unaffected carriers”), all with NMD+ BMPPR2 mutations, were subjected to expression analysis (see Table 1 for patient characteristics and Figure E1 for BMPR2 expression levels of these samples). We limited our analysis to a single type of mutation (NMD+) to minimize the mutation-specific effects that are the case with NMD− mutations. Furthermore, because the patients and carriers have the same mutation type, the effects of the mutation are normalized. Our goal was to identify genes that were differentially expressed between patients and unaffected carriers. This analysis showed that only a limited number of genes are differentially expressed among the two groups (Figure 1; Table 2). These data were confirmed by real-time PCR analysis (Figure E2). We then used a Pearson correlation coefficient to confirm this finding. We found high correlation (r = 0.998), indicating that the two groups are similar at the expression level (Figure E3). This result is not surprising because the two groups were so closely matched. Overall, we identified a signature (the “PAH penetrance signature”) of 35 genes that were differentially regulated, of which 12 were up-regulated and 23 were down-regulated in patients (Table 2) compared with unaffected mutation carriers. A member of the BMP family, BMP8, was up-regulated in the patient group.
TABLE 1.
CLINICAL CHARACTERISTICS OF THE BMPR2 MUTATION CARRIERS
| Patients* | Carriers† | |
| Subjects, n | 10 | 10 |
| Women, n (%) | 7 (70.0) | 4 (40.0) |
| Age at diagnosis, yr (affected subject, AMCs) | 41.5 ± 13.1‡ | n/a |
| Current age or age at death, yr (healthy BMPR2 carrier subject, UMCs) | n/a | 62.7 ± 16.5 |
| Baseline hemodynamic data at diagnosis, mean (affected subject) | ||
| Right atrial pressure, mm Hg | 8.25 ± 6.6 | n/a |
| Mean PAP, mm Hg | 55.7 ± 5.6 | n/a |
| Cardiac index, L/min/m2 | 2.08 ± 1.1 | n/a |
| Indexed PVR, U/m2 | 17.67 ± 14.2 | n/a |
Definition of abbreviations: n/a = not available; PAP = pulmonary artery pressure; PVR = pulmonary vascular resistance.
Patients: BMPR2 mutation carriers with disease.
Carriers: BMPR2 mutation carriers without disease.
Data are presented as mean ± SD unless otherwise stated.
Figure 1.
Volcano plot showing up- and down-regulated genes. Raw data were analyzed by the Partek software package at the Vanderbilt Functional Genomics Shared Resource.
TABLE 2.
HEREDITARY PULMONARY ARTERIAL HYPERTENSION PENETRANCE SIGNATURE IN NONSENSE-MEDIATED DECAY–POSITIVE MUTATION CARRIERS
| Probe ID | Fold Change (Log base 2) | Entrez ID | Gene Name* |
| 3577443 | −2.80786447 | 51676 | Ankyrin repeat and SOCS box-containing 2 (ASB2) |
| 3403414 | −2.23944727 | 359787 | Developmental pluripotency associated 3 (DPPA3) |
| 3920850 | −1.74975727 | 3772 | Potassium inwardly rectifying channel, subfamily J, member 15 (KCNJ15) |
| 3734355 | −1.43369476 | 55890 | G-protein–coupled receptor, family C, group 5, member C (GPRC5C) |
| 3415046 | −1.33007436 | 283401 | Hypothetical protein FLJ33996 |
| 3268895 | −1.28707489 | 2849 | G-protein–coupled receptor 26 (GPR26) |
| 2488038 | −1.28256327 | 55577 | N-acetylglucosamine kinase (NAGK) |
| 2835662 | −1.24426623 | 85027 | MSTP150 |
| 3960042 | −1.19884239 | 6753 | Somatostatin receptor 3 (SSTR3) |
| 3377385 | −1.18904359 | 10004 | N-acetylated α-linked acidic dipeptidase-like 1 (NAALADL1) |
| 3619116 | −1.18891988 | 11245 | G-protein–coupled receptor 176 (GRP176) |
| 3442941 | −1.15759035 | 719 | Complement component 3a receptor 1 (C3AR1) |
| 3348940 | −1.1248184 | 83875 | β-carotene oxygenase 2 (BCO2) |
| 3789680 | −1.12328802 | 51046 | ST8 α-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (ST8SIA3) |
| 3146723 | −1.11646583 | 169166 | Sorting nexin 31 (SNX31) |
| 3216195 | −1.1164567 | 3293 | Hydroxysteroid (17-beta) dehydrogenase 3 (HSD17B3) |
| 2633737 | −1.10829105 | 84873 | G-protein–coupled receptor 128 (GRP128) |
| 3624003 | −1.09905562 | 1588 | Cytochrome P450, family 19, subfamily A, polypeptide 1 (CYP19A1) |
| 3549264 | −1.09331377 | 57578 | KIAA1409 |
| 3023295 | 1.137712078 | 5605 | Mitogen-activated protein kinase kinase 2 (MAP2K2) |
| 2964139 | 1.141819416 | 2570 | γ-aminobutyric acid (GABA) receptor, rho 2 (GABRR2) |
| 2408062 | 1.15072843 | 656 | Bone morphogenetic protein 8b (BMP8b) |
| 3056226 | 1.15109356 | 6804 | Syntaxin 1A (brain) (STX1A) |
| 2377020 | 1.20453073 | 50604 | Interleukin 20 (IL20) |
| 2434716 | 1.282768294 | 29956 | LAG1 homolog, ceramide synthase 2 (CERS2) |
| 2356425 | 1.473341637 | 5174 | PDZ domain containing 1 (PDZK1) |
| 2939469 | 1.699583803 | 221749 | Chromosome 6 open reading frame 145 (C6orf145) |
Up- and down-regulated genes are shown.
