Abstract
In the formation of the spinal network, various transcription factors interact to develop specific cell types. Using a gene trap technique, we established a stable line of zebrafish in which the red fluorescent protein (RFP) was inserted in the pax8 gene. RFP insertion marked putative pax8-lineage cells with fluorescence and inhibited pax8 expression in homozygous embryos. Pax8 homozygous embryos displayed defects in the otic vesicle, as previously reported in studies using morpholinos. The pax8 homozygous embryos survived to adulthood in contrast to mammalian counterparts that die prematurely. RFP is expressed in the dorsal spinal cord. Examination of the axon morphology revealed that RFP (+) neurons include Commissural Bifurcating Longitudinal (CoBL) interneurons, but other inhibitory neurons such as Commissural Local (CoLo) interneurons and Circumferential Ascending (CiA) interneurons do not express RFP. We examined the effect of inhibiting pax2a/pax8 expression on interneuron development. In pax8 homozygous fish, the RFP (+) cells undergo differentiation similar to that of pax8 heterozygous fish, and the swimming behavior remained intact. In contrast, the RFP (+) cells of pax2a/pax8 double mutants displayed altered cell fates. CoBLs were not observed. Instead, RFP (+) cells exhibited axons descending ipsilaterally: a morphology resembling that of V2a/V2b interneurons.
Keywords: Spinal cord, Transcription factor, Differentiation, Gene trap, Cell fate, Paired box gene
Introduction
Pax8 is a transcription factor that belongs to the Pax gene family. Pax genes are homologues of the Drosophila gene paired (Treisman et al., 1991), which is involved in the pair-rule segmentation. The Pax gene is characterized by the existence of a paired domain, which is comprised of two sub-domains, with each of these domains having the ability to bind to DNA. Genes in this family play important roles in various aspects of development (Chi and Epstein, 2002). Pax2, Pax5 and Pax8 constitute a subfamily of the Pax genes. Pax8 has been cloned from zebrafish (Pfeffer et al., 1998; Mackereth et al., 2005), and its expression was detected in similar organs as in mammals (Bouchard et al., 2004).
A null mutant of pax8 was never isolated in a large-scale mutagenesis screening of zebrafish. To date several studies have used morpholinos (MOs) to knockdown Pax8 (Hans et al., 2004; Mackereth et al., 2005). In these studies, injection of pax8 MOs resulted in a decrease in size of the otic vesicle at 30 hours post-fertilization (hpf).
The expression of transcription factors and the regulation of cell fates in the spinal cord have been studied extensively in mammals (Burrill et al., 1997; Jessell, 2000; Goulding and Pfaff, 2005). Several genes of the Pax family have been found to be involved in these processes. Pax2 is expressed in postmitotic cells in the lateral portion of the spinal cord (Burrill et al., 1997). Pax6 establishes distinct ventral cell populations and controls identity of motor neurons and interneurons (Ericson et al., 1997). Studies in zebrafish are less comprehensive compared to that of mammals. However, in several studies, the identity of neurons in the spinal cord was clearly linked to the expression of transcription factors. Engrailed-1 -expressing neurons are ipsilateral ascending glycinergic inhibitory neurons (Higashijima et al., 2004) and alx-expressing neurons are ipsilateral descending glutamatergic neurons (Kimura et al., 2006). Transcription factors including islet1, islet2, and nkx6 are important for the development of primary motor neurons (Segawa et al., 2001; Hutchinson and Eisen, 2006; Hutchinson et al., 2007).
Gene trap screening, widely used in Drosophila and mouse, is a genetic technique used to identify gene expression and function (O’Kane and Gehring, 1987; Friedrich and Soriano, 1991). It has only recently been applied to zebrafish (Kawakami et al., 2004). In our gene trap screening, using the red fluorescent protein (RFP), we identified a line in which the RFP gene was inserted in the pax8 gene. The insertion of RFP occurred in the first intron of pax8, leading to a fusion transcript between pax8 and RFP. We used this fish to analyze RFP (+) neurons in developing zebrafish. Embryos homozygous for the insertion had a very low pax8 transcript level, rendering the fish a pax8 hypomorph. Analysis of RFP (+) neurons in the spinal cord revealed that a subpopulation of inhibitory interneurons arose from the putative pax8-lineage. The analysis of pax2/pax8 double mutants revealed an interaction of pax8 and pax2a in the differentiation of putative pax8-lineage neurons.
Material and Methods
Maintenance of zebrafish lines
Wild type zebrafish were obtained from Aquatic Ecosystems, and maintained in stand-alone self-circulating AHAB systems (Aquatic Ecosystems, Apopka, FL) following the guidelines of IACUC at the University of Florida (protocol #D464) and NIH/NIAAA (protocol #LMP-FO-11). Both protocols conformed to the NIH Guidelines for Use of Zebrafish. A founder fish harboring the RFP insertion in the pax8 gene (F0) was out-crossed with wild type fish for several generations. F3 and later generations were used for experiments. Embryos were maintained at 28.5°C. Noi mutant (noib593) (Erickson et al., 2007), a mutant of pax2a, was obtained from the Zebrafish International Resource Center.
Gene trap screening and mapping of the insertion site
The GFP sequence between the BamHI site and the NotI site of the clone pT2KSAG (Kawakami et al., 2004) was cut out and the sequence of DsRedExpress (BD, Franklin Lakes, NJ) was inserted in its place. The plasmid, named Tol2-DsRed, was used for injection into zebrafish eggs.
Transposase mRNA was prepared from pCS-TP (Kawakami et al., 2004) using the mMessage mMachine SP6 kit (Ambion, Austin, TX). Tol2DsRed plasmid and transposase was diluted to a final concentration of 50 ng/μl. Injection into fertilized zebrafish eggs was performed as previously described (Ono et al., 2001). Surviving embryos were raised to adulthood. They were in-crossed or out-crossed with wild type fish to screen for RFP (+) fish. Screening of fish was done between 72 hpf to 120 hpf.
Inverse PCR was performed following the protocol described in Kawakami et al. (2004) with slight modifications. To determine the site of insertion, amplified products were directly sequenced and used to search the zebrafish genomic database (Sanger) using BLAST.
Real-time PCR (qPCR)
RNA was extracted from 24 hpf embryos and 10 days post fertilizaton (dpf) larvae, using RNeasy mini kit (Qiagen, Valencia, CA). RNA samples were treated with TurboDNAse (Ambion). Total RNA was reverse transcribed using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA), followed by RNase treatment. Pax8 primers and probe were designed to amplify the junction of exons 5 and 6; forward 5′-AAG TGC AGC AGC CAT TTA ACC TCC-3′, reverse 5′-TTG ATG GAG TAA GTG GAG CCC AGA-3′, FAM labeled probe/56-FAM/TAA AGG CCT GAG CCC AGG ACA C/3BHG_1/(Integrated DNA Technologies, Coralville, IA). Exons 5 and 6 are present in all reported splicing isoforms of pax8 (Mackereth et al., 2005).
qPCR was carried out with ABI StepOnePlus (ABI, Foster City, CA). Data was analyzed using the comparative Ct method (Ct) with elongation factor (elf) 1-alpha (NM_131263.1) as endogenous reference. Primers and probe for elf1-alpha were; forward 5′-TTG ATG CCC TTG ATG CCA TTC TGC-3′, reverse 5′-ACA ACC ATA CCA GGC TTG AGG ACA-3′ and probe/56-FAM/ATT GGC ACT GTA CCT GTG GGT CGT GT/3BHQ_1/.
Optical studies
Screening of RFP signals in zebrafish embryos was performed on the Olympus MVX fluorescent stereomicroscope. Images were recorded with Olympus DP70 Color CCD camera. Confocal microscopy imaging was done on a Zeiss confocal microscope 510 Meta with 25X or 40X water immersion objective or Olympus FV-300 with 40X water immersion objective. For double imaging of RFP and GFP, sequential scannings were performed. Images were imported into Photoshop (Adobe Systems, San Jose, CA) and analyzed. Size, contrast and brightness were adjusted in Photoshop.
Retrograde labeling of spinal interneurons and reticulo-spinal neurons
To visualize the spinal interneurons in pax8 heterozygous (pax8+/m) or pax8 homozygous (pax8m/m) fish, retrograde labeling with Alexa Fluor 488 conjugated dextran (3,000 MW, Invitrogen, Eugene, OR) was performed as previously described (Hale et al., 2001). In brief, fish were anesthetized in 0.1 g/l MS-222 (tricaine methanesulfonate, Sigma, St. Louis, MO) and placed onto an agar plate. Injection electrodes, with a tip diameter of ~15 μm, were filled with 50% (v/v) Alexa Fluor 488 conjugated dextran solution and inserted into the spinal cord. After allowing a few seconds for the dye to diffuse, the electrode was withdrawn. Fish were allowed to recover in a physiologically conditioned solution for at least 3 hours prior to confocal imaging.
For the retrograde labeling of reticulo-spinal neurons, fish were anesthetized and embedded in 2% low melting point agarose (melting point 25 ± 5 °C, Fisher Scientific, Pittsburgh, PA). A section of the fish trunk was exposed by removal of the surrounding agarose. This trunk section was then cut using a scalpel blade coated with dextran fluorescein (10,000 MW, Invitrogen). Following this procedure the fish were left in the agarose for 5 minutes to allow for the intake of dextran into the spinal cord. Post surgical recovery and confocal imaging were carried out as described previously.
Stochastic labeling of spinal interneurons
Stochastic labeling of the spinal interneurons in pax8+/m or pax8m/m fish was obtained by the injection of a(n) eng1-EGFP, HuC-GFP, or EF-GVP-Ulyn-UH2B construct at 1–64 cell stage. The EF-GVP-Ulny-UH2B construct is comprised of three genes: Gal4-VP16 following the Xenopus EF-1α-promoter, lynEGFP following the 14xUAS-E1b promoter and H2B-EYFP following the 14xUAS-E1b promoter (Koster and Fraser, 2001). LynEGFP encodes EGFP localized to the membrane, and H2B-EYFP encodes EYFP localized to the nucleus. At 3 or 4 dpf, live larvae were imaged using confocal microscopy. Larvae were then fixed with paraformaldehyde (PFA) in preparation for immunohistochemistry.
