Movement is a fundamental characteristic of all living things. Many researchers are therefore interested in muscle, the tissue that produces movement in animals. Muscle contraction is a result of the relative sliding movement between actin and myosin filaments, fueled by the chemical energy of ATP hydrolysis. In 1969, based on an x-ray diffraction study of muscle, H. E. Huxley (1) proposed a “tilting crossbridge model” in which the myosin head, attached to an actin filament, tilts and pulls on the actin filament. In 1971, A. F. Huxley and Simmons (2) provided the theoretical framework for this model to explain the mechanical properties of muscle. Since then, many investigations of muscle contraction have been performed with this model as a working hypothesis. The mechanism, however, has not yet been fully worked out. In an article in this issue of PNAS, Rief et al. (3) demonstrate that an unconventional myosin, myosin V, allows the detailed mechanical kinetics to be measured directly at the single-molecule level, which is very advantageous for solving the remaining problems.
At present, the subject is roughly divided into two areas of research. One concerns the problem of whether structural changes predicted by the “myosin head tilting model” indeed take place. This area has progressed largely through the clarification of the atomic structure of muscle myosin, based on its crystal structure (4) and the development of in vitro motility assays (5). X-ray crystallography demonstrated that the angle of the base of a myosin head changes relative to the main body (catalytic domain) of the head, depending on the form of bound nucleotides. The tilting crossbridge model has been refined, on the basis of this finding, into the “lever-arm model.” In this model, small structural changes that are coupled to the ATPase cycle in the catalytic domain of the myosin head are magnified by pivoting of the 8-nm-long light-chain domain (neck), which acts as a lever arm. The translation caused by this pivoting of the lever arm would be about 5 nm (Fig. 1a; refs. 6 and 7). However, there is no guarantee that the structural changes seen in crystals indeed take place while myosin generates motion. New technologies for manipulating single actin filaments with microneedles, optical traps, and optical sensors that can resolve objects at nanometer scales have allowed the displacement of single myosin molecules to be measured directly in vitro, and the lever-arm model has been tested with these technologies. The displacements reported, however, have varied considerably (8). Some reports have shown myosin displacements of about 5 nm, which is consistent with the lever-arm model. However, others have shown that if myosin is oriented correctly relative to the actin filament axis as in muscle, the value increases to 10–15 nm, which is significantly larger than the values expected from the lever-arm model. A major difficulty in the precise determination of myosin displacement is that the scale of the motion is smaller than the Brownian motions of microneedles or optically trapped beads, the average amplitudes of which are 30–40 nm (9).
Figure 1.
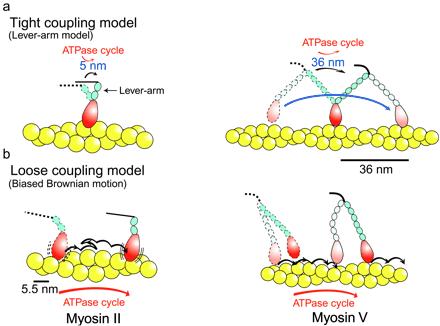
Models of an actomyosin motor. (a) Tight coupling model. A typical tight coupling model is a lever-arm model. In this model, a lever-arm tilting (conformational change) is coupled to each ATPase cycle in a one-to-one fashion. Displacement per ATPase cycle is expected to be proportional to the length of a neck domain (lever arm). (b) Loose coupling model. Each mechanical event is not coupled to the ATPase cycle. A typical loose coupling model is a biased Brownian ratchet model, in which the myosin head thermally diffuses along an actin filament, say, according to asymmetric potentials (8). One of the two heads of myosin II is not drawn here, because the two heads are thought to operate independently. On other hand, the two heads of myosin V are expected to operate in a hand-over-hand fashion to travel long distances processively.