GSEA Confirms Enrichment of Key Cellular Pathways Associated with Growth and Metabolism
To gain further pathway-specific biological insights into HPAH penetrance, the expression signatures were analyzed using GSEA. One major advantage of this method is that it capitalizes on established gene associations, and thus relatively small individual gene expression changes can be combined to create strong correlations between a gene set and a particular phenotype. In our analysis, we used the Gene Ontology Biological Process (c5) from the MSigDB database (http://www.broadinstitute.org/gsea/msigdb/index.jsp). The top pathways enriched in CLs from patients and unaffected mutation carriers are shown in Tables 3 and 4, respectively. CLs from patients were enriched in pathways involved in apoptosis, cell division, cell cycle regulation, regulation of G-protein–coupled signaling pathways, and chromatin remodeling (Table 3). CLs from unaffected mutation carriers were enriched in pathways involved in the regulation of metabolic processes, phosphorylation, proliferation, and G-protein–coupled signaling, among others (Table 4).
TABLE 3.
TOP-ENRICHED PATHWAYS IN CULTURED LYMPHOCYTES FROM NONSENSE-MEDIATED DECAY–POSITIVE BMPR2 MUTATION CARRIERS WITH PULMONARY ARTERIAL HYPERTENSION
| Pathway | Genes (n) | Nes | Nom P Value |
| Apoptotic nuclear changes | 19 | 1.671 | 0.010 |
| Cell division | 17 | 1.600 | 0.019 |
| Cytokinesis | 15 | 1.591 | 0.015 |
| Establishment of organelle localization | 16 | 1.576 | 0.034 |
| Regulation of G-protein–coupled receptor protein signaling pathway | 22 | 1.569 | 0.044 |
| M phase of mitotic cell cycle | 83 | 1.569 | 0.041 |
| Nuclear organization and biogenesis | 30 | 1.565 | 0.019 |
| Chromatin remodeling | 23 | 1.562 | 0.032 |
| Mitosis | 81 | 1.548 | 0.051 |
| Mitotic cell cycle | 147 | 1.548 | 0.047 |
| Cell cycle process | 188 | 1.539 | 0.053 |
| Organelle localization | 23 | 1.535 | 0.051 |
| Cell cycle phase | 167 | 1.525 | 0.059 |
| M phase | 112 | 1.516 | 0.058 |
| Interphase | 66 | 1.498 | 0.064 |
Definition of abbreviations: Nes, normalized error squared; Nom, nominal.
TABLE 4.
TOP-ENRICHED PATHWAYS IN CULTURED LYMPHOCYTES CELLS FROM NONSENSE-MEDIATED DECAY–POSITIVE BMPR2 MUTATION CARRIERS WITHOUT DISEASE
| Pathway | Genes (n) | Nes | Nom P Value |
| Negative regulation of metabolic process | 46 | −1.711 | 0.006 |
| Negative regulation of cellular protein metabolic process | 43 | −1.689 | 0.008 |
| Innate immune response | 23 | −1.572 | 0.014 |
| Protein autoprocessing | 32 | −1.532 | 0.031 |
| Protein amino acid autophosphorylation | 31 | −1.485 | 0.045 |
| Protein amino acid dephosphorylation | 61 | −1.440 | 0.059 |
| Cytokine and chemokine mediated signaling pathway | 20 | −1.435 | 0.041 |
| Regulation of immune response | 32 | −1.421 | 0.057 |
| Homophilic cell adhesion | 15 | −1.419 | 0.075 |
| Cellular defense response | 48 | −1.415 | 0.035 |
| Reproductive process | 145 | −1.410 | 0.034 |
| Leukocyte activation | 66 | −1.404 | 0.031 |
| T-cell proliferation | 18 | −1.404 | 0.023 |
| G protein signaling coupled to IP3 Sec. mess. phospholipase C activating | 40 | −1.403 | 0.038 |
| Cell activation | 73 | −1.396 | 0.036 |
For definition of abbreviations, see Table 3.