Antibody characterization
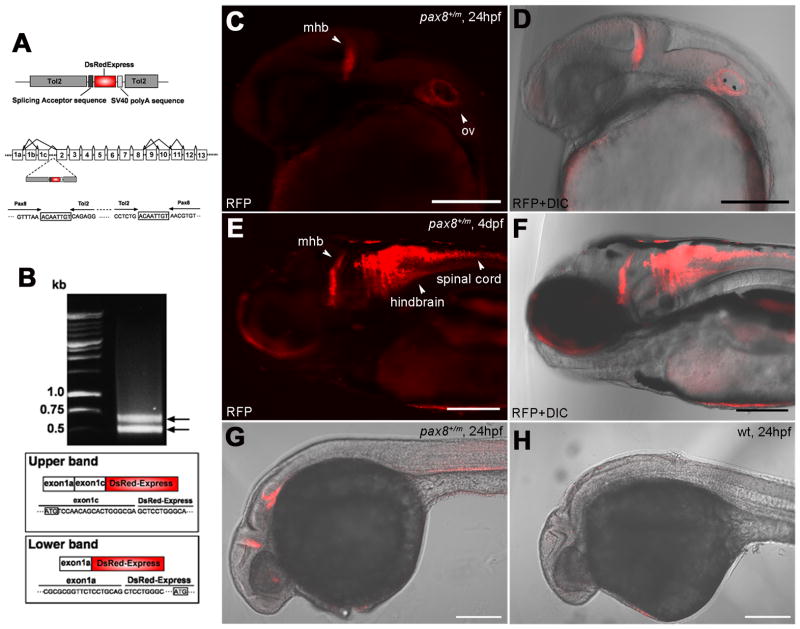
Anti-DsRed rabbit polyclonal antibody (Clontech, Mountain View, CA, #632496) was raised against DsRed-Express, a variant of Discosoma sp. red fluorescent protein. The quality and performance of this antibody was tested by Western blot analysis using lysates of HEK 293 cells stably expressing DsRed-Express, DsRed-Monomer, or AcGFP1 and untransfected HEK293 cells. A specific band corresponding to 30–38 kDa was observed only in lanes loaded with lysates of cells expressing DsRed-Express or DsRed-Monomer (manufacturer’s technical information). In zebrafish, treatment of wild type fish with this antibody showed no positive signal (Fig. 1H).
Figure 1.
Insertion of RFP in the pax8 gene leads to RFP expression. A: A schematic diagram of the DNA construct used in the gene trap screening (top). The construct has a splicing acceptor site, DsRedExpress sequence and the SV40 polyA sequence. Tol2 sequences are at both ends. Exon composition of the pax8 gene in zebrafish is shown with possible alternative splicing combinations (middle). The insertion of the Tol2 sequence was detected in the first intron between exons 1c and 2. The nucleic acid sequence surrounding junctions is shown (bottom). Note the repeat of eight nucleic acids at both ends of the insertion, a phenomenon commonly observed with the Tol2 insertion. B: An electrophoresis image of RACE PCR products. Two distinct bands were detected. The upper band corresponded to the splicing of exons1a+1c and DsRed, while the lower band corresponded to the splicing between exon1a and DsRed. Presumptive translation initiation site for each transcript is shown with a box. C, D: Confocal image of RFP signal in pax8+/m embryos at 24 hpf (C). The signal is detected at the midbrain-hindbrain boundary (mhb) and the otic vesicle (ov). D shows the merged image with transmitted light. Scale: 200 μm. E, F: At 4 dpf, the signal is detected also in the hindbrain and the spinal cord. F is the merged image with transmitted light. Scale: 200 μm. G, H: RFP antibody stains the mhb, otic vesicle and the kidney at 24 hpf (G). H is a wild type embryo control. Scale: 200 μm.
A rabbit polyclonal Pax2 antibody (Invitrogen, #71-6000) was raised from a GST-Pax2 fusion protein derived from the C-terminal domain (aa188–385) of the murine Pax2 protein (Dressler and Douglas, 1992). Specificity of anti-Pax2 antiserum with zebrafish was discussed in detail in a previous study (Puschel et al., 1992). This antibody detects a protein of approximately 46 kDa in extracts from 24 hpf zebrafish embryos, as well as in extracts from mouse embryos.
A mouse monoclonal anti-GFP antibody (Invitrogen, #A-11120) was purified from clone 3E6 and recognizes native GFP (manufacturer’s technical information). The specificity of this GFP antibody was confirmed by using Isl1-GFP fish (Higashijima et al., 2000). Immunostaining using this GFP antibody and the Alexa 598 anti-mouse IgG (1/500, Invitrogen) showed a complete overlap between the GFP fluorescence and the Alexa598 signal (data not shown).
Information on antibodies is summarized in Table 1.
Table 1.
Antibody characteristics
| Antigen | Immunogen | Manufacturer | Dilution |
|---|---|---|---|
| DsRed | DsRed-Express, a variant of Discosoma sp. red fluorescent protein | Clontech (Mountain View), rabbit polyclonal, #632496 | 1:2,000 |
| Pax2 | GST-Pax2 fusion protein derived from the C-terminal domain (aa188–385) of the murine Pax2 protein | Invitrogen (Eugene), rabbit polyclonal, #71-6000 | 1:400 |
| Green Fluorescent Protein (GFP) | Green fluorescent protein purified from A. victoria | Invitrogen (Eugene), mouse monoclonal, clone 3E6, #A- 11120 | 1:1,000 |
Histology and immunohistochemistry
Fish were anesthetized with MS-222 and fixed with 4% PFA in 0.1 M phosphate buffer (PB, pH 7.2) at 4 °C for 5–7 hours. Samples were washed overnight with 30% sucrose in PB, followed by embedding with Tissue Tek OCT Compound (Sakura Finetech, Torrance, CA). Samples were then frozen and sectioned transversely (25μm) on a cryostat. Sections were mounted on Superfrost Plus slides (Fisher) and dried. Slides were then washed in 0.1M PB for 30 minutes and coverslipped with Fluoromount-G (Southern Biotechnology Associates, Birmingham, AL).
Immunohistochemistry with anti-RFP was performed to emphasize DsRedExpress signals from fixed embryos. 24 hpf embryos were fixed with 4% PFA in 0.1 M PB overnight and then washed in 0.1 M PB for 5–7 hours. Samples were washed 2 times (10 minutes each) in phosphate buffered saline (PBS) containing 0.5% Triton X-100 (PBST) and then incubated in 100% ethanol followed by Acetone, at −20 °C. After washing in PBST (2 times, 5 minutes each), samples were incubated in 2% normal goat serum (NGS) overnight at 4 °C and in anti-DsRed antibody (1/2000) for 24 hours at 4 °C. Samples were washed 3 times (2–3 hours each) in PBST and incubated overnight with secondary antibody, Alexa 568 goat anti-rabbit IgG (1/500, Invitrogen), at 4 °C. After 3 washes in PBST, samples were observed with the confocal microscope.
For Pax2 immunohistochemistry, ventral trunk muscles of 3 dpf larvae were excised with a razor blade and treated with 1% collagenase (Sigma) for 20–30 minutes. Muscle cells and other tissues were removed to expose the spinal cord. The obtained spinal-fish were fixed with 4% PFA in 0.1 M PB and washed in 0.1 M PB as described previously. Samples were washed in PBST for 30 minutes and incubated in 2% NGS for 2–5 hours at room temperature. Samples were treated with rabbit polyclonal anti-Pax2 (1/400) for 12–60 hours at 4 °C. In specimens expressing GFP, anti-GFP antisera (1/1000) were also added to the solution. Samples were washed 3 times (2–3 hours each) in PBST and incubated overnight with secondary antibodies, Alexa 488 goat anti-mouse IgG and Alexa 647 goat anti-rabbit IgG (1/500, Invitrogen). After 3 washes in PBST, samples were observed with the confocal microscope.
In situ hybridization
The pax8 in situ probe was designed to target a 798bp region of the pax8 transcript spanning exons 5 through 9 (Pfeffer et. al., 1998). The RFP probe was designed against DsRedExpress (700bp). In situ hybridization was carried out as previously described (Toyama et al.1995; Kudoh et. al 2001) with minor modifications. In brief, RNA probes were synthesized using a DIG RNA labeling Kit (SP6/T7) (Roche, Indianapolis, IN) following manufactures instructions. DIG labeled probes were purified with illustra ProbeQuant G-50 Micro Columns (GE Healthcare, Piscataway, NJ). In situ samples were fixed in 4% PFA in PBS overnight at 4°C. Following fixation, samples were washed with PBS, and transferred to methanol and stored at −20°C until processing. For processing, samples were rehydrated and treated with proteinase K (10mg/ml) for 7 minutes. Samples were then washed in PBS with 0.1% Tween-20 (PBTw) followed by fixation for 30 minutes at room temperature with 4% PFA in PBS. Samples were washed as before and then transferred to hybridization buffer (50% formamide, 5xSSC, 5mM EDTA, 0.1% Tween-20, 0.1% CHAPS, 50ug/ml heparin, 1mg/ml tRNA) and incubated at 65°C for 3 hours. Hybridization buffer was replaced with hybridization buffer containing ~1ng/ul of probe and samples were incubated overnight. The optimal hybridization temperature for the Pax8 probe was 60°C and 62°C for the RFP probe. Following probe incubation samples were subjected to several rounds of washes with 4 separate buffers. Washing was followed by treatment with 2% blocking solution (Roche) containing 5% lamb serum (heat inactivated) for 1hour. Samples were then incubated overnight with anti-DIG-AP-Fab fragment antibody (Roche) at 4°C, optimal antibody dilution was determined for each probe. Color reaction was carried out using BM Purple AP Substrate (Roche) for 24h. Reaction was stopped with PBS and samples were fixed as before. Samples were then cleared and mounted in 75% glycerol and imaged with an Olympus MVX10 microscope.