Class V myosin is alleviating this obstacle, and research is soon likely to take a large step. Myosin V is one of 15 known classes of actin-based molecular motors and is implicated in several forms of organelle transport. Unlike muscle myosin (class II), myosin V does not make a filament but is believed to function as an independent molecule. Organelles are transported by only a small amount of myosin V; thus, one molecule is expected to travel long distances continuously without dissociation from an actin filament, much as one molecule of kinesin moves progressively along a microtubule. Furthermore, because the head of myosin V has a long neck domain (23 nm), which is about three times as long as that of muscle myosin (8 nm), displacements may also be large (Fig. 1a). If one molecule of myosin V moves progressively along an actin filament with large steps, it will be possible to determine precisely the size of steps and the stepping kinetics. Fortunately, myosin V has not betrayed these expectations and has become very valuable for research into the myosin motor mechanism.
Metha et al. (10) observed the sliding movements of actin filaments on a glass surface on which myosin V was adsorbed at very low densities. They applied a statistical method in the analysis of their data, which was used to demonstrate that one molecule of kinesin can move progressively along a microtubule, and concluded that myosin V is a progressive motor. Biochemical data showed that myosin V has a high affinity for actin during the ATPase cycle, also supporting the progressivity of myosin V (11, 12). Ando and his colleagues (13) have obtained harder evidence for the progressivity of myosin V. They demonstrated that single fluorescently labeled myosin V molecules can travel progressively a long distance along an actin filament fixed on a glass surface (Fig. 2). Moreover, Metha et al. (10) showed in an optical trapping assay that an actin filament was pulled continuously for several steps, the size of which were 30–40 nm, by one molecule of myosin V.
Figure 2.
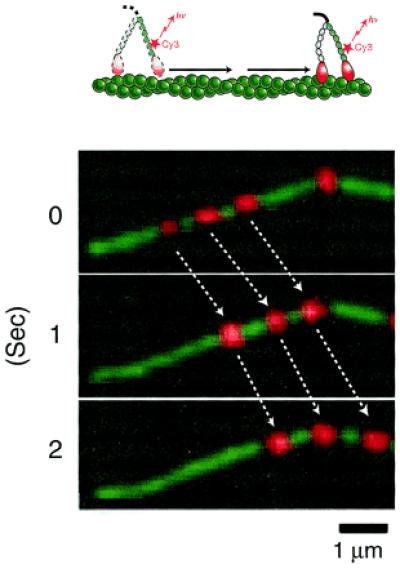
Direct observation of movement of single myosin V molecules along an actin filament (13). Myosin V is fluorescently labeled, exchanging one of the endogenous calmodulins bound to the neck domains by a fluorescently labeled one with Cy-3. Movement of single labeled myosin V molecules along an actin filament is observed by total internal reflection fluorescence microscopy (5). Red spots indicate Cy-3-labeled myosin V. An actin filament (green), which is labeled with rhodamine green phalloidine, is visualized separately and superimposed. Myosin V and an actin filament are artificially colored for convenience.
Because the size of the observed steps of myosin V coincides with the actin helical repeat (36 nm), myosin V is thought to walk along this repeat. Negatively stained electron micrographs show that the two long heads of myosin V span adjacent actin helixes, in agreement with this view (14). The observed large steps of myosin V are consistent with the lever-arm model, because in this model, the size of the steps is expected to increase in proportion to the length of neck (Fig. 1a).
The other major area of research into the mechanism of the myosin motor is concerned with how mechanical events are coupled to the ATPase cycle. Since 1985, when we suggested that a single mechanical event was not coupled tightly to each ATPase cycle (tight coupling) but rather that many mechanical events could occur within one ATPase cycle (loose coupling; ref. 15), this mechanochemical coupling has been studied extensively and remains controversial. In the lever-arm model, one conformational change (lever-arm tilting) is coupled to each ATPase cycle in a one-to-one fashion, and thus this model assumes tight coupling (Fig. 1a). In loose coupling, each mechanical event is not coupled to the ATPase cycle; thus, models of motion such as the biased thermal ratchet model, which is fundamentally different from the lever-arm model, should be considered (Fig. 1b). Clearly, the form of the mechanochemical coupling is essential for understanding how molecular motors convert the chemical energy obtained from the ATPase cycle into mechanical energy (8).