Functional Analysis of the Gene Signature
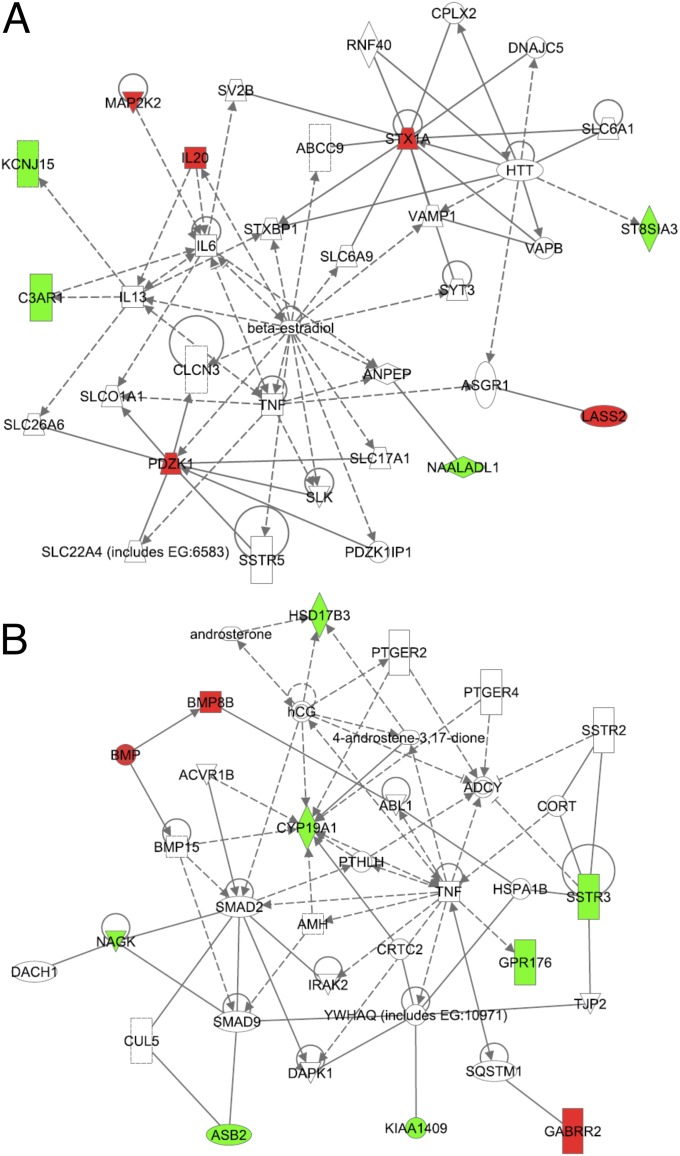
We used Ingenuity Pathway Analysis (IPA version 8.7) to determine whether the genes in our PAH penetrance signature belonged to any canonical pathways and to further delineate the network of genes connected to this signature. Using a threshold value of 0.01, we found three significant canonical pathways—estrogen signaling, BMP signaling, and IL-6 signaling—to be enriched in our signature (Table 5). The IPA identified two networks with very significant P values (1.0 × 10−21) (Figure 2). Network 1 included genes that have been shown to be involved in pulmonary hypertension or pathways related to pulmonary hypertension, including IL6, TNF, MAP2K2, solute carrier family members, and potassium channel family members (Figure 2A). Network 2 included SMAD as well as BMP family member genes, both known to be important in PAH pathogenesis. Network 2 also included another BMP family member, BMP15, in direct regulatory relationships with SMAD9 and SMAD2 as well as an inhibitory relationship with P450 aromatase (CYP19A1). Both networks included TNF, which may indicate that it has a role in PAH pathogenesis. Furthermore, this network analysis identified several small molecules in both networks (e.g., β-estradiol in Network 1) (Figure 2A), and androsterone and androstenedione in Network 2 (Figure 2B)—suggesting their functional role in PAH. Functional annotation of these networks showed that the top functions of these networks are important in cell–cell signaling, cell–cell interaction, cancer, and connective tissue disorders.
TABLE 5.
TOP CANONICAL PATHWAYS IDENTIFIED BY PATHWAY ANALYSIS
| Ingenuity Canonical Pathway | P Value |
| Estrogen-dependent signaling | 3.39 × 10−3 |
| BMP signaling pathway | 4.57 × 10−3 |
| IL-6 signaling | 6.61 × 10−3 |
Definition of abbreviation: BMP, bone morphogenetic protein.
Figure 2.
Networks generated by Ingenuity Pathway Analysis of the expression signature. A network is a graphical representation of the molecular relationships between genes and gene products. Genes or gene products are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). Red and green nodes represent genes positively and negatively correlated genes with the phenotypic categories in the signature, respectively. Solid lines represent protein–protein interactions; dashed lines represent regulatory relationships. All lines are supported by at least one reference from the literature, from a textbook, or from canonical information stored in the Ingenuity Pathways Knowledge Base.
cMap Analysis of the PAH Penetrance Signature
To identify pathways that might have been missed by GSEA, we queried the Broad Institute's cMap database with the PAH penetrance signature to identify drugs that produce an expression signature similar to the PAH penetrance signature. The cMap system uses a pattern comparison algorithm that is similar to the Kolmogorov-Smirnov test (24), which calculates the maximal distance between cumulative functions of two distributions. The given normalized similarity score indicated the comparative similarity between the PAH penetrance signature and the drug-treated cell line expression profiles in the cMap database. cMap analysis identified a number of candidate drugs with a similar expression signature to the PAH penetrance signature (Table 6). A review of the literature suggested that of the top 20 drugs (Table 6), 9 have been repeatedly identified in the literature as affecting ROS production directly or indirectly. This suggested to us that there might be differences in ROS production between patients and unaffected carriers.