Video imaging
Swimming analysis was performed as described in a previous study (Epley et al., 2008). In brief, high-speed video imaging was recorded with a 1024 PCI camera (TechImaging, Salem, MA) mounted on a Zeiss stereomicroscope Stemi 2000-C. Images were taken at the rate of 1000 frames/second. Sequential images were stored, and later processed in Photoshop. Swim pattern angles were analyzed using NIH ImageJ (NIH, Bethesda, MD). Rostral midlines were drawn in sequential images, within ImageJ, from these lines head tip and the angle were measured at individual time points.
Results
RFP gene was inserted in the pax8 gene
Gene trap screening was performed using DsRedExpress for detection of insertion into the zebrafish genome. DsRedExpress is a mutant form of DsRed with altered amino acids at nine locations (Matz et al., 1999). While both DsRed and DsRedExpress require tetramerization to function as a RFP, the latter features a better solubility and a faster maturation of the chromophore (Bevis and Glick, 2002). The DsRedExpress sequence was positioned downstream of a splicing acceptor site, such that it would fuse to the endogeneous transcript in the splicing process. DsRedExpress, with a splicing acceptor site, was flanked by Tol2. In the presence of transposase the flanking Tol2 sequences facilitate integration into the zebrafish genome (Fig. 1A). Fertilized zebrafish eggs were injected with a mixture of the Tol2-DsRedExpress construct and transposase mRNA, and were raised to adulthood. These adult fish were either in-crossed or out-crossed with wild type fish to screen for embryos that expressed RFP. A founder fish (F0) was identified that gave rise to offspring expressing RFP after ~20 hpf (Fig. 1C, G). The positive rate of embryos from this F0 fish was 1.5 % (3 embryos out of 206 embryos). Positive embryos were raised to adulthood, and out-crossed to generate F2 embryos. All experiments were done with F3 or later generations.
Using RFP (+) embryos as a template for inverse PCR, the site of RFP insertion was determined to be in the pax8 gene. Further analysis mapped the insertion in the intron region between exons 1 and 2 (Fig. 1A). Previous studies have shown that pax8 in zebrafish undergoes extensive alternative splicing (Mackereth et al., 2005). The zebrafish pax8 gene has 13 exons, and alternative splicing gives rise to 10 different isoforms, with two predicted translation initiation sites.
We examined if the fusion of the DsRedExpress transcript and the pax8 transcript occurred in the splicing process as predicted. We extracted mRNA from RFP (+) embryos, reverse-transcribed, and performed RACE PCR using reverse primers in the DsRedExpress sequence. From this PCR two amplicon bands were detected (Fig. 1B). Sequencing of these amplicons revealed the fusion of pax8 and DsRedExpress. The inserted DsRedExpress sequence fused to either exon1a or 1c. When exon1a fuses to the DsRedExpress, the translation initiation site is the first methionine of DsRedExpress. Therefore, mRNA will lead to the expression of RFP. On the other hand, when exon1c fuses to the DsRedExpress, the translation initiation site in exon1c does not generate the DsRedExpress protein. In either case, fusion transcripts do not produce the Pax8 protein. Since the generation of the pax8-RFP fusion transcript is controlled by the innate transcription mechanism of pax8, RFP production is expected to serve as a faithful marker for pax8-lineage cells, as was reported in other gene trap lines (Tian et al., 2009).
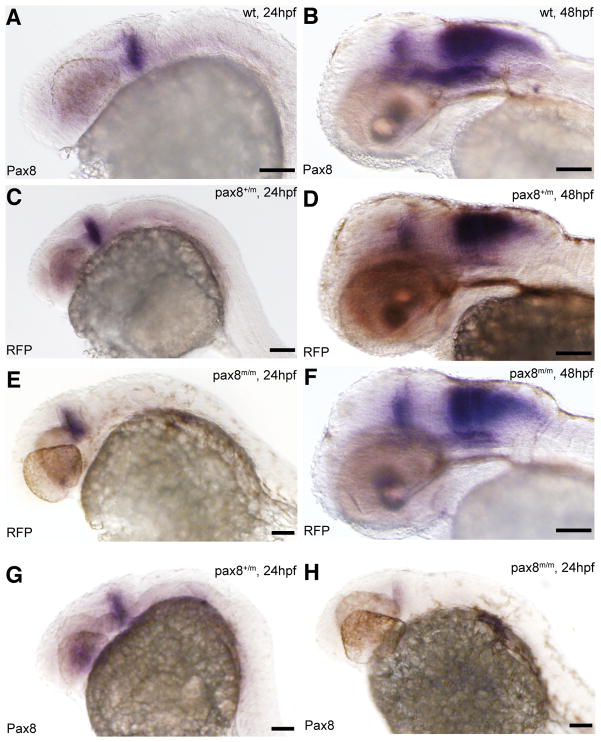
To confirm that RFP expression follows the expression pattern of pax8, we performed in situ hybridization using probes for pax8 and RFP and compared the staining pattern. The expression of pax8 in wild type embryos (Fig. 2A, B) closely matched that of RFP in RFP (+) embryos, both at 24 and 48 hpf (Fig. 2C–F). At 24 hpf, the expression of RFP in pax8+/m and pax8m/m was detected in the midbrain-hindbrain boundary (mhb) region (Fig. 2C, E). At 48 hpf, the expression was observed also in the hindbrain region (Fig. 2D, F). This expression pattern was also observed in wild type samples probed with pax8 (Fig. 2A, B). This similarity in expression pattern shows that RFP transcription indeed follows the intrinsic transcriptional regulation of the pax8 gene and that the RFP fluorescence can serve as a marker of pax8 expression in developing zebrafish embryos.
Figure 2.
Pax8 expression is identical to RFP expression. Transcripts were detected using in situ hybidization with either pax8 (A, B, G, H) or RFP (C, D, E, F) probes in 24hpf (A, C, E, G, H) or 48hpf (B, D, F) embryos. Expression pattern, at 24hpf, of pax8 in wild type (pax8+/+, A) was same to that of RFP in pax8+/m (C) and pax8m/m (E). Signal was detected in the mhb region of the embryos. RFP showed no signal in wild type samples (data not shown). At 48hpf the staining pattern of pax8 in wild type (B) resembled that of RFP expression in pax8+/m (D) and pax8m/m (F) embryos. Pax8+/m embryos showed positive staining also with the pax8 probe (G). Pax8 transcripts were also detected, at a much lower intensity, in pax8m/m embryos (H). Scale: 100 μm.
At 24 hpf, RFP expression was detected in the mhb and the otic vesicles (Fig. 1C, D). By 4 dpf, expression occurred in the mhb, hindbrain and the spinal cord (Fig. 1E, F). RFP expression in otic vesicles was hard to detect at this stage. Previous studies, employing in situ hybridization, showed the presence of pax8 in the nephric duct (Pfeffer et al., 1998). However, due to the auto-fluorescence of the yolk sac, RFP signal in the surrounding area was obscured. This was alleviated by using immunohistochemistry with an anti-RFP antibody. Results revealed positive expression of RFP in the nephric duct (Fig. 1G)(Diep et al., in press). Wild type embryos treated with the same antibody do not show a positive signal in the same area (Fig. 1H). RFP immunohistochemistry also detected RFP expression in the eye (Fig. 1G). In the cross section of the eye, RFP expression was in the center of the retina and a positive signal was observed extending toward the brain (Fig. S1A), raising the possibility that retinal ganglion cells express RFP. However, confocal observation of a live fish indicated that a strong RFP signal was observed encircling the HuC-Cameleon-expressing optic nerve (Fig. S1B–G). HuC-Cameleon expressing retinal ganglion cells did not express RFP. This suggests that glial cells express pax8.
Fish homozygous for RFP insertion are Pax8 hypomorph
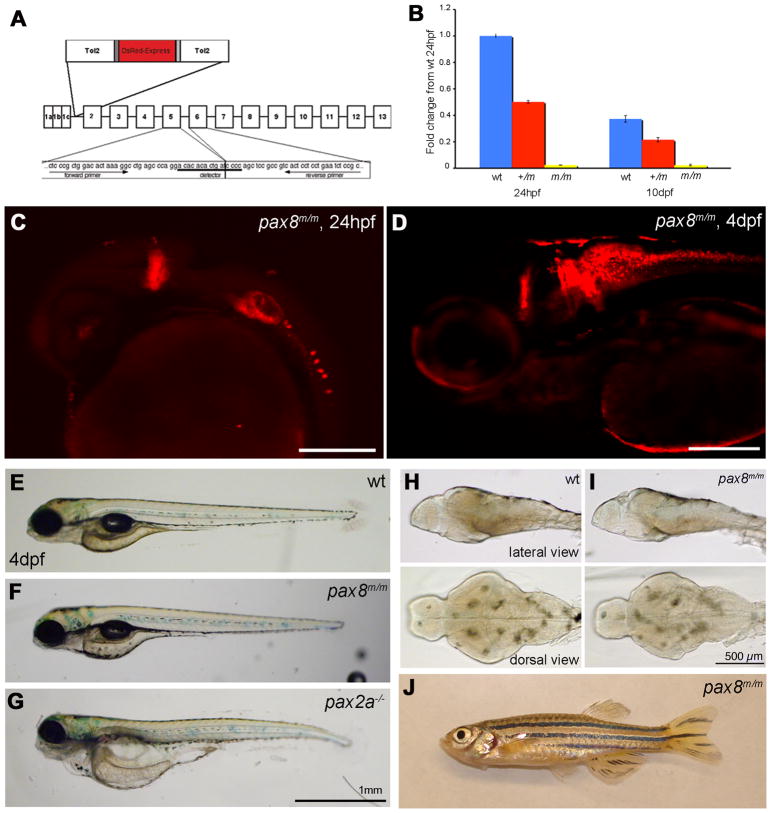
Using qPCR we examined if the insertion of the Tol2-RFP sequence was able to interfere with the endogenous expression of the pax8 gene. Primers were designed to the junction between exons 5 and 6, to quantify normally-spliced transcripts in pax8+/m or pax8m/m fish (Fig. 3A). The generation of amplicon was detected using a Taqman probe corresponding to the sequence encompassing the junction. At 1 dpf, the normal transcript in pax8+/m fish was reduced to 52.8 ± 9.9 % of wild type fish (n=3, Fig. 3B). In pax8m/m fish it was further reduced to 3.1 ± 1.2 % of wild type fish (n=3). At 10 dpf, the amount of pax8 transcript in wild type larvae was 41.9 ± 4.9 % compared to 1 dpf wild type embryos (n=3). Pax8+/m or pax8m/m larvae also displayed reduced pax8 levels: 29.5 ± 4.2 % and 2.1 ± 0.6 %, respectively (n=3; all values are relative to wild type transcript at 1 dpf; Average ± SEM). An additional set of primers and probe, encompassing exons 7–8, was also tested and gave similar results (data not shown). In order to confirm the qPCR result, we compared in situ hybridization for pax8 in pax8+/+, pax8+/m and pax8m/m embryos (Fig. 2). The signal was weaker in pax8m/m (Fig. 2A, G, H), and the decrease of transcript seems to happen uniformly among pax8-expressing cells. The in situ hybridization signal, however, was stronger than expected from the qPCR, presumably because the pax8 probe can bind to the pre-mRNA.
