Abstract
A central focus of clinical proteomics is to search for biomarkers in plasma for diagnostic and therapeutic use. We studied a set of plasma proteins accessed from the HIP2 database, a larger set of curated human proteins, and a subset of inflammatory proteins, for overlap with sets of known protein biomarkers, drug targets, and secreted proteins. Most inflammatory proteins were found to occur in plasma, and over three times the level of biomarkers were found in inflammatory plasma proteins and their interacting protein neighbors compared to the sets of plasma and curated human proteins. Percentage overlaps with Gene Ontology terms were similar between the curated human set and plasma protein set, yet the set of inflammatory plasma proteins had a distinct ontology-based profile. Most of the major hub proteins within protein-protein interaction networks of tissue specific sets of inflammatory proteins were found to occur in disease pathways. The present study presents a systematic approach for profiling a plasma subproteome’s relationship to both its potential range of clinical application and its overlap with complex disease.
Keywords: Mass spectrometry, plasma proteome, inflammatory response, biomarkers, drug targets
1 Introduction
The identification of protein biomarkers for the early diagnosis, subtyping, and monitoring of treatment for chronic diseases, including cardiovascular diseases, cancer, arthritis, Alzheimer’s disease, pulmonary disease, and autoimmune diseases, is now a central research focus in clinical proteomics [1, 2]. While biomedical researchers have traditionally sought to study molecular and immunological responses in tissues collected from patients, a recent trend has been to identify sets of proteins that may respond to changes in disease states or drug treatments, and be detected in easy-to-access biofluids such as human plasma. For decades, biomedical researchers and clinicians have used plasma to isolate and measure proteins that can be useful for the diagnosis or monitoring of disease [3, 4]. With the recent completion of the human genome and increasing availability of sensitive and reproducible analytical platforms, e.g., protein microarrays and liquid chromatography coupled tandem mass spectrometers (LC-MS/MS), researchers can now study hundreds or even thousands of proteins expressed in patient tissues or biofluids in parallel, therefore creating new opportunities for developing and managing disease [5–7]. An emerging focus has been to use panel biomarkers consisting of more than one protein and peptide marker to achieve a higher level of sensitivity and specificity [8]. Interpreting patterns of protein expressions in the human plasma proteome—sometimes very complex due to noisy signals and incomplete data sets—as a function of disease states often involves high-throughput data management, non-trivial statistical data analysis, and computational interpretation of data in the context of physiological and molecular pathways. A general process for biomarker discovery in the plasma proteome can be viewed in three steps: 1) defining the detectable “human plasma proteome” based on a specific analytical platform such as mass spectrometry or antibodies; 2) selecting differentially expressed proteins between different experimental conditions; and 3) identifying candidate protein disease biomarker profiles to achieve decent detection sensitivity and specificity [9].
Understanding the constituents, variability, and detectability of the human plasma proteome has been an elusive and challenging effort even today. With the release of the Human Proteome Organization (HUPO) Plasma Proteome Project (PPP) core dataset of 3020 plasma proteins in 2005, annotation of the human plasma proteome was initially performed by several groups [10–13]. Ping et al. [10] studied the HUPO PPP core datasets of 3020 proteins, and described plasma proteins to be comprised of a diverse group of proteins from the human proteome, including glycoproteins, DNA binding proteins, coagulation pathway, cardiovascular, liver, inflammation, and monocular phagocyte proteins. In this study, liver was the dominant tissue-based source of proteins, although many of the proteins detected are also expressed in other tissues. Berhane et al. [11] observed 354 proteins of particular interest for cardiovascular research to be found in the core dataset of HUPO. They classified these proteins into eight categories: protein markers of inflammation in cardiovascular disease, vasoactive and coagulation proteins, signal transduction pathway proteins, growth and differentiation-associated proteins, cytoskeletal proteins, transcription proteins, channel and receptor proteins, and heart failure and remodeling-related proteins. Muthusamy et al. [12] identified 3778 gene-based plasma proteins in the literature to generate a plasma proteome database and GO profile, and compared this set of proteins with the contents of the HUPO PPM core data set of 3020 proteins (mapping to 2446 genes). Liu et al. [13] performed a gene ontology annotation of cellular components in the plasma proteome generated from IMS-MS of 9087 plasma proteins identified by independent study and found differences between GO annotation percentages of human versus plasma proteomes. Recently, Saha et al. [14] reported a comprehensive catalogue of plasma proteins from healthy individuals called HIP2 (Healthy Human Individual’s Integrated Plasma Proteome), which collected set of 11,588 unique human plasma proteins detectable with different shotgun mass spectrometry methods [15]. In this study, while each human plasma protein has peptide-based evidence underlying its identification, only 106 high-abundant proteins were common among four different shotgun proteomics experiments, revealing an ongoing challenge in improving reproducibility among low-abundant proteins. Sub-proteomic analyses of the plasma proteome have also begun to evaluate biological processes of potential biomedical significance such as inflammatory pathways [10, 11]. Chronic inflammation is especially of interest for human plasma protein studies, because it is characterized by a response of tissue destruction by inflammatory cells like macrophages and plasma cells, and it has been found to be a factor associated with a wide variety of chronic diseases such as cancer [2, 16–18]. While inflammatory proteins are generally recognized as being present across many varieties of chronic diseases including cancer [19], the extent to which inflammatory proteins may be used in disease biomarker studies has not yet been studied systematically.
In this study, we dissected the human plasma proteome by examining its overlap with other datasets of interest: especially concerning proteins associated with drug treatment and biomarkers, and the subset of inflammatory proteins in the plasma proteome. We first analyzed the prevalence of candidate cancer biomarkers, drug targets and secreted proteins in human plasma, inflammatory proteins and expanded inflammatory proteins. Second, statistical comparisons of Gene Ontology (GO) were performed between the set of human Uniprot proteins and the plasma proteome, and between the plasma proteome and the set of inflammatory protein. Third, tissue specific protein-protein interactions (PPIs) of inflammatory proteins were calculated for each inflammatory protein subset present in each of five major tissues: brain, heart, lung, kidney and liver. We observed many of the interacting protein partners to be disease specific. Fourth, the hub proteins (10 or more interacting protein partners) in the PPIs networks were searched in the pathway database and found to be related to disease biology pathways.
Our overall objective is to foster within the biomedical research community, for the first-time, a systematic survey of the human plasma proteome relating to the human proteome for those interested in plasma proteomics applications. For purpose of application, we further sought to inspect detail of how the general inflammatory response may differentiate across tissues, biomolecular interaction networks and pathways to provide insight concerning the non-trivial dynamics that may be impeding efforts to establish sensitive and specific panels of biomarkers.
2 Materials and methods
2.1 Datasets
We used different datasets of proteins in our study. The description of each database and dataset used in our study is below:
HIP2 database
Human plasma proteins were collected from the HIP2 database, which currently contains 11,588 non-redundant International Protein Index (IPI) number protein entries [14]. This database is a comprehensive collection of healthy human plasma proteins, and has protein data mappings of supporting peptide evidence from several high-quality and high-throughput mass-spectrometry (MS) experimental data sets.
Human Plasma Proteome and Filtered Human Plasma Proteome sets
We defined the set of 10,138 human plasma proteins as the human plasma proteome, and the plasma proteins from HIP2 with two or more unique peptide sequences associated with their identification as the filtered human plasma proteome (n=7817). With peptide count thresholds of ≥3, ≥5 and ≥10, we defined additional filtered sets of plasma proteins (respectively, n={5392, 2718, 747}) and further gathered UniProt names corresponding to a high-confidence plasma protein set reported by States et al. [20]. The high-confidence plasma protein set was a subset of HUPO, and was based on a confidence level of at least 95% as determined by multiple hypothesis-testing techniques and coding region lengths of genes.
Curated Human Proteome set
For a comprehensive list of curated human proteins, 17,807 human proteins were extracted from UniProtKB/Swiss-Prot Release 54.6 of 04-Dec-2007 with the sequence retrieval system (SRS; http://srs.ebi.ac.uk/). We defined the 17,807 human Uniprot proteins as the curated human proteome. We generally considered the coverage of the plasma proteome compared to the curated human proteome to be sufficiently high (9995/17,807=56%) and reasonable for an initial analysis. Out of 10,138 human plasma proteins from HIP2, 9995 proteins have distinct Uniprot names; In our set-to set comparison, we used Uniprot names as identifiers.