TABLE 6.
DRUGS AND AGENTS IDENTIFIED BY CONNECTIVITY MAP ANALYSIS WITH EXPRESSION SIGNATURES SIMILAR TO THE PENETRANCE SIGNATURE
| Rank | Cmap name | Score | P Value | Pathways Affected |
| 1 | Viomycin | 0.914 | 0.00004 | Increased ROS by possible effect on mitochondria |
| 2 | Isoflupredone | 0.94 | 0.00032 | Transcription, phosphatidylinositol 3-kinase and protein kinase AKT stimulation, affects endothelial nitric oxide synthase (eNOS), and nitric oxide dependent vaso relaxation. SOD mimetic. |
| 3 | Nadolol | 0.871 | 0.00036 | Activation of adenylate cyclase through the action of G proteins |
| 4 | Heptaminol | 0.8 | 0.00078 | Increased cAMP |
| 5 | Isoniazid | 0.799 | 0.00078 | Mitochondrial energy transfer, increased ROS |
| 6 | Metronidazole | 0.784 | 0.00104 | Decrease ROS/energy and electron transport |
| 7 | CP-320650-01 | 0.638 | 0.00112 | |
| 8 | Fludrocortisone | 0.604 | 0.00243 | Oxidative uncoupling/increased ROS |
| 9 | Tetrahydroberberine | 0.808 | 0.00251 | K channel blocker/decreased ROS |
| 10 | Chenodeoxycholic acid | 0.781 | 0.0043 | Liver enzymes/increased ROS |
| 11 | Finasteride | 0.644 | 0.00598 | Inhibits activation of testosterone leading to increased estrogenic actions/increased ROS |
| 12 | PHA-00745360 | 0.564 | 0.00608 | |
| 13 | Vincamine | 0.642 | 0.00618 | Mitochondrial function/decreases ROS, sodium channels |
| 14 | Pindolol | 0.694 | 0.00663 | Activation of adenylate cyclase through the action of G proteins |
| 15 | Harmol | 0.754 | 0.00698 | Mitochondrial based apoptosis/decreases ROS |
| 16 | Fusidic acid | 0.753 | 0.00718 | Antimicrobial |
| 17 | Carbimazole | 0.845 | 0.00723 | Action on NF-κB and inhibition of apoptosis |
| 18 | Etiocholanolone | 0.634 | 0.00733 | Testosterone metabolite |
| 19 | Pentoxifylline | 0.685 | 0.00783 | Inhibits cAMP phosphodiesterase activity/decreased ROS |
| 20 | Pivmecillinam | 0.741 | 0.00863 | Induces hypocarnitemia which affects mitochondrial energy metabolism |
| 21 | Cefotiam | 0.74 | 0.00869 | GABA receptor blockage |
| 22 | Felbinac | 0.735 | 0.00975 | Typical NSAID effects also GABA and mitochondrial effect |
| 23 | Sulmazole | 0.822 | 0.01122 | Phosphodiesterase inhibitors, increased calcium |
| 24 | Meteneprost | 0.724 | 0.01156 | Anti-progestrone PGE2 analog |
| 25 | Adiphenine | 0.65 | 0.01456 | NAChR and sodium channel inhibition |
| 26 | 3-Acetamidocoumarin | 0.705 | 0.01574 | ROS scavenging |
| 27 | Pralidoxime | 0.705 | 0.01574 | Increased acetyl choline |
| 28 | Thiamazole | 0.586 | 0.01702 | |
| 29 | Paclitaxel | 0.586 | 0.01738 | Increased ROS |
Definition of abbreviations: Cmap = connectivity map; ROS = reactive oxygen species.
Affected BMPR2 Mutation Carriers Produce Higher Levels of ROS than Unaffected Mutation Carriers
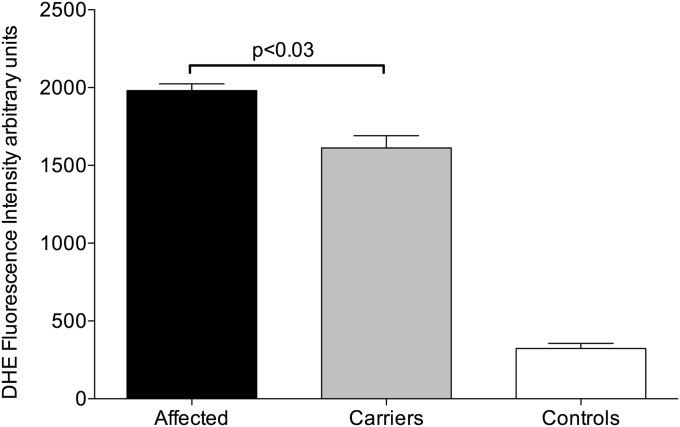
To determine whether the expression differences in carrier and PAH-affected CLs revealed by microarray analyses become manifest as a detectable change in ROS formation, CL pairs were subjected to ROS measurement using the dihydroethidium staining and flow cytometric analysis as well as a second method using the ROS-sensitive dye 2′,7′-dichlorofluorescein diacetate. CLs from patients with HPAH had higher ROS production than CLs from unaffected mutation carriers with both methods (Figure 3; Figure E4).
