Fanconi anemia (FA) is a cancer predisposition syndrome driven by defects in the FA pathway, which controls DNA interstrand cross-link (ICL) repair. In this review, Kim and D'Andrea provide a comprehensive survey of ICL repair by ubiquitin- and SUMO-activated pathways and the coordination of multiple repair activities.
Keywords: Fanconi anemia, ubiquitination, DNA interstrand cross-link, DNA repair, translesion DNA synthesis, homologous recombination
Abstract
The maintenance of genome stability is critical for survival, and its failure is often associated with tumorigenesis. The Fanconi anemia (FA) pathway is essential for the repair of DNA interstrand cross-links (ICLs), and a germline defect in the pathway results in FA, a cancer predisposition syndrome driven by genome instability. Central to this pathway is the monoubiquitination of FANCD2, which coordinates multiple DNA repair activities required for the resolution of ICLs. Recent studies have demonstrated how the FA pathway coordinates three critical DNA repair processes, including nucleolytic incision, translesion DNA synthesis (TLS), and homologous recombination (HR). Here, we review recent advances in our understanding of the downstream ICL repair steps initiated by ubiquitin-mediated FA pathway activation.
The Fanconi anemia (FA) pathway
FA is a chromosomal instability disorder characterized by multiple congenital abnormalities, progressive bone marrow failure, and cancer predisposition (Joenje and Patel 2001; Kennedy and D'Andrea 2005; Kee and D'Andrea 2010). Although it is a rare autosomal recessive disease (incidence of one to five per 1,000,000 births), FA is an important model for studying DNA repair, cancer pathogenesis, and ubiquitin signaling (Moldovan and D'Andrea 2009). Clinically, most FA patients manifest anemia and bone marrow failure during childhood and are at risk of developing acute myelogenous leukemia, squamous cell carcinoma of head and neck, and hepatocellular carcinoma (D'Andrea 2010). Mechanistically, FA is caused by germline mutations in genes that cooperate in a DNA repair pathway specialized for resolving DNA interstrand cross-link (ICL), a fatal lesion blocking both DNA replication and transcription (Deans and West 2011). Accordingly, FA patient-derived cells are hypersensitive to DNA cross-linking agents, such as mitomycin C (MMC) or diepoxybutane (DEB), with a dramatic increase of chromosome aberrations and quadradials, a phenotype widely used as a diagnostic test for FA.
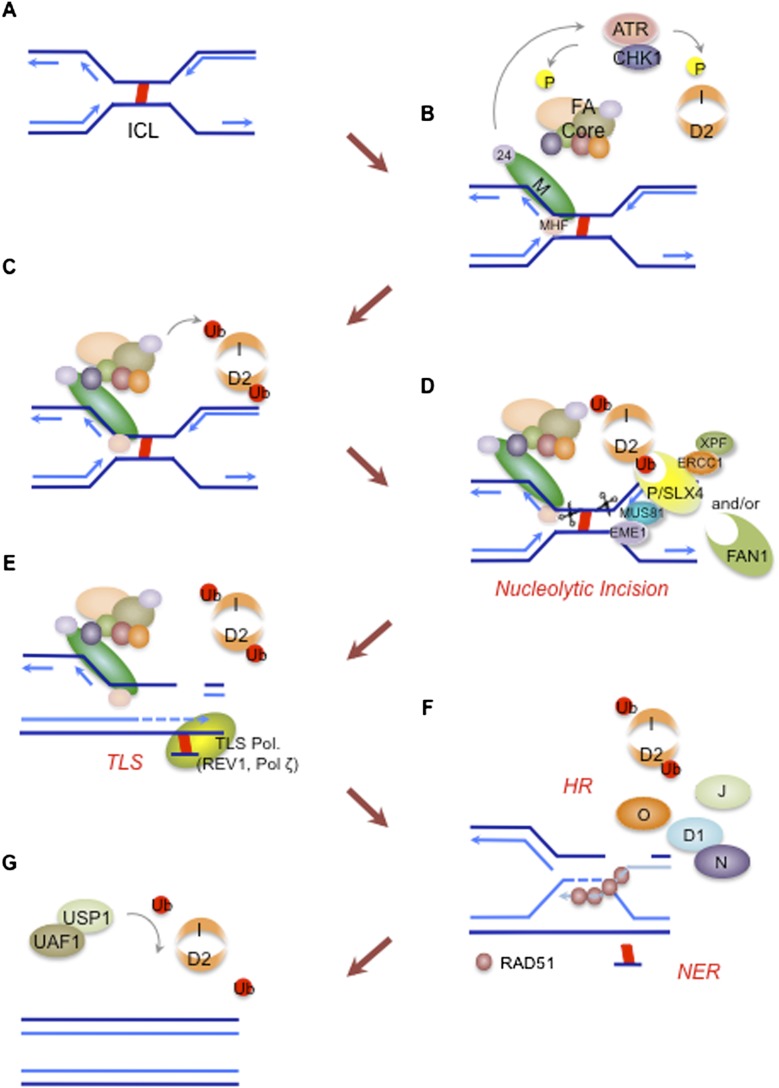
At least 15 FA gene products constitute a common DNA repair pathway, the FA pathway, which resolves ICLs encountered during replication (Fig. 1A). Specifically, eight FA proteins (FANCA/B/C/E/F/G/L/M) form a multisubunit ubiquitin E3 ligase complex, the FA core complex, which activates the monoubiquitination of FANCD2 and FANCI after genotoxic stress or in S phase (Wang 2007). The FANCM subunit initiates the pathway (Fig. 1B). It forms a heterodimeric complex with FAAP24 (FA-associated protein 24 kDa), and the complex resembles an XPF–ERCC1 structure-specific endonuclease pair (Ciccia et al. 2008). The FANCM–FAAP24 complex plays multiple roles in pathway activation by recognizing the DNA lesion and recruiting the FA core complex, stabilizing the stalled replication fork, and initiating ATR (ataxia-telangiectasia and Rad3-related)-mediated checkpoint signaling (Ciccia et al. 2007; Collis et al. 2008; Schwab et al. 2010). Histone fold protein 1 (MHF1) and MHF2 maintain the stable association of FANCM with chromatin and augment efficient pathway activation (Singh et al. 2010; Yan et al. 2010). Beside the FANCM–FAAP24–MHF1/2 complex, the MutS mismatch repair complex plays a redundant role in DNA damage sensing and pathway activation by further promoting the recruitment of the FA core complex to chromatin (Huang et al. 2011; Williams et al. 2011b). During activation, multiple FA proteins undergo phosphorylation by ATR-CHK1 checkpoint kinases, representing a close interconnection of the FA pathway with DNA damage response signaling (Cohn and D'Andrea 2008).
Figure 1.
Interaction of the 15 FA proteins in a common ICL repair pathway in S phase. (A) Two replication forks converge on the DNA ICL that covalently links the two strands of DNA. (B) The FANCM–FAAP24–MHF1/2 complex recognizes the stalled replication fork structure and recruits the FA core complex to the ICL region. The translocase activity of FANCM prevents the collapse of replication fork independent of FA pathway activation. FANCM also initiates ATR-CHK1-dependent checkpoint response, which in turn phosphorylates multiple FA proteins, including FANCA/E/D2/I. (C) The FA core complex, a ubiquitin E3 ligase, monoubiquitinates FANCD2 and FANCI, and the ID heterodimeric complex is recruited to the DNA lesion. (D) FANCD2-Ub acts as a platform to recruit multiple nucleases to coordinate nucleolytic incisions flanking the ICL. FANCP/SLX4, which interacts with ERCC1–XPF and MUS81–EME1 structure-specific nucleases, and FAN1 5′-flap endonuclease are good candidates for this process. Both SLX4 and FAN1 contain the UBZ4 UBM essential for FANCD2-Ub-dependent recruitment to the DNA lesion. (E) Unhooking leaves cross-linked nucleotides tethered to the complementary strand, which is bypassed by TLS, mediated by specialized TLS polymerases such as REV1 and Pol ζ. (F) Incision creates a DSB, which is repaired by HR. Downstream FA proteins promote RAD51-dependent strand invasion and the resolution of recombinant intermediates. NER removes remaining adducts and fills the gap. (G) The USP1–UAF1 DUB complex removes monoubiquitin from FANCD2-I and completes the repair.
Monoubiquitination of FANCD2 and FANCI by the FA core complex is the key regulatory step in the pathway. The RING domain-containing FANCL subunit is a ubiquitin E3 ligase, and in concert with the UBE2T E2 enzyme, it conjugates a single ubiquitin to Lys 561 and Lys 523 of human FANCD2 and FANCI, respectively (Fig. 1C; Cole et al. 2010; Garner and Smogorzewska 2011). Monoubiquitination also occurs during S-phase progression, and phosphorylation-dependent FANCM degradation regulates the localization of the FA core complex to chromatin (Kee et al. 2009). The monoubiquitinated FANCD2-I (ID) heterodimeric complex is relocalized to DNA lesions, where it coordinates cross-link repair activities together with downstream FA proteins (D1/J/N/O/P).
Multiple DNA nucleases function in ICL repair, and the monoubiquitinated FANCD2 (FANCD2-Ub) acts as a landing pad for recruiting nucleases such as FAN1 (FA-associated nuclease 1) and the newly identified FA complementation group P/SLX4 protein to the ICL lesion in order to initiate nucleolytic incision (Fig. 1D; Crossan and Patel 2012). These proteins contain a unique ubiquitin-binding domain (UBD) called the UBZ4 (ubiquitin-binding zinc finger 4) that can specifically recognize the ubiquitin moiety of FANCD2 (Huang and D'Andrea 2010; Yamamoto et al. 2011). SLX4-associated MUS81–EME1 and XPF–ERCC1 nucleases promote cross-link unhooking (Ciccia et al. 2008). The unhooking process converts a stalled replication fork into a double-strand break (DSB), and translesion DNA synthesis (TLS) allows the bypass of the unhooked cross-linked oligonucleotides and the restoration of a nascent strand (Fig. 1E). The DSB is then repaired by homologous recombination (HR), while nucleotide excision repair (NER) excises the remaining adducts, and the gap is filled by DNA polymerases (Fig. 1F). The downstream FA proteins D1/J/N/O play an important role in the HR process. These proteins facilitate RAD51 loading and the resolution of recombination intermediates. Finally, the modified ID complex is deubiquitinated by the deubiquitinating (DUB) enzyme USP1 (ubiquitin-specific peptidase 1), associated with its activating partner, UAF1 (USP1-associated factor 1) (Fig. 1G; Nijman et al. 2005; Cohn et al. 2007). Of note, genetic disruption of murine Usp1 results in a phenotype similar to FA, and both Usp1 and Uaf1 knockout chicken DT40 cells exhibit hypersensitivity to DNA cross-linking agents (Oestergaard et al. 2007; Kim et al. 2009; Murai et al. 2011). Therefore, USP1-dependent deubiquitination constitutes another critical repair step in the completion of DNA ICL repair.