Inflammatory protein and inflammatory plasma sets
The inflammatory protein data set was obtained by using gene ontology queries against the human Uniprot protein database. The gene product overlaps to inflammatory response (GO:0006954) and its descendants in the gene ontology were found with GO::TermFinder [21], and identified as the set of inflammatory proteins. This set was defined based on presence within both the human plasma proteome and the curated human proteome to identify 204 and 291 Swiss-Prot accession number proteins respectively. We defined the set of 291 proteins as the inflammatory protein set (I), and the set of 204 proteins as the inflammatory plasma protein set (Ip).
Cancer Biomarker Data set
The cancer biomarker list of 1261 proteins, mapping to 1049 Uniprot names, was from Anderson laboratory [22]. This data was compiled from literature and other sources and believed to be differentially expressed in human cancer [22]. In this data set, of the 34 biomarkers with more than 1000 citations, 79% were reported to be plasma proteins and 56% were reported as being used for clinical diagnosis (89% of these were reported as being plasma proteins). Of the 28 biomarkers with between 500 and 1000 citations, 57% were reported as being plasma proteins, but only 7% were reported as being used clinically.
DrugBank Data set
A list of 2,396 drug target proteins, mapping to 837 human Uniprot names, was obtained by parsing entries from the DrugBank database that combines detailed drug/chemical data with comprehensive drug target or protein information where each entry contains >80 data fields with half of the information being devoted to drug chemical structure and the other half devoted to drug target proteins [23]. The drug target protein source was from different species; in our study we worked on human proteins (837 Uniprot proteins).
Secreted Protein data set
The human secreted protein list of 1191 proteins, mapping to 1187 Uniprot names, was obtained from the Secreted Protein Database (SPD), which consists of a core dataset and a reference dataset. The core dataset of SPD contains 18,152 secreted proteins retrieved from Swiss-Prot/TrEMBL, RefSeq and CBI-Gene of human, mouse and rat [24]. All the entries of SPD were ranked according to the prediction confidence, and contain both experimental and computationally predicted secreted proteins. For our analysis using SPD, we used human proteins coming from Rank0 that consist of the manually curated set of Swiss-Prot proteins.
2.2 Gene Ontology annotations
We studied the three major GO vocabularies – biological process, molecular function and cellular component – using GO::TermFinder, a tool for accessing and evaluating GO annotations, and calculated significance for comparisons between sets of proteins [21]. We used the Gene Ontology project Open Biomedical Ontologies (OBO) file version 1.2 format revision 5.640, and the Gene Ontology project human annotation file revision 1.75 [25]. 9576 proteins of the 10,138 plasma proteins were observed to have one or more associated GO terms. Of the 7817 proteins in the filtered human plasma proteome, 7354 had GO annotations. Of the 17,807 proteins in the curated human proteome, 16,196 had GO annotations. We defined the filtered subset of 7354 proteins in the human plasma proteome with GO annotations as P. We defined the filtered subset of 16,196 proteins in the curated human proteome with GO annotations as H.
To account for the relative differences in sizes of protein sets and frequencies of GO annotations, we based the significance of annotation frequencies on hypergeometric calculation of an adjusted p value using a simulation-based correction based on the fraction (padj) of 1000 independent null-hypothesis simulations (samplings drawn from the larger data set) having any node with a p value equal or better than the p value for that node in the smaller data set, where the node is the GO term to which elements are annotated [21]. Comparisons were conducted between H and P, and between P and Ip. In addition to our criterion for statistical significance (padj≤0.001), initial selection of Gene Ontology categories was based on those GO terms whose shortest distance to the root of the GO hierarchy was 4 (i.e., GO level 4). In general, a GO level is the shortest distance to the root of the direct acyclic graph GO hierarchy, where level 0 is the root of the hierarchy, level 1 are terms “biological process”, “cellular component” and “molecular function” and higher number levels relate to increased qualitative specificity.
2.3 Protein-protein interactions (PPIs) study and visualization
A tool for visually exploring biological networks, Cytoscape version 2.5.1 with the APID2NET plugin, was used to expand and study the interacting proteins associated with the initial set of 204 proteins [26]. Cytoscape uses a December 2006 release of the Biomolecular Interaction Network Database (BIND) for protein-protein interactions data. BIND records are based on interactions as they have been shown experimentally and published in at least one peer-reviewed journal [27]. Two search filters of APID2NET were used: i) connection level; and ii) number of experimental methods. A connection level set to 1 collected first neighbors of the initial set of 710 proteins/nodes, and a connection level set to 2 collected first and second neighbors of the initial set of 2866 proteins/nodes. We set the number of experimental methods to be 2, meaning that the PPIs have two or more types of experimental evidence to avoid false positives from single experiments. Network views of protein interactions were generated with APID2NET [28], and showed proteins as nodes, and interactions as edges on the network. PPI networks (connection level=1) were built for five tissues: heart, brain, lung, kidney and liver. Tissue-specificities for inflammatory proteins were based on UniProtKB/Swiss-Prot entry comments sections as accessed from the ExPASy Proteomics Server [29].
2.4 Pathway database searching
We conducted a pathway analysis of proteins in the five tissue-based PPIs for those hub proteins whose degree is ≥10. We studied the presence of these hub proteins in pathways based on four pathway databases: BioCarta (http://cgap.nci.nih.gov/Pathways/BioCarta_Pathways/), NCI-Curated (http://pid.nci.nih.gov), Reactome [30] and ProteinLounge (http://www.proteinlounge.com).
3 Results and discussion
For practical application, an issue for our investigation was to evaluate the plasma proteins and inflammatory proteins inferred by mass spectrometry for diagnostic and therapeutic use. We analyzed the presence of cancer biomarkers, drug targets and secreted proteins across different data sets: i) curated human proteome, ii) human plasma proteome, iii) inflammatory proteins and inflammatory plasma proteins, and iv) inflammatory plasma expanded proteins. Extensive GO cross-comparison analyses were performed across these sets. To check the specificity of inflammatory proteins present in the plasma, tissue specific PPIs were studied in five major tissues: brain, heart, lung, kidney and liver. The hub proteins in the network analysis coming from the inflammatory protein seed list were then searched in four pathway databases: NCI-curated, Reactome, BioCarta, and ProteinLounge.
3.1 Prevalence of Biomarkers, Drug Targets, and Secreted Proteins
Protein biomarkers and drug targets are studied here in the context of curated human proteome and human plasma proteome, because we believe they indicate a range of clinical application opportunities. In particular, protein biomarkers in the plasma may have especially significant utility for clinical diagnosis. Ideally, however, a tissue-specific drug target protein should not be present in the plasma in high concentration, because the drug would then bind to the proteins in the plasma and not reach the specific tissue(s) [31]. Tissue-specific proteins that are secreted outside of the cell membrane may eventually reach the plasma, so we made an attempt to study the relationship of plasma proteins to secreted proteins. As a means for studying potential association with chronic disease, we surveyed both plasma and non-plasma inflammatory proteins of the curated human proteome for biomarkers, drug targets and secreted proteins.
3.1.1 Curated human proteome, human plasma proteome and filtered human plasma proteome comparison
We compared the curated human proteome and human plasma proteome by examining its percentage of overlap in each set for biomarkers, drug targets and secreted proteins. As shown in Table 1, the human plasma proteome and filtered human plasma proteome have slightly higher percentages of biomarkers, drug targets and secreted proteins relative to the curated human proteome. Percentage differences of overlap with biomarkers and drug targets were <0.5% between the filtered human plasma proteome and the human plasma proteome. The set of secreted proteins had the smallest percentage-wise distinction of overlap between the curated human proteome and the human plasma proteome, and was the only category to decrease in overlap between the unfiltered and filtered human plasma proteomes. All percentage overlaps were less than 10%, and these results highlight the need for using additional classifications of protein subsets within these proteomes to uncover higher amounts of overlap with biomarkers and drug targets. When adjusting the filter from peptide count ≥2, to thresholds of 3 and 5, changes to the overall percentage overlaps were within ±2%. With a threshold peptide count ≥10, the changes to percentage overlaps were also similarly minor for candidate biomarkers and drug targets, but the percentage overlap more than doubled for secreted proteins. We also conducted the percentage overlap comparison with a high-confidence set of plasma proteins from States et al. [20], and found these overlaps to be distinctly different from what was found for each of the specific peptide count thresholds. Since our study relates primarily to usage of actual output and performance of existing clinical proteomics technologies, we opted to use the peptide count threshold ≥2 as reported in [10, 13].
Table 1.