Figure 3.
Reactive oxygen species measurement using dihydroethidium (DHE) staining and flow cytometry. Aggregate data from CLs derived from eight affected and eight carrier individuals are presented. The error bars represent SEM. P values were two-tailed and were calculated using Student's t test. Statistical analysis was done using the Prism V software package for Mac.
Discussion
One of the most perplexing features of BMPR2-associated HPAH is its reduced penetrance. Nearly 80% of mutation carriers never develop the disease, but they can produce offspring that develop HPAH (11). We used gene expression profiling and layered bioinformatics analyses, including the use of cMap, to identify the cellular pathways involved in HPAH penetrance. Gene expression profiling identified a unique expression signature, the “PAH penetrance signature,” that characterized HPAH. Furthermore, cMap analysis of these data predicted that ROS production was different between patients with HPAH and unaffected mutation carriers. This hypothesis was confirmed by direct measurement of ROS in the CL cell lines used in the expression studies. Our study is unique on several levels in that this is the first expression interrogation of HPAH secondary to NMD+ BMPR2 mutations, the first to use the cMap database in respiratory disease, and the first to show that there is increased ROS production in patients with HPAH compared with unaffected mutation carriers. Thus, our studies demonstrate the potential role of ROS as a modifier of HPAH penetrance and show the unique and important role of cMap analysis as a discovery tool in pulmonary research.
Studies of gene expression in pulmonary hypertension in the past had several fundamental limitations, including the use of only idiopathic PAH and the use of diseased tissue, which resulted in expression signatures overwhelmed with end-stage and adaptive responses, making it impossible to discern cause from effect (25). A gap in our knowledge is the paucity of expression studies in familial PAH. Geraci and colleagues (26) and West and colleagues (27) analyzed a small number of patients with familial PAH; however, these two studies were done on patients and carriers with NMD− BMPR2 mutations. No studies have been reported where the expression signatures of NMD+ mutation carriers were analyzed. This study was designed to address this gap in our knowledge.
We used CLs free of disease and drug effects so that we could focus on the underlying genetic differences. Furthermore, we decided to use patient and carrier samples containing only one type of BMPR2 mutation (NMD+) so that any potential direct effects of the BMPR2 mutation on the expression profiles would be normalized. Our PAH penetrance signature included several G protein–coupled receptor genes, genes important in immune regulation, a potassium channel gene, and BMP8b. The GSEA of the raw data showed that affected individuals had enrichment of apoptosis, cytokinesis, G protein–coupled receptor signaling, and chromatin remodeling pathways. The carriers were enriched in negative regulation of metabolic processes, immune response, protein phosphorylation, and dephosphorylation and cytokine signaling pathways. These analyses suggest that the penetrance may be determined by subtle genetic differences in the known HPAH pathways, in particular the G-protein–coupled receptor pathways because that pathway was present in the final signature and in the GSEA. These findings are not surprising because G protein–coupled receptors have been implicated in pulmonary vasoconstriction pathways in several studies (28–35).
Our group has recently published two other papers examining CLs from patients with PAH. In the first of these, we assessed risk factors separating carriers from noncarriers in NMD− patients possessing a point mutation in the ligand-binding domain of BMPR2 (26). Differences in estrogen metabolism were the central risk factor identified a finding that matches the current data well (Figure 2; Table 4). However, the previous study did not identify metabolic differences as risk factors in NMD− patients. This may be because the previous study used a smaller sample size (three patients and four control subjects) or because of some fundamental difference between NMD− and NMD+ patients. We have also recently published a study concentrating on identifying risk factors in patients with idiopathic PAH (36). Because of its design, carriers and noncarriers with BMPR2 mutations were not well separated, but in that study patients with idiopathic and heritable PAH were distinguished by alterations in gene expression related to metabolism and oxidative stress, in agreement with the current study's assessment of metabolism as a central risk factor for development of PAH.
To gather additional information from our expression data and expand our understanding of the differences in the cellular processes at work in the NMD+ affected and carrier-derived cells, we subjected our penetrance signature to cMap analysis. Although the cMap database can be used for drug discovery, we decided to test the utility of the database in hypothesis generation in HPAH. Our goals were to determine whether cMap analysis can identify cellular pathways and processes that might have been missed using the standard expression signature and GSEA analysis. cMap analysis of the PAH penetrance signature indicated that ROS production might be different in the affected individuals compared with the carrier individuals, a hypothesis confirmed by direct ROS measurement in the CLs. These data represent a validation of our approach of using cMap analysis and suggest that ROS production may be an important modifier of the observed penetrance in BMPR2-related HPAH.
