Abstract
Molecular processes depending on protein-protein interactions can utilize consensus recognition sequences that possess defined secondary structures. Left-handed polyproline II (PPII) helices are a class of secondary structure commonly involved with cellular signal transduction. However, unlike α-helices, for which a substantial body of work exists regarding applications of ring-closing metathesis (RCM), there are few reports on the stabilization PPII helices by RCM methodologies. The current study examined the effects of RCM macrocyclization on left-handed PPII helices involved with the SH3 domain-mediated binding of Sos1 to Grb2. Starting with the Sos1-derived peptide “Ac-V1-P2-P3-P4-V5-P6-P7-R8-R9-R10-amide”, RCM macrocyclizations were conducted utilizing alkenyl chains of varying lengths originating from the pyrrolidine rings of the Pro4 and Pro7 residues. The resulting macrocyclic peptides showed increased helicity as indicated by circular dichroism and enhanced abilities to block Grb2-Sos1 interactions in cell lysate pull-down assays. The synthetic approach may be useful in RCM macrocyclizations where maintenance of proline integrity at both ring junctures is desired.
Keywords: peptide macrocyclization, ring-closing metathesis, polyproline type II helix, Grb2, Sos1, SH3 domain
INTRODUCTION
Stabilization of α-helices through ring closing metathesis (RCM) macrocyclization1–6 has been shown to be impart biological advantages to certain peptides.7, 8 Application of RCM macrocyclization to other classes of helices is less common, with stabilization of 310 helices being a rare example.9, 10 Polyproline type II (PPII) helices represent physiologically important motifs that are characterized by an extended left-handed helical conformation having a translation of 3.12 Å per residue, trans-amide bonds, three residues per turn and typical backbone dihedral values ϕ − 75° and ψ + 145°.11 PPII helices are widely involved in protein-protein interactions related to cellular signaling.12–14 This is exemplified by the growth factor receptor bound protein 2 (Grb2), which is composed of a central SH2 domain that recognizes phosphotyrosyl-containing sequences, flanked by amino- and carboxy-terminal SH3 domains that bind to PPII and 310 helices.15 Grb2 is a prototypical adaptor protein mediating receptor tyrosine kinase signaling, whose involvement in oncogenic pathways makes it a potential target for anticancer therapeutic development.16, 17 Because inhibitors of SH3 domain binding may down-regulate Grb2 function, efforts have been undertaken to increase the effectiveness of SH3 domain-inhibitor interactions.18–22 Artificial induction of PPII helices through non-covalent conformational stabilization has been reported using modified proline residues,23–26 however, only a single example has been reported of an RCM approach to PPII stabilization, and in this case the requisite alkenyl chains were appended to peptide backbone amide nitrogens.27 The objective of the current study was to examine the effects of RCM macrocyclization on Grb2 SH3 domain-binding peptides by employing alkenyl chains originating from the pyrrolidine rings of key proline residues.
RESULTS
Macrocycle Design
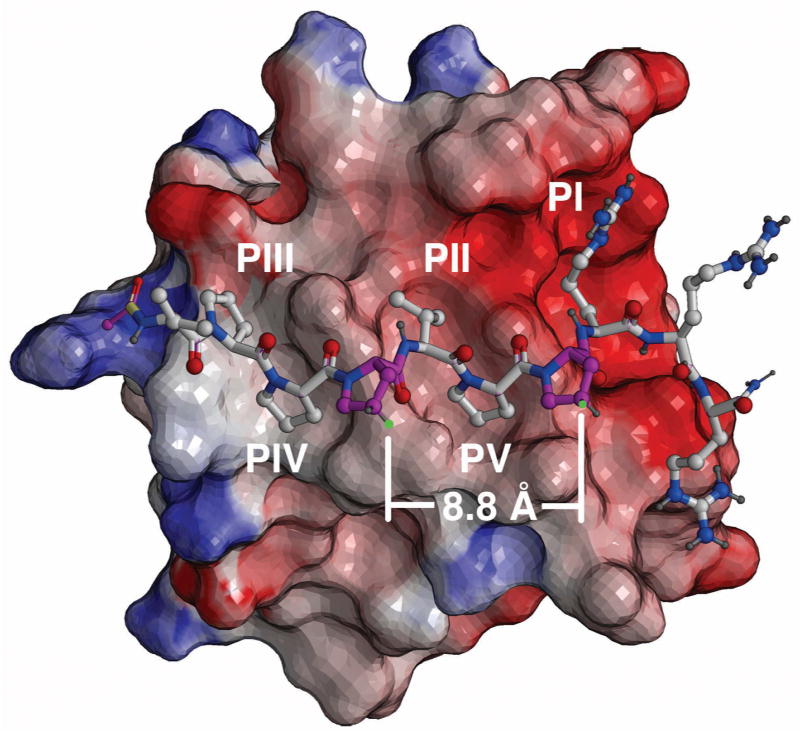
Two classes of binding modes (Class I and Class II) that differ on their binding orientations have been shown for the interaction of PPII helices with SH3 domains.28 The Sos1-derived sequence, “Ac-V1-P2-P3-P4-V5-P6-P7-R8-R9-R10-amide” (1) has been shown to bind with high affinity to the Grb2 N-terminal domain SH3 domain and to bind to the SH3 C-terminal domain in a same orientation and mode, but with lower affinity.29, 30 (The affinities for the C-terminal and N-terminal domains have been reported as 2.6 μM and 40 μM, respectively.)31 NMR solution structures of 1 bound to the Grb2 N-terminal SH3 domain show the peptide binds in a Class II orientation with the region P3-P4-V5-P6-P7-R8 encompassing a prototypical PPII Pro-x-x-Pro motif, in which the P3, V5 and P6 residues bind in hydrophobic recognition pockets designated PIV, PII and PV, respectively (Figure 1).28, 32 The neighboring P4 and P7 residues, which are situated one turn apart (approximately 9 Å) and lie on the side of the helix opposite to the protein-binding P3 and P6 residues, were chosen as sites for introduction of RCM ring-closing functionality (Figure 1).
Figure 1.
NMR solution structure of Ac-V-P-P-P-V-P-P-R-R-R-amide (1) bound to the Grb2 SH3 N-domain (PDB 3GBQ, reference 32). The peptide binds in a Class II orientation, with interaction pockets identified as PI – PV (reference 28). The protein surface is shown as electrostatic potential (blue – positive; red = negative). Atom coloring: blue = nitrogen; red = oxygen; light grey = carbon and dark grey = hydrogen. Key proline residues of the “P-x-x-P” recognition motif are rendered in solid color (turquoise). Proline residues involved with RCM modification are rendered in magenta, with sites of alkenyloxy attachment shown in green.