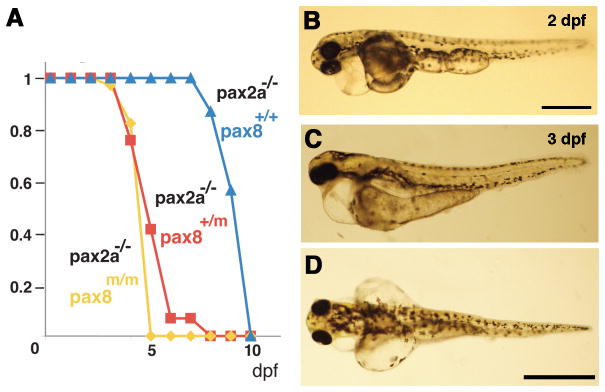
Figure 3.
Homozygous embryos are Pax8 hypomorph. A: The exon composition of zebrafish pax8 indicating the insertion site of Tol2. Primers for the qPCR were designed so that the amplicon spans the junction between exons 5 and 6. B: qPCR of the pax8 transcript in wild type (wt), pax8+/m, and pax8m/m fish at 1 dpf and 10 dpf. Wild type at 1dpf was used as a reference sample to obtain relative transcript amounts. Error bars represent the SEM of the average from triplicates of a single run (n=3). C, D: Confocal images of RFP signals in pax8m/m fish at 24 hpf (C) and 4 dpf (D). Scale: 200 μm. E, F, G: The gross morphology of larva at 4 dpf. E is wild type, F is pax8m/m, and G is pax2a−/−. The pax2a−/− larva is deformed, and dies at 7–10 dpf. Scale: 1 mm. H, I: The brain morphology is normal in the pax8m/m larva (I) compared to the WT larva (H). Top is the lateral view and bottom is the dorsal view. Scale: 500 μm. J: Adult pax8m/m fish.
In spite of the pax8 ablation, the RFP expression pattern in pax8m/m fish (Fig. 3C, D) seemed similar to that of pax8+/m fish (Fig. 1C, E). The overall structure of the brain was also normal in pax8m/m larvae (Fig. 3H, I). This was in contrast to a mutant of pax2a, a gene closely linked to pax8. Pax2a homozygous embryos exhibited a grossly deformed brain at 1 dpf, most notably the lack of isthmus and cerebellum (Brand et al., 1996). The pax8m/m fish did not display an obvious morphological defect (Fig. 3E, F, G) and followed a normal course of development to adulthood. (Fig. 3J). This was unexpected because pax8 knockout mice present severe phenotypes and die soon after weaning (Mansouri et al., 1998).
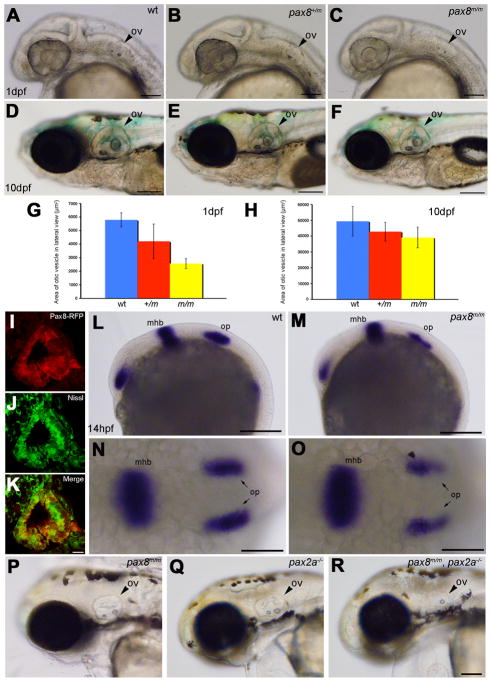
The unexpected normal growth of pax8m/m fish raised a question whether Pax8 protein was indeed decreased as predicted by the mRNA measurements of qPCR (Fig. 3B). Previous studies of zebrafish reported that blocking of Pax8 by MO injection leads to an abnormal otic vesicle development (Hans et al., 2004; Mackereth et al., 2005). We looked at the formation of otic vesicles in pax8m/m fish. The size of otic vesicles in pax8m/m fish was slightly reduced at 1 dpf (Fig. 4A–C). The average area of otic vesicle was 5.79 ± 0.51 × 103μm2 for wild type, 4.21 ±1.27 × 103μm2 for pax8+/m and 2.57 ± 0.38 × 103μm2 for pax8m/m (n=6; one-way ANOVA, F(2,15)=22.9, p<0.001) (Fig. 4G). The reduction in the otic vesicle size became less pronounced as the larva gets older (Fig. 4D–F). The average area of otic vesicle at 10 dpf was 49.4 ± 9.3 × 103 μm2 for wild type, 42.8 ±5.9 × 103μm2 for pax8+/m, and 39.1 ± 6.5 × 103μm2 for pax8m/m (n=6; one-way ANOVA, F(2,15)=2.9, p=0.08) (Fig. 4H).
Figure 4.
Otic vesicle formation defects in pax2a/pax8 mutants. A–F: The otic vesicle (ov) size is slightly reduced in pax8m/m fish. Lateral view of the head region at 1 dpf (A, B, C) and 10 dpf (D, E, F). A and D are the wild type (wt), B and E are pax8+/m, and C and F are pax8m/m. Scale: 100 μm (A, B, C), 200 μm (D, E, F). G, H: The area of the otic vesicle as measured in the lateral view at 1 dpf (G) and 10 dpf (H). Wild type (Blue), pax8+/m (Red), and pax8m/m fish (Yellow). The average and the standard deviation are shown (n=6 each). I, J, K: C. RFP (+) cells (I) line the cavity of the otic vesicle, as visualized by the Nissle counterstain (J) at 1dpf. The merged picture is shown in K. Scale 10 μm. L–O: In situ hybridization with pax2a probes in wild type (L, N) and pax8m/m embryos (M, O) at 14 hpf. Pax2a expression is detected in retina, mhb and the otic placode (op) in wild type. Note the signal in the otic placode is weaker in pax8m/m embryo. The difference is more obvious in the higher magnification of the dorsal view (N and O). Scale: 200 μm (L, M), 100 μm (N, O). P, Q, R: The reduction of the otic vesicle size is more pronounced in the pax2a/pax8 double mutant. The lateral view of the head is shown for pax8m/m (P), pax2a−/− (Q) and pax2a/pax8 double mutant embryos at 2 dpf(R). Scale: 100 μm. Magenta/green images for panels I, J and K are provided in suppl. figure. 2.
To examine whether the smaller size of the otic vesicle results from the lack of pax8, we observed if pax8 is expressed in the otic vesicle at 1dpf. Because in situ hybridization signal of pax8 at this stage was too weak to perform analysis (Fig. 2A), we used RFP as the marker to show putative pax8 expressing cells. The otic vesicle had a cavity as revealed by Nissl staining. RFP expressing cells formed a layer that lined this cavity (Fig. 4I–K), which likely determines the size of the otic vesicle. Several groups reported that pax8 regulates pax2a in the otic placode formation and together regulate the formation of otic vesicles (Hans et al., 2004; Mackereth et al., 2005). In situ hybridization of pax8m/m embryos with a pax2a probe, indeed revealed a reduced pax2a expression in the otic placodes at 14 hpf (Fig. 4L–O).
Finally, we generated a double mutant of pax2a and pax8, by making double-homozygous embryos for pax8 and no isthmus (noi) (Brand et al., 1996). Pax2a/pax8 double mutant embryos displayed a severely reduced otic vesicle size compared to single mutants of either pax8 or pax2a (Fig. 4P–R). This again agrees with previous MO studies (Hans et al., 2004; Mackereth et al., 2005). Taken together, these results on the inner ear formation confirm the qPCR findings and that Pax8 function is inhibited in pax8m/m.