We have previously reported that patients with BMPR2 mutations, whether affected or carriers, produce higher levels of ROS, as assessed by oxidized lipids in urine, although in that study we did not distinguish between NMD+ and NMD− patients (37). The current study suggests that, at least in NMD+ patients, levels of ROS distinguish between affected patients and carriers. There are two possible explanations for this. First, it may just be a matter of degree: On a cellular level, patients with PAH may produce more ROS than carriers, who produce more than healthy control subjects. However, the difference may also lie in the ability to adapt to oxidative stress or the metabolic problems that underlie it, either directly through increased activity of systems that counteract oxidative stress (e.g., we have recently shown increased metallothioneins in carriers) or indirectly through increased function of salvage pathways in energy metabolism (36).
Our data cannot distinguish between the molecular mechanisms behind this observed increase in ROS production. For example, the increase in ROS production could be due to differences in mitochondrial metabolism (38–41) (which have recently been implicated in PAH) or could be symptomatic of a broader organismal metabolic phenotype. These areas warrant further investigation. Nevertheless, these differences are likely related to genetically determined differences in ROS production rather than adaptive to disease state or drug effect because the cells used in this study were in culture and thus had been removed from the disease environment.
Our study design has several limitations. This study uses only one cohort of patients, and thus its conclusion would need to be confirmed in another independent cohort. In this study, we used patient-derived CLs and not effector cells in the lungs (which can only be obtained in limited amounts and in special cases in affected individuals and cannot be obtained at all from unaffected carriers). Thus, caution is needed in interpreting data generated through the use these cells because the functions of the effector cells in the lungs (pulmonary microvascular endothelial smooth muscle cells) are likely different from CLs. Nevertheless, we have consistently shown in several previously published studies that CLs can be used for discovery approaches and to generate specific hypotheses about the role of BMPR2 in HPAH pathogenesis (5, 8, 11, 26, 42–43). Another potential limitation of this study is our use of CLs for ROS determination because ROS production may be different in effector lung cells compared with the CLs. However, ROS in effector cells derived from patients and unaffected carriers cannot be measured. Despite these limitations, we believe our study demonstrates that a layered bioinformatics approach using cMap analysis can generate meaningful data and testable hypotheses.
In summary, we present the first multifaceted bioinformatics analysis of the phenomenon of reduced penetrance in BMPR2-associated HPAH. This analysis was unique not only in terms of the subset of patients studied and the approach of using sequential bioinformatics tools including cMap analysis. cMap analysis offers a novel method to identify the role of undiscovered pathways in the pathogenesis of pulmonary disease. In addition, cMap analysis has the potential to identify possible FDA-approved pharmacological agents that may be useful for treating pulmonary diseases for which current treatment options are severely limited (e.g., PAH, pulmonary fibrosis, etc.). Because the cMap database contains only FDA approved agents, any therapies or preventive treatments identified in this way could be implemented immediately if shown to be effective.
Supplementary Material
Acknowledgments
The authors thank Melissa Stauffer of Scientific Editing Solutions for help in editing this manuscript.
Footnotes
This work was supported by NHLBI grant 1R01HL102020 (R.H.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2011-0251OC on February 3, 2012
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.McLaughlin VV, Shillington A, Rich S. Survival in primary pulmonary hypertension: the impact of epoprostenol therapy. Circulation 2002;106:1477–1482 [DOI] [PubMed] [Google Scholar]
- 2.Rai PR, Cool CD, King JA, Stevens T, Burns N, Winn RA, Kasper M, Voelkel NF. The cancer paradigm of severe pulmonary arterial hypertension. Am J Respir Crit Care Med 2008;178:558–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabinovitch M. Molecular pathogenesis of pulmonary arterial hypertension. J Clin Invest 2008;118:2372–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cogan JD, Pauciulo MW, Batchman AP, Prince MA, Robbins IM, Hedges LK, Stanton KC, Wheeler LA, Phillips III JA, Loyd JE, et al. High frequency of BMPR2 exonic deletions/duplications in familial pulmonary arterial hypertension. Am J Respir Crit Care Med 2006;174:590–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cogan JD, Vnencak-Jones CL, Phillips JA, III, Lane KB, Wheeler LA, Robbins IM, Garrison G, Hedges LK, Loyd JE. Gross BMPR2 gene rearrangements constitute a new cause for primary pulmonary hypertension. Genet Med 2005;7:169–174 [DOI] [PubMed] [Google Scholar]