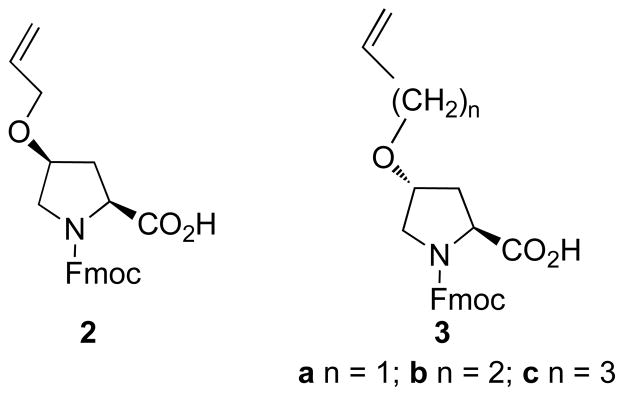
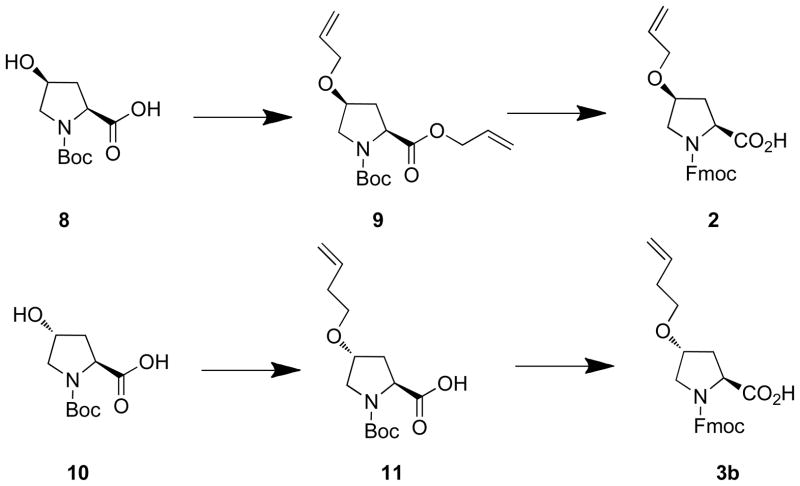
Alkenyl ether groups were introduced at the P4 and P7 proline γ-methylenes (pyrrolidine C4). To minimize perturbation of the protein-bound helix geometry conformation, the alkenyloxy groups originate from the pyrrolidine rings of the P4 and P7 residues with (4S) and (4R) configurations, respectively (Figure 1). The distance between these positions in the protein-bound solution structure was calculated to be approximately 8.8 Å (Figure 1). However, because the optimal lengths for the ring-closing segments were not obvious, three macrocycles were prepared that contained ring-closing segments of increasing length, from four to six carbons. This required the synthesis of the N-Fmoc-protected proline analogues 2 and 3a – 3c (Figure 2).
Figure 2.
Structures of reagents for the solid-phase introduction of alkenyloxy-containing proline residues.
Macrocycle Synthesis
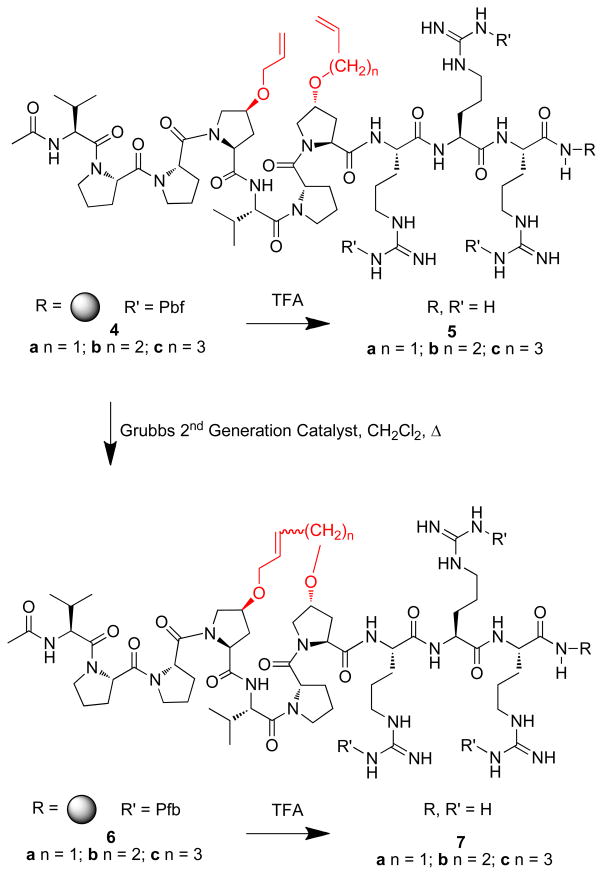
Solid-phase peptide synthesis was conducted on NovaSyn®TGR resin using standard Fmoc solid-phase protocols and active ester coupling. The alkenyloxy-containing proline analogues 2 and 3a – 3b were employed to prepare the resin-bound metathesis precursors 4a – 4c, respectively. Portions of the resins were cleaved to provide the corresponding open-chain peptides 5a – 5c (Scheme 1). On-resin ring closure of 4a – 4c was then conducted using Grubbs 2nd Generation33 and the resulting peptide resins (6a – 6c) were cleaved by treatment with TFA and purified by HPLC to yield the final macrocyclic peptides 7a – 7c (Scheme 1). Scrambling of the wild-type peptide (WT) (1) provided Ac-P-R-V-P-P-P-P-V-R-R-NH2 (13) as a negative control for binding experiments.
Scheme 1.
Synthesis peptides described in the text. Shown in red are alkenyloxy groups involved with RCM ring closure.