Expression of RFP in the nervous system
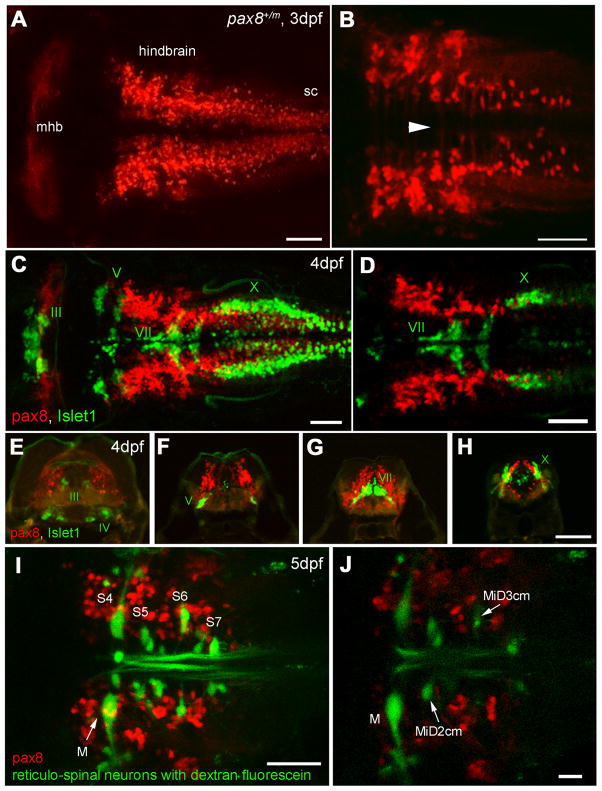
RFP expressing cells in the nervous system were examined in greater detail. Dorsal view of the hindbrain exhibited RFP (+) cells bilaterally (Fig. 5A). At higher magnification, axon-like structures crossing the midline were observed (Fig. 5B). In contrast to retinal expression (Fig. S1), RFP expressing cells also express HuC-Cameleon (data not shown). These observations suggest that RFP (+) cells in the hindbrain are mainly neurons. RFP (+) fish were also crossed with other transgenic lines to identify possible overlaps with RFP signals. Isl1-GFP fish express GFP under the promoter of Islet-1, labeling GFP in cranial motor neurons (Higashijima et al., 2000). GFP/RFP double positive larvae showed that RFP (+) cells were distinct from GFP (+) motor neurons (Fig. 5C). They never overlapped, as shown clearly in a single confocal plane (Fig. 5D). After in vivo observation, GFP (+)/RFP (+) larvae were further examined in fixed slices (Fig. 5E–H). At the level of the IIIrd, Vth, VIIth, and Xth cranial motor nuclei, RFP (+) cells occupied distinct domains relative to motor neurons. When the reticulospinal neurons were stained with dextran fluorescein, there was no overlap with RFP expressing cells (Fig. 5I, J). RFP (+) cells were distributed more caudal than the 3rd segment. RFP (+) axons ascending to more rostral brain regions were not observed (data not shown). These results suggest that RFP (+) cells in the hindbrain include interneurons.
Figure 5.
RFP expression in the central nervous system. A, B: Confocal images displaying RFP (+) cells in the hindbrain. A is a dorsal view of the hindbrain and B is a higher magnification of the ventral region. Note the rostral end of the hindbrain lacks RFP (+) cells (A). In the ventral region (B), axon-like structures were detected crossing the midline (arrowhead). sc, spinal cord. Scale: 50 μm. C, D: RFP (+) cells (red) in the hindbrain compared to the islet-1 GFP expressing motor neurons (green). C is a stack of confocal images. D displays a single confocal plane near the Xth (X) motor nuclei. Note that two populations (red and green) do not overlap. Scale: 50 μm. E-H: A longitudinal series of the hindbrain sections are shown from the level of IIIrd, IVth (E), Vth (F), VIIth (G), and Xth motor nucleus (H). RFP (+) cells are red and islet-1 expressing motor neurons are green. Scale: 100 μm. I, J: RFP (+) cells (red) in the hindbrain were compared to the reticular neurons stained by the tracing dye, dextran fluorescein (green). I is a stack of confocal images and J is a single plane at the level of the Mauthner cell (M). S4 through S7 indicates segment number. Scale: 50 μm (I), 20 μm (J). Magenta/Green images are provided in suppl. figure. 3.
Pax2 and pax8 in spinal interneurons
Though pax8 is expressed in the zebrafish spinal cord, the role it plays in the spinal cord had remained unexplored, until recently. Batista and Lewis (2008) studied the expression of pax8 along with pax2a in the spinal cord. While their findings suggested that the expression of these two genes largely overlaps, further characterization of pax8-expressing neurons was not performed. The better sensitivity of RFP fluorescence compared to in situ hybridization (Fig. 1, 2) enabled us to characterize putative pax8-lineage cells in the spinal cord.
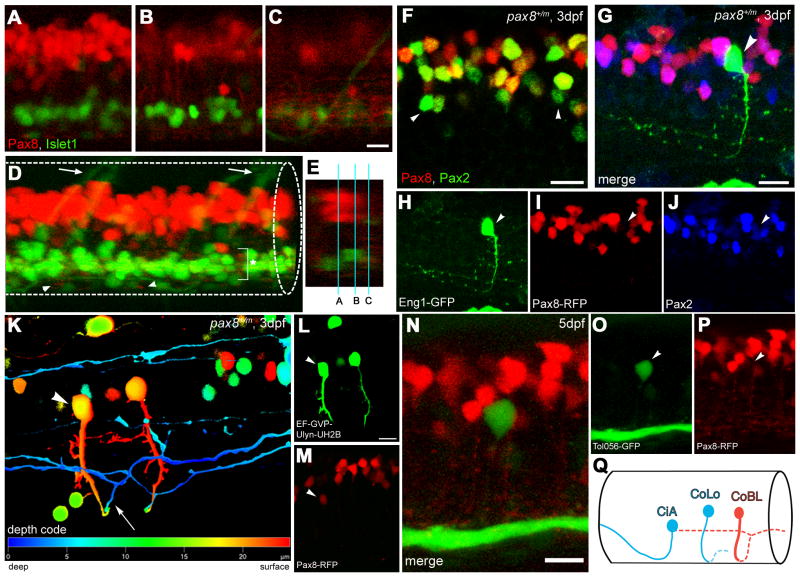
The lateral view of a spinal cord in a pax8+/m fish expressing isl1-GFP showed that RFP (+) cells were located in the dorsal spinal cord, distant from the motor neurons labeled by GFP (Fig. 6A). From the cell bodies of these RFP (+) neurons, axons extending ventrally could be visualized (Fig. 6B). In the ventro-lateral region, a plexus-like structure was observed, presumably arising from processes of RFP (+) cells (Fig. 6C). At the midline, RFP (+) fibers were observed near the ventral edge, that seemed to represent axons of commissural neurons (Fig. 6D).
Figure 6.
Interneurons in the spinal cord expressing pax2a or pax8. A-E: RFP (+) neurons are detected in the dorsal portion of the spinal cord; RFP (red), islet-1:GFP (green). Single place confocal images (A, B, C) show the position of RFP(+) cells and islet-1 expressing secondary motor neurons. Scale: 10 μm. D is the 3D reconstruction. Arrowheads indicate commissural axons labeled by RFP and arrows indicate motor neuron axons labeled by GFP. The area with an asterisk has a plexus-like structure of RFP (+) fibers. An optical cross section (E) displays the level of confocal planes corresponding to A, B and C. F: Pax8+/m fish stained with anti-Pax2; RFP (red), anti-Pax2 (green). Arrowheads are Pax2 (+) cells without RFP. Scale: 10 μm. G–J: CiA neurons express Pax2 but lack RFP expression. G is a stack of confocal images with eng1-GFP (green), RFP (red) and Pax2 (blue). H–J are single plane images to show that the GFP (+) cell is Pax2 (+) but RFP (-). Scale: 10 μm. K, L, M:.CoBL neurons express RFP. K is a stack of confocal images in depth coding. The cell with an arrow has the characteristic axon pattern of CoBL. In single planes, the GFP (+) cell is also RFP (+) (L and M). Scale: 10 μm. N, O, P: CoLo neurons labeled with GFP (green) in Tol-056 fish do not express RFP (red). N is a merged image. O and P show that the marked cell expresses GFP but not RFP. Scale: 10 μm. Q: Schema of CiA, CoLo and CoBL interneurons in the spinal cord. The position of the cell body and the axon projection are shown. Rostral to the left. Note that CiA and CoLo (blue) do not express RFP, while CoBL (red) express RFP. Magenta/Green images are provided in suppl. figure. 4.
We examined the overlap of Pax2a and RFP expression. We stained pax8+/m fish at 3 dpf with anti-Pax2 antibody. There were some cells expressing both Pax2a and RFP, which agrees with a report by Batista and Lewis (2008), but the overlap was not complete. In these stains there were also populations of neurons expressing only Pax2a or RFP (Fig. 6F). The Pax2a-expressing cells were overall shifted to the ventral side compared to the RFP (+) cells.
Because Circumferential Ascending interneurons (CiAs) express Pax2 (Batista and Lewis, 2008), we examined if CiAs also express RFP. To visualize CiA neurons in pax8+/m embryos, we injected a BAC clone with the engrailed-1 (eng1) promoter driving GFP (Higashijima et al., 2004). Eng1 is expressed exclusively in CiAs, and GFP driven by the eng1 promoter labels CiAs efficiently. Among the GFP positive cells, CiA neurons were identified by their characteristic axon projection pattern (Fig. 6G). These CiA neurons were also further examined for RFP expression. Out of 11 CiA neurons identified by GFP, none displayed the expression of RFP (Fig. 6H, I). On the other hand, Pax2a was indeed expressed in CiA neurons (Batista and Lewis, 2008), evidenced by the anti-Pax2 signal in CiA (Fig. 6H, J). Therefore, CiA neurons express pax2a, but do not express RFP (pax8).
We next tried to determine the morphology of RFP (+) cells. Fibers visualized by RFP are too dense to allow morphological analysis of individual neurons (Fig. 6B). In order to visualize the morphology of RFP (+) neurons in isolation, a dye was injected into the spinal cord of pax8+/m fish (Hale et al., 2001). However, the efficiency of labeling RFP (+) cells with the dye was very low (data not shown). This data suggested that RFP (+) neurons do not extend long axons longitudinally. We therefore turned to a genetic method by injecting HuC-GFP or EF-GVP-Ulyn-UH2B constructs into pax8+/m embryos (Koster and Fraser, 2001). GFP expressed in a stochastic fashion enabled visualization of the axon morphology. By looking for cells positive for both GFP and RFP, we determined the morphology of RFP (+) cells. RFP (+) cells included Commisural Bifurcating Longitudinal interneurons (CoBLs), which extended axons contralaterally before bifurcating longitudinally (n = 11; Fig. 6K–M). We occasionally observed RFP (+) cells with shorter axons, which do not extend longitudinal after crossing midlines (n=2) or even do not cross the midline and remain in the ipsilateral side (n = 2) (data not shown). These cells may represent an undifferentiated form of CoBLs.