Myosin V has also allowed detailed stepping kinetics to be measured directly from single molecules, because it makes large steps continuously. In general, the stepping kinetics vary depending on the force. However, in conventional optical trapping nanometry, the force exerted on the motors increases with the displacement of the motors. To avoid this effect, Rief et al. (3) used “feedback-enhanced optical trapping nanometry” to observe steps of myosin V at a constant force. They observed more regular and more steadily repeating steps. Dwell times between adjacent steps were measured at various concentrations of ATP and ADP. The data analysis agrees with the idea that each step corresponds to a single ATPase cycle, and the cycle time of the steps is limited by ADP release. The heads of myosin V bound with ADP have a high affinity for actin; thus, it is thought that either of the two heads of a myosin V molecule binds to an actin filament during the step cycle and that the molecule can thus move progressively along the actin filament without dissociation. Rief et al. (3) have proposed a model to suit the stepping kinetics data, based on the lever-arm model and the hand-over-hand model (Fig. 1a), which is used to explain the progressivity of kinesin movement (16).
Myosin V is thus expected to advance rapidly the study of myosin motors and lead it to the final stage. Recently, however, I fear that some investigators may have focused on the lever-arm model so much that important points have been overlooked. Below are some studies that challenge the tight coupling and lever-arm model.
Ando and his colleagues (13) have argued loose coupling for myosin V. They measured the sliding velocity (v) of single myosin V molecules along actin filaments in solution (Fig. 2). This condition is almost the same as that for the measurement of the ATPase cycle rate of an acto-myosin V suspension in solution; thus, the sliding velocity and the ATPase cycle rate (Va) can be compared. They obtained the travel distance per ATPase cycle (d) as d = v/Va = (0.5–1 μm/s)/0.24/s = 200–400 nm. This result suggests that myosin V undergoes 5–10 steps, the sizes of which are about 40 nm, within each ATPase cycle. Rief et al. (3) have mentioned that this scenario is not suited to the mechanical stepping kinetics of myosin V. Now it is possible to measure simultaneously individual ATPase cycles and mechanical steps of a single myosin molecule (17); thus, this problem will soon be solved. For skeletal muscle myosin, optical trapping nanometry cannot obtain individual displacements but can obtain the averaged values of many events caused by the large Brownian motion of the beads (9). Therefore, if the average displacement is 5 nm, some of the displacements may be much larger, say, 30 nm. To overcome this problem, we have developed a more direct assay for manipulating a single myosin head and measuring the displacements with a scanning probe (18). This assay allowed measurement of individual displacements of single myosin heads with high resolution. The data showed that a myosin head (class II) moved along an actin filament with single mechanical steps of 5.5 nm; groups of two to five rapid steps in succession often produce displacements of 11–30 nm. Similar multiple substeps were observed at high loads (3 pN), although the number of substeps per displacement decreased. Because multiple steps are produced within one ATPase cycle, the mechanochemical coupling is not tight but loose. Substeps of similar size were also observed in smooth muscle myosin (9). Each 5.5-nm-long substep could be explained by tilting of a lever arm. However, because the size of the substeps coincides with the repeat of actin monomers (5.5 nm) and is constant, independent of force, and because some substeps are backward, it is more likely that a myosin head may step along the actin monomer repeat by biased Brownian motion (Fig. 1b; ref. 8).
Concerning the lever-arm model, some questions arise. The actin translocation velocity of multiple monomeric myosin V constructs with a short neck domain, which has only one light-chain binding site, is similar to that of intact two-headed myosin V with a long neck domain (11). If the rate of lever-arm tilting were constant, the actin translocation velocity would be proportional to the length of the neck domain (lever arm) as previously argued for myosin II by Spudich and his colleagues (19). Tanaka et al. (20) have reported that the displacement of myosin II is not affected by deleting the neck domain from it, in disagreement with the lever-arm model. The question of whether the myosin step size is proportional to the neck length as expected from the lever-arm model would be more easily tested with deletion mutants of myosin V with short necks, and a clearer answer would thereby be obtained.