Determination of Grb2 SH3 N-domain Binding Affinities
Grb2 forms a complex with Sos1 through interactions of its C- and N-terminal SH3 domains with “P-x-x-P-x-R” sequences in Sos1, with the N-terminal SH3 domain-binding affinity of WT peptide 1 having been reported as Kd = 39 μM by isothermal calorimetry.32 In our current work, in vitro Grb2 SH3 N-domain binding affinities of synthetic peptides were determined using a Trp-fluorescence assay.34 The WT peptide 1, as well as the open-chain metathesis precursors 5a – 5c, showed similar binding affinities (Kd ≈ 11 – 14 μM; Table 1). In comparison, the affinities of the macrocyclic peptides were lower and ranged from Kd ≈ 33 μM for the smallest macrocycle (7a) to Kd ≈ 18 μM for the largest macrocycle (7c), with the intermediate sized 7b exhibiting Kd ≈ 24 μM (Table 1).
Table 1.
Grb2 SH3 N-Domain in vitro binding affinities1
| Peptide | Kd ± SD (μM)2 |
|---|---|
| 1 | 11.1 ± 1.31 |
| 5a | 12.6 ± 0.98 |
| 5b | 12.2 ± 0.40 |
| 5c | 14.5 ± 0.75 |
| 7a | 33.5 ± 1.63 |
| 7b | 24.2 ± 1.21 |
| 7c | 18.6 ± 1.11 |
| 13 | 173.1 ± 1.16 |
Data were obtained using a Trp-fluorescence assay as described in the Materials and Methods.
Dissociation constant ± standard deviation.
Blockade of Grb2 – Sos1 Association in Cell Lysates
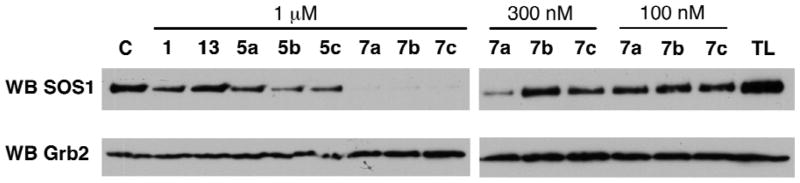
The peptides were also examined for their ability to block the binding of Sos1 to full-length Grb2. Non-ionic detergent lysates were prepared from human pheochromocytoma-derived PC12 cells, which express high levels of both Grb2 and Sos1.35 Synthetic peptides at different concentrations were added to the lysates and measured for their ability to compete for Grb2 pull down using our recently reported biotinylated Grb2 capture reagent immobilized to streptavidin-coated microbeads.36 This reagent binds selectively to the Grb2 SH2 domain with high affinity.37 At 1 μM concentrations, the scrambled negative control peptide 13 showed little inhibition of Grb2-Sos1 association. In contrast, the WT peptide 1 and the open-chain RCM precursor peptide 5a exhibited similar but modest inhibition (Figure 3). Relative to 1 and 5a, the open-chain analogues 5b and 5c showed slightly higher inhibitory potencies. At 1 μM concentrations the macrocyclic peptides 7a – 7c were able to effectively block Grb2 – Sos1 interactions, while at 300 nM concentrations these peptides displayed clear differential inhibition, in the potency order: 7a > 7c > 7b (Figure 3). At the lowest concentrations tested (100 nM), only 7a showed visible inhibition. While the IC50 values for 7b and 7c were in the range of 1 μM – 300 nM, the most effective analogue (7a) had an apparent IC50 value between approximately 100 nM and 300 nM.
Figure 3.
SDS-PAGE gels showing the effects of synthetic peptides on Grb2-Sos1 association in PC12 cell lysates. Experiments were performed as summarized in the Materials and Methods with protein detection by means of Western blotting. Shown is the amount of Sos1 (top gel) bound to Grb2 (bottom gel) in the presence of the indicated concentrations of peptides. Control (C) was run in the absence of peptide with total cell lysate shown as TL.
Examination of PPII Helicity Using Circular Dichroism
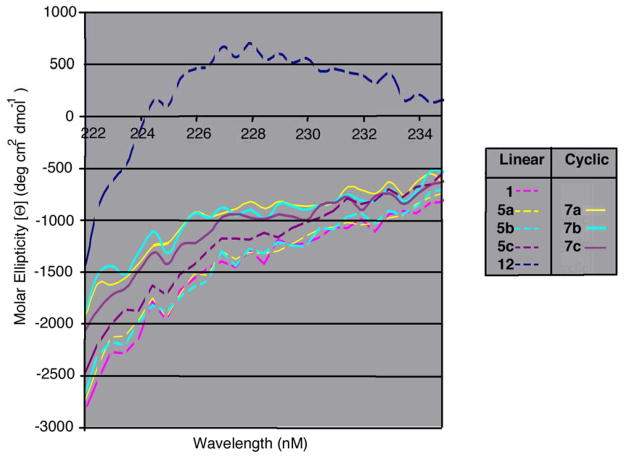
From the above data it was clear that introduction of alkenyloxy functionality modestly enhanced inhibitory potency, while macrocyclization resulted in significantly more effective binding antagonists. Evaluation of PPII helicity was undertaken by measuring circular dichroism (CD) peak intensities at 226 nm.38 An overlay of linear and cyclic peptide spectra expanded about λ226 showed that none of the linear peptides 5a – 5c or WT peptide 1 showed appreciable PPII helical character (Figure 4). All three of the macrocyclic peptides (7a – 7c) showed slightly increased PPII helical character relative to the WT peptide 1 and open-chain metathesis precursors 5a – 5c.
Figure 4.
Expanded CD spectra comparing linear and cyclic peptides.