Taken together, the FA pathway regulates three critical DNA repair processes; namely, nucleolytic incision, TLS, and HR. A Xenopus egg extract system, using a plasmid with a single site-specific ICL, has established a role of FANCD2-Ub in multiple ICL repair steps, including nucleolytic incision and TLS (Knipscheer et al. 2009). However, understanding the molecular details of the FA pathway and its coordination of nucleases, helicases, and polymerases is far from complete. Recent biochemical, cellular, and genetic studies have provided invaluable insights into the DNA repair network, and the cross-talk among the DNA repair pathways is now emerging. Moreover, new players have been added to the repertoire of the FA pathway. Here, we discuss recent advances in understanding the downstream ICL repair steps regulated by the FA pathway.
Regulation of nucleolytic incision by the FA pathway
FANCP/SLX4, the scissors in the FA pathway
The DNA ICL is a complex lesion, capable of blocking replication fork progression. A new FA gene, FANCP/SLX4, is an essential player in the nucleolytic incision of ICL. Slx4 in Saccharomyces cerevisiae was initially identified through a synthetic-lethal screen as one of six genes required for the survival of cells lacking Sgs1, a member of the RecQ family of DNA helicases (Mullen et al. 2001). Slx4 was also a hit in a genome-wide screen for proteins that confer resistance to DNA ICL-inducing agents (Wu et al. 2004; Lee et al. 2005). The Drosophila melanogaster Slx4 homolog MUS312 was shown to be required for ICL repair (Yildiz et al. 2002). Slx4 interacts with its catalytic subunit, Slx1, and the Slx1–Slx4 complex acts as a structure-specific 5′-flap endonuclease directed toward branched DNA structures and Holliday junctions (HJs) (Fricke and Brill 2003; Coulon et al. 2004). Slx4 also interacts with the Rad1–Rad10 complex, a member of the yeast XPF/MUS81 structure-specific nuclease family required for NER and ICL repair (Flott et al. 2007; Ciccia et al. 2008). The Slx4–Rad1–Rad10 complex is required for DSB repair during single-strand annealing (SSA) (Li et al. 2008; Lyndaker et al. 2008). Taken together, Slx4 coordinates multiple DNA repair pathways and recombination events.
Efforts to identify a vertebrate ortholog of yeast Slx4, using a conserved C-terminal DNA-binding motif in a bioinformatic search, led to the discovery of the human BTBD12/SLX4 protein (Svendsen and Harper 2010; Cybulski and Howlett 2011). Unlike its yeast counterpart, the human protein contains a BTB/POZ (Broad Complex, Tramtrack, and Bric a brac/POxvirus and Zinc finger) protein–protein interaction domain and two UBZ4 UBDs. The protein is an ATM/ATR kinase substrate, consistent with the known Slx4 phosphorylation by Tel1 and Mec1 following DNA damage in yeast (Flott et al. 2007; Matsuoka et al. 2007). SLX4 interacts with XPF–ERCC1 and MUS81–EME1 structure-specific endonucleases and with SLX1 and cleaves 5′-flap, 3′-flap, and replication fork structures in vitro (Andersen et al. 2009; Fekairi et al. 2009; Munoz et al. 2009; Svendsen et al. 2009). In addition, SLX1–SLX4 functions as a HJ resolvase that processes recombination intermediates. Importantly, depletion of SLX4 leads to hypersensitivity to MMC and cisplatin, but not to ultraviolet (UV) irradiation or ionizing radiation (IR), and reduces the efficiency of HR repair, implicating SLX4 in the FA pathway (Fekairi et al. 2009; Munoz et al. 2009). Overall, SLX4 acts as a scaffold for multiple nucleases regulating ICL repair and HR.
Two independent groups identified biallelic SLX4 mutations in unassigned FA patients (Kim et al. 2011; Stoepker et al. 2011). Introduction of wild-type SLX4 cDNA complemented most of the cellular phenotypes, confirming that SLX4 is a new FA complementation group, FANCP. Importantly, each mutant allele encodes a truncated protein, thus providing valuable insights into the structural domains of the SLX4 protein.
First, some patient-derived mutations result in in-frame deletion of the UBZ4 domains at the N terminus (Kim et al. 2011; Stoepker et al. 2011). Cells expressing these truncated products are hypersensitive to MMC, despite intact interactions with the nuclease complexes XPF–ERCC1 and MUS81–EME1. The interaction of SLX4 with ubiquitin is therefore essential for SLX4 function in ICL repair. In chicken DT40 cells, the recruitment of SLX4 to DNA damage foci requires its UBZ4 domain and FANCD2-Ub, and cells expressing UBZ4-deficient SLX4 are selectively sensitive to ICL-inducing agents (Yamamoto et al. 2011). Therefore, the FA pathway appears to channel SLX4 into a subset of HR processes to resolve ICLs through FANCD2-Ub.
Second, independent frameshift mutations produce N-terminal fragments that only interact with ERCC1 (Kim et al. 2011). These mutant proteins, predicted to lose their interaction with EME1–MUS81, do not fully rescue MMC sensitivity. In contrast, cells expressing a low level of an N-terminal truncated protein show reduced chromatin localization of XPF–ERCC1 and MMC sensitivity (Stoepker et al. 2011). A survival assay using Slx4−/− mouse embryonic fibroblasts (MEFs) also demonstrated that the interaction of ERCC1with SLX4 is required for ICL repair (Crossan et al. 2011). These observations emphasize the scaffolding role of SLX4 in the recruitment of multiple nucleases to sites of ICL repair. Interestingly, Ercc1−/− MEFs exhibit additional sensitivity to UV irradiation, in contrast to Sxl4−/− MEFs, which are UV-resistant (Crossan et al. 2011). The ERCC1 mutant, which cannot interact with the NER factor XPA, is defective in complementing UV sensitivity but not MMC sensitivity (Orelli et al. 2010). Therefore, the recruitment of a specific pool of XPF–ERCC1 to ICLs may be one of the critical functions of SLX4 in the FA pathway. The association of SLX4 with XPF–ERCC1 may alter the catalytic activity of XPF nuclease, thereby promoting an ICL-specific function.
Intriguingly, the FA pathway and SLX4 are not epistatic in MMC or cisplatin sensitivity in DT40 cells (Yamamoto et al. 2011). In addition, SLX4 binds to Lys 63 polyubiquitin chains in vitro via its UBZ4 domains (Kim et al. 2011; Yamamoto et al. 2011). SLX4 may therefore have FANCD2-Ub-independent roles in ICL processing. Alternatively, the role of SLX4 in coordinating nucleases in human and chicken cells may be different, since chicken cells do not express Mus81 and Ercc1.
Last, Slx4 knockout mice recapitulate key cellular and clinical phenotypes of FA patients, including growth retardation, developmental defects, and hematological dysfunction associated with genome instability (Crossan et al. 2011). Primordial germ cell failure is also evident, as in other FA-deficient mice (Parmar et al. 2009). Therefore, the FA-P mouse establishes a new disease model for FA and will be a valuable tool for studying FA-associated cancer predisposition.
FANCD2-Ub coordinates the nuclease event
The identification of nuclease-associated proteins with UBZ4 domains established an important role of FANCD2-Ub in ICL repair; namely, the recruitment of structure-specific nucleases to the site of repair. The Xenopus egg extract system showed that FANCD2-Ub is required for the incision at the site of ICLs (Knipscheer et al. 2009). Blocking the incision step by depletion of either FANCD2-Ub or SLX4-associated nucleases can prevent downstream TLS and DSB repair by HR. However, the endonuclease responsible for the initial incision event during ICL repair remains undetermined. ICL-induced DSBs can be generated without ERCC1 or XPF (De Silva et al. 2000; Niedernhofer et al. 2004), whereas MUS81–EME1 promotes the conversion of an ICL into a DSB in S phase of mouse embryonic stem cells (Hanada et al. 2006). Collectively, initiation of incision appears to be mediated by MUS81–EME1, followed by a second incision by XPF–ERCC1 5′ to the ICL. Nevertheless, it is also possible that XPF–ERCC1 alone is sufficient to initiate ICL incisions, as shown by its ability to perform both 5′ and 3′ incisions against psolaren-induced Y-shaped DNA mimicking a stalled fork structure (Fisher et al. 2008). Indeed, the hSNM1A exonuclease, a mammalian homolog of Pso2, was shown to collaborate with XPF–ERCC1 to initiate ICL repair by creating a favorable substrate for TLS, while MUS81–EME1 acts in reserve (Wang et al. 2011). Intriguingly, FANCD2 recruitment to chromatin and foci formation is impaired in the absence of XPF–ERCC1, indicating that a specific DNA structure generated by incision may help stabilize the FANCD2-Ub association to the lesion, thereby further promoting the recruitment of nucleases via SLX4 (Bhagwat et al. 2009).
FAN1 is also a strong candidate for the incision at ICL, since (1) it is recruited to FANCD2-Ub via its UBZ4 domain to the site of DNA repair, and (2) it exhibits 5′-flap endonuclease as well as 5′–3′ exonuclease activity (Kratz et al. 2010; Liu et al. 2010; MacKay et al. 2010; Smogorzewska et al. 2010; Yoshikiyo et al. 2010). However, FAN1 knockdown does not affect ICL-induced DSB formation, suggesting that it may have nuclease functions downstream from the ICL unhooking step (Kratz et al. 2010). Biallelic mutations of FAN1 have not been identified in unassigned FA patients yet, so it is not considered a bona fide FA gene. FANCJ and BLM helicases may further augment nuclease events in which they synergistically unwind a damaged DNA duplex substrate (Suhasini et al. 2011). FANCD2 may directly process DNA, as it was shown to have intrinsic nuclease activity (Pace et al. 2010). Depletion of each nuclease component from Xenopus egg extracts or complementation with SLX4 mutant proteins lacking specific nuclease complex interaction sites may address the identity of endonucleases responsible for the initial incision step.