- 6.Lane KB, Machado RD, Pauciulo MW, Thomson JR, Phillips JA, III, Loyd JE, Nichols WC, Trembath RC. Heterozygous germlie mutations in BMPR2, encoding a TGF-beta receptor, cause familial primary pulmonary hypertension: the international pph consortium. Nat Genet 2000;26:81–84 [DOI] [PubMed] [Google Scholar]
- 7.Deng Z, Morse JH, Slager SL, Cuervo N, Moore KJ, Venetos G, Kalachikov S, Cayanis E, Fischer SG, Barst RJ, et al. Familial primary pulmonary hypertension (PPH) is caused by mutations in the bone morphogenetic protein receptor-II gene. Am J Hum Genet 2000;67:737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cogan JD, Pauciulo MW, Batchman AP, Prince MA, Robbins IM, Hedges LK, Stanton KC, Wheeler LA, Phillips JA, III, Loyd JE, et al. High frequency of BMPR2 exonic deletions/duplications in familial pulmonary arterial hypertension. Am J Respir Crit Care Med 2006;174:590–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez CI, Bhattacharya A, Wang W, Peltz SW. Nonsense-mediated mRNA decay in saccharomyces cerevisiae. Gene 2001;274:15–25 [DOI] [PubMed] [Google Scholar]
- 10.Newman JH, Phillips JA, III, Loyd JE. Narrative review: the enigma of pulmonary arterial hypertension. New insights from genetic studies. Ann Intern Med 2008;148:278–283 [DOI] [PubMed] [Google Scholar]
- 11.Hamid R, Cogan JD, Hedges LK, Austin E, Phillips JA, III, Newman JH, Loyd JE. Penetrance of pulmonary arterial hypertension is modulated by the expression of normal BMPR2 allele. Hum Mutat 2009;30:649–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Austin ED, Hamid R, Ahmad F. Somatic mutations in pulmonary arterial hypertension: primary or secondary events? Am J Respir Crit Care Med 2010;182:1094–1096 [DOI] [PubMed] [Google Scholar]
- 13.Michnick SW. The connectivity map. Nat Chem Biol 2006;2:663–664 [DOI] [PubMed] [Google Scholar]
- 14.Lamb J. The connectivity map: a new tool for biomedical research. Nat Rev Cancer 2007;7:54–60 [DOI] [PubMed] [Google Scholar]
- 15.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 2006;313:1929–1935 [DOI] [PubMed] [Google Scholar]
- 16.Machado RD, Pauciulo MW, Thomson JR, Lane KB, Morgan NV, Wheeler L, Phillips JA, III, Newman J, Williams D, Galie N, et al. BMPR2 haploinsufficiency as the inherited molecular mechanism for primary pulmonary hypertension. Am J Hum Genet 2001;68:92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh HM, Oh JM, Choi SC, Kim SW, Han WC, Kim TH, Park DS, Jun CD. An efficient method for the rapid establishment of epstein-barr virus immortalization of human B lymphocytes. Cell Prolif 2003;36:191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bird AG, McLachlan SM, Britton S. Cyclosporin a promotes spontaneous outgrowth in vitro of epstein-barr virus-induced b-cell lines. Nature 1981;289:300–301 [DOI] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang HC, Zheng S, VanBuren V, Zhao Z. Discovering disease-specific biomarker genes for cancer diagnosis and prognosis. Technol Cancer Res Treat 2010;9:219–230 [DOI] [PubMed] [Google Scholar]
- 21.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002;415:530–536 [DOI] [PubMed] [Google Scholar]
- 22.Zhao H, Joseph J, Fales HM, Sokoloski EA, Levine RL, Vasquez-Vivar J, Kalyanaraman B. Detection and characterization of the product of hydroethidine and intracellular superoxide by HPLC and limitations of fluorescence. Proc Natl Acad Sci USA 2005;102:5727–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noguchi Y, Young JD, Aleman JO, Hansen ME, Kelleher JK, Stephanopoulos G. Effect of anaplerotic fluxes and amino acid availability on hepatic lipoapoptosis. J Biol Chem 2009;284:33425–33436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myles Hollander DAW. Nonparametric statistical methods. Hoboken, NJ: Wiley; 1999 [Google Scholar]
- 25.Menon S, Fessel J, West J. Microarray studies in pulmonary arterial hypertension. Int J Clin Pract Suppl 2011;65:19–28 [DOI] [PubMed] [Google Scholar]
- 26.West J, Cogan J, Geraci M, Robinson L, Newman J, Phillips JA, Lane K, Meyrick B, Loyd J. Gene expression in BMPR2 mutation carriers with and without evidence of pulmonary arterial hypertension suggests pathways relevant to disease penetrance. BMC Med Genomics 2008;1:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geraci MW, Moore M, Gesell T, Yeager ME, Alger L, Golpon H, Gao B, Loyd JE, Tuder RM, Voelkel NF. Gene expression patterns in the lungs of patients with primary pulmonary hypertension: a gene microarray analysis. Circ Res 2001;88:555–562 [DOI] [PubMed] [Google Scholar]