Percentage profile of candidate biomarkers, drug target and secreted proteins for the curated human proteome set, unfiltered and filtered plasma proteome sets.
| Curated human proteome (%) n=17,807 |
Human plasma proteome (%) peptides ≥1 n=10,138 |
Filtered human plasma proteome (%) peptides ≥2 n=7817 |
Filtered-3 human plasma proteome (%) peptides ≥3 n=5392 |
Filtered-5 human plasma proteome (%) peptides ≥5 n=2718 |
Filtered-10 human plasma proteome (%) peptides ≥10 n=747 |
States et al 2006 set of plasma proteins n=436 |
|
|---|---|---|---|---|---|---|---|
| Candidate biomarkers | 5.9 | 8.4 | 8.7 | 9.1 | 10.4 | 12.7 | 8.2 |
| Drug target proteins | 4.7 | 6.9 | 7.2 | 7.4 | 6.8 | 7.5 | 4.3 |
| Secreted proteins | 6.7 | 7.9 | 7.6 | 7.7 | 9.5 | 20.00 | 13.7 |
3.1.2 Inflammatory proteins
We next examined inflammatory proteins and their interacting protein neighbors for overlap with biomarkers, drug targets and secreted proteins. The examined datasets were the set of 291 inflammatory proteins, the subset of 204 inflammatory plasma proteins, and datasets based on expansions with interacting protein neighbors. As shown in Table 2, restricting the initial set of 291 inflammatory proteins to the 204 proteins in the plasma proteome produced only marginal changes (≤1%) in percentage overlap with biomarkers, drug targets and secreted proteins. Overlap with biomarkers increased by 4% with a first neighbor-based protein interaction expansion on the set of inflammatory plasma proteins, yet an overall percentage-wise decrease was found for the second neighbor-based protein interaction expansion. We calculated p-value based on Fisher’s exact test to test whether inflammatory proteins present in plasma (n=204), first neighbor expansion proteins (n=710) and second neighbor expansion proteins (n=2866) are biomarkers or non biomarkers. We got p-value of 0.1977 between inflammatory proteins present in plasma and first neighbor expansion proteins; p-value of 0.0006 between first neighbor expansion proteins and second neighbor expansion proteins; p-value of 0.0141 between inflammatory proteins present in plasma and second neighbor expansion proteins (See Table S1–S3). Protein interaction expansions to the set of inflammatory plasma proteins produced consecutive reductions in overlap with drug targets and secreted proteins, most significantly for secreted proteins and the contingency tables of Fisher’s exact test are shown in Table S4–S9. Based on protein interactions, both first neighbor and second neighbor expansions were found to be successful in overlapping with higher numbers of biomarkers, drug targets and secreted proteins than observed for the curated human proteome or human plasma proteome.
Table 2.
Overlapping analysis between inflammatory proteins/expanded network proteins and different sets of proteomes of interests.
| Inflammatory proteins (%) | Inflammatory proteins present in plasma proteome (%) | Inflammatory plasma expanded proteins* (%) | Inflammatory plasma expanded proteins** (%) | |
|---|---|---|---|---|
| Plasma proteome | 70 | 100 | 80 | 75 |
| Candidate biomarkers | 27 | 26 | 30 | 20 |
| Drug target proteins | 14 | 15 | 12 | 8.0 |
| Secreted proteins | 39 | 39 | 19 | 8.7 |
Degree of neighborhood=1
Degree of neighborhood=2
For an additional survey of biomarkers, drug targets and secreted proteins, we next distinguished for plasma versus non-plasma proteins in the set of inflammatory proteins and its related set of expanded proteins. The subsets of non-plasma proteins in the inflammatory and expanded sets of inflammatory proteins had elevated percentages of overlap for biomarkers and secreted proteins as shown in Table 3. We found the greatest increases between plasma and non-plasma protein sets to occur for percentage overlaps with inflammatory protein biomarkers. We found this trend to be consistent with the presence of inflammatory protein biomarkers outside of plasma. The inflammatory response proteins and their interacting protein partners thus implicate possibilities for clinical application related to cancer detection and therapy. Investigating the relationship of these prospective sets of proteins with functional annotations, disease biology and pathways may therefore yield insight on the range and activity of these proteins in cellular and organism physiology.
Table 3.
Comparison counts with a distinction for plasma proteins between inflammatory-related proteins and expanded proteins with candidate biomarkers, drug targets and secreted proteins.§
| Comparison Set | Ip* | Ip′* | Ep* | Ep′* |
|---|---|---|---|---|
| Candidate biomarkers | 54 (26%) | 25 (29%) | 119 (30%) | 40 (31%) |
| Drug target proteins | 31 (15%) | 10 (11%) | 49 (12%) | 10 (7.8%) |
| Secreted proteins | 79 (39%) | 35 (40%) | 48 (12%) | 22 (17%) |
Ip∪Ip′ = I; Ep∪Ep′ = E; Ip∩Ep = ∅; Ip′∩Ep′ = ∅;
- denotes intersection with comparison set;
Ip: n=204; Ip′: n=87; Ep: n=399; Ep′: n=128;
Ip*: subset of the inflammatory protein set in the plasma proteome that intersects with comparison set;
Ip′*: subset of the inflammatory protein set, not in the plasma proteome, that intersects with comparison set;
Ep*: subset of the expanded inflammatory protein set (degree of neighbor = 1) in the plasma proteome that intersects with comparison set;
Ep′*: subset of the expanded inflammatory protein set (degree of neighbor = 1), not in the plasma proteome, that intersects with comparison set.
3.2 Comparative gene functional category analysis
We use gene ontology as a primary tool to annotate and compare protein functions between the annotated subsets of the curated human proteome (H) and the filtered human plasma proteome (P) to make comparisons. We conducted two sets of comparisons involving: i) the annotated subsets of the curated human proteome and filtered human plasma proteome (H vs P); and ii) the filtered human plasma proteome and inflammatory plasma proteins (P vs Ip). To account for the relative difference in sizes between protein sets and their frequency ranges of GO annotations, we determined significance based on an adjusted p-value as calculated from 1000 null-hypothesis simulations of hypergeometric comparisons based on random selection (padj≤0.001).
Following our initial selection of statistically significant, level 4 GO terms, we made further refinements to the set of GO terms to be used for analysis. The set of biological process GO terms was pruned for general redundancies with inflammatory response resulting in the removal of 35 GO terms from our analytical comparisons. Calculation of percentages was based on the sets of proteins annotated to each of three selected sets of GO terms: 11 cellular component terms, 18 molecular function terms and 14 biological process terms.
3.2.1 Curated human proteome and filtered human plasma proteome
We studied the annotations of 43 GO terms as they are applied to the curated human proteome and filtered human plasma proteome. Differences between the GO annotation percentages of the H and P sets were <2%, and the overall distributions were identical as shown in Figure 1. Across the three GO vocabularies, for the 13 GO terms in H and P with <2% annotation frequencies, the P set uniformly had higher percentages except for the indexed P8 category (response to bacterium, GO:0009617). For the 8 cellular component and 10 molecular function GO terms in H and P with >2% annotation frequencies, the H set had only a slightly higher number of GO terms with greater percentages than the P set. For biological process, the P set had a higher percentage than the H set for 8 of the 12 GO terms with >2% annotation frequencies. While the differences of percentages between GO annotations in the H and P sets were low (<2%), 34 of the 43 GO terms were statistically significant for the overall comparison as presented in Tables 4–6.
Figure 1.
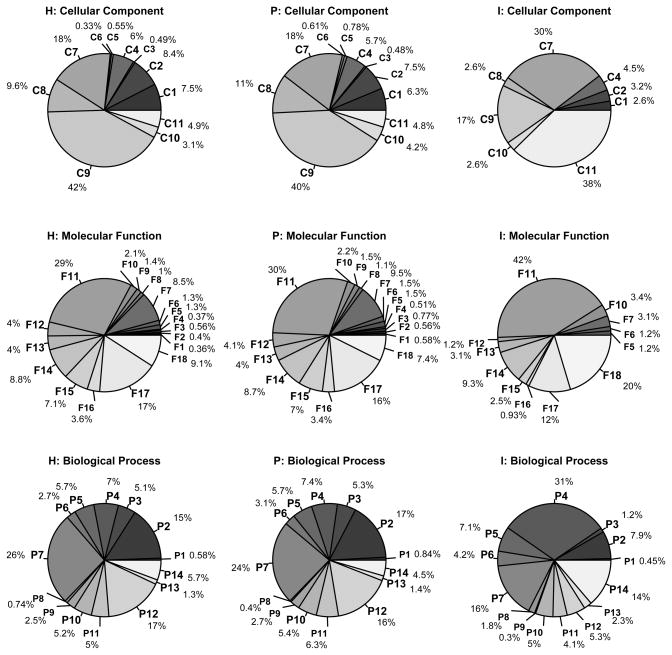
Gene ontology (GO) percentages for the curated human proteome (H), the filtered plasma proteome (P), and the inflammatory response plasma protein set (I). The indexed labels C1,…,C11; F1,…,F18; and P1,…,P14 are defined in Tables 4–6, and correspond to cellular component, molecular function, and biological process vocabularies respectively.