Regulation of TLS by the FA pathway
TLS
Following ICL unhooking, the lesion must be bypassed by a TLS polymerase, thereby extending the leading strand. The leading strand is subsequently used as a template of HR, followed by restoration of the replication fork (Fig. 1E,F). Therefore, TLS constitutes a crucial step in ICL repair. TLS is one of the cellular mechanisms for DNA damage tolerance, or post-replication repair. TLS, through the activity of error-prone polymerases, allows cells to replicate over the replication-blocking lesion without correcting it (Lehmann et al. 2007; Chang and Cimprich 2009). Many types of DNA lesions impede replication fork progression, resulting in replication fork collapse and DSB formation. Thus, this DNA damage tolerance mechanism prevents prolonged replication stalling and ensures the completion of DNA replication in a timely manner under conditions of stress, at the expense of creating mutations across the genome.
TLS uses specialized low-fidelity DNA polymerases to directly bypass the lesion. Unlike the B family replicative polymerases (Pols) such as Pol α, Pol δ, and Pol ɛ, TLS polymerases lack 3′–5′ proofreading activity and contain an unconstrained active site that can accommodate distorted bases and base pair mismatches (Sale et al. 2012). In mammalian cells, the Y family TLS polymerases include Pol ι, Pol η, Pol κ, and Rev1, while Pol ζ belongs to the B family. Although the nature of lesion bypass is mutagenic, there are specific lesions (referred to as cognate lesions) that are fixed in a relatively error-free manner, such as removal of UV-induced cis–syn thymine dimers by Pol η (Johnson et al. 1999; Washington et al. 1999). Mutation of the Pol η gene (POLH) is associated with a cancer-prone disease, the variant form of xeroderma pigmentosum (XPV) (Masutani et al. 1999).
TLS polymerases are recruited and regulated by a post-translational modification of PCNA (proliferating cell nuclear antigen). PCNA is a polymerase processivity factor that encircles DNA and functions as a moving platform for DNA synthesis (Moldovan et al. 2007). Following replication arrest, the E2–E3 ubiquitin ligase complex RAD6–RAD18 associates with RPA (replication protein A)-bound ssDNA and monoubiquitinates PCNA at Lys 164, thus recruiting a TLS polymerase to the lesion (Hoege et al. 2002; Davies et al. 2008; Ulrich 2009). Many TLS polymerases, including Pol ι, Pol η, and Pol κ, interact with PCNA through their PIP (PCNA-interacting protein) box motif. Moreover, specialized UBDs, such as the UBM (ubiquitin-binding motif) of REV1 or the UBZ3 motif of Pol η, further provide specificity for the recognition of PCNA monoubiquitin (PCNA-Ub) (Xu et al. 2001; Kannouche et al. 2004; Guo et al. 2006a,b; Plosky et al. 2006; Parker et al. 2007). In S. cerevisiae, PCNA is also polyubiquitinated at Lys 164 via the Lys 63-linked chain by the Ubc13/Mms2–Rad5 E2–E3 ligase complex (Hoege et al. 2002; Chiu et al. 2006). This modification mediates template switching, an error-free process that uses a newly synthesized and undamaged template to temporarily replace a damaged DNA template via either fork reversal or D-loop formation (Chang and Cimprich 2009). In humans, the Rad5 orthologs HLTF (helicase-like transcription factor) and SHPRH (SNF2 histone linker PHD RING helicase) E3 ligases are responsible for the Lys 63-linked polyubiquitination of PCNA (Unk et al. 2006, 2008). In contrast, SUMO conjugation at Lys 164 of PCNA recruits the anti-recombinogenic DNA helicase Srs2, thereby releasing RAD51 and restricting recombination in yeast (Pfander et al. 2005).
TLS polymerases in ICL repair
Rev1 and Pol ζ, a heterodimer of Rev3 and Rev7, function together in the TLS step of ICL repair. The yeast rev3 strain exhibits hypersensitivity to ICL-inducing agents, which is epistatic with Rev1 (McHugh et al. 2000; Lawrence 2002; Wu et al. 2004; Sarkar et al. 2006). Similarly, chicken DT40 or human cells deficient in Pol ζ and/or Rev1 are hypersensitive to ICL-inducing agents (Sonoda et al. 2003; Niedzwiedz et al. 2004; Okada et al. 2005; Wittschieben et al. 2006; Hicks et al. 2010). In addition, DT40 genetic studies indicate that both Rev1 and Rev3 are epistatic with FANCC in cisplatin sensitivity (Niedzwiedz et al. 2004). Furthermore, depletion of Pol ζ abolishes the extension of a nascent strand beyond the ICL in Xenopus egg extracts, arguing for a direct role of Pol ζ in replication-dependent ICL repair (Raschle et al. 2008).
Rev1 is the major TLS polymerase in eukaryotes required for the introduction of DNA mutations. Rev1 is responsible for >90% of base pair substitutions induced by UV irradiation (Lawrence 2002). Consequently, Rev1 deficiency leads to a dramatic reduction in both spontaneous and damage-induced mutations and greatly increases the incidence of chromosome aberrations (Simpson and Sale 2003). Rev1 is not a polymerase per se, but instead functions as a deoxycytidyl transferase. It inserts a deoxycytidine (dC) across a template G or an abasic site, using its conserved arginine residue in the catalytic domain (Nelson et al. 1996; Haracska et al. 2002). Mammalian Rev1 is required for the bypass of UV-induced damage and cross-links, but its transferase activity seems dispensable, since a catalytically dead mutant can complement cisplatin and UV sensitivity in Rev1 knockout DT40 cells (Ross et al. 2005). Rather, protein–protein interaction seems essential, as the extreme C terminus of Rev1 interacts with various TLS polymerases including Pol ζ, Pol ι, Pol η, and Pol κ (Guo et al. 2003). The crystal structure of human REV3–REV7 also demonstrated a functional collaboration between Pol ζ and REV1; the disruption of REV1–REV7 interaction sensitizes cells to DNA cross-linking agents (Hara et al. 2010). Rev1 is also recruited to PCNA-Ub via its UBM, thus allowing Rev1 to regulate the loading of different TLS polymerases to the lesion (Guo et al. 2006a,b). Therefore, a model has evolved in which Rev1 functions as a scaffold to recruit and coordinate TLS polymerases, rather than directly bypassing the DNA lesion. This mechanism may facilitate so-called “polymerase switching,” in which Rev1 exerts a spatiotemporal regulatory role in the action of TLS polymerases specialized for nucleotide insertion and extension beyond the lesion (Friedberg et al. 2005; Waters et al. 2009). Rev1 may also potentiate the enzymatic activity of the recruited TLS polymerases, as Rev1 was shown to stimulate Pol ζ extension in vitro (Acharya et al. 2006). Nevertheless, the enzymatic activity of Rev1 may still be required in other contexts. For instance, an altered mutation spectrum was observed with the Rev1 catalytically dead mutant in both yeast and vertebrate cells (Otsuka et al. 2005; Ross et al. 2005). In addition, the catalytic activity of Rev1 is required for coping with specific adducts, such as N2-dG, formed by 4-NQO (4-nitroquinoline-1-oxide) in S. cerevisiae (Wiltrout and Walker 2011).
Several lines of evidence indicate that other DNA polymerases may be involved in the TLS step of cross-link repair. Pol κ was shown to catalyze accurate bypass of N2-N2-guanine ICL in vitro, and Pol κ-depleted cells show chromosome instability and decreased survival following MMC exposure (Minko et al. 2008). The A family Pol θ (POLQ) can extend DNA from mismatched bases in vitro (Seki and Wood 2008). POLQ-deficient cells exhibit spontaneous and MMC-induced chromosomal abnormalities, although they are not hypersensitive to cross-linking agents per se (Shima et al. 2004). Another A family Pol ν (POLN) can perform nonmutagenic bypass of psoralen-induced DNA cross-links, albeit with low efficiency, suggesting a role in TLS during ICL repair (Zietlow et al. 2009). However, the biological significance of these in vitro data remains to be established.
Function of the UBZ domain in DNA repair
Ubiquitin plays a crucial role in the regulation of DNA repair processes (Hofmann 2009). Ubiquitin signaling is mediated by UBDs that recognize various ubiquitin modifications. As expected from diverse ubiquitin signaling networks, UBDs are structurally and functionally distinct modules, and more than 20 different UBD classes exist to interact with various ubiquitin modifications (Dikic et al. 2009).
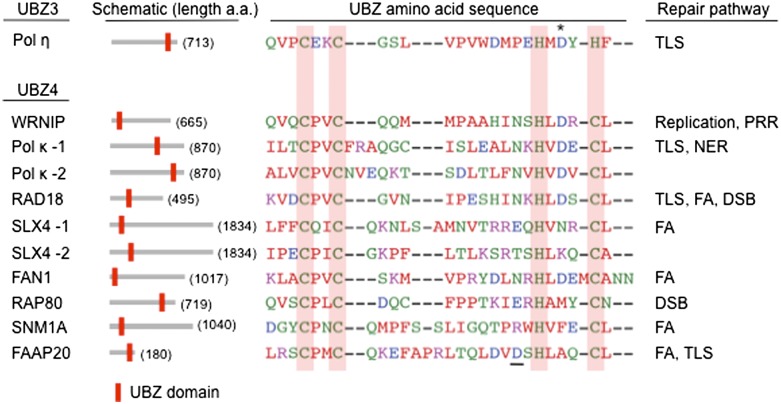
Among those, a UBZ family is particularly intriguing, since it exclusively appears in proteins involved in DNA ICL repair and TLS (Hofmann 2009). The UBZ domain, along with UBM, was originally identified through yeast two-hybrid screening and bioinformatic analysis (Bienko et al. 2005). Two subfamilies of UBZ are especially relevant: UBZ3 contains a highly conserved C2H2 short mononucleate zinc finger, whereas UBZ4 is defined as a RAD18-like C2HC zinc finger (Fig. 2). Yeast Rad30 and its mammalian homolog, pol η, are the only UBZ3-containing proteins identified so far. The NMR structure of the pol η UBZ3 motif revealed similarities to the DNA-binding zinc finger, in which the ubiquitin binding α-helical surface is on the opposite side of the zinc-coordinating residues (Bomar et al. 2007). In contrast, the UBZ4 domain has been found in multiple proteins involved in post-replication repair, such as Pol κ, WRNIP1 (Werner helicase-interacting protein 1), and RAD18 (Hofmann 2009). Pol κ interacts with ubiquitinated PCNA, while WRNIP and RAD18 bind to Lys 48– and Lys 63–polyubiquitin chains (Ogi et al. 2005; Bish and Myers 2007; Crosetto et al. 2008; Guo et al. 2008). Additionally, SNM1A, a mammalian ortholog of yeast Pso2 that plays a critical role in ICL repair, was shown to target a stalled replication fork by recognizing PCNA-Ub via its UBZ4 domain (Yang et al. 2010). Interestingly, the recently identified FAN1 nuclease and the FANCP/SLX4 nuclease scaffold protein also contain UBZ4 domains that specifically recognize FANCD2-Ub and function in the downstream ICL repair process, emphasizing a specialized role of UBZ4-containing proteins in mediating ICL repair (Huang and D'Andrea 2010; Cybulski and Howlett 2011). In summary, UBZ is a variant form of the zinc finger motif that has evolved to mediate the DNA repair signaling pathways through its ability to recognize ubiquitin. In this sense, the UBZ domain is a key signature of DNA repair proteins that require interaction with ubiquitinated targets or polyubiquitin chains at chromatin to fulfill their functions.