Recently, single-molecule detection techniques, used not only for mechanical events as shown here but also for ATPase cycles and conformational changes, are progressing rapidly (5). These techniques can be applied to myosin V, and the remaining problems will be solved in the near future. Research on the mechanism of myosin motors is reaching the goal.
Footnotes
See companion article on page 9482.
References
- 1.Huxley H E. Science. 1969;164:1356–1366. [PubMed] [Google Scholar]
- 2.Huxley A F, Simmons R M. Nature (London) 1971;233:533–538. doi: 10.1038/233533a0. [DOI] [PubMed] [Google Scholar]
- 3.Rief M, Rock R S, Mehta A D, Mooseker M S, Cheney R E, Spudich J A. Proc Natl Acad Sci USA. 2000;97:9482–9486. doi: 10.1073/pnas.97.17.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rayment I, Rypniewski W R, Schmidt-Base K, Smith R, Tomchick D R, Benning M M, Winkelmann D A, Wesenberg G, Holden H M. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 5.Ishii Y, Yanagida T. Single Mol. 2000;1:5–16. [Google Scholar]
- 6.Goldman E Y. Cell. 1998;93:1–4. doi: 10.1016/s0092-8674(00)81137-x. [DOI] [PubMed] [Google Scholar]
- 7.Cooke R. Physiol Rev. 1997;77:671–697. doi: 10.1152/physrev.1997.77.3.671. [DOI] [PubMed] [Google Scholar]
- 8.Yanagida T, Kitamura K, Tanaka H, Iwane A H, Esaki S. Curr Opin Cell Biol. 2000;12:20–25. doi: 10.1016/s0955-0674(99)00052-6. [DOI] [PubMed] [Google Scholar]
- 9.Veigel C, Coluccio L M, Jontes J D, Sparrow J C, Milligan R A, Molloy J E. Nature (London) 1999;398:530–533. doi: 10.1038/19104. [DOI] [PubMed] [Google Scholar]
- 10.Mehta A D, Rock R S, Rief M, Spudich J A, Mooseker M S, Cheney R E. Nature (London) 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 11.Trybus K M, Krementsova E, Freyzon Y. J Biol Chem. 1999;274:27448–27456. doi: 10.1074/jbc.274.39.27448. [DOI] [PubMed] [Google Scholar]
- 12.De La Cruz E M, Wells A L, Rosenfield S S, Ostrap E M, Sweeney H L. Proc Natl Acad Sci USA. 1999;96:13726–13731. doi: 10.1073/pnas.96.24.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakamoto T, Amitani I, Yokota E, Ando T. Biochem Biophys Res Commun. 2000;272:586–590. doi: 10.1006/bbrc.2000.2819. [DOI] [PubMed] [Google Scholar]
- 14.Walker M L, Burgess S A, Sellers J R, Wang F, Hammer J A, Trinick J, Knight P J. Nature (London) 2000;405:804–807. doi: 10.1038/35015592. [DOI] [PubMed] [Google Scholar]
- 15.Yanagida T, Arata T, Oosawa F. Nature (London) 1985;316:366–369. doi: 10.1038/316366a0. [DOI] [PubMed] [Google Scholar]
- 16.Vale R D, Milligan R A. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 17.Ishijima A, Kojima H, Funatsu T, Tokunaga M, Higuchi H, Tanaka H, Yanagida T. Cell. 1998;92:161–171. doi: 10.1016/s0092-8674(00)80911-3. [DOI] [PubMed] [Google Scholar]
- 18.Kitamura K, Tokunaga M, Iwane A H, Yanagida T. Nature (London) 1999;397:129–134. doi: 10.1038/16403. [DOI] [PubMed] [Google Scholar]
- 19.Uyeda T Q P, Abramson P D, Spudich J A. Proc Natl Acad Sci USA. 1996;93:4459–4464. doi: 10.1073/pnas.93.9.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka H, Iwane A H, Yanagida T. Biophys J. 1999;78:234. [Google Scholar]