Table 4.
Selected cellular component GO terms applied to comparisons between H versus P protein sets and P versus Ip protein sets. The results of comparisons are shown in the last two columns; significance (Y) is based on an adjusted p-value as calculated from 1000 null-hypothesis simulations of hypergeometric comparisons based on random selection, padj≤0.001; n/a is when there are not any representatives of the GO term in the Ip set.
| Index | Level | GO ID | GO term | H vs P | P vs Ip |
|---|---|---|---|---|---|
| C1 | 6 | GO:0005783 | endoplasmic reticulum | N | N |
| C2 | 6 | GO:0005739 | mitochondrion | N | N |
| C3 | 6 | GO:0005765 | lysosomal membrane | N | n/a |
| C4 | 6 | GO:0005794 | golgi apparatus | N | N |
| C5 | 5 | GO:0016459 | myosin complex | Y | n/a |
| C6 | 4 | GO:0005581 | collagen | Y | n/a |
| C7 | 5 | GO:0044459 | plasma membrane part | Y | N |
| C8 | 6 | GO:0005856 | cytoskeleton | Y | N |
| C9 | 6 | GO:0005634 | nucleus | Y | N |
| C10 | 3 | GO:0031012 | extracellular matrix | Y | N |
| C11 | 4 | GO:0005615 | extracellular space | N | Y |
Table 6.
Selected biological process GO terms applied to comparisons between H versus P protein sets and P versus Ip protein sets. The results of comparisons are shown in the last two columns; significance (Y) is based on an adjusted p-value as calculated from 1000 null-hypothesis simulations of hypergeometric comparisons based on random selection, padj≤0.001; n/a is when there are not any representatives of the GO term in the Ip set.
| Index | Level | GO ID | GO term | H vs P | P vs Ip |
|---|---|---|---|---|---|
| P1 | 5 | GO:0043062 | extracellular structure organization and biogenesis | Y | n/a |
| P2 | 4 | GO:0007275 | multicellular organismal development | Y | N |
| P3 | 4 | GO:0007049 | cell cycle | Y | N |
| P4 | 4 | GO:0006950 | response to stress | Y | Y |
| P5 | 4 | GO:0016265 | death | Y | Y |
| P6 | 4 | GO:0006928 | cell motility | Y | Y |
| P7 | 4 | GO:0007154 | cell communication | Y | Y |
| P8 | 5 | GO:0009617 | response to bacterium | N | Y |
| P9 | 4 | GO:0022402 | cell cycle process | Y | N |
| P10 | 4 | GO:0008283 | cell proliferation | Y | N |
| P11 | 4 | GO:0007155 | cell adhesion | Y | N |
| P12 | 4 | GO:0006810 | transport | Y | N |
| P13 | 4 | GO:0002520 | immune system development | N | Y |
| P14 | 4 | GO:0006955 | immune response | N | Y |
Interestingly, our findings show the percentage ontology classifications of the H and P sets to be much more similar than has been previously reported in studies where the proteome coverage was low or the data came from a single instrument type [10, 12, 13]. Possible differences likely arose from our use of a more expansive and up-to-date reference set for the plasma proteome, our choice of a reference set of human proteins, and our use of more recent ontology and annotation files. Another source of variation between our results and the other proteomic analyses may have been our inclusion of all evidence codes including the inferred from electronic annotation code (IEA). There was some notable, almost identical, similarity however between some of the percentages in our study, and percentages from a study based on a different plasma proteome database [12]. Similarities were found for cell communication (P7: 24% versus 23.7% respectively), immune response (P14: 4.5% versus 4.3% respectively) and hydrolase activity (F14: 8.8% versus 9% respectively). The similarities are surprisingly close considering that our percentages are calculated based on a different overall set of GO terms and, unlike Muthusamy et al. [12], we did not include a category for proteins without annotation. Significant differences (two-fold) were observed however for transport (P12) and signal transducer activity (F18). We found consistency of sorted rankings for percentages of different GO terms, although percentages and proteomic comparisons between Ping et al. [10], Liu et al. [13], and our study varied. Nucleus (C9) and protein binding (F11) were the top-ranked GO categories of cellular component and molecular function for both our study (see Figure 1) and Ping et al. [10]. Nucleus was the second highest ranking category for Liu et al. [13]. For biological process terms across both human and plasma protein sets, transport was in the top three GO terms in our study and the most prevalent GO term in Ping et al. [10]. When comparing the human and plasma protein sets, Liu et al. [13] found a noticeable increase in extracellular component (+5%) and a decrease in nuclear proteins (-12%). We found similar directions of change, but of a reduced magnitude; for extracellular matrix and space (C10 and C11), there was a 1% increase, and for nucleus (C9), there was a 2% decrease. The biological processes of cell proliferation, immune response and cell motility in our analysis matched the percentage frequency rank order of Ping et al., as did the molecular functions of protein binding, ion binding and nucleotide binding. We did not find a similarly high magnitude of percentage differences between plasma and human protein sets. In general, our study did not find the many two-fold or more changes in percentages common to most of Ping et al. (for instance, from a human protein set to plasma protein set comparison, they found changes of 2% to 4%, and 2% to 6% for extracellular matrix and space respectively). We did however find, consistent with findings reported in Ping et al., plasma proteins to be increasingly over-represented for low frequency GO annotations relative to human proteins.
Our findings may also help further resolve proposed interpretations concerning the percentage differences between the plasma and the entire human proteome. Intriguingly, with plasma, Liu et al. and Ping et al. report similar percentages for their respective nuclear and nucleus component categories (18%). Liu et al. find a rise of the percentage (to 30%) observed in a human protein set, yet Ping et al. find an unexpected reduction (to <14%) which they suggested may be the result of the secretion of cellular breakdown products into circulation. We found 40% of the human plasma proteome to consist of nucleus proteins, with a minor rise in value to 42% for the curated human proteome. Liu et al. suggest that mimicry of percentages in the human proteome by the plasma map would indicate that it is composed of random assignments, and they find evidence against random assignment based on significant differences in percentage. We did not find significant differences in GO term percentages but our tests for statistical significance would reject a scenario of completely random assignment.
3.2.2 Filtered human plasma proteome and inflammatory plasma proteins
We compared GO annotations between the human plasma proteome and inflammatory plasma proteins. Across the set of comparisons between 43 GO terms shown in Tables 4–6 and Figure 1, Ip did not have any representative proteins for 3 of the cellular component terms: lysosomal membrane, myosin complex and collagen (C3, C5 and C6), 6 of the molecular function terms: structural constituent of cytoskeleton, chromatin binding, helicase activity, extracellular matrix structural constituent, RNS polymerase II transcription factor activity and GTPase regulator activity (F1, F2, F3, F4, F8 and F9) and a single biological process term: extracellular structure organization and biogenesis (P1). The distribution of GO term percentages on the Ip set was broadly distinguishable from the P set. Differences between the GO term percentages of the P and Ip sets were generally >2%. Differences were >10% for 3 of the 8 GO terms within the P versus Ip cellular component comparison, and 2 of the GO terms within each of the 12 molecular function and 13 biological process comparisons. For larger differences among cellular component GO terms (>10%), the Ip set had higher percentages for plasma membrane part (C7) and extracellular space (C11), and a smaller percentage for nucleus (C9). For larger differences among molecular function GO terms (>10%), the Ip set had higher percentages for protein binding (F11) and signal transducer activity (F18). For larger differences among biological process GO terms (>10%), the Ip set had a higher percentage for response to stress (P4) and a smaller percentage for transport (P12). For a percentage difference threshold of >8%, the Ip set also had a higher percentage for immune response (P14) and a smaller percentage for cell communication (P7). Statistically significant comparisons between the P and Ip sets were observed for 1 cellular component GO term : extracellular space (C11), 1 molecular function GO term: signal transducer activity (F18) and 7 biological process GO terms: response to stress, death, cell motility, cell communication, response to bacterium, immune system development and immune response (P4, P5, P6, P7, P8, P13 and P14). Extracellular space and signal transducer activity were exclusively significant for the P vs Ip comparison. The statistically significant distinctions of biological process with the inflammatory protein set characterize a system of cellular communication, motility, immune response and system development, death, and response to stress. For biological process, the response to bacterium, immune response and immune system development terms were exclusively significant for the P vs Ip comparison. The Ip set is based on the biological process GO term of inflammatory response, and the interdependency among concepts within the biological process GO vocabulary suggested that over-representation may occur for other biological process GO terms, as was indeed found.