Figure 2.
UBZ is a signature domain of DNA repair proteins. Various amino acid sequences of known UBZ domains are shown, and their relative positions are marked as a red bar in the protein schematic. Amino acid residues are colored according to their physicochemical properties, and the conserved residues that comprise a zinc-binding core are shaded in the red box. UBZ domains are defined as a C2H2 (UBZ3) or C2HC (UBZ4) zinc finger module, and these two classes appear in DNA damage response proteins related to ICL repair (the FA pathway) and post-replication repair (PRR) (TLS). The major difference between the two domains is the fourth zinc ligand residue, which is a cysteine in the UBZ4 motif instead of histidine in the UBZ3. In addition, the aspartate residue within the H2 zinc-binding dyad of the UBZ3 (indicated as asterisk) is not absolutely conserved in the UBZ4 group, although alanine substitution was shown to disrupt the interaction with ubiquitin in WRNIP (Crosetto et al. 2008). In FAAP20, the aspartate residue outside the HC zinc-binding dyad (underlined) is also required for binding to ubiquitin, possibly compensating for the lack of aspartate in the conserved position (Ali et al. 2012). PCNA-Ub and FANCD2-Ub were proposed as primary targets for some of these UBZ domains, but several in vitro data also suggest that these UBZ domains are able to bind to the Lys 48- or Lys 63-linked polyubiquitin chains. The main function of the UBZ domain is to recruit UBZ-containing DNA repair factors to the site of DNA lesion by recognizing DNA damage-specific ubiquitin conjugation, including monoubiquitinated targets or polyubiquitin chains.
The role of RAD18 in the FA pathway
Increasing evidence suggests interactions between the FA and TLS pathways in ICL repair. RAD18, a primary E3 ligase that regulates TLS, promotes FA pathway activation. By monoubiquitinating PCNA, RAD18 activates FANCD2 following cellular treatment with cisplatin or BPDE (benzo[a]pyrene diol-epoxide), which generates bulky DNA adducts (Geng et al. 2010; Song et al. 2010). PCNA-Ub may stabilize the association of the FA core complex to chromatin, or binding of TLS polymerases to PCNA-Ub may augment FA pathway activation. RAD18 also promotes FANCD2 monoubiquitination independent of monoubiquitinating PCNA following MMC or camptothecin (CPT) treatment (Palle and Vaziri 2011; Williams et al. 2011a). However, the catalytic activity of RAD18 is essential, indicating that RAD18 may have unknown substrates that functionally interact with the FA pathway. In contrast, RAD18 participates in the HR repair of DSBs induced by IR in an E3 ligase-independent manner (Huang et al. 2009). Instead, the UBZ4 domain of RAD18 is required for RAD51C accumulation and RAD51 loading at the DSBs. In this regard, RAD18 may regulate RAD51C recruitment during the HR process of ICL repair, given that RAD51C/FANCO is one of the FA genes necessary for ICL repair (Vaz et al. 2010).
Regulation of REV1 by the FA pathway
Although TLS constitutes an essential step in the FA pathway, how the REV1–Pol ζ polymerase complex is recruited to a stalled replication fork is not well understood. Several lines of evidence implicate the FA core complex in regulating the TLS step. Genetic studies in DT40 cells indicate that Rev1 and Rev3 act downstream from the FA core complex (Niedzwiedz et al. 2004). One interesting feature of FA patient-derived cells is their hypomutability, similar to the phenotype of TLS-deficient cells (Papadopoulo et al. 1990a,b). FancC-deficient DT40 cells have reduced mutational repair in response to endogenously generated abasic sites in the IgV gene locus (Niedzwiedz et al. 2004). FancG knockout hamster CHO cells also show reduced generation of viable Hprt mutations, indicating impaired TLS at the hprt locus during DNA replication (Hinz et al. 2006). These data imply that the FA pathway normally promotes error-prone point mutagenesis through TLS and protects cells from large chromosomal insertions and deletions through HR. Mechanistically, the FA core complex is required for efficient REV1 foci formation following UV irradiation and cisplatin treatment, a process essential for its TLS activity (Mirchandani et al. 2008; Hicks et al. 2010). Accordingly, FA core-deficient patient cells are not as efficient as cDNA-corrected cells in generating spontaneous and UV-induced point mutations. FANCD2 monoubiquitination by the FA core complex is dispensable in this process, suggesting that the FA core complex regulates the TLS step independent of its E3 ligase activity (Mirchandani et al. 2008).
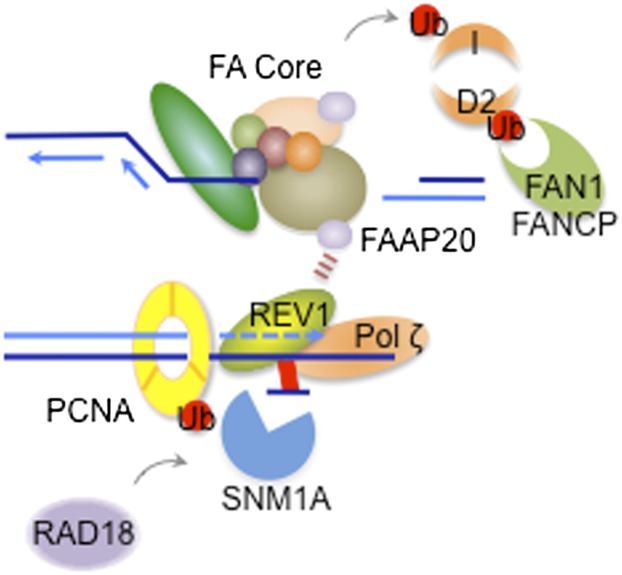
Recently, a new subunit of the FA core complex, FAAP20, was shown to link the FA core complex with REV1-dependent TLS (Kim et al. 2012). FAAP20 was identified by several groups as an integral subunit of the FA core complex (Ali et al. 2012; Kim et al. 2012; Leung et al. 2012). FAAP20 interacts with FANCA through its N terminus, maintains the integrity of the core complex, and allows FANCD2 monoubiquitination. Importantly, the C terminus of FAAP20 contains a UBZ4 domain, which regulates the localization of monoubiquitinated REV1 to the DNA lesion (Kim et al. 2012). Therefore, the FA core complex controls both the incision step of cross-link repair by monoubiquitinating FANCD2 and the TLS step by recruiting REV1 to the site of the lesion (Fig. 3). In addition, the FA core complex may promote the enzymatic activity of REV1. The role of PCNA-Ub in recruiting REV1 to the ICL lesion is less clear. PCNA is only weakly monoubiquitinated in response to cross-linking agents such as MMC, and cross-linkers do not generate long stretches of ssDNA required for RAD18 recruitment (Ho and Schärer 2010). Indeed, RAD18 does not play a major role in resolving replication-associated DSBs during ICL repair (Hicks et al. 2010). Furthermore, Rad18 and FancC double-knockout DT40 cells show greater sensitivity to cisplatin than single knockouts, indicating that FA-dependent TLS in the cross-link repair may be RAD18-independent (Hirano et al. 2005). The FAAP20–REV1 interaction may stabilize a pre-existing nonmonoubiquitinated PCNA–REV1 complex at a stalled replication fork. Consistent with this notion, Rev1 is required in DT40 cells for maintaining fork progression following damage, acting independently of PCNA-Ub (Edmunds et al. 2008). Conversely, FANCD2 depletion in Xenopus egg extracts impairs both incision and TLS, indicating that FANCD2-Ub may directly control both processes (Knipscheer et al. 2009). Perhaps the incision step is a prerequisite for the subsequent TLS step. Also, FANCD2 activation may regulate the activity of certain TLS polymerases (e.g., Pol η) under specific conditions of genotoxic stress (Park et al. 2010).
Figure 3.
Regulation of TLS in replication-associated ICL repair. The FA core complex not only regulates the incision step by monoubiquitinating FANCD2, but also contributes to the recruitment of a TLS polymerase REV1 to the lesion, thus promoting bypass of the ICL intermediate. FAAP20 is an important factor in both steps, as it stabilizes FANCA (therefore keeping the integrity of the FA core complex) and interacts with monoubiquitinated REV1 via its UBZ4 domain to promote stable association of the PCNA/REV1 DNA damage bypass complex at the stalled replication fork. REV1 controls the polymerase switching of TLS polymerases to restore the leading strand synthesis mediated by Pol ζ. It is not clear whether RAD18-dependent PCNA monoubiquitination is a prerequisite for the REV1 recruitment to the ICL region. In contrast, PCNA-Ub enhances the recruitment of UBZ4-containing SNM1A to the ICL site. The 5′–3′ exonuclease activity of SNM1A trims the unhooked intermediate to generate a favorable substrate for TLS (Wang et al. 2011).