We also studied the expression of RFP in another class of inhibitory interneurons: Commisural Local interneurons (CoLos). These interneurons are glycinergic and extend axons that make connections onto contralateral motor neurons (Liao and Fetcho, 2008). An enhancer trap line expressing GFP in all CoLos, Tol-056, was recently reported (Satou et al., 2009). We crossed the RFP (+) fish with this enhancer trap line. GFP and RFP never overlapped in these fish (Fig. 6N–P). In summary, RFP (+) cells were limited to a subclass of inhibitory neurons: CoBLs express RFP, but CiAs or CoLos do not.
The formation of spinal networks in pax8 mutants
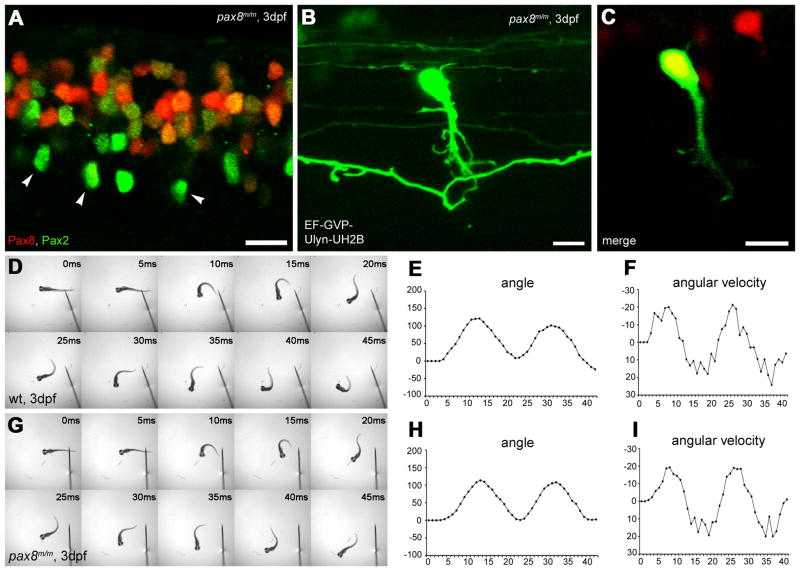
We examined the effect of inhibiting Pax8 on RFP (+) cells in the spinal cord, using pax8m/m fish. We first examined the expression of Pax2 and RFP in the spinal cord. The location and the density of Pax2 (+) cells or RFP (+) cells did not display an obvious change from those in pax8+/m fish (Fig. 7A).
Figure 7.
Spinal neurons in pax8 mutant. A: Spinal cord of a pax8m/m fish stained with anti-Pax2; RFP (red), anti-Pax2 (green). Scale: 10 μm. B, C: CoBL cell in pax8m/m fish. B is a stack of confocal images and C is a single confocal plane; GFP (green), RFP (red). Scale: 10 μm. D–I: Touch response of wild type (top, D, E, F) and pax8m/m fish (bottom, G, H, I) at 3 dpf. High-speed images of escape response are shown from 0 ms to 45 ms (D, G). In E, F, H, I, the head angle (E, H) and the angular velocity (F, I) were plotted against time. Magenta/Green images of panels A, B and C are provided in suppl. figure. 5.
The morphology of RFP (+) neurons was examined in pax8m/m fish using the stochastic expression of GFP. RFP (+) neurons in these fish also included CoBLs. Axon morphology of the CoBL neurons (n = 8) did not show any obvious abnormalities (Fig. 7B).
The ostensible normal differentiation of CoBL neurons was further examined at the functional level. We compared the touch response behavior of pax8m/m larvae with that of wild type embryos. Larvae at 3 dpf were touched on the tail with a tungsten needle and the escape response was recorded under a high-speed camera. The escape response showed no obvious differences between wild type and pax8m/m larvae (Fig. 7D, G). For a greater detail comparison, movements of the rostral midline were measured (Fig. 7E, F, H, I) (Epley et al., 2008). The maximum angle for wild type (Fig. 7E) and pax8m/m (Fig. 7H) larvae was 138 ± 20 and 140 ± 24 respectively (n=5, average ± SD, p=0.80, student’s t-test). The peak angular velocity was 21.6 ± 1.4 and 21.4 ± 3.3 (p=0.90) (Fig. 7F, I). The duration was 11.2 ± 1.1 and 11.8 ± 1.3 (p=0.45). The normal escape response of pax8m/m fish, the smooth coordinated alternation of left-right excitation in particular, suggests that the networking of the spinal cord is maintained.
Spinal network formation in the pax2a/pax8 double mutants
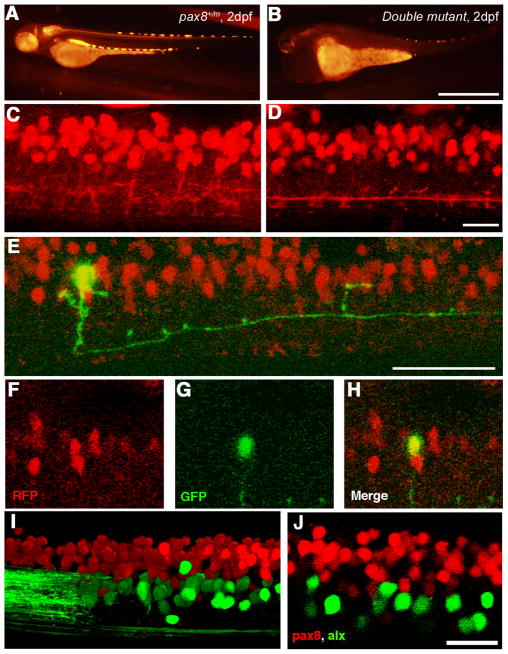
Lastly, we examined the spinal cord of pax2a/pax8 double mutants (pax2a−/−pax8m/m fish). Double mutants displayed a severe systemic phenotype. At 2 dpf, the edema started to develop (Fig. 8B). At 3 dpf, the edema became severe, most evident from the dorsal view (Fig. 8C, D). The survival of pax2a−/−fish with different pax8 genotypes (pax8+/+, pax8+/m and pax8m/m) was plotted against dpf (Fig. 8A). Pax2a single mutants die at ~10 dpf, as previously reported (Lun and Brand, 1998). When one or two mutant alleles of pax8 were introduced, the survival of the fish was severely shortened.
Figure 8.
Pax2a/pax8 double mutant. A: Survival curve of pax2a mutants with different pax8 genotypes. Blue, red and yellow represents pax8+/+, pax8+/m, pax8m/m fish, respectively. B: Lateral view of a double mutant embryo at 2 dpf. Scale: 500 μm. C, D: Lateral (C) and dorsal (D) view of a 3 dpf double mutant larvae. Scale: 1 mm.
In the double mutant, the RFP signal in the mhb was not observable at 1 dpf. This agrees with previous reports that pax2 regulates the expression of pax8 in the mhb (Liu and Joyner, 2001). In the spinal cord, RFP signal was visible but considerably weaker than that of pax8 single-mutant fish (Fig. 9B). Due to the severe systemic abnormality at 3 dpf, the spinal cord of double mutant fish was observed at 2 dpf. At this stage, RFP (+) cells in the spinal cord were observed in the dorsal domain (Fig. 9D). However, the overall pattern of the RFP signal seemed different from control embryos. The network of processes arising from RFP (+) cells looked less dense. In order to analyze the specific change occurring at the single cell level, we turned to the stochastic expression of GFP; driven by eng1-GFP, HuC-GFP or EF-GVP-Ulyn-UH2B. We did not observe typical CoBLs in the double mutant. Instead, some RFP (+) cells differentiated into neurons that extended ipsilateral axons caudally (Fig. 8E–H). The morphology of these cells resembles that of Circumferential Ipsilateral Descending (CiD) interneurons or Ventral Lateral Descending (VeLD) interneurons (Batista et al., 2008). CiDs and VeLDs correspond to V2a and V2b neurons, respectively, in mammalian spinal cords. 3 out of 7 cells with ipsilateral descending axons were RFP (+). In wild type fish, nearly all CiD neurons arise from cells expressing the transcription factor alx (Kimura et al., 2006). We crossed RFP (+) fish with a stable transgenic line expressing alx-GFP, in which the promoter of alx drives the expression of GFP and marks CiD with a high fidelity (Kimura et al., 2006). In these fish, the RFP signal and the GFP signal never overlapped (Fig. 9I, J). This shows that CiDs do not arise from RFP (+) cells in normal fish. Stochastically labeled neurons with ipsilateral descending neurons (n=10) were never observed to express RFP in pax8+/m fish. This suggests that VeLDs do not arise from the putative pax8-lineage either. The generation of RFP (+) neurons with ipsilateral descending axons in double mutant embryos therefore represents a change of cell fate in putative pax8-lineage neurons.
Figure 9.
Spinal neurons in pax2a/pax8 double mutant. A, B: RFP fluorescence of a pax8+/m embryo (A) and a double mutant embryo (B) under the same optical condition. The RFP signal is reduced in the double mutant. Scale: 1 mm. C, D: The RFP signal in the spinal cord of a pax8+/m embryo (C) and a double mutant embryo (D) near the 15th body segment. 3D images were constructed from confocal slices. Scale: 20 μm. E–H: Stochastic labeling by GFP (green) identified a neuron with an ipsilateral descending axon in the double mutant. E is a 3D reconstruction. F (RFP), G (GFP), and H (merged) are single plane images. Scale: 50 μm. I, J: The spinal cord of a pax8+/m larva obtained from crossing with the Alx-GFP transgenic line. The image is from an embryo at 4 dpf. I is a 3D reconstruction and J is a single confocal plane. Note that RFP and GFP do not overlap. Scale: 20 μm. Magenta/Green images are provided in suppl. figure. 6.