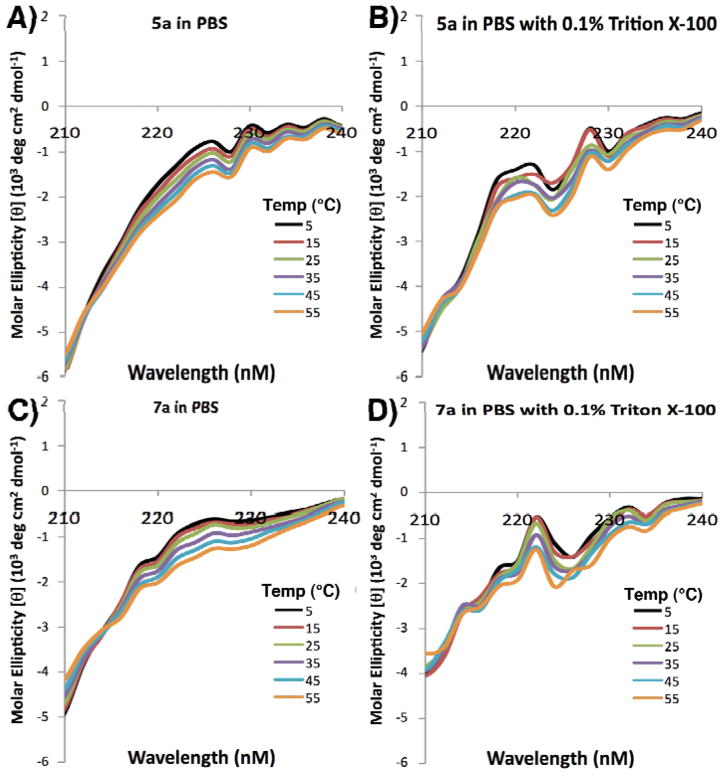
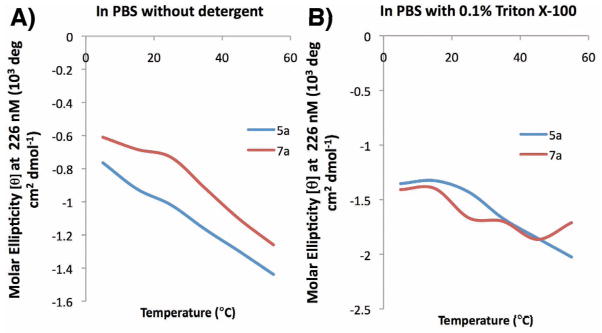
In order to gain greater understanding of the effect of macrocyclization on PPII helical conformation, cryogenic studies were conducted by running far UV CD spectra of 7a and the corresponding linear precursor 5a at different termperatures in pH 7.4 PBS buffer, with and without detergent Triton X-100 (Figure 5). The changes in [θ]max = 226 nm as a function of temperature for peptides 5a and 7a in pH 7.4 PBS buffer without detergent were also plotted (Figure 6). Overall, the data indicated Triton X-100 did have some effects in disturbing the peptide solution conformation, and a negative correlation between molar ellipticity at 226 nm and temperature confirmed the existence of PPII helix. However the plot of the molar ellipticity of 5a and 7a at 226 nm against temperature indicated that macrocyclization has minimal effects on the stability of PPII helical structure against increasing temperature.
Figure 5.
Far-UV CD run at varying temperatures of peptides 5a and 7a in pH 7.4 PBS buffer in the absence (panels A nd C, respectively) and presence of 0.1% Triton X detergent (Panels B and D, respectively).
Figure 6.
Change of CD [θ]max = 226 nm as a function of temperature for peptide 5a and 7a in pH 7.4 PBS in the absence (panel A) and presence (panel B) of Triton X detergent.
DISCUSSION
In the in-vitro system with purified components, the addition of carbon macrocycles to the main peptide backbone (7a – 7c) has the effect of slightly reducing the binding affinity to the Grb2 SH3 N-domain relative to that of the WT Sos peptide (1). Linear peptides with uncoupled hydrocarbon chains display affinities equivalent to the WT peptide. A likely explanation for this observation is that the macrocyclic structures introduce constraint into the peptide backbone, resulting in a less flexible form or even a marginally altered structure compared to the WT peptide. This is consistent with the results of the circular dichroism analysis, which revealed a slightly higher helical content. Such a conformational difference, albeit minor, could impart different binding characteristics. Furthermore, with increasing macrocycle size, the binding affinity increases and approaches that of the wild-type peptide. Presumably this is related to a corresponding increased relaxation of the constraint imposed, as a function of macrocycle chain length. Finally, as the modified linear peptides (5a – 5c) would not be expected to constrain the native backbone configuration, they should not substantially alter the mode of docking (unless they hinder or enhance it somehow). This is reflected in their Kd values being similar to that of the WT Sos peptide (1).
The in-vitro fluorescence assay results, although internally consistent, are not in keeping with the competition assay using whole cell lysates, as revealed by Western blot analysis. Here the cyclic peptides at 1 μM concentration show a more potent inhibition of Sos binding. This could be due to the fact that the effective concentrations of peptides accessing the target may differ markedly between species. In addition, the affinities of the WT sequence and linear modified peptides for other SH3 domains present in the lysate may also be greater than those of the cyclized peptides, potentially leading to off-target binding that could reduce the available pool of peptide for specific Grb2 binding. It is also possible that the added hydrophobic character introduced by the hydrocarbon linker of the macrocyclic structures, could confer an increased tendency of these peptides to interact hydrophobic binding sites on the Sos protein substrate, thereby augmenting their inhibitory nature. The possibility of less specific binding by such hydrophobic structures is indicated by the fact that even the scrambled peptide (13) displays a low affinity for the Grb2 SH3 N-domain in the fluorescence assay. It should also be considered that differences in buffers used in assays could contribute to differences in apparent affinities. For example, the cell lysis buffer contains detergent unlike in the Trp-fluorescence assay, which may influence the solubility or aggregation state of either the peptide or proteins, and give rise to varying interaction behavior.
CONCLUSIONS
The current study represents only the second application of RCM macrocyclization to biologically active PPII helical peptides. The synthetic strategy employed is characterized by its use of proline γ-methylenes (pyrrolidine C4 positions) for introduction of ring-closing alkenyloxy functionality, with the locations of the ring-closing proline residues and the stereochemistry of the alkenyloxy substituents being based on the NMR solution structure of a linear WT peptide bound to the target Grb2 N-terminal SH3 domain protein. Alkenyloxy-substituted proline residues have been used for RCM macrocyclization onto alkenylglycine residues of random coil-constrained mimics of the membrane-proximal domain of FcεRIα.38 Our current approach extends this technology by allowing RCM macrocyclization with maintenance of proline residues at both ring junctures. Within our current system we have found that RCM macrocyclization can increase PPII helicity and enhance the ability of peptides to block Grb2 – Sos1 interactions.