USP1, a master regulator of the FA and TLS pathways
USP1 is a DUB enzyme that regulates the level of FANCD2-Ub (Nijman et al. 2005). The UAF1 protein associates with USP1 stoichiometrically and stimulates its enzymatic activity (Cohn et al. 2007). Depletion of USP1 or UAF1 causes hyperaccumulation of both FANCD2-Ub and PCNA (Huang et al. 2006). Thus, USP1 controls the monoubiquitination status of two key proteins working in the FA and TLS pathways, indicating that the USP1–UAF1 DUB complex coordinates at least two DNA repair processes. USP1 is degraded following UV irradiation, and cell cycle-dependent USP1 degradation by APC/CCdh1 ubiquitin E3 ligase allows rapid PCNA monoubiquitination for post-replication repair in G1 phase (Huang et al. 2006; Cotto-Rios et al. 2011). The murine Usp1 knockout model displays several important FA phenotypes, and Usp1−/− cells are hypersensitive to DNA cross-linking agents, emphasizing that timely deubiquitination of FANCD2 is required for the resolution of ICLs (Kim et al. 2009). USP1 depletion also increases spontaneous and UV-induced point mutation frequency by elevating PCNA-Ub (Huang et al. 2006; Hendel et al. 2011). Loss of USP1 results in aberrant engagement of Pol κ to DNA, leading to a slowdown of replication fork progression and interference of DNA synthesis (Jones et al. 2012). This may explain the intrinsic genome instability observed upon USP1 down-regulation and further highlights the role of USP1 in maintaining genome stability by coordinating both FA and TLS pathways.
SUMO-like signaling network in the FA and TLS pathways
Similar to ubiquitin–UBD interaction, SUMO binds to a SUMO-interacting motif (SIM) (Bergink and Jentsch 2009). SIM was initially identified through yeast two-hybrid screening as a short consensus sequence composed of hydrophobic amino acids plus an acidic/polar residue at position 2 or 3 ([V/I]-X-[V/I]-[V/I]), flanked by acidic residues on either side (Minty et al. 2000; Song et al. 2004; Hecker et al. 2006). The combination of α-helix and β-sheet conformation in SIM allows a noncovalent interaction with the hydrophobic pocket on the SUMO surface (Gareau and Lima 2010). One of the important functions of the SUMO–SIM interaction is to properly target an enzyme to its substrate. For instance, RNF4, a member of STUbL (SUMO-targeted ubiquitin E3 ligase), contains four SIMs at the N terminus that bind and target SUMOylated substrates for proteasomal degradation (Tatham et al. 2008). SUMO-like domains (SLDs) were found in a primary sequence of certain proteins such as RENi (Rad60–Esc2–NIP45) family (Novatchkova et al. 2005). However, the functional importance of integrated SUMO structures in SUMO signaling remains unclear.
Recently, SLD–SIM interactions were shown to target the USP1–UAF1 DUB complex to its monoubiquitinated substrates. UAF1 harbors two tandem SLDs (SLD1 and SLD2) at its C terminus, and SLD2 is required for USP1-mediated FANCD2 and PCNA deubiquitination in vivo (Yang et al. 2011). The USP1–UAF1 complex is targeted to FANCD2-Ub and PCNA-Ub via the SIM-like (SLIM) sequence of FANCI, a partner of FANCD2, and the SLIM of hELG1, a partner of PCNA (Lee et al. 2010; Yang et al. 2011). Disruption of each SLIM–SLD interaction increases the level of FANCD2-Ub and PCNA-Ub, respectively, rendering cells DNA repair-deficient. Therefore, UAF1 not only regulates the enzymatic activity of USP1, but also determines the substrate targeting of USP1. Intriguingly, the SLD2 of UAF1 does not bind to a canonical SIM sequence. Furthermore, SUMO isoforms do not interact with the SLIM sequences of FANCI and hELG1, illustrating the specificity of the SLD2–SLIM interaction in substrate recognition (Yang et al. 2011). Thus, the FA and TLS pathways are coordinated by a concerted action of ubiquitin (FANCD2 and PCNA monoubiquitination) and SUMO (FANCD2 and PCNA deubiquitination) signaling networks. Beside USP1, UAF1 interacts with other DUBs, such as USP12 and USP46 (Cohn et al. 2009). Similar SUMO-like delivery networks may be used for substrate recognition by these enzyme complexes as well.
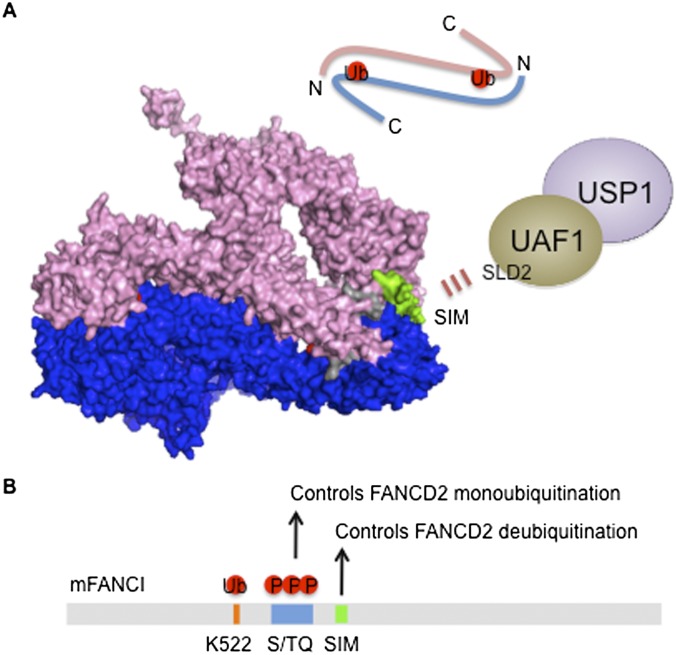
A recently solved three-dimensional structure of the ID complex further elucidates USP1 targeting in vivo. The ID complex has the shape of two juxtaposed saxophones, with the monoubiquitination sites localized at the ID interface, but allowing the access of a ubiquitin tail through a solvent-accessible tunnel (Fig. 4A; Joo et al. 2011). In contrast to the embedded ubiquitinated lysine residues, the SIM sequence of FANCI, necessary for interacting with SLD2 of UAF1, is exposed, thereby allowing the recognition of SIM by the USP1–UAF1 DUB complex. How the active site of USP1 gains access to the sequestered lysine–ubiquitin isopeptide bond at the ID interface is not known. In addition, FANCD2-Ub is located opposite the FANCI SIM, making it difficult for USP1 to access its substrate. Thus, the ubiquitinated ID complex, associated with a damaged DNA structure, may adapt a different conformation from the one predicted in the model. Alternatively, USP1–UAF1 may play an active role in inducing a structural rearrangement of the ID complex and promoting the deubiquitination process upon docking to the FANCI SIM. Whether deubiquitination precedes the dissociation of the ID complex or vice versa remains unclear.
Figure 4.
The FANCI–FANCD2 (ID) complex is targeted by USP1–UAF1. (A) The structure of the mouse ID complex generated by PyMOL software using Protein Data Bank ID 3S4W is shown where a trough-like shape of FANCI (pink) and FANCD2 (blue) are juxtaposed in an anti-parallel manner (schematic on top). The monoubiquitination sites of FANCD2 (Lys 559) and FANCI (Lys 522) are embedded in the ID interface just wide enough to create a tunnel to accommodate the C-terminal ubiquitin tail (shown in red). FANCI contains S/TQ clusters following the monoubiquitination site, and FANCI phosphorylation promotes FANCD2 monoubiquitination (shown in gray). FANCD2 and FANCI may be monoubiquitinated as a monomer, which increases the affinity for chromatin and allows heterodimerization at the DNA lesion. Ubiquitination may stabilize the heterodimerization and induces a conformational change that can fully expose the modified lysine for recruiting DNA repair factors. Alternatively, the ID complex may be loosely associated with chromatin, and ubiquitination may open the ID interface, relocating the ID complex to damage-induced foci at the lesion. FANCD2 and FANCI by themselves have been shown to recognize several DNA structures. Thus, the ubiquitinated ID complex at the ICL may adopt a different conformation compared with the one modeled here. Of note, the SIM of FANCI is exposed outside (shown in green), allowing the targeting of USP1–UAF1 via the SLD2 domain of UAF1. The interaction may trigger the exposure of the ubiquitinated lysine to provide access to the USP1 active site for isopeptide cleavage. (B) By heterodimerizing with FANCD2, FANCI regulates both FANCD2 ubiquitination and deubiquitination through phosphorylation and SIM-mediated DUB targeting, respectively.
The overall similarity between FANCD2 and FANCI suggests that they have evolved from a common ancestor. However, FANCI contains multiple S/TQ phosphorylation sites that regulate FANCD2 monoubiquitination and localization to damage-induced foci (Ishiai et al. 2008). In addition, there is a unique FANCD2 interaction domain in FANCI, which undergoes substantial conformational change upon FANCD2 binding and stabilizes the association with FANCD2 (Joo et al. 2011). FANCI is also required for restricting monoubiquitination to the correct lysine residue (Lys 561) of FANCD2 (Alpi et al. 2008). Thus, FANCI may have acquired a regulatory role by forming heterodimers with FANCD2 instead of homodimerizing to control both FANCD2 monoubiquitination (via phosphorylation) and deubiquitination (via SIM-mediated DUB targeting) (Fig. 4B).
Regulation of HR by the FA pathway
HR factors in the FA pathway
Following an incision flanking the region of the ICL, a DSB is created as an intermediate in the ICL repair process. HR is a primary mechanism for resolving this replication-associated DSB, as it can use the homologous template restored by TLS. The FA pathway not only promotes HR, but also suppresses NHEJ (nonhomologous end-joining), another mechanism for repairing DSBs. NHEJ ligates two broken DSB ends without requiring homology of the template (Bunting and Nussenzweig 2010). Intriguingly, FA phenotypes can be rescued by inhibition of the NHEJ pathway (e.g., deleting NHEJ factors Ku70, DNA-PKcs, or Lig4), suggesting that the FA pathway channels DSBs into HR by suppressing NHEJ in order to prevent inappropriate DNA repair by the NHEJ machinery (Adamo et al. 2010; Pace et al. 2010). However, recent mouse knockout studies demonstrated that 53BP1 or Ku80 deletion exacerbates the genomic instability of FANCD2-deficient cells, suggesting that FANCD2 has an essential role in ICL repair that cannot be bypassed simply by targeting the NHEJ pathway (Bunting et al. 2012).