Overall, we found inflammatory proteins to be annotated with functions, processes and cellular components in ways that would distinctly separate this class of proteins from the human plasma proteome in general. Beyond this observed distinction in functional annotation for inflammatory proteins, objectives for direct clinical application may be further advanced by investigating the specificity of inflammatory proteins to multiple aspects of different complex diseases. In particular, we next sought to complement the findings of our ontology analysis by investigations of tissue specificity, interactions between proteins, and biological pathways.
3.3 Tissue specific PPIs
Inflammation processes are tissue specific [32], thus we investigated overlap of inflammatory plasma proteins in tissue. We observed in which tissues those inflammatory proteins were expressed, and found these proteins to be expressed in heart, brain, lung, kidney, pancreas, skin, colon, testis and ovary, liver and spleen. For our study, we evaluated five main organs such as heart, brain, lung, kidney and liver for which the inflammatory proteins were also present in amounts ≥ 20. The UniProt IDs of the proteins expressed in these tissues has been shown in Table 7. For the visualization of PPIs, we used plugin software APID2NET in the Cytoscape with settings described in Section 2.2 of this paper. All the GO terms mentioned in the tissue PPIs analysis are related to cancer prognosis [33–35].
Table 7.
Expression of inflammatory proteins (present in plasma) in different tissue organs.
| Uniprot name | Uniprot Primary Accession number | Protein name | Gene name | Organs |
|---|---|---|---|---|
| A1AG1_HUMAN | P02763 | Alpha-1-acid glycoprotein 1 | ORM1 | Liver |
| A1AG2_HUMAN | P19652 | Alpha-1-acid glycoprotein 2 | ORM2 | Liver |
| A2AP_HUMAN | P08697 | Alpha-2-antiplasmin | SERPINF2 | Liver |
| AACT_HUMAN | P01011 | Alpha-1-antichymotrypsin | SERPINA3 | Brain, Liver |
| ADO_HUMAN | Q06278 | Aldehyde oxidase | AOX1 | Brain, Heart, Lung, Kidney, Liver |
| ADPRH_HUMAN | P54922 | ADP-ribosylarginine hydrolase | ADPRH | Heart, Lung, Kidney, Liver |
| AOC3_HUMAN | Q16853 | Membrane copper amine oxidase | AOC3 | Heart, Lung, Kidney |
| APOL2_HUMAN | Q9BQE5 | Apolipoprotein-L2 | APOL2 | Brain, Lung, Kidney, Liver |
| ATRN_HUMAN | O75882 | Attractin | ATRN | Liver |
| B4GT1_HUMAN | P15291 | Beta-1,4-galactosyltransferase 1 | B4GALT1 | Brain |
| BMP2_HUMAN | P12643 | Bone morphogenetic protein 2 | BMP2 | Brain, Heart, Lung, Liver |
| C163A_HUMAN | Q86VB7 | Scavenger receptor cysteine-rich type 1 protein M130 | CD163 | Liver |
| C3AR_HUMAN | Q16581 | C3a anaphylatoxin chemotactic receptor | C3AR1 | Brain, Heart, Lung |
| CCL17_HUMAN | Q92583 | C-C motif chemokine 17 | CCL17 | Lung |
| CCL18_HUMAN | P55774 | C-C motif chemokine 18 | CCL18 | Lung |
| CCL21_HUMAN | O00585 | C-C motif chemokine 21 | CCL21 | Heart, Liver |
| CCL23_HUMAN | P55773 | C-C motif chemokine 23 | CCL23 | Lung, Liver |
| CCL26_HUMAN | Q9Y258 | C-C motif chemokine 26 | CCL26 | Heart |
| CCL8_HUMAN | P80075 | C-C motif chemokine 8 | CCL8 | Brain, Heart, Lung, Liver |
| CDO1_HUMAN | Q16878 | Cysteine dioxygenase type 1 | CDO1 | Brain, Heart, Liver |
| CEBPB_HUMAN | P17676 | CCAAT/enhancer-binding protein beta | CEBPB | Lung, Kidney |
| CFAH_HUMAN | P08603 | Complement factor H | CFH | Liver |
| CHST1_HUMAN | O43916 | Carbohydrate sulfotransferase 1 | CHST1 | Brain |
| CP4FB_HUMAN | Q9HBI6 | Cytochrome P450 4F11 | CYP4F11 | Heart, Kidney Liver |
| CXCR4_HUMAN | P61073 | C-X-C chemokine receptor type 4 | CXCR4 | Brain, Heart, Lung, Kidney, Liver |
| CXL13_HUMAN | O43927 | C-X-C motif chemokine 13 | CXCL13 | Liver |
| EDG3_HUMAN* | Q99500 | Sphingosine 1-phosphate receptor 3 | S1PR3 | Heart, Kidney, Liver |
| EPCR_HUMAN | Q9UNN8 | Endothelial protein C receptor | PROCR | Heart, Lung, Kidney, Liver |
| FETUA_HUMAN | P02765 | Alpha-2-HS-glycoprotein | AHSG | Liver |
| FHR1_HUMAN | Q03591 | Complement factor H-related protein 1 | CFHR1 | Liver |
| FHR5_HUMAN | Q9BXR6 | Complement factor H-related protein 5 | CFHR5 | Liver |
| FPRL1_HUMAN** | P25090 | N-formyl peptide receptor 2 | FPR2 | Lung |
| GCR_HUMAN | P04150 | Glucocorticoid receptor | NR3C1 | Heart |
| HDAC9_HUMAN | Q9UKV0 | Histone deacetylase 9 | HDAC9 | Brain, Heart |
| ICBR_HUMAN | P57730 | Caspase-1 inhibitor Iceberg | ICEBERG | Heart |
| IL1AP_HUMAN | Q9NPH3 | Interleukin-1 receptor accessory protein | IL1RAP | Lung, Liver |
| IL1F6_HUMAN | Q9UHA7 | Interleukin-1 family member 6 | IL1F6 | Brain |
| IRAK2_HUMAN | O43187 | Interleukin-1 receptor-associated kinase- like 2 | IRAK2 | Lung, Kidney, Liver |
| LT4R1_HUMAN | Q15722 | Leukotriene B4 receptor 1 | LTB4R | Brain, Heart, Liver |
| MEFV_HUMAN | O15553 | Pyrin | MEFV | Brain, Heart, Lung, Kidney, Liver |
| MGLL_HUMAN | Q99685 | Monoglyceride lipase | MGLL | Brain, Heart, Lung, Kidney, Liver |
| MMP25_HUMAN | Q9NPA2 | Matrix metalloproteinase-25 | MMP25 | Lung |
| NDST1_HUMAN | P52848 | Bifunctional heparan sulfate N- deacetylase/N-sulfotransferase 1 | NDST1 | Heart, Liver |
| NFAC3_HUMAN | Q12968 | Nuclear factor of activated T-cells, cytoplasmic 3 | NFATC3 | Heart, Kidney |
| NFAC4_HUMAN | Q14934 | Nuclear factor of activated T-cells, cytoplasmic 4 | NFATC4 | Lung, Kidney |
| NFAM1_HUMAN | Q8NET5 | NFAT activation molecule 1 | NFAM1 | Lung |
| NMI_HUMAN | Q13287 | N-myc-interactor | NMI | Brain, Liver |
| NOD1_HUMAN | Q9Y239 | Nucleotide-binding oligomerization domain-containing protein 1 | NOD1 | Heart, Lung, Kidney, Liver |
| NOX4_HUMAN | Q9NPH5 | NADPH oxidase 4 | NOX4 | Brain, Heart, Kidney |
| PA24C_HUMAN | Q9UP65 | Cytosolic phospholipase A2 gamma | PLA2G4C | Heart |
| PTAFR_HUMAN | P25105 | Platelet-activating factor receptor | PTAFR | Heart, Lung |
| SAA_HUMAN | P02735 | Serum amyloid A protein | SAA1 | Liver |
| SAA4_HUMAN | P35542 | Serum amyloid A-4 protein | SAA4 | Liver |
| SN_HUMAN | Q9BZZ2 | Sialoadhesin | SIGLEC1 | Brain, Lung, Liver |
| SPR1_HUMAN | Q15743 | Sphingosylphosphorylcholine receptor | GPR68 | Brain, Lung |
| STAB1_HUMAN | Q9NY15 | Stabilin-1 | STAB1 | Liver |
| STAT3_HUMAN | P40763 | Signal transducer and activator of transcription 3 | STAT3 | Brain, Lung, Kidney, Liver |
| THRB_HUMAN | P00734 | Prothrombin | F2 | Liver |