MATERIALS AND METHODS
(2S,4S)-2-Allyl 1-Tert-butyl 4-(Allyloxy)pyrrolidine-1,2-dicarboxylate (9)
To a suspension of sodium hydride (60% in mineral oil, 380 mg, 9.50 mmol) in DMF (6 mL) at 0° C, was added a solution of (4S)-N-Boc- 4-hydroxy-L-proline (8, Scheme 2) (1.0 g, 4.32 mmol) [Sigma-Aldrich, Cat. 654019] in DMF (6 mL) dropwise (over 5 minutes). The mixture was kept at 0° C (15 minutes) then allylbromide (1.00 mL, 11.2 mmol) was added and the mixture was allowed to come to room temperature and stirred (overnight). The reaction was quenched by the addition of saturated aqueous NH4Cl (50 mL) and extracted (EtOAc, 150 mL). The organic layer was washed, dried and purified silica gel chromatography (hexanes : EtOAc) to yield 9 (Scheme 2) as a colorless oil (1.25 g, 93% yield). 1H NMR (400 MHz, CDCl3) δ 5.87 – 5.71 (m, 2 H), 5.28 – 5.06 (m, 4 H), 4.60 – 4.45 (m, 2 H), 4.36 (dd, J = 8.4, 4.0 Hz, 0.4 H), 4.25 (dd, J = 8.4, 4.0 Hz, 0.6 H), 4.00 (m, 1 H), 3.87 – 3.82 (m, 2 H), 3.58 (dd, J = 7.6, 5.6 Hz, 0.6 H), 3.53 (dd, J = 7.6, 5.2 Hz, 0.4 H), 3.44 (dd, J = 7.6, 3.2 Hz, 0.6 H), 3.40 (dd, J = 7.6, 2.8 Hz, 0.4 H), 2.30 – 2.15 (m, 2 H), 1.40 (s, 3.5 H), 1.34 (s, 5.5 H). 13C NMR (100 MHz, CDCl3) δ 171.9, 171.5, 154.2, 153.8, 134.2, 132.0, 131.8, 118.4, 117.9, 116.8, 80.0, 76.6, 75.6, 69.7, 65.5, 57.7, 57.3, 51.9, 51.2, 36.1, 35.0, 28.3, 28.2. ESI (+VE) m/z: 334.4 (M + Na)+.
Scheme 2.
Synthesis of reagents 2 and 3b.
(2S,4S)-1-((2-(9H-Fluoren-9-yl)ethoxy)carbonyl)-4-(allyloxy)pyrrolidine-2-carboxylic Acid [(4S)-N-Fmoc-4-allyloxyproline] (2)
A mixture 9 (Scheme 2, 1.25 g, 4.00 mmol) and LiOH monohydrate (336 mg, 8.00 mmol) in THF (5 mL) and H2O (5 mL) was stirred at room temperature (3 h). Volatile organics were removed by rotary evaporation under reduced pressure and the aqueous phase was then washed (ether, 2 × 50 mL), acidified to pH 3 – 4 (1N aqueous HCl) and extracted (EtOAc, 150 mL). The EtOAc layer was washed, dried (NaSO4) and evaporated to yield a colorless oil. This was treated with a mixture of TFA (10 mL) and dichloromethane (10 mL) (room temperature, 2 h). The solvent was removed by rotary evaporation under reduced pressure and the residue was placed under high vacuum (2 h) and then dissolved in dioxane (15 mL) and H2O (15 mL). To this was added NaHCO3 (1.68 g, 20 mmol) and Fmoc-OSu (1.52 g, 4.50 mmol), and the mixture was stirred (room temperature, overnight). Dioxane was removed by rotary evaporation under reduced pressure and the remaining aqueous phase was washed (ether, 50 mL × 2), acidified to pH 3 – 4 (1N aqueous HCl) and extracted (EtOAc, 200 mL). The organic layer was washed (brine), dried (NaSO4) and evaporated to yield pure 2 as a white wax (1.52 g, 96% yield). 1H NMR (400 MHz, CDCl3) δ 11.0 (brs, 1 H), 7.73 (d, J = 7.6 Hz, 1 H), 7.68 (d, J = 7.6 Hz, 1 H), 7.62 – 7.53 (m, 2 H), 7.40 – 7.23 (m, 4 H), 5.80 (m, 1 H), 5.25 – 5.05 (m, 2 H), 4.54 – 4.45 (m, 2 H), 4.40 – 4.30 (m, 2 H), 4.23 (m, 0.5 H), 4.15 (m, 0.5 H), 4.08 (m, 0.5 H), 4.03 (m, 0.5 H), 3.89 (m, 1 H), 3.62 – 3.58 (m, 2 H), 2.40 (m, 1 H), 2.22 (m, 1 H). 13C NMR (100 MHz, CDCl3) δ 176.3, 175.8, 155.5, 155.0, 143.9, 143.6, 141.3, 134.0, 127.7, 127.1, 125.1, 125.0, 120.0, 117.1, 76.4, 75.5, 69.6, 67.9, 66.9, 57.8, 57.5, 52.2, 47.1, 36.0, 34.5. ESI (+VE) m/z: 416.2 (M + Na)+. ESI-HRMS cacld for C23H24NO5+(M + H)+: 394.1649, Found 394.1657.
(2S,4R)-1-((2-(9H-Fluoren-9-yl)ethoxy)carbonyl)-4-(allyloxy)pyrrolidine-2-carboxylic Acid [(4R)-N-Fmoc-4-allyloxyproline] (3a)
The synthesis of 3a has been reported.39
(2S,4R)-4-(But-3-en-1-yloxy)-1-(tert-butoxycarbonyl)pyrrolidine-2-carboxylic Acid [(4R)-N-Boc-4-(but-3-en-1-yloxy)proline] (11)
A mixture of (4R)-N-Boc- 4-hydroxy-L-proline (8) (1.0 g, 4.32 mmol) [Sigma Aldrich: Cat. 518360] (2.0 g, 8.64 mmol), 4-bromo-1-butene (3.5 mL, 34.6 mmol) and potassium hydroxide (5 g) in a mixture of dioxane (5 mL) and H2O (5 mL) was stirred room temperature, 48 h), acidified to pH 3 – 4 (1N aqueous HCl) and extracted (EtOAc). The organic layer was washed (brine), dried (NaSO4) and evaporated to yield 11 (Scheme 2) as a colorless oil (100 mg). 1H NMR (400 MHz, CDCl3) δ 5.75 (m, 1 H), 5.07 - 4.99 (m, 2 H), 4.11 (dd, J = 8.0, 2.4 Hz, 0.6 H), 4.31 (t, J = 8.0 Hz, 0.4 H), 4.03 (m, 1 H), 3.58 (m, 1 H), 3.47 – 3.40 (m, 3 H), 2.36 (m, 1 H), 2.30 – 2.22 (m, 2 H), 2.21 – 2.05 (m, 1 H), 1.45 (s, 5.5 H), 1.39 (s, 3.5 H).