Many factors in the FA pathway are involved in promoting HR directly or indirectly. Deletion of some HR genes renders cells hypersensitive to ICL-inducing agents, and the FA core- or FANCD2/I-deficient patient cells are defective in efficient HR repair (Nakanishi et al. 2005; Zhang et al. 2007; Smogorzewska et al. 2010). In DT40 cells, FANCC was shown to be epistatic with XRCC2, a RAD51 paralog required for the HR process, in repairing DNA cross-links (Niedzwiedz et al. 2004). One of the key steps in HR is the loading of RAD51 onto newly resected DNA and the RAD51-mediated strand invasion of the sister chromatid. Downstream FA gene products are directly involved in this process. BRCA2, mutated in the FA-D1 subtype, interacts with RAD51 and promotes its loading to RPA-coated ssDNA (Moynahan et al. 2001; Howlett et al. 2002). PALB2 (partner and localizer of BRCA2), mutated in the FA-N subtype, binds to BRCA2 and regulates its intranuclear localization and stability (Xia et al. 2006). BRCA1 also promotes RAD51 loading via its interaction with FANCN and FANCD1 (Zhang et al. 2009). BRIP1 (BRCA1-interacting protein C-terminal helicase 1), mutated in the FA-J subtype, works downstream from RAD51 to complete HR repair by preventing untimely or promiscuous recombination via its helicase activity (Litman et al. 2005; Sommers et al. 2009). RAD51C/FANCO facilitates RAD51 loading and the resolution of HJs, recombination intermediates in the later step of HR (French et al. 2002; Liu et al. 2007; Vaz et al. 2010). Heterozygous mutations in these downstream FA genes are associated with increased risk of developing breast and ovarian cancer, emphasizing the connection between FA and breast cancer, the FA–BRCA network, associated with HR repair dysfunction. Of note, a homozygous XRCC2 mutation was recently found in a Saudi FA patient (Shamseldin et al. 2012). As XRCC2 is also a breast cancer susceptibility gene (Park et al. 2012), these results further support the linkage of the FA pathway with HR repair and cancer predisposition. Pol ν may be involved in the DNA synthesis step of HR during ICL repair, and cells with POLN knockdown are hypersensitive to cross-linking agents (Moldovan et al. 2010).
Last, FANC proteins may prevent NHEJ factors from accessing DSB ends, thus providing a favorable environment for DSB resection and the downstream HR process. BRCA1 plays a similar role by excluding 53BP1 accumulation and preventing aberrant NHEJ (Cao et al. 2009; Bouwman et al. 2010; Bunting et al. 2010).
Replication-associated HR process in the FA pathway
The stalled replication fork adjacent to the ICL initiates the incision step, leading to the generation of a DSB in one sister chromatid (Fig. 1D,E). The FA pathway next promotes replication-dependent homology-directed DSB repair. Accordingly, inhibition of RAD51 activity in Xenopus egg extracts resulted in the ablation of replication-coupled ICL repair, thus proving a functional link between the FA pathway and the HR machinery (Long et al. 2011). RAD51 may play a distinct role in HR at the stalled replication fork, as it protects newly synthesized DNA from extensive MRE11-dependent degradation, a process that normally facilitates end resection in two-ended DSB repair (Hashimoto et al. 2010). Additional insights into replication-associated DSB repair were provided by the development of the TR-GFP assay, a modified version of the DR-GFP HR assay system (Nakanishi et al. 2011). The TR-GFP assay uses a DNA template with a site-specific ICL at sequences that are complemented to triplex-forming oligonucleotide conjugated with psoralen (pso-TFO). The construct also contains an origin of replication of Epstein-Barr virus (EBV) for replication in human cells. ICL-induced HR was substantially compromised in the absence of FA proteins, providing experimental evidence that the FA pathway is specifically involved in replication-coupled HR repair.
BRCA1 plays a unique role in the HR process of ICL repair, as it promotes chromatin loading of FANCD2-Ub at an early step and RAD51 loading at a later step of ICL repair. Since the FANCD2-Ub-dependent incision step is a prerequisite for the generation of DSBs in ICL repair, depletion of an NHEJ factor such as 53BP1 in Brca1-deficient cells can rescue cellular hypersensitivity to PARP1 inhibitor but cannot rescue cross-link hypersensitivity. (Bunting et al. 2012).
The deubiquitination of FANCD2 may also play an active role in ICL repair by facilitating HR. Usp1 knockout MEFs as well as both Usp1 and Uaf1 knockout DT40 cells show decreased HR efficiency, indicating that the USP1–UAF1 DUB promotes the HR process (Kim et al. 2009; Murai et al. 2011). Interestingly, disruption of the NHEJ pathway improves the cellular sensitivity and HR efficiency of the Usp1 and Uaf1 knockout DT40 cells (Murai et al. 2011). Suppressing NHEJ may therefore be one mechanism by which USP1–UAF1 promotes HR, consistent with previous studies indicating that the FA pathway suppresses the NHEJ pathway.
Negative regulation of HR in the FA pathway
Uncontrolled HR causes inappropriate hyperrecombination and the accumulation of toxic recombination intermediates (Heyer et al. 2010). HR is activated strictly in the S and G2 phases. In yeast, Srs2 negatively regulates HR by disassembling ATP-bound RAD51 filaments, reversing the early HR step (Pfander et al. 2005). In humans, the RecQ family helicases BLM and RECQL5 can disrupt RAD51 filaments in vitro and further attenuate HR (Bugreev et al. 2007; Hu et al. 2007). In addition, RTEL1 (regulator of telomere elongation helicase 1), a RAD3-type helicase, antagonizes HR at a later step by removing RAD51 from D-loop recombination intermediates (Barber et al. 2008). Down-regulation of negative HR regulators leads to hyperrecombination and DNA damage sensitivity phenotypes, emphasizing the role of restricting inappropriate HR in preserving genome stability. PARI (PCNA-associated recombination inhibitor) was recently identified as an Srs2 ortholog in higher eukaryotes (Moldovan et al. 2012). Importantly, PARI depletion reverses the HR defect and sensitivity to PARP1 inhibitor of FANCD1/BRCA2-deficient cells, indicating that increasing HR efficiency by antagonizing an inhibitory factor in DNA repair-deficient cells may be beneficial for FA patients.
Clinical perspective
Cancer cells become resistant to conventional chemotherapy by relying on certain DNA repair pathways. Thus, inhibiting the FA pathway may provide a strategy for resensitizing resistant cancer cells. Given their critical role in the FA pathway, TLS polymerases are possible targets for augmenting the effect of DNA cross-linking chemotherapeutic agents. Depletion of Rev1 or Rev3 causes pronounced sensitivity to cisplatin in lymphoma and non-small-cell lung cancer (NSCLC) models and prevents the development of acquired drug resistance, indicating that inhibition of TLS can not only kill tumor cells, but also antagonize TLS-mediated generation of resistance-causing mutations (Doles et al. 2010; Xie et al. 2010). Thus, selectively inhibiting TLS polymerases may sensitize cancer cells to DNA-damaging agents.
Recent studies indicate that small molecules that perturb ubiquitin-mediated signal transduction can modulate the FA pathway. The anti-cancer agent Bortezomib (Orlowski and Kuhn 2008) reduces intracellular pools of free ubiquitin and thereby blocks FANCD2 monoubiquitination (Jacquemont and Taniguchi 2007). Thus, Bortezomib may also function as a FA pathway inhibitor, capable of sensitizing cancer cells to cisplatin. Inhibitors of DUB enzymes (Chen et al. 2011) or protein neddylation (Kee et al. 2012) may also block the FA pathway and function as novel cisplatin sensitizers.
As FA cells exhibit spontaneous chromosome aberrations, there may be endogenous sources of DNA damage resolved by the FA pathway. Genetic knockout studies have revealed that aldehyde metabolites may be one of the relevant sources. Mice deficient in Fancd2 and Aldh2 (encodes an enzyme that detoxifies acetaldehyde to acetate) are embryonic-lethal, and exposure of newborn animals to ethanol (an intermediate precursor of acetaldehyde) results in bone marrow failure and severe anemia, a hallmark of FA (Langevin et al. 2011). Synthetic lethality between the FA pathway and formaldehyde catabolism was also shown in DT40 cells, emphasizing the importance of the coordinated activity of aldehyde detoxification and the FA pathway for cellular survival (Rosado et al. 2011). Increasing aldehyde detoxification may therefore alleviate some of the symptoms of FA patients. Nevertheless, the absence of HR (e.g., Xrcc2−/− and Xrcc3−/−) or TLS (e.g., Rev1−/− and Rev3−/−) factors does not lead to hypersensitivity to formaldehyde in DT40 cells (Rosado et al. 2011). This suggests that aldehyde metabolites may cause cross-link lesions other than DNA ICLs, such as protein–DNA cross-links.
Concluding remarks and future directions
Since the discovery of FANCD2-Ub as a surrogate marker for FA pathway activation (Garcia-Higuera et al. 2001), the FA field has witnessed several conceptual and technical advances. New players in the FA pathway, such as FAN1, SLX4, and FAAP20, have further elucidated the regulatory mechanisms of nucleolytic incision and TLS. Ubiquitin modifications in the FA and TLS pathways are recognized by a specialized UBZ4 UBM, and signaling is terminated by the concerted action of the USP1–UAF1 DUB enzyme complex. SUMO signaling is emerging as a new regulatory mechanism that fine-tunes the ubiquitin-mediated FA signaling pathway. Structural determination of the ID complex and new technologies, such as the Xenopus cell-free replication system and the TR-GFP HR assay, have further accelerated discovery.
Despite this progress, several questions regarding the FA pathway remain unresolved. First, the biochemical role of each structure-specific nuclease in the pathway is unclear. It will be important to determine which endonuclease initiates the ICL nucleolytic incision and whether each enzyme has a specialized role in the processing of different classes of ICL. Second, the sequence and orchestration of the nucleolytic incision, trimming, and TLS steps are unknown. The Xenopus system has already proven particularly valuable in determining the order of these repair events. Third, the relative importance of post-translational modifications of FANCD2 and FANCI, such as phosphorylation, ubiquitination, and sumoylation, has not been resolved. Monoubiquitinated FANCD2 and FANCI may bind to additional (unknown) partners with UBZ4 domains. Fourth, additional (unknown) proteins may be components of the FA core complex, the ID complex, and the HR complex (i.e., BRCA1, BRCA2, FANCO, and FANCJ complex), and these protein components may correspond to new FANC genes. Fifth, the relative importance of monoubiquitination and deubiquitination in controlling DNA repair structures during S phase and following DNA damage remains unresolved. Finally, small molecule inhibitors, or activators, of the FA pathway may find clinical utility in the treatment of cancer or of the HR deficiency of FA patients, respectively.