| TIRAP_HUMAN | P58753 | Toll/interleukin-1 receptor domain- containing adapter protein | TIRAP | Brain, Heart, Lung, Kidney, Liver |
| TLR10_HUMAN | Q9BXR5 | Toll-like receptor 10 | TLR10 | Lung |
| TLR2_HUMAN | O60603 | Toll-like receptor 2 | TLR2 | Lung, Liver |
| TLR7_HUMAN | Q9NYK1 | Toll-like receptor 7 | TLR7 | Brain, Lung |
| TLR8_HUMAN | Q9NR97 | Toll-like receptor 8 | TLR8 | Brain, Heart, Lung, Liver |
| TLR9_HUMAN | Q9NR96 | Toll-like receptor 9 | TLR9 | Lung, Liver |
| TRFE_HUMAN | P02787 | Serotransferrin | TF | Liver |
| VPS45_HUMAN | Q9NRW7 | Vacuolar protein sorting-associated protein 45 | VPS45 | Brain, Heart, Lung, Kidney, Liver |
| X3CL1_HUMAN | P78423 | Fractalkine | CX3CL1 | Brain, Heart, Lung, Kidney |
3.3.1 Brain
We observed 25 inflammatory proteins expressed in brain tissue, and the interaction network was expanded to 129 nodes and 278 edges. We found six major hub proteins – NMI(17), CXCR4 (26), HDAC (17), AACT (18) and BMP2 (14), STAT3 (103) as shown in Fig. S1. In the expanded list of 129 proteins, we studied the GO terms of biological process, and observed that eight proteins are from the JAK-STAT cascade (GO:7259)- STAT4, SOCS3, CCR2, STAT4, STAT3, STA5A, STA5B, NMI; four proteins are from the anti-apoptosis (GO:6916)- TF65, SOCS3, NFKB1, HDAC3; two proteins are from the I-kappaB kinase/NF-kappaB cascade (GO:7249)- STAT1 TLR8; two proteins are from activation of NF-kappaB-inducing kinase (GO:7250)- TLR4 M3K7. Interestingly, we observed other proteins to be associated with GO development processes such as a protein in eye development (GO:1654) - BMR1B; two proteins in nervous system development (GO:7399) - STAT3, HDAC4; and two proteins in muscle contraction (GO:6936)- LT4R1, DAG1.
3.3.2 Heart
We observed 29 inflammatory proteins expressed in heart tissue, and the interaction network was expanded to 116 nodes and 234 edges. We found five major hub proteins, HDAC9 (17), NOD1 (13), GCR (98), BMP2 (14), CXCR4 (26) as shown in Fig. S2. In the expanded list of 116 proteins, we studied the GO terms of biological process, and observed that five proteins are from the JAK-STAT cascade (GO:7259) - STA5A, CCR2, STAT3, STA5A, STA5B; five proteins are associated with anti-apoptosis (GO:6916)- HDAC3, BAG1, NFKB1, TF65, SOCS3; one protein is associated with activation of NF-kappaB transcription factor (GO:51092)- TF65; and one protein is from the I-kappaB kinase/NF-kappaB cascade(GO:7249)- TLR8.
3.3.3 Lung
We observed 34 inflammatory proteins expressed in lung tissue, and the interaction network was expanded to 153 nodes and 353 edges. We found eight major hub proteins – IRAK2 (12), IL1AP (13), CXCR4 (26), NOD1 (13), TLR2 (18), CEBPB (60), BMP2 (14), STAT3 (103) as shown in Fig. S3. Six proteins are from the JAK-STAT cascade pathway (GO:7259)- INAR1, SOCS3, STA5A, CCR2, NMI, STAT3; five proteins are associated with anti-apoptosis (GO:6916)- FOXO1, NFKB1, SOCS3, TF65, ENPL; three proteins are from the I-kappaB kinase/NF-kappaB cascade pathway (GO:7249)- IRAK2, STAT1, TLR8; three proteins are from the activation of NF-kappaB-inducing kinase pathway (GO:7250)- TLR4, M3K7, TRAF6. All the GO terms mentioned were found to relate to cancer prognosis [33–35].
3.3.4 Kidney
We observed 21 inflammatory proteins expressed in kidney tissue, and the interaction network was expanded to 106 nodes and 281 edges. We found six major hub proteins – IRAK2 (12), CXCR4 (26), NOD1 (13), CEBPB (60), BMP2 (14), STAT3 (103) as shown in Fig. S4. We observed that five proteins are from the JAK-STAT cascade pathway (GO:7259)- INAR1, SOCS3, STA5A, NMI, STAT3; four proteins are associated with anti-apoptosis (GO:6916)- SOCS3, NFKB1, TF65, FOXO1; two proteins are from the I-kappaB kinase/NF-kappaB cascade (GO:7249)- STAT1, IRAK2; and three proteins are associated with activation of NF-kappaB-inducing kinase (GO:7250)- TLR4, M3K7, TRAF6.
3.3.5 Liver
We observed 43 inflammatory proteins expressed in liver tissue, and the interaction network was expanded to 165 nodes and 318 edges. We found fourteen major hub proteins – IRAK2 (12), NMI (25), IL1AP (13), CXCR4 (26), NOD1 (13), TLR2 (18), AACT (18), BMP2 (14), THRB (48), FETUA (10), TRFE (20), CFAH (11), A2AP (15), STAT3 (103) as shown in Fig S5. We observed that eight proteins are from the with JAK-STAT cascade (GO:7259)- STAT4, STA5B, INAR1, SOCS3, STA5A, CCR2, NMI, STAT3; four proteins are associated with anti-apoptosis (GO:6916)- NFKB1, SOCS3, TF65, ENPL; three proteins are from the I-kappaB kinase/NF-kappaB cascade (GO:7249)- IRAK2, TLR8, STAT1; three proteins are associated with in activation of NF-kappaB-inducing kinase (GO:7250) - TLR4, M3K7, TRAF6.
3.4 Pathway database searching for network hub proteins
Seventeen hub proteins expressed in five tissues were analyzed with known pathways as shown in Table 8. It was observed that many inflammatory proteins were expressed in many tissues. For example, BMP2_HUMAN protein is expressed in all the five organs studied (heart, brain, lung, kidney, and liver) and it is secreted protein [24]. NOD1_HUMAN, AACT_HUMAN, FETUA_HUMAN, TRFE_HUMAN, CFAH_HUMAN, A2AP_HUMAN were the six hub proteins not found in our set of pathway databases. All of those pathways containing our proteins related to complex disease. For instance, HDAC9_HUMAN is expressed in heart, brain, and liver and is associated with pathways of cardiac hypertrophy, notch signaling, and the p53 signaling pathway. GCR_HUMAN is expressed only in the heart and is found in chromatin remodelling[36]; defects in the GCR gene cause a hypertensive, hyperandrogenic disorder characterized by increased serum cortisol concentrations [37]. AACT_HUMAN is expressed only in brain and liver tissue, and defects in the AACT gene are a proposed cause of chronic obstructive pulmonary disease [38]. TLR2_HUMAN is expressed in liver and is observed in Toll-like receptors pathway and TLR-TRIF pathway. THRB_HUMAN is a part of thrombin signalling, thrombopoietin pathway. Six hub proteins, BMP2_HUMAN, AACT_HUMAN, CXCR4_HUMAN, FETUA_HUMAN, TRFE_HUMAN and THRB_HUMAN were found in the cancer biomarker list as reported in [22]. Four hub proteins, GCR_HUMAN, TLR2_HUMAN, TRFE_HUMAN and THRB_HUMAN were found in the drug target list [23].
Table 8.