- 28.Chandra SM, Razavi H, Kim J, Agrawal R, Kundu RK, de Jesus Perez V, Zamanian RT, Quertermous T, Chun HJ. Disruption of the apelin-apj system worsens hypoxia-induced pulmonary hypertension. Arterioscler Thromb Vasc Biol 2011;31:814–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Damron DS, Kanaya N, Homma Y, Kim SO, Murray PA. Role of PKC, tyrosine kinases, and rho kinase in alpha-adrenoreceptor-mediated pasm contraction. Am J Physiol Lung Cell Mol Physiol 2002;283:L1051–L1064 [DOI] [PubMed] [Google Scholar]
- 30.Foletta VC, Lim MA, Soosairajah J, Kelly AP, Stanley EG, Shannon M, He W, Das S, Massague J, Bernard O. Direct signaling by the BMPR2 type II receptor via the cytoskeletal regulator limk1. J Cell Biol 2003;162:1089–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janssen LJ, Lu-Chao H, Netherton S. Excitation-contraction coupling in pulmonary vascular smooth muscle involves tyrosine kinase and rho kinase. Am J Physiol Lung Cell Mol Physiol 2001;280:L666–L674 [DOI] [PubMed] [Google Scholar]
- 32.Machado RD, Rudarakanchana N, Atkinson C, Flanagan JA, Harrison R, Morrell NW, Trembath RC. Functional interaction between BMPR-II and TCTEX-1, a light chain of dynein, is isoform-specific and disrupted by mutations underlying primary pulmonary hypertension. Hum Mol Genet 2003;12:3277–3286 [DOI] [PubMed] [Google Scholar]
- 33.Meiri D, Greeve MA, Brunet A, Finan D, Wells CD, LaRose J, Rottapel R. Modulation of rho guanine exchange factor IFC activity by protein kinase A-mediated phosphorylation. Mol Cell Biol 2009;29:5963–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagaoka T, Morio Y, Casanova N, Bauer N, Gebb S, McMurtry I, Oka M. Rho/rho kinase signaling mediates increased basal pulmonary vascular tone in chronically hypoxic rats. Am J Physiol Lung Cell Mol Physiol 2004;287:L665–L672 [DOI] [PubMed] [Google Scholar]
- 35.Wong WK, Knowles JA, Morse JH. Bone morphogenetic protein receptor type II c-terminus interacts with c-src: implication for a role in pulmonary arterial hypertension. Am J Respir Cell Mol Biol 2005;33:438–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Austin ED, Menon S, Hemnes AR, Robinson LR, Talati M, Fox KL, Cogan JD, Hamid R, Hedges LK, Robbins I, et al. Idiopathic and heritable PAH perturb common molecular pathways, correlated with increased MSX 1 expression. Pulm Circ 2011;1:389–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lane KL, Talati M, Austin E, Hemnes AR, Johnson JA, Fessel JP, Blackwell T, Mernaugh RL, Robinson L, Fike C, et al. Oxidative injury is a common consequence of BMPR2 mutations. Pulm Circ 2011;1:72–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piao L, Marsboom G, Archer SL. Mitochondrial metabolic adaptation in right ventricular hypertrophy and failure. J Mol Med 2010;88:1011–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rehman J, Archer SL. A proposed mitochondrial-metabolic mechanism for initiation and maintenance of pulmonary arterial hypertension in fawn-hooded rats: the warburg model of pulmonary arterial hypertension. Adv Exp Med Biol 2010;661:171–185 [DOI] [PubMed] [Google Scholar]
- 40.Archer SL, Gomberg-Maitland M, Maitland ML, Rich S, Garcia JG, Weir EK. Mitochondrial metabolism, redox signaling, and fusion: a mitochondria-ros-hif-1alpha-kv1.5 o2-sensing pathway at the intersection of pulmonary hypertension and cancer. Am J Physiol Heart Circ Physiol 2008;294:H570–H578 [DOI] [PubMed] [Google Scholar]
- 41.Bonnet S, Michelakis ED, Porter CJ, Andrade-Navarro MA, Thebaud B, Haromy A, Harry G, Moudgil R, McMurtry MS, Weir EK, et al. An abnormal mitochondrial-hypoxia inducible factor-1alpha-kv channel pathway disrupts oxygen sensing and triggers pulmonary arterial hypertension in fawn hooded rats: similarities to human pulmonary arterial hypertension. Circulation 2006;113:2630–2641 [DOI] [PubMed] [Google Scholar]
- 42.Meyrick BO, Friedman DB, Billheimer DD, Cogan JD, Prince MA, Phillips JA, III, Loyd JE. Proteomics of transformed lymphocytes from a family with familial pulmonary arterial hypertension. Am J Respir Crit Care Med 2008;177:99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phillips JA, III, Poling JS, Phillips CA, Stanton KC, Austin ED, Cogan JD, Wheeler L, Yu C, Newman JH, Dietz HC, et al. Synergistic heterozygosity for TGFbeta1 snps and BMPR mutations modulates the age at diagnosis and penetrance of familial pulmonary arterial hypertension. Genet Med 2008;10:359–365 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.