So far, comprehensive studies of the FA pathway have revealed a complex interaction of nucleolytic incision, TLS, and HR repair steps initiated from a ubiquitin signaling pathway. FANCD2-Ub is a requisite gateway to the ICL repair process, connecting upstream signaling with downstream enzymatic repair steps. The biochemical and genetic analyses of the pathway have also provided a rationale for platinum-based chemotherapies in cancer treatment. Overall, a better understanding of the FA pathway and its regulation of DNA repair will allow improvement in therapy for both FA and non-FA cancer patients.
Acknowledgments
We thank Kalindi Parmar, George-Lucian Moldovan, Jenny Xie, and Grace Hsieh for critical reading of the manuscript. H.K. is a recipient of the Leukemia and Lymphoma Society Career Development Fellowship. Research in the D'Andrea laboratory related to this work is supported by NIH grants R01DK43889, R01HL52725, and 2P01HL048546.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.195248.112.
References
- Acharya N, Johnson RE, Prakash S, Prakash L 2006. Complex formation with Rev1 enhances the proficiency of Saccharomyces cerevisiae DNA polymerase ζ for mismatch extension and for extension opposite from DNA lesions. Mol Cell Biol 26: 9555–9563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, Martinez-Perez E, Boulton SJ, La Volpe A 2010. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell 39: 25–35 [DOI] [PubMed] [Google Scholar]
- Ali AM, Pradhan A, Singh TR, Du C, Li J, Wahengbam K, Grassman E, Auerbach AD, Pang Q, Meetei AR 2012. FAAP20: A novel ubiquitin-binding FA nuclear core complex protein required for functional integrity of the FA–BRCA DNA repair pathway. Blood 119: 3285–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpi AF, Pace PE, Babu MM, Patel KJ 2008. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol Cell 32: 767–777 [DOI] [PubMed] [Google Scholar]
- Andersen SL, Bergstralh DT, Kohl KP, LaRocque JR, Moore CB, Sekelsky J 2009. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol Cell 35: 128–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O'Neil NJ, Petalcorin MIR, Martin JS, Collis SJ, Cantor SB, Auclair M, et al. 2008. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell 135: 261–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S, Jentsch S 2009. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458: 461–467 [DOI] [PubMed] [Google Scholar]
- Bhagwat N, Olsen AL, Wang AT, Hanada K, Stuckert P, Kanaar R, D'Andrea A, Niedernhofer LJ, McHugh PJ 2009. XPF–ERCC1 participates in the Fanconi anemia pathway of cross-link repair. Mol Cell Biol 29: 6427–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. 2005. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310: 1821–1824 [DOI] [PubMed] [Google Scholar]
- Bish RA, Myers MP 2007. Werner helicase-interacting protein 1 binds polyubiquitin via its zinc finger domain. J Biol Chem 282: 23184–23193 [DOI] [PubMed] [Google Scholar]
- Bomar MG, Pai M-T, Tzeng S-R, Li SS-C, Zhou P 2007. Structure of the ubiquitin-binding zinc finger domain of human DNA Y-polymerase η. EMBO Rep 8: 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, et al. 2010. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol 17: 688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Yu X, Egelman EH, Mazin AV 2007. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev 21: 3085–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Nussenzweig A 2010. Dangerous liaisons: Fanconi anemia and toxic nonhomologous end joining in DNA crosslink repair. Mol Cell 39: 164–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen H-T, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callèn E, Kozak ML, Kim JM, Wong N, López-Contreras AJ, Ludwig T, Baer R, Faryabi RB, Malhowski A, et al. 2012. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell 46: 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Xu X, Bunting SF, Liu J, Wang R-H, Cao LL, Wu JJ, Peng T-N, Chen J, Nussenzweig A, et al. 2009. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol Cell 35: 534–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DJ, Cimprich KA 2009. DNA damage tolerance: When it's OK to make mistakes. Nat Chem Biol 5: 82–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Dexheimer TS, Ai Y, Liang Q, Villamil MA, Inglese J, Maloney DJ, Jadhav A, Simeonov A, Zhuang Z 2011. Selective and cell-active inhibitors of the USP1/UAF1 deubiquitinase complex reverse cisplatin resistance in Non-small cell lung cancer cells. Chem Biol 18: 1390–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu RK, Brun J, Ramaekers C, Theys J, Weng L, Lambin P, Gray DA, Wouters BG 2006. Lysine 63-polyubiquitination guards against translesion synthesis-induced mutations. PLoS Genet 2: e116 doi: 10.1371/journal.pgen.0020116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani EH, Joenje H, McDonald N, de Winter JP, et al. 2007. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol Cell 25: 331–343 [DOI] [PubMed] [Google Scholar]
- Ciccia A, McDonald N, West SC 2008. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem 77: 259–287 [DOI] [PubMed] [Google Scholar]
- Cohn MA, D'Andrea AD 2008. Chromatin recruitment of DNA repair proteins: Lessons from the Fanconi anemia and double-strand break repair pathways. Mol Cell 32: 306–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, D'Andrea AD 2007. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell 28: 786–797 [DOI] [PubMed] [Google Scholar]
- Cohn MA, Kee Y, Haas W, Gygi SP, D'Andrea AD 2009. UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J Biol Chem 284: 5343–5351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole AR, Lewis LPC, Walden H 2010. The structure of the catalytic subunit FANCL of the Fanconi anemia core complex. Nat Struct Mol Biol 17: 294–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collis SJ, Ciccia A, Deans AJ, Horejsi Z, Martin JS, Maslen SL, Skehel JM, Elledge SJ, West SC, Boulton SJ 2008. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol Cell 32: 313–324 [DOI] [PubMed] [Google Scholar]
- Cotto-Rios XM, Jones MJK, Busino L, Pagano M, Huang TT 2011. APC/CCdh1-dependent proteolysis of USP1 regulates the response to UV-mediated DNA damage. J Cell Biol 194: 177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon S, Gaillard P-HL, Chahwan C, McDonald WH, Yates JR, Russell P 2004. Slx1–Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in fission yeast. Mol Biol Cell 15: 71–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetto N, Bienko M, Hibbert RG, Perica T, Ambrogio C, Kensche T, Hofmann K, Sixma TK, Dikic I 2008. Human Wrnip1 is localized in replication factories in a ubiquitin-binding zinc finger-dependent manner. J Biol Chem 283: 35173–35185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossan GP, Patel KJ 2012. The Fanconi anaemia pathway orchestrates incisions at sites of crosslinked DNA. J Pathol 226: 326–337 [DOI] [PubMed] [Google Scholar]
- Crossan GP, van der Weyden L, Rosado IV, Langevin F, Gaillard P-HL, McIntyre RE, Gallagher F, Kettunen MI, Lewis DY, Brindle K, et al. 2011. Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat Genet 43: 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski KE, Howlett NG 2011. FANCP/SLX4: A Swiss army knife of DNA interstrand crosslink repair. Cell Cycle 10: 1757–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea AD 2010. Susceptibility pathways in Fanconi's anemia and Breast cancer. N Engl J Med 362: 1909–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD 2008. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol Cell 29: 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans AJ, West SC 2011. DNA interstrand crosslink repair and cancer. Nat Rev Cancer 11: 467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva IU, McHugh PJ, Clingen PH, Hartley JA 2000. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol 20: 7980–7990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I, Wakatsuki S, Walters KJ 2009. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol 10: 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doles J, Oliver TG, Cameron ER, Hsu G, Jacks T, Walker GC, Hemann MT 2010. Suppression of Rev3, the catalytic subunit of Polζ, sensitizes drug-resistant lung tumors to chemotherapy. Proc Natl Acad Sci 107: 20786–20791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds CE, Simpson LJ, Sale JE 2008. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol Cell 30: 519–529 [DOI] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong M-Q, Ruse C, Yates JR III, Russell P, et al. 2009. Human SLX4 Is a holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138: 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher LA, Bessho M, Bessho T 2008. Processing of a psoralen DNA interstrand cross-link by XPF–ERCC1 complex in vitro. J Biol Chem 283: 1275–1281 [DOI] [PubMed] [Google Scholar]
- Flott S, Alabert C, Toh GW, Toth R, Sugawara N, Campbell DG, Haber JE, Pasero P, Rouse J 2007. Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol Cell Biol 27: 6433–6445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CA, Masson J-Y, Griffin CS, O'Regan P, West SC, Thacker J 2002. Role of Mammalian RAD51L2 (RAD51C) in recombination and genetic stability. J Biol Chem 277: 19322–19330 [DOI] [PubMed] [Google Scholar]
- Fricke WM, Brill SJ 2003. Slx1–Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1–Top3. Genes Dev 17: 1768–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Lehmann AR, Fuchs RP 2005. Trading places: How do DNA polymerases switch during translesion DNA synthesis? Mol Cell 18: 499–505 [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell 7: 249–262 [DOI] [PubMed] [Google Scholar]
- Gareau JR, Lima CD 2010. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 11: 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner E, Smogorzewska A 2011. Ubiquitylation and the Fanconi anemia pathway. FEBS Lett 585: 2853–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng L, Huntoon CJ, Karnitz LM 2010. RAD18-mediated ubiquitination of PCNA activates the Fanconi anemia DNA repair network. J Cell Biol 191: 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC 2003. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J 22: 6621–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Sonoda E, Tang TS, Parker JL, Bielen AB, Takeda S, Ulrich HD, Friedberg EC 2006a. REV1 protein interacts with PCNA: Significance of the REV1 BRCT domain in vitro and in vivo. Mol Cell 23: 265–271 [DOI] [PubMed] [Google Scholar]
- Guo C, Tang TS, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC 2006b. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol Cell Biol 26: 8892–8900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Tang T-S, Bienko M, Dikic I, Friedberg EC 2008. Requirements for the interaction of mouse Pol κ with ubiquitin and its biological significance. J Biol Chem 283: 4658–4664 [DOI] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R 2006. The structure-specific endonuclease Mus81–Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J 25: 4921–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Hashimoto H, Murakumo Y, Kobayashi S, Kogame T, Unzai S, Akashi S, Takeda S, Shimizu T, Sato M 2010. Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase ζ and REV1. J Biol Chem 285: 12299–12307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Prakash S, Prakash L 2002. Yeast Rev1 protein is a G template-specific DNA polymerase. J Biol Chem 277: 15546–15551 [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Chaudhuri AR, Lopes M, Costanzo V 2010. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol 17: 1305–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker C-M, Rabiller M, Haglund K, Bayer P, Dikic I 2006. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem 281: 16117–16127 [DOI] [PubMed] [Google Scholar]