Major inflammatory hub proteins in Tissue-based PPIs (>=10) and their role in known biological pathways.
| Major HUB proteins in Tissue-based PPIs (>=10) and GO | Degree | Tissues expressed | MS platforms | Pathways involved | Source |
|---|---|---|---|---|---|
| HDAC9_HUMAN | 17 | Heart, Brain | IMS-MS/MS_TOF | DNA Methylation and Transcriptional Repression, NFAT and Cardiac Hypertrophy, Notch Signaling, p53 Signaling | Protein Lounge, HDAC9, NCI-curated |
| NOD1_HUMAN | 13 | Heart, Lung, kidney Liver | IMS-MS/MS_TOF | -- | |
| GCR_HUMAN | 98 | Heart | IMS-MS/MS_TOF | Chromatin remodelling, Defects in this gene cause a hypertensive, hyperandrogenic disorder characterized by increased serum cortisol concentrations. | BioCarta, NCI-curated |
| BMP2_HUMAN | 14 | Heart, Brain, Lung, Kidney, Liver | IMS-MS/MS_TOF | BMP Pathway, JAK/STAT Pathway, Mitochondrial Apoptosis, PAK Pathway,, MIF Regulation of Innate Immune Cells, MIF Mediated Glucocorticoid Regulation, TGF-Beta Pathway, MAPK Family Pathway, PKR Pathway, Rac1 Pathway, Rho Family GTPases | Protein Lounge, BioCarta |
| NMI_HUMAN | 25 | Brain, Liver | IMS-MS/MS_TOF | Prolactin Signaling | Protein Lounge, BioCarta |
| CXCR4_HUMAN | 26 | Brain, Lung, Kidney, Liver | IMS-MS/MS_TOF | CXCR4 Pathway, Signaling by Slit, NF-kappaB Activation by Viruses, EphB-EphrinB Signaling, Ephrin-Eph Signaling, Apoptotic Pathways Triggered By HIV1 | Protein Lounge, BioCarta, NCI-curated |
| AACT_HUMAN | 18 | Brain, Liver | ESI-MS/MS_LTQ | Defects may be a cause of chronic obstructive pulmonary disease | |
| IRAK2_HUMAN | 12 | Lung, Kidney, Liver | IMS-MS/MS_TOF | Toll-Like Receptors Pathway, TLR-TRIF Pathway, NF-KappaB Family Pathway, IL-1 Pathway, NF-KappaB (p50/p65) Pathway | Protein Lounge, BioCarta |
| IL1AP_HUMAN | 13 | Lung, Liver | IMS-MS/MS_TOF and ESI-MS/MS_LTQ | IL-1 Pathway, IL-10 Pathway | Protein Lounge |
| CEBPB_HUMAN | 60 | Lung, Kidney | IMS-MS/MS_TOF | Glucocorticoid Receptor Signaling, Prolactin Signaling, Growth Hormone Signaling | Protein Lounge, BioCarta, NCI-curated |
| TLR2_HUMAN | 18 | Lung, Liver | IMS-MS/MS_TOF | Toll-Like Receptors Pathway, TLR-TRIF Pathway | Protein Lounge |
| STAT3_HUMAN | 103 | Brain, Lung, Kidney, Liver | IMS-MS/MS_TOF | STAT3 Pathway | Protein Lounge |
| FETUA_HUMAN | 10 | Liver | ESI-MS/MS_LTQ, IMS-MS/MS_TOF | ||
| TRFE_HUMAN | 20 | Liver | ESI-MS/MS_LCQ, IMS-MS/MS_TOF, ESI-MS/MS_QTOF, ESI-MS/MS_DECAXP | ||
| CFAH_HUMAN | 11 | Liver | ESI-MS/MS_LCQ, IMS-MS/MS_TOF | ||
| A2AP_HUMAN | 15 | Liver | ESI-MS/MS_LTQ, IMS-MS/MS_TOF | ||
| THRB_HUMAN | 48 | Liver | IMS-MS/MS_TOF, ESI-MS/MS_LTQ | Thrombin Signaling, Thrombopoietin Pathway | Protein Lounge |
4 Conclusions
Characterizing the components of plasma proteome systematically is closely related to future clinical applications of MS proteomics emerging from a wave of promising new technologies such as SELDI [39], shotgun proteomics [40], multiple reaction monitoring (MRM) [41] and glycoprotein-mass spectrometry coupled proteomics [42]. In order to measure the consequence of these advancements which may add significant quality to the biomarker discovery process, there is a need for inspecting how current proteomic surveys generally relate to systems of complex disease. In this work, we examined proteomic and inflammatory protein sets for clinically important overlaps with biomarkers and drug targets in the healthy human plasma proteome recently integrated in the HIP2 database [14]. We analyzed the functional annotations of these protein sets with gene ontology, and investigated patterns of protein interaction, tissue specificity and overlap with pathways. The human plasma proteome and filtered human plasma proteome were found to have higher percentages of biomarkers and drug targets than was found in the curated human proteome. Plasma is readily accessible and, based on the presence of biomarkers in plasma, this suggests that it is useful for objectives of clinical diagnosis and prognosis. As we found, especially for GO level 4, tests for statistical significance identified differences between the curated human proteome and the filtered human plasma proteome across all three GO vocabularies. Distinct differences of percentages for GO term overlaps were not observed however between the curated human proteome and the human plasma proteome, although we did find higher percentage representations of P compared to H for GO term annotation percentages <2%. The higher percentage representations for low frequency GO term annotations in plasma can be generally interpreted to represent how the plasma proteome specifically deviates from the overall human proteome. Although the inflammatory process is tissue specific, 70% of inflammatory proteins are also found in plasma, and inflammation has been found to have some overlap with complex disease. We found inflammatory plasma proteins to have a higher chance to be biomarkers than the plasma proteome. Additionally, we found inflammatory plasma protein interacting partners to have a higher chance to be biomarkers than the plasma proteome. Compared to the plasma proteome, the inflammatory plasma protein set also presented a distinctive profile of statistically significant GO terms and percentages of overlap. Our visualizations of PPI network topologies showed differences between tissues, most distinctively for the higher centrality observed in brain and heart PPI networks. Six hub proteins in the PPIs expanded inflammatory network were found to be potential cancer biomarkers: BMP2_HUMAN, AACT_HUMAN, CXCR4_HUMAN, FETUA_HUMAN, TRFE_HUMAN and THRB_HUMAN. We identified a specificity issue in terms of how many inflammatory proteins are expressed in many tissues, and may thus be difficult to use as biomarkers of tissue specific cancer. Despite decades of effort, single biomarkers have not been found that can reach the levels of specificity and sensitivity that are required for routine clinical use for the detection or monitoring of the most common cancers. Alternative approaches that measure sensitivity and specificity based on multiple protein and peptide markers may therefore be necessary to achieve a higher level of diagnostic specificity. Overall, we found that a systematic evaluation of plasma, inflammatory proteins and interacting protein partners facilitates the study of complex disease and opportunities for clinical application.
Supplementary Material
Table 5.
Selected molecular function GO terms applied to comparisons between H versus P protein sets and P versus Ip protein sets. The results of comparisons are shown in the last two columns; significance (Y) is based on an adjusted p-value as calculated from 1000 null-hypothesis simulations of hypergeometric comparisons based on random selection, padj≤0.001; n/a is when there are not any representatives of the GO term in the Ip set.
| Index | Level | GO ID | GO term | H vs P | P vs Ip |
|---|---|---|---|---|---|
| F1 | 4 | GO:0005200 | structural constituent of cytoskeleton | Y | n/a |
| F2 | 4 | GO:0003682 | chromatin binding | Y | n/a |
| F3 | 4 | GO:0004386 | helicase activity | Y | n/a |
| F4 | 4 | GO:0005201 | extracellular matrix structural constituent | Y | n/a |
| F5 | 4 | GO:0016563 | transcription activator activity | Y | N |
| F6 | 4 | GO:0003712 | transcription cofactor activity | Y | N |
| F7 | 4 | GO:0000166 | nucleotide binding | Y | N |
| F8 | 4 | GO:0003702 | RNA polymerase II transcription factor activity | Y | n/a |
| F9 | 4 | GO:0030695 | GTPase regulator activity | Y | n/a |
| F10 | 4 | GO:0008289 | lipid binding | Y | N |
| F11 | 4 | GO:0005515 | protein binding | Y | N* (p=0.06) |
| F12 | 4 | GO:0022892 | substrate-specific transporter activity | Y | N |
| F13 | 4 | GO:0003700 | transcription factor activity | Y | N |
| F14 | 4 | GO:0016787 | hydrolase activity | Y | N |
| F15 | 4 | GO:0016740 | transferase activity | Y | N |
| F16 | 4 | GO:0022857 | transmembrane transporter activity | Y | N |
| F17 | 4 | GO:0043167 | ion binding | Y | N |
| F18 | 4 | GO:0004871 | signal transducer activity | N | Y |
Acknowledgments
This work was supported by a grant from the National Cancer Institute (U24CA126480-01), part of NCI’s Clinical Proteomic Technologies Initiative (http://proteomics.cancer.gov). This initiative is designed to advance the field of clinical cancer proteomics by addressing the challenges to the measurement of peptides and/or proteins in clinical specimens. A component of this initiative is the set of Clinical Proteomic Technology Assessment for Cancer (CPTAC) teams that includes the Broad Institute of MIT and Harvard, Memorial Sloan-Kettering Cancer Center, Purdue University, University of California, San Francisco, and Vanderbilt University School of Medicine.