Discussion
In this study, we established a mutant zebrafish line that has an insertion of a RFP gene in the pax8 gene. The novelties of this study are as follows: First, we used the gene trap technique in zebrafish to label a population of neurons as well as to inhibit the gene expression in the labeled neurons. Second, due to the time-dependent nature of the MO knockdown technique, the long-term effect of pax8 loss in zebrafish was previously unknown. Thanks to the gene inhibition at the genomic level, we showed that pax8-less zebrafish survive to adulthood, unlike pax8 knockout mice that die prematurely. Third, putative pax8-lineage neurons in the spinal cord were revealed to include CoBLs, but other inhibitory neurons including CiAs or CoLos do not arise from this population. Fourth, the pax8/pax2 double mutant displayed a specific cell fate change of putative pax8-lineage neurons; they do not differentiate into CoBLs and instead give rise to neurons with ipsilateral descending axons.
The insertion of RFP in the pax8 gene and the loss of Pax8 expression
The insertion of the RFP gene in pax8 resulted in a fusion transcript. Due to the stop codon and the polyA addition signal that follows the RFP open reading frame, the expression of a functional Pax8 is inhibited. qPCR showed the expression level of pax8 was, in fact, reduced to 2.5 % of wild type fish (Fig. 3B). This suppression of endogenous pax8 is pronounced not only at 1 dpf but also at 10 dpf. The persistent suppression throughout development is due to the genomic nature of the RFP insertion. This is in contrast to earlier studies using MOs (Hans et al., 2004; Mackereth et al., 2005).
In spite of the nearly complete inhibition of the normal transcript in the pax8 hypomorphic mutant, its phenotype was surprisingly mild. This mild phenotype was probably the reason why a random mutagenesis screening failed to isolate a mutant of pax8. Some teleost genomes have two duplicate genes corresponding to a single gene in mammals. It therefore raises the possibility that the zebrafish pax8 gene has a duplicate copy that remains functional in pax8m/m fish. However, this is unlikely for four reasons. First, only a single gene homologous to pax8 can be detected in the zebrafish genome database. Second, in the genome of fugu rubripes, another teleost species whose genome is completely sequenced, only a single copy of pax8 exists (data not shown). Third, pax8m/m embryos presented a smaller otic vesicle size and reduced pax2a expression in the otic placode (Fig. 4). Fourth, pax2a/pax8 double mutant displayed a much-reduced otic vesicle size (Fig. 4P–R) and an enhanced mortality rate compared to pax2a single mutant (Fig. 8A). Therefore, it is unlikely that a duplicate gene compensates for the inhibited pax8 gene.
Pax8 expression in developing CNS
Previous studies in zebrafish used in situ hybridization to determine the timing and location of pax8 expression (Pfeffer et al., 1998; Mackereth et al., 2005). However, some regions with a low level of pax8 or RFP expression failed to exhibit in situ hybridization signals. For example, the otic vesicle at 24 hpf showed fluorescence but did not stain with in situ hybridization (Fig. 1C, 2A). Similarly, RFP fluorescence detected in the spinal cord lacked matching in situ hybridization signals (Fig. 2D, 9A). The better sensitivity of the fluorescence signal enabled us to clarify the anatomical identity of pax8-expressing cells in the spinal cord for the first time.
On the other hand, there are some factors that need to be considered when interpreting the data. First, the time course of RFP may be delayed compared to the endogenous Pax8 expression, because RFP requires tetramerization and maturation before it starts to emit fluorescence. However, DsRedExpress, used in this study, has a much expedited maturation process compared to wild type DsRed (Bevis and Glick, 2002). Indeed, when compared to in situ hybridization with pax8 or RFP specific probes, we did not observe a major delay in the expression of RFP fluorescence. Second, we needed to use pax8+/m fish to observe RFP. In these fish, one allele of pax8 is inhibited, as evidenced by qPCR (Fig. 3B). Therefore, there is a possibility that the morphology of cells expressing RFP in heterozygous fish may be different from that of pax8-lineage cells in wild type fish. However, the normal morphology of CiAs, CoBLs and CoLos in pax8+/m or pax8m/m fish (Fig. 6, 7) suggests it is unlikely. Third, while the nature of the RFP insertion in the pax8 gene and the similarity of pax8 and RFP in situ signals strongly support the validity of using RFP as the marker of pax8-lineage cells, a direct analysis of pax8 expression using in situ hybridization in the spinal cord was impossible due to the weak signal. We refer to RFP (+) cells, used for the morphology analysis, as “putative” pax8-lineage cells to acknowledge this point.
The expression of Pax8 was detected in retina, mhb, hindbrain, spinal cord and nephric duct. In mammals, Pax8 was also found in the thyroid gland. Pax8 knock-out mice die prematurely due to thyroid gland failure (Mansouri et al., 1998). Given the normal development of pax8m/m fish, it will be an interesting question, in the future, to examine the function of thyroid hormones in zebrafish (Yonkers and Ribera, 2008).
Pax8 and Pax2a in the spinal cord
In the zebrafish genome, pax2 has two duplicated copies: pax2a and pax2b. Only a mutant of pax2a (noi) was isolated in a large-scale mutagenesis screening. The interaction of pax2a and pax8 in the zebrafish spinal cord was first studied by Batista and Lewis (2008). Based on the number of cells stained with single or double in situ hybridization, it was concluded that the expression of pax2a, pax2b and pax8 largely overlap. It was also shown that pax2a neurons include CiAs. However, the identity of pax8-expressing interneurons was not studied. Our current study showed that the putative pax8-lineage neurons include CoBLs, but not CiAs or CoLos.
In mammalian spinal cord, Pax2 (+) domains include V0, V1, dl4 and dl6 (Burrill et al., 1997; Pillai et al., 2007). Alx (+) areas and eng1 (+) areas in zebrafish correspond to V2a and V1, respectively (Higashijima et al., 2004, Kimura et al., 2006). Areas of putative pax8 (+) cells in zebrafish spinal cord are dorsal to alx (+) neurons (Fig. 9I), and partially overlap with the pax2a (+) domain (Fig. 6F). Some of the cell bodies, observed by RFP, without obvious axon extensions may be glias.
Our results from pax8m/m embryos showed that CoBLs develop normally in pax8 single mutants (Fig. 7). A MO study by Lewis et al. suggested that spinal neurons develop normally in pax2a single mutants (Batista and Lewis, 2008). We also observed CoBL neurons expressing RFP in pax2a single mutants (data not shown). In contrast to pax8 or pax2a single mutants, pax2a/pax8 double mutants showed a clear change of fate for RFP (+) interneurons in the spinal cord. The expression of transcription factors have been linked to specific populations of spinal neurons (Lewis, 2006); the knocking out of transcription factors leads to different effects in mammalian spinal cord. The knockout of Dbx1 prevents the formation of V0 neurons and leads to abnormal left-right alternation (Lanuza et al., 2004). In engrailed1 knockout mice, a subpopulation of V1 neurons showed a subtler defect of reduced synaptic connection to target neurons (Sapir et al., 2004). Although RFP (+) cells in pax2a/pax8 double mutants are positioned roughly in the correct domain of the spinal cord, their cell fate was clearly changed. First, the overall pattern of RFP (+) fibers was altered. Second, more remarkably, double mutants lead to the generation of neurons with ipsilateral descending axons from RFP (+) cells. We failed to identify CoBLs in double mutants, which we routinely encounter in control fish. Together with the altered pattern of plexus, it is likely that the CoBLs are absent in the double mutant. It also agrees with the previous report that in pax2/pax8 knockdowns the development of inhibitory neurons is inhibited (Bastista and Lewis, 2008). RFP (+) neurons with ipsilateral descending axons that newly arose in the double mutant may represent CiDs, VeDLs or neither. The identity of these neurons awaits a further study.
An interesting question is how the motor output in the double mutant isaffected by the cell fate change. This is difficult to address in the current study due to the poor health of the double mutants. The severe edema likely comes from defects in the mhb or the nephric duct. In the future, it will be interesting to partially rescue double mutants with mhb or nephric duct specific promoters (Gosgnach et al., 2006) and examine the effect of pax2a/pax8 knockdown in the spinal cord in viable fish.
The analysis of the spinal interneuron network in zebrafish started from the anatomical classification based on the cell body location and the axon extension pattern (Bernhardt et al., 1990; Hale et al., 2001). Combination of genetic markers and electrophysiology made it possible to clarify functions that particular cell types display (Higashijima et al., 2004; Kimura et al., 2006; Liao and Fetcho, 2008; Satou et al., 2009). Use of MOs addressed the cell function from a different angle, by disturbing the development of specific interneurons (Batista and Lewis, 2008). The current study showed that the gene trap technique is a very powerful tool for such analyses; unequivocally labeling classes of interneurons expressing one type of transcription factor with a high sensitivity, while allowing the inhibition of its function in development. Generating more gene trap lines and combining them with other genetic techniques will further extend our understanding of the spinal cord network development.
Supplementary Material
Acknowledgments
We thank Drs Shinichi Higashijima and Joseph Fetcho for kindly providing alx–GFP fish, HuC-Cameleon fish, Tol-056 enhancer trap line, HuC-GFP construct, and engrailed1-GFP construct. We thank Drs Scott Fraser and Reiko Toyama for the EF-GVP-Ulyn-UH2B and pax2a clone respectively. We appreciate the technical advice from Drs Yu Katsuyama and Reiko Toyama on in situ hybridization. Dr. Kimberly Epley and Ms. Alison Delargy provided valuable help in maintaining zebrafish colonies. We thank Dr. Stephen Ikeda for critical reading of the manuscript.
Footnotes
Supporting grant information: This research was supported by NINDS Grant 1R01NS050388-01A1 to FO, Muscular Dystrophy Association Grant MDA3818 to FO, and the intramural research program of the NIH/NIAAA.