(2S,4R)-1-((2-(9H-Fluoren-9-yl)ethoxy)carbonyl)-4-(but-3-en-1-yloxy)pyrrolidine-2-carbo xylic Acid Acid [(4R)-N-Boc-4-(but-3-en-1-yloxy)proline] (3b)
A solution of 11 (100 mg) in TFA (3 mL) and dichloromethane (3 mL) was stirred (room temperature, 2 h). The solvent was removed by rotary evaporation under reduced pressure and the residue was placed under high vacuum (2 h) and then dissolved in dioxane (3 mL) and H2O (3 mL). To this was added NaHCO3 (353 mg, 4.2 mmol) and Fmoc-OSu (142 mg, 0.42 mmol), and the mixture was stirred (room temperature, overnight). Dioxane was removed by rotary evaporation under reduced pressure and the remaining aqueous phase was washed (ether, 30 mL × 2), acidified to pH 3 – 4 (1N aqueous HCl) and extracted (EtOAc, 100 mL). The organic layer was washed (brine), dried (NaSO4) and evaporated to yield pure 3b (Scheme 2) as a white wax (140 mg, 98% yield). 1H NMR (400 MHz, CDCl3) δ 9.40 (brs, 1 H), 7.75 (d, J = 7.6 Hz, 1 H), 7.67 (d, J = 7.6 Hz, 1 H), 7.57 – 7.48 (m, 2 H), 7.40 – 7.23 (m, 4 H), 5.77 (m, 1 H), 5.10 – 4.99 (m, 2 H), 4.50 – 4.22 (m, 4 H), 4.10 (m, 1 H), 3.70 – 3.57 (m, 2 H), 3.47 – 3.37 (m, 2 H), 2.45 – 2.00 (m, 4 H). 13C NMR (100 MHz, CDCl3) δ 177.6, 176.3, 155.7, 154.7, 143.7, 141.3, 134.7, 127.7, 127.1, 125.0, 120.0, 116.7, 76.9, 76.2, 68.6, 67.9, 67.8, 58.0, 57.5, 51.9, 51.7, 47.1, 47.0, 36.8, 35.1, 34.1. ESI (+VE) m/z: 430.2 (M + Na)+. ESI-HRMS cacld for C24H26NO5+(M + H)+: 408.1806, Found, 408.1811.
(2S,4R)-1-((2-(9H-Fluoren-9-yl)ethoxy)carbonyl)-4-(pent-4-en-1-yloxy)pyrrolidine-2-carbo xylic Acid [(4R)-N-Boc-4-(pent-4-en-1-yloxy)proline] (3c)
The synthesis of 3c has been reported.39
Peptide Synthesis
Peptides were synthesized on NovaSyn®TGR resin (Novabiochem, cat. no. 01-64-0060) by standard Fmoc solid-phase protocols using active ester coupling (1-hydroxybenzotriazole (HOBT) and N, N′-diisopropylcarbodiimide (DIC) as coupling reagents (single couple, 2 hours). The guanidine functionality of arginine was protected using the 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl (Pbf) group. On resin ring-closing metathesis was conducted using dried resin (200 mg) mixed with anhydrous dichloromethane (3 mL) and de-gassed under argon (3 minutes). Grubbs 2nd Generation Catalyst [benzylidene[1,3-bis(2,4,6-trimethylphenyl)-2-imidazolidinylidene]dichloro(tricyclohexylphosphine) ruthenium]33 (15 mg) in anhydrous dichloromethane (4 mL) DCM) was added, and the mixture was agitated in a sealed (room temperature, 24 h). The resulting resin was washed with N, N-dimethylformamide, methanol, dichloromethane and ether and then dried under vacuum (overnight). Peptides were cleaved from resin (200 mg) by treatment with TFA: triisopropylsilane : H2O (90 : 5 : 5) (5 mL, 4 h). The resin was removed by filtration and the filtrate was concentrated under vacuum, then precipitated with ether and the precipitate washed with ether. The resulting solid was dissolved in 50% aqueous acetonitrile (5 mL) and purified by reverse phase preparative HPLC using a Phenomenex C18 column (21 mm dia × 250 mm, cat. no: 00G-4436-P0) with a linear gradient from 0% aqueous acetonitrile (0.1% TFA) to 65% acetonitrile (0.1% TFA) over 30 minutes at a flow rate of 10.0 mL/minute (detection at 220 nm). Lyophilization provided products as white powders. Mass spectral data are provided in Table 2.
Table 2.
Peptide ESI Mass Spectral Data.
| No | Expected (M + H)+ | Observed (M + H)+ | HRMS Expected (M + H)+ | HRMS Observed (M + H)+ |
|---|---|---|---|---|
| 1 | 1211.7 | 1211.5 | ||
| 8 | 1211.7 | 1211.7 | ||
| 9 | 1207.7 | 1207.5 | ||
| 5a | 1323.8 | 1323.8 | 1323.8008 | 1323.7892 |
| 5b | 1337.8 | 1337.8 | 1337.8165 | 1337.8144 |
| 5c | 1351.8 | 1351.8 | 1351.8321 | 1351.8306 |
| 7a | 1295.8 | 1295.8 | 1295.7695 | 1295.7672 |
| 7b | 1309.8 | 1309.8 | 1309.7852 | 1309.7824 |
| 7c | 1323.8 | 1323.9 | 1323.8008 | 1323.7986 |
Determination of Grb2 SH3 N-Domain-Binding Constants
Purification of Grb2 SH3-N domain
GST-Grb2SH3N fusion protein was expressed in E. coli strain BL21(DE3) from the pGEX-2T vector by inducing cultures overnight at 20°C with 50 μM IPTG. Cells were lysed by sonication in TPE lysis buffer (1% Triton X-100, PBS and 100 mM EDTA) with a protease inhibitor cocktail added. Insoluble material was centrifuged at 48,000 g for 1 hour and the soluble fraction was incubated overnight with glutathione sepharose beads. The next day beads were washed extensively with cold wash buffer (50 mM Tris pH 7.5, 100 mM EDTA, 0.1% Tween20) before overnight incubation with elution buffer (100 mM reduced glutathione brought to pH 8.0 with a concentrated Tris buffer stock). The eluted GST-Grb2SH3N was dialysed against 5 mM Tris pH 7.5 and then cleaved for 24 hours by incubation with thrombin protease before final purification using size exclusion chromatography (Superdex 75 column, GE Healthcare) in gel filtration buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1 mM DTT). Purified Grb2 SH3-N domain was concentrated to give a stock of >1 mM.