References
- 1.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 2.Aggarwal BB. Nuclear factor-kappaB: the enemy within. Cancer cell. 2004;6:203–208. doi: 10.1016/j.ccr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Alper C, Taggart HA. Plasma protein patterns in disease. Am Pract Dig Treat. 1954;5:349–353. [PubMed] [Google Scholar]
- 4.Parfentjev IA, Johnson ML. The plasma protein pattern and its significance in geriatrics and cancer diagnosis. Geriatrics. 1955;10:232–238. [PubMed] [Google Scholar]
- 5.Trusheim MR, Berndt ER, Douglas FL. Stratified medicine: strategic and economic implications of combining drugs and clinical biomarkers. Nature reviews. 2007;6:287–293. doi: 10.1038/nrd2251. [DOI] [PubMed] [Google Scholar]
- 6.Williams SA, Slavin DE, Wagner JA, Webster CJ. A cost-effectiveness approach to the qualification and acceptance of biomarkers. Nature reviews. 2006;5:897–902. doi: 10.1038/nrd2174. [DOI] [PubMed] [Google Scholar]
- 7.Veenstra TD, Conrads TP, Hood BL, Avellino AM, et al. Biomarkers: mining the biofluid proteome. Mol Cell Proteomics. 2005;4:409–418. doi: 10.1074/mcp.M500006-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Petricoin EF, Belluco C, Araujo RP, Liotta LA. The blood peptidome: a higher dimension of information content for cancer biomarker discovery. Nat Rev Cancer. 2006;6:961–967. doi: 10.1038/nrc2011. [DOI] [PubMed] [Google Scholar]
- 9.Liotta LA, Petricoin EF. Putting the “bio” back into biomarkers: orienting proteomic discovery toward biology and away from the measurement platform. Clinical chemistry. 2008;54:3–5. doi: 10.1373/clinchem.2007.097659. [DOI] [PubMed] [Google Scholar]
- 10.Ping P, Vondriska TM, Creighton CJ, Gandhi TK, et al. A functional annotation of subproteomes in human plasma. Proteomics. 2005;5:3506–3519. doi: 10.1002/pmic.200500140. [DOI] [PubMed] [Google Scholar]
- 11.Berhane BT, Zong C, Liem DA, Huang A, et al. Cardiovascular-related proteins identified in human plasma by the HUPO Plasma Proteome Project pilot phase. Proteomics. 2005;5:3520–3530. doi: 10.1002/pmic.200401308. [DOI] [PubMed] [Google Scholar]
- 12.Muthusamy B, Hanumanthu G, Suresh S, Rekha B, et al. Plasma Proteome Database as a resource for proteomics research. Proteomics. 2005;5:3531–3536. doi: 10.1002/pmic.200401335. [DOI] [PubMed] [Google Scholar]
- 13.Liu X, Valentine SJ, Plasencia MD, Trimpin S, et al. Mapping the human plasma proteome by SCX-LC-IMS-MS. Journal of the American Society for Mass Spectrometry. 2007;18:1249–1264. doi: 10.1016/j.jasms.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saha S, Harrison SH, Shen C, Tang H, et al. HIP2: An online database of human plasma proteins from healthy individuals. BMC medical genomics. 2008;1:12. doi: 10.1186/1755-8794-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shadforth I, Bessant C. Genome annotating proteomics pipelines: available tools. Expert review of proteomics. 2006;3:621–629. doi: 10.1586/14789450.3.6.621. [DOI] [PubMed] [Google Scholar]
- 16.Balkwill F, Charles KA, Mantovani A. Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer cell. 2005;7:211–217. doi: 10.1016/j.ccr.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Theodoropoulos G, Papaconstantinou I, Felekouras E, Nikiteas N, et al. Relation between common polymorphisms in genes related to inflammatory response and colorectal cancer. World J Gastroenterol. 2006;12:5037–5043. doi: 10.3748/wjg.v12.i31.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oremek GM, Sauer-Eppel H, Bruzdziak TH. Value of tumour and inflammatory markers in lung cancer. Anticancer research. 2007;27:1911–1915. [PubMed] [Google Scholar]
- 19.Aggarwal BB, Shishodia S, Sandur SK, Pandey MK, Sethi G. Inflammation and cancer: how hot is the link? Biochemical pharmacology. 2006;72:1605–1621. doi: 10.1016/j.bcp.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 20.States DJ, Omenn GS, Blackwell TW, Fermin D, et al. Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nature biotechnology. 2006;24:333–338. doi: 10.1038/nbt1183. [DOI] [PubMed] [Google Scholar]
- 21.Boyle EI, Weng S, Gollub J, Jin H, et al. GO::TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics (Oxford, England) 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polanski M, Anderson LN. Biomarker Insights. 2006:1–48. [PMC free article] [PubMed] [Google Scholar]
- 23.Wishart DS, Knox C, Guo AC, Shrivastava S, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic acids research. 2006;34:D668–672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, Zhang Y, Yin Y, Gao G, et al. SPD--a web-based secreted protein database. Nucleic acids research. 2005;33:D169–173. doi: 10.1093/nar/gki093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashburner M, Ball CA, Blake JA, Botstein D, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon P, Markiel A, Ozier O, Baliga NS, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alfarano C, Andrade CE, Anthony K, Bahroos N, et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic acids research. 2005;33:D418–424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernandez-Toro J, Prieto C, De las Rivas J. APID2NET: unified interactome graphic analyzer. Bioinformatics (Oxford, England) 2007;23:2495–2497. doi: 10.1093/bioinformatics/btm373. [DOI] [PubMed] [Google Scholar]
- 29.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, et al. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic acids research. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vastrik I, D’Eustachio P, Schmidt E, Joshi-Tope G, et al. Reactome: a knowledge base of biologic pathways and processes. Genome biology. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shargel L. Applied Biopharmaceutics & Pharmacokinetics. McGraw-Hill; New York: 2005. [Google Scholar]
- 32.Luster AD, Alon R, von Andrian UH. Immune cell migration in inflammation: present and future therapeutic targets. Nature immunology. 2005;6:1182–1190. doi: 10.1038/ni1275. [DOI] [PubMed] [Google Scholar]
- 33.Spano JP, Milano G, Rixe C, Fagard R. JAK/STAT signalling pathway in colorectal cancer: a new biological target with therapeutic implications. Eur J Cancer. 2006;42:2668–2670. doi: 10.1016/j.ejca.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 34.Monks NR, Biswas DK, Pardee AB. Blocking anti-apoptosis as a strategy for cancer chemotherapy: NF-kappaB as a target. Journal of cellular biochemistry. 2004;92:646–650. doi: 10.1002/jcb.20080. [DOI] [PubMed] [Google Scholar]
- 35.Luo JL, Kamata H, Karin M. IKK/NF-kappaB signaling: balancing life and death--a new approach to cancer therapy. The Journal of clinical investigation. 2005;115:2625–2632. doi: 10.1172/JCI26322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fryer CJ, Archer TK. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 37.Malchoff DM, Brufsky A, Reardon G, McDermott P, et al. A mutation of the glucocorticoid receptor in primary cortisol resistance. The Journal of clinical investigation. 1993;91:1918–1925. doi: 10.1172/JCI116410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poller W, Faber JP, Scholz S, Weidinger S, et al. Mis-sense mutation of alpha 1-antichymotrypsin gene associated with chronic lung disease. Lancet. 1992;339:1538. doi: 10.1016/0140-6736(92)91301-n. [DOI] [PubMed] [Google Scholar]
- 39.Tang N, Tornatore P, Weinberger SR. Current developments in SELDI affinity technology. Mass spectrometry reviews. 2004;23:34–44. doi: 10.1002/mas.10066. [DOI] [PubMed] [Google Scholar]
- 40.Hu L, Ye M, Jiang X, Feng S, Zou H. Advances in hyphenated analytical techniques for shotgun proteome and peptidome analysis--a review. Analytica chimica acta. 2007;598:193–204. doi: 10.1016/j.aca.2007.07.046. [DOI] [PubMed] [Google Scholar]
- 41.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Abbott KL, Aoki K, Lim JM, Porterfield M, et al. Targeted glycoproteomic identification of biomarkers for human breast carcinoma. Journal of proteome research. 2008;7:1470–1480. doi: 10.1021/pr700792g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.