Literature cited
- Batista MF, Lewis KE. Pax2/8 act redundantly to specify glycinergic and GABAergic fates of multiple spinal interneurons. Dev Biol. 2008;323:88–97. doi: 10.1016/j.ydbio.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista MF, Jacobstein J, Lewis KE. Zebrafish V2 cells develop into excitatory CiD and Notch signalling dependent inhibitory VeLD interneurons. Dev Biol. 2008;322:263–275. doi: 10.1016/j.ydbio.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Bernhardt RR, Chitnis AB, Lindamer L, Kuwada JY. Identification of spinal neurons in the embryonic and larval zebrafish. J Comp Neurol. 1990;302:603–616. doi: 10.1002/cne.903020315. [DOI] [PubMed] [Google Scholar]
- Bevis BJ, Glick BS. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed) Nat Biotechnol. 2002;20:83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- Bouchard M, Souabni A, Busslinger M. Tissue-specific expression of cre recombinase from the Pax8 locus. Genesis. 2004;38:105–109. doi: 10.1002/gene.20008. [DOI] [PubMed] [Google Scholar]
- Brand M, Heisenberg CP, Jiang YJ, Beuchle D, Lun K, Furutani-Seiki M, Granato M, Haffter P, Hammerschmidt M, Kane DA, Kelsh RN, Mullins MC, Odenthal J, van Eeden FJ, Nusslein-Volhard C. Mutations in zebrafish genes affecting the formation of the boundary between midbrain and hindbrain. Development. 1996;123:179–190. doi: 10.1242/dev.123.1.179. [DOI] [PubMed] [Google Scholar]
- Burrill JD, Moran L, Goulding MD, Saueressig H. PAX2 is expressed in multiple spinal cord interneurons, including a population of EN1+ interneurons that require PAX6 for their development. Development. 1997;124:4493–4503. doi: 10.1242/dev.124.22.4493. [DOI] [PubMed] [Google Scholar]
- Chi N, Epstein JA. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 2002;18:41–47. doi: 10.1016/s0168-9525(01)02594-x. [DOI] [PubMed] [Google Scholar]
- Diep C, Ma D, Arora N, Wingert R, Bollig F, Djordjevic G, Lichman B, Zhu H, Ikenaga T, Ono F, Englert C, Hukriede N, Handin R, Davidson A. Identification of adult renal progenitor cells capable of nephron formation and regeneration in zebrafish. Nature. doi: 10.1038/nature09669. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler GR, Douglas EC. Pax2 is a DNA-binding protein expressed in embryonic kidney and Wilms tumor. Proc Natl Acad Sci U S A. 1992;89:1179–1183. doi: 10.1073/pnas.89.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epley KE, Urban JM, Ikenaga T, Ono F. A modified acetylcholine receptor {delta}-subunit enables a null mutant to survive beyond sexual maturation. J Neurosci. 2008;28:13223–13231. doi: 10.1523/JNEUROSCI.2814-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–180. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- Erickson T, Scholpp S, Brand M, Moens CB, Waskiewicz AJ. Pbx proteins cooperate with Engrailed to pattern the midbrain-hindbrain and diencephalic-mesencephalic boundaries. Dev Biol. 2007;301:504–517. doi: 10.1016/j.ydbio.2006.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- Gosgnach S, Lanuza GM, Butt SJ, Saueressig H, Zhang Y, Velasquez T, Riethmacher D, Callaway EM, Kiehn O, Goulding M. V1 spinal neurons regulate the speed of vertebrate locomotor outputs. Nature. 2006;440:215–219. doi: 10.1038/nature04545. [DOI] [PubMed] [Google Scholar]
- Goulding M, Pfaff SL. Development of circuits that generate simple rhythmic behaviors in vertebrates. Curr Opin Neurobiol. 2005;15:14–20. doi: 10.1016/j.conb.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Hale ME, Ritter DA, Fetcho JR. A confocal study of spinal interneurons in living larval zebrafish. J Comp Neurol. 2001;437:1–16. doi: 10.1002/cne.1266. [DOI] [PubMed] [Google Scholar]
- Hans S, Liu D, Westerfield M. Pax8 and Pax2a function synergistically in otic specification, downstream of the Foxi1 and Dlx3b transcription factors. Development. 2004;131:5091–5102. doi: 10.1242/dev.01346. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J Neurosci. 2000;20:206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima S, Masino MA, Mandel G, Fetcho JR. Engrailed-1 expression marks a primitive class of inhibitory spinal interneuron. J Neurosci. 2004;24:5827–5839. doi: 10.1523/JNEUROSCI.5342-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson SA, Eisen JS. Islet1 and Islet2 have equivalent abilities to promote motoneuron function and to specify motoneuron subtype identity. Development. 2006;133:2137–2147. doi: 10.1242/dev.02355. [DOI] [PubMed] [Google Scholar]
- Hutchinson SA, Cheesman SE, Hale LA, Boone JQ, Eisen JS. Nkx6 proteins specify one zebrafish primary motoneuron subtype by regulating late islet1 expression. Development. 2007;134:1671–1677. doi: 10.1242/dev.02826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Okamura Y, Higashijima S. alx, a zebrafish homolog of Chx10, marks ipsilateral descending excitatory interneurons that participate in the regulation of spinal locomotor circuits. J Neurosci. 2006;26:5684–5697. doi: 10.1523/JNEUROSCI.4993-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster RW, Fraser SE. Tracing transgene expression in living zebrafish embryos. Dev Biol. 2001;233:329–346. doi: 10.1006/dbio.2001.0242. [DOI] [PubMed] [Google Scholar]
- Kudoh T, Tsang M, Hukriede NA, Chen X, Dedekian M, Clarke CJ, Kiang A, Schultz S, Epstein JA, Toyama R, Dawid IB. A gene expression screen in zebrafish embryogenesis. Genome Res. 2001;11:1979–1987. doi: 10.1101/gr.209601. [DOI] [PubMed] [Google Scholar]
- Lanuza GM, Gosgnach S, Pierani A, Jessell TM, Goulding M. Genetic identification of spinal interneurons that coordinate left-right locomotor activity necessary for walking movements. Neuron. 2004;42:375–386. doi: 10.1016/s0896-6273(04)00249-1. [DOI] [PubMed] [Google Scholar]
- Liao JC, Fetcho JR. Shared versus specialized glycinergic spinal interneurons in axial motor circuits of larval zebrafish. J Neurosci. 2008;28:12982–12992. doi: 10.1523/JNEUROSCI.3330-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun K, Brand M. A series of no isthmus (noi) alleles of the zebrafish pax2.1 gene reveals multiple signaling events in development of the midbrain-hindbrain boundary. Development. 1998;125:3049–3062. doi: 10.1242/dev.125.16.3049. [DOI] [PubMed] [Google Scholar]
- Mackereth MD, Kwak SJ, Fritz A, Riley BB. Zebrafish pax8 is required for otic placode induction and plays a redundant role with Pax2 genes in the maintenance of the otic placode. Development. 2005;132:371–382. doi: 10.1242/dev.01587. [DOI] [PubMed] [Google Scholar]
- Mansouri A, Chowdhury K, Gruss P. Follicular cells of the thyroid gland require Pax8 gene function. Nat Genet. 1998;19:87–90. doi: 10.1038/ng0598-87. [DOI] [PubMed] [Google Scholar]
- Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- O’Kane CJ, Gehring WJ. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci U S A. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono F, Higashijima S, Shcherbatko A, Fetcho JR, Brehm P. Paralytic zebrafish lacking acetylcholine receptors fail to localize rapsyn clusters to the synapse. J Neurosci. 2001;21:5439–5448. doi: 10.1523/JNEUROSCI.21-15-05439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer PL, Gerster T, Lun K, Brand M, Busslinger M. Characterization of three novel members of the zebrafish Pax2/5/8 family: dependency of Pax5 and Pax8 expression on the Pax2.1 (noi) function. Development. 1998;125:3063–3074. doi: 10.1242/dev.125.16.3063. [DOI] [PubMed] [Google Scholar]
- Pillai A, Mansouri A, Behringer R, Westphal H, Goulding M. Lhx1 and Lhx5 maintain the inhibitory-neurotransmitter status of interneurons in the dorsal spinal cord. Development. 2007;134:357–366. doi: 10.1242/dev.02717. [DOI] [PubMed] [Google Scholar]
- Puschel AW, Westerfield M, Dressler GR. Comparative analysis of Pax-2 protein distributions during neurulation in mice and zebrafish. Mech Dev. 1992;38:197–208. doi: 10.1016/0925-4773(92)90053-m. [DOI] [PubMed] [Google Scholar]
- Sapir T, Geiman EJ, Wang Z, Velasquez T, Mitsui S, Yoshihara Y, Frank E, Alvarez FJ, Goulding M. Pax6 and engrailed 1 regulate two distinct aspects of renshaw cell development. J Neurosci. 2004;24:1255–1264. doi: 10.1523/JNEUROSCI.3187-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou C, Kimura Y, Kohashi T, Horikawa K, Takeda H, Oda Y, Higashijima S. Functional role of a specialized class of spinal commissural inhibitory neurons during fast escapes in zebrafish. J Neurosci. 2009;29:6780–6793. doi: 10.1523/JNEUROSCI.0801-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa H, Miyashita T, Hirate Y, Higashijima S, Chino N, Uyemura K, Kikuchi Y, Okamoto H. Functional repression of Islet-2 by disruption of complex with Ldb impairs peripheral axonal outgrowth in embryonic zebrafish. Neuron. 2001;30:423–436. doi: 10.1016/s0896-6273(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Tian T, Zhao L, Zhao X, Zhang M, Meng A. A zebrafish gene trap line expresses GFP recapturing expression pattern of foxj1b. J Genet Genomics. 2009;36:581–589. doi: 10.1016/S1673-8527(08)60150-2. [DOI] [PubMed] [Google Scholar]
- Toyama R, O’Connell ML, Wright CV, Kuehn MR, Dawid IB. Nodal induces ectopic goosecoid and lim1 expression and axis duplication in zebrafish. Development. 1995;121:383–391. doi: 10.1242/dev.121.2.383. [DOI] [PubMed] [Google Scholar]
- Treisman J, Harris E, Desplan C. The paired box encodes a second DNA-binding domain in the paired homeo domain protein. Genes Dev. 1991;5:594–604. doi: 10.1101/gad.5.4.594. [DOI] [PubMed] [Google Scholar]
- Yonkers MA, Ribera AB. Sensory neuron sodium current requires nongenomic actions of thyroid hormone during development. J Neurophysiol. 2008;100:2719–2725. doi: 10.1152/jn.90801.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.