Intrinsic tryptophan fluorescence measurements
Tryptophan fluorescence measurements to determine peptide-binding affinities were carried out with a Perkin Elmer LS50B spectrofluorimeter essentially as described previously.34 Briefly, the excitation wavelength employed was 295 nm and the emission wavelength was 342 nm, at the maximum fluorescence intensity, using excitation and emission slit widths, respectively, set to 5 and 15 nm. Titration experiments, each in triplicate, were performed in filtered, degassed PBS buffer containing 1 mM DTT and the purified Grb2 SH3-N domain (6 μM) with continual stirring. The cuvette was maintained at a constant 21°C in its holder by a cycling water-cooling system linked to a thermostat-controled water bath. Peptides were titrated in until no increase in the fluorescence intensity was observed, typically giving 12 to 15 data points per experiment. A single-site binding model enabled an accurate curve to be fit to the data using Origin (v5.0) software, from which mean Kd values and standard deviations could be determined. Data is shown in Table 1.
Inhibition of Grb2 – SOS1 Association in Cell Lysates
Peptides were evaluated for their ability to block Grb2 – Sos1 binding interactions in cell lysates. Non-ionic detergent lysates were prepared from human pheochromocytoma-derived PC12 cells as described.35 The detergent used was Triton X-100 (1%) and the composition of the buffer is as follow: Cell lysis buffer 100 mL: 1.2 g HEPES or 10 mL 500 mM HEPES (50 mM final), 0.4 g NaF, 0.05 g Na3VO4 (sodium orthovanadate, high purity), 0.45 g Na4PO2 (sodium pyrophosphate), 4 mL of 100 mM EDTA (4 mM final) and 10 mL 10% Triton X-100 (TX-100; high purity, Pierce). These cells were selected because they contain relatively high levels of both Grb2 and Sos1. SH3 domain binding antagonists were added to the lysates at the indicated concentrations together with a biotinylated Grb2 capture reagent that had been immobilized to streptavidin-coated microbeads.36 The synthetic biotinylated Grb2 capture reagents binds selectively to the Grb2 SH2 domains with high affinity.37 After 1 hour, the beads were removed by centrifugation, washed 3 times with lysis buffer [50 mM HEPES pH 7.4, 5 mM NaF, 1 mM Na3VO4, 4 mM EDTA, 5 mM Na4PO2, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1 ug/mL aprotinin] and bound proteins were extracted with boiling Laemmli buffer. Samples were resolved by reducing SDS-PAGE, electrophoretically transferred to PVDF membranes and immuno-detected using primary antibodies for Sos1 (Millipore Corp.) or Grb2 (Santa Cruz Biotechnology) and secondary detection reagents for chemiluminescence. Results are shown in Figure 3.
CD Spectral Data
CD spectra of peptides 1, 5a – 5c and 7a – 7c were collected on a Circular Dichroism Spectrometer AVIV Model 202 at 200 μM concentration in H2O, pH 7.4 PBS buffer, and pH 7.4 PBS buffer with 0.1% Triton X-100. The CD spectrum at each condition was collected three times independently; the final plot was based on the average data of three independent runs. The spectrum of Ac-P-P-P-P-P-P-P-R-R-amide (12) is included as a reference exhibiting PPII helicity in solution. Results are shown in Figures 4 – 6.
Acknowledgments
Appreciation is expressed to Drs. Marzena Dyba and Sergey Tarasov of the Biophysics Resource in the Structural Biophysics Laboratory, Molecular Discovery Program, CCR, NCI for providing assistance in obtaining CD spectra. This work was supported in part by the Intramural Research Program of the NIH, Center for Cancer Research, NCI-Frederick and the National Cancer Institute, National Institutes of Health and by a Cancer Research UK project grant and a Heads Up programme grant to SF. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
References
- 1.Schafmeister CE, Po J, Verdine GL. J Am Chem Soc. 2000;122:5891–5892. [Google Scholar]
- 2.Walensky LD, Pitter K, Morash J, Oh KJ, Barbuto S, Fisher J, Smith E, Verdine GL, Korsmeyer S. J Mol Cell. 2006;24:199–210. doi: 10.1016/j.molcel.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 3.Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. J Am Chem Soc. 2007;129:2456–2457. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henchey LK, Jochim AL, Arora PS. Curr Opin Chem Biol. 2008;12:692–697. doi: 10.1016/j.cbpa.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang D, Lu M, Arora PS. Angew Chem Int Ed. 2008;47:1879–1882. doi: 10.1002/anie.200704227. [DOI] [PubMed] [Google Scholar]
- 6.Kutchukian PS, Yang JS, Verdine GL, Shakhnovich EI. J Am Chem Soc. 2009;131:4622–4627. doi: 10.1021/ja805037p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D, Liao W, Arora PS. Angew Chem Int Ed. 2005;44:6525–6529. doi: 10.1002/anie.200501603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arora PS, Ansari AZ. Nature. 2009;462:171–173. doi: 10.1038/462171a. [DOI] [PubMed] [Google Scholar]
- 9.Chapman R, Kulp JL, III, Patgiri A, Kallenbach NR, Bracken C, Arora PS. Biochemistry. 2008;47:4189–4195. doi: 10.1021/bi800136m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobsen O, Klaveness J, Ottersen OP, Amiry-Moghaddam MR, Rongved P. Org Biomol Chem. 2009;7:1599–1611. doi: 10.1039/b823559g. [DOI] [PubMed] [Google Scholar]
- 11.Kapitán J, Gallo D, Goasdoué N, Nicaise M, Desmadril M, Hecht L, Leclercq G, Barron LD, Jacquot Y. J Pep Sci. 2009;15:455–464. doi: 10.1002/psc.1136. [DOI] [PubMed] [Google Scholar]
- 12.Jacobsen O, Klaveness J, Rongved P. Molecules. 2010;15:6638–6677. doi: 10.3390/molecules15096638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siligardi G, Drake AF. Biopolymers. 1995;37:281–292. doi: 10.1002/bip.360370406. [DOI] [PubMed] [Google Scholar]
- 14.Kay BK, Williamson MP, Sudol P. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 15.Harkiolaki M, Tsirka T, Lewitzky M, Simister PC, Joshi D, Bird LE, Jones EY, O’Reilly N, Feller SM. Structure. 2009;17:809–822. doi: 10.1016/j.str.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 16.Dharmawardana PG, Peruzzi B, Giubellino A, Burke TR, Jr, Bottaro DP. Anti-Cancer Drugs. 2005;17:13–20. doi: 10.1097/01.cad.0000185180.72604.ac. [DOI] [PubMed] [Google Scholar]
- 17.Giubellino A, Burke TR, Jr, Bottaro DP. Exp Opin Therap Targ. 2008;12:1021–1033. doi: 10.1517/14728222.12.8.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kardinal C, Konkol B, Lin H, Eulitz M, Schmidt EK, Estrov Z, Talpaz M, Arlinghaus RB, Feller SM. Blood. 2001;98:1773–1781. doi: 10.1182/blood.v98.6.1773. [DOI] [PubMed] [Google Scholar]
- 19.Lewitzky M, Kardinal C, Gehring NH, Schmidt EK, Konkol B, Eulitz M, Birchmeier W, Schaeper U, Feller SM. Oncogene. 2001;20:1052–1062. doi: 10.1038/sj.onc.1204202. [DOI] [PubMed] [Google Scholar]
- 20.Feller SM, Lewitzky M. Curr Pharm Des. 2006;12:529–548. doi: 10.2174/138161206775474369. [DOI] [PubMed] [Google Scholar]
- 21.Gril B, Vidal M, Assayag F, Poupon M-F, Liu W-Q, Garbay C. Int J Cancer. 2007;121:407–415. doi: 10.1002/ijc.22674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye Y-B, Lin J-Y, Chen Q, Liu F, Chen H-J, Li J-Y, Liu W-Q, Garbay C, Vidal M. Biochem Pharmacol. 2008;75:2080–2091. doi: 10.1016/j.bcp.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 23.Ruzza P, Siligardi G, Donella-Deana A, Calderan A, Hussain R, Rubini C, Cesaro L, Osler A, Guiotto A, Pinna LA, Borin G. J Peptide Sci. 2006;12:462–471. doi: 10.1002/psc.750. [DOI] [PubMed] [Google Scholar]
- 24.Jacquot Y, Broutin I, Miclet E, Nicaise M, Lequin O, Goasdoue N, Joss C, Karoyan P, Desmadril M, Ducruix A, Lavielle S. Bioorg Med Chem. 2007;15:1439–1447. doi: 10.1016/j.bmc.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Flemer S, Wurthmann A, Mamai A, Madalengoitia JS. J Org Chem. 2008;73:7593–7602. doi: 10.1021/jo8012258. [DOI] [PubMed] [Google Scholar]
- 26.Tuchscherer G, Grell D, Tatsu Y, Durieux P, Fernandez-Carneado J, Hengst B, Kardinal C, Feller S. Angew Chem Int Ed. 2001;40:2844–2848. doi: 10.1002/1521-3773(20010803)40:15<2844::AID-ANIE2844>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 27.Liu F, Stephen AG, Waheed AA, Freed EO, Fisher RJ, Burke TR., Jr Bioorg Med Chem Lett. 2010;20:318–321. doi: 10.1016/j.bmcl.2009.10.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon J, Schreiber SL. Chem Biol. 1995;2:53–60. doi: 10.1016/1074-5521(95)90080-2. [DOI] [PubMed] [Google Scholar]
- 29.Yuzawa S, Yokochi M, Hatanaka H, Ogura K, Kataoka M, Miura K-i, Mandiyan V, Schlessinger J, Inagaki F. J Mol Biol. 2001;306:527–537. doi: 10.1006/jmbi.2000.4396. [DOI] [PubMed] [Google Scholar]
- 30.McDonald Caleb B, Seldeen Kenneth L, Deegan Brian J, Farooq A. Biochemistry. 2009;48:4074–4085. doi: 10.1021/bi802291y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cussac D, Vidal M, Leprince C, Liu W-Q, Cornille F, Tiraboschi G, Roques BP, Garbay C. Faseb J. 1999;13:31–38. doi: 10.1096/fasebj.13.1.31. [DOI] [PubMed] [Google Scholar]
- 32.Wittekind M, Mapelli C, Lee V, Goldfarb V, Friedrichs MS, Meyers CA, Mueller L. J Mol Biol. 1997;267:933–952. doi: 10.1006/jmbi.1996.0886. [DOI] [PubMed] [Google Scholar]
- 33.Kingsbury JS, Harrity JPA, Bonitatebus PJ, Hoveyda AH. J Am Chem Soc. 1999;121:791–799. [Google Scholar]
- 34.Posern G, Zheng J, Knudsen BS, Kardinal C, Muller KB, Voss J, Shishido T, Cowburn D, Cheng G, Wang B, Kruh GD, Burrell SK, Jacobson CA, Lenz DM, Zamborelli TJ, Adermann K, Hanafusa H, Feller SM. Oncogene. 1998;16:1903–1912. doi: 10.1038/sj.onc.1201714. [DOI] [PubMed] [Google Scholar]
- 35.Atabey N, Gao Y, Yao Z-J, Breckenridge D, Soon L, Soriano JV, Burke TR, Jr, Bottaro DP. J Biol Chem. 2001;276:14308–14314. doi: 10.1074/jbc.M010202200. [DOI] [PubMed] [Google Scholar]
- 36.Shi Z-D, Liu H, Zhang M, Worthy KM, Bindu L, Yang D, Fisher RJ, Burke TR., Jr Bioor Med Chem. 2005;15:4200–4208. doi: 10.1016/j.bmc.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 37.Giubellino A, Shi Z-D, Miller Jenkins LM, Worthy KM, Bindu LK, Athauda G, Peruzzi B, Fisher RJ, Appella E, Burke TR, Jr, Bottaro DP. J Med Chem. 2008;51:7459–7468. doi: 10.1021/jm800523u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helbecque N, Loucheux-Lefebvre MH. Int J Peptide Protein Res. 1982;19:94–101. doi: 10.1111/j.1399-3011.1982.tb03028.x. [DOI] [PubMed] [Google Scholar]
- 39.Peters C, Bacher M, Buenemann CL, Kricek F, Rondeau JM, Weigand K. ChemBioChem. 2007;8:1785–1789. doi: 10.1002/cbic.200700362. [DOI] [PubMed] [Google Scholar]