- Hendel A, Krijger PHL, Diamant N, Goren Z, Langerak P, Kim J, Reissner T, Lee KY, Geacintov NE, Carell T, et al. 2011. PCNA ubiquitination is important, but not essential for translesion DNA synthesis in mammalian cells. PLoS Genet 7: e1002262 doi: 10.1371/journal.pgen.1002262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W-D, Ehmsen KT, Liu J 2010. Regulation of homologous recombination in eukaryotes. Annu Rev Genet 44: 113–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks JK, Chute CL, Paulsen MT, Ragland RL, Howlett NG, Gueranger Q, Glover TW, Canman CE 2010. Differential roles for DNA polymerases η, ζ, and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol Cell Biol 30: 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz JM, Nham PB, Salazar EP, Thompson LH 2006. The Fanconi anemia pathway limits the severity of mutagenesis. DNA Repair (Amst) 5: 875–884 [DOI] [PubMed] [Google Scholar]
- Hirano S, Yamamoto K, Ishiai M, Yamazoe M, Seki M, Matsushita N, Ohzeki M, Yamashita YM, Arakawa H, Buerstedde J-M, et al. 2005. Functional relationships of FANCC to homologous recombination, translesion synthesis, and BLM. EMBO J 24: 418–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TV, Schärer OD 2010. Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ Mol Mutagen 51: 552–566 [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Hofmann K 2009. Ubiquitin-binding domains and their role in the DNA damage response. DNA Repair (Amst) 8: 544–556 [DOI] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, de Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, et al. 2002. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297: 606–609 [DOI] [PubMed] [Google Scholar]
- Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, et al. 2007. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev 21: 3073–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, D'Andrea AD 2010. A new nuclease member of the FAN club. Nat Struct Mol Biol 17: 926–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TT, Nijman SMB, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D'Andrea AD 2006. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol 8: 341–347 [DOI] [PubMed] [Google Scholar]
- Huang J, Huen MSY, Kim H, Leung CCY, Glover JNM, Yu X, Chen J 2009. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol 11: 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Kennedy R, Ali AM, Moreau LA, Meetei AR, D'Andrea AD, Chen CC 2011. Human MutS and FANCM complexes function as redundant DNA damage sensors in the Fanconi Anemia pathway. DNA Repair (Amst) 10: 1203–1212 [DOI] [PubMed] [Google Scholar]
- Ishiai M, Kitao H, Smogorzewska A, Tomida J, Kinomura A, Uchida E, Saberi A, Kinoshita E, Kinoshita-Kikuta E, Koike T, et al. 2008. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol 15: 1138–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont C, Taniguchi T 2007. Proteasome function is required for DNA damage response and Fanconi anemia pathway activation. Cancer Res 67: 7395–7405 [DOI] [PubMed] [Google Scholar]
- Joenje H, Patel KJ 2001. The emerging genetic and molecular basis of Fanconi anaemia. Nat Rev Genet 2: 446–457 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L 1999. Efficient bypass of a thymine–thymine dimer by yeast DNA polymerase, Pol η. Science 283: 1001–1004 [DOI] [PubMed] [Google Scholar]
- Jones MJK, Colnaghi L, Huang TT 2012. Dysregulation of DNA polymerase κ recruitment to replication forks results in genomic instability. EMBO J 31: 908–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo W, Xu G, Persky NS, Smogorzewska A, Rudge DG, Buzovetsky O, Elledge SJ, Pavletich NP 2011. Structure of the FANCI–FANCD2 complex: Insights into the Fanconi anemia DNA repair pathway. Science 333: 312–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR 2004. Interaction of human DNA polymerase η with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol Cell 14: 491–500 [DOI] [PubMed] [Google Scholar]
- Kee Y, D'Andrea AD 2010. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev 24: 1680–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, Kim JM, D'Andrea AD 2009. Regulated degradation of FANCM in the Fanconi anemia pathway during mitosis. Genes Dev 23: 555–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, Huang M, Chang S, Moreau L, Park E, Smith PG, D'Andrea AD 2012. Inhibition of the Nedd8 system sensitizes cells to DNA interstrand crosslinking agents. Mol Cancer Res 10: 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy RD, D'Andrea AD 2005. The Fanconi anemia/BRCA pathway: New faces in the crowd. Genes Dev 19: 2925–2940 [DOI] [PubMed] [Google Scholar]
- Kim JM, Parmar K, Huang M, Weinstock DM, Ruit CA, Kutok JL, D'Andrea AD 2009. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell 16: 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A 2011. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet 43: 142–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Yang K, Dejsuphong D, D'Andrea AD 2012. Regulation of Rev1 by the Fanconi anemia core complex. Nat Struct Mol Biol 19: 164–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC 2009. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326: 1698–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz K, Schöpf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannav ÛE, Sartori AA, Hengartner MO, Jiricny J 2010. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell 142: 77–88 [DOI] [PubMed] [Google Scholar]
- Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ 2011. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 475: 53–58 [DOI] [PubMed] [Google Scholar]
- Lawrence CW 2002. Cellular roles of DNA polymerase ζ and Rev1 protein. DNA Repair (Amst) 1: 425–435 [DOI] [PubMed] [Google Scholar]
- Lee W, St.Onge RP, Proctor M, Flaherty P, Jordan MI, Arkin AP, Davis RW, Nislow C, Giaever G 2005. Genome-wide requirements for resistance to functionally distinct DNA-damaging agents. PLoS Genet 1: e24 doi: 10.1371/journal.pgen.0010024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Yang K, Cohn MA, Sikdar N, D'Andrea A, Myung K 2010. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through its interactions with PCNA and USP1. J Biol Chem 285: 10362–10369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM 2007. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 6: 891–899 [DOI] [PubMed] [Google Scholar]
- Leung JWC, Wang Y, Fong KW, Huen MSY, Li L, Chen J 2012. Fanconi anemia (FA) binding protein FAAP20 stabilizes FA complementation group A (FANCA) and participates in interstrand cross-link repair. Proc Natl Acad Sci 109: 4491–4496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Dong J, Pan X, Oum J-H, Boeke JD, Lee SE 2008. Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates. Mol Cell 30: 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, Andreassen PR, Cantor SB 2005. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell 8: 255–265 [DOI] [PubMed] [Google Scholar]
- Liu Y, Tarsounas M, O'Regan P, West SC 2007. Role of RAD51C and XRCC3 in genetic recombination and DNA repair. J Biol Chem 282: 1973–1979 [DOI] [PubMed] [Google Scholar]
- Liu T, Ghosal G, Yuan J, Chen J, Huang J 2010. FAN1 acts with FANCI–FANCD2 to promote DNA interstrand cross-link repair. Science 329: 693–696 [DOI] [PubMed] [Google Scholar]
- Long DT, Räschle M, Joukov V, Walter JC 2011. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science 333: 84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyndaker AM, Goldfarb T, Alani E 2008. Mutants defective in Rad1–Rad10–Slx4 exhibit a unique pattern of viability during mating-type switching in Saccharomyces cerevisiae. Genetics 179: 1807–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay C, Dèclais AC, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, et al. 2010. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell 142: 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F 1999. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399: 700–704 [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- McHugh PJ, Sones WR, Hartley JA 2000. Repair of intermediate structures produced at DNA interstrand cross-links in Saccharomyces cerevisiae. Mol Cell Biol 20: 3425–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minko IG, Harbut MB, Kozekov ID, Kozekova A, Jakobs PM, Olson SB, Moses RE, Harris TM, Rizzo CJ, Lloyd RS 2008. Role for DNA polymerase κ in the processing of N2-N2-guanine interstrand cross-links. J Biol Chem 283: 17075–17082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minty A, Dumont X, Kaghad M, Caput D 2000. Covalent modification of p73α by SUMO-1. J Biol Chem 275: 36316–36323 [DOI] [PubMed] [Google Scholar]
- Mirchandani KD, McCaffrey RM, D'Andrea AD 2008. The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA Repair (Amst) 7: 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, D'Andrea AD 2009. How the Fanconi anemia pathway guards the genome. Annu Rev Genet 43: 223–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S 2007. PCNA, the maestro of the replication fork. Cell 129: 665–679 [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Madhavan MV, Mirchandani KD, McCaffrey RM, Vinciguerra P, D'Andrea AD 2010. DNA polymerase POLN participates in cross-link repair and homologous recombination. Mol Cell Biol 30: 1088–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Dejsuphong D, Petalcorin MIR, Hofmann K, Takeda S, Boulton SJ, D'Andrea AD 2012. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol Cell 45: 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Pierce AJ, Jasin M 2001. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell 7: 263–272 [DOI] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ 2001. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157: 103–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz IM, Hain K, Declais A-C, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, et al. 2009. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell 35: 116–127 [DOI] [PubMed] [Google Scholar]
- Murai J, Yang K, Dejsuphong D, Hirota K, Takeda S, D'Andrea AD 2011. The USP1/UAF1 complex promotes double-strand break repair through homologous recombination. Mol Cell Biol 31: 2462–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Yang Y-G, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, Wang Z-Q, Jasin M 2005. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci 102: 1110–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Cavallo F, Perrouault L, Giovannangeli C, Moynahan ME, Barchi M, Brunet E, Jasin M 2011. Homology-directed Fanconi anemia pathway cross-link repair is dependent on DNA replication. Nat Struct Mol Biol 18: 500–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC 1996. Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382: 729–731 [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NGJ, Beverloo HB, Hoeijmakers JHJ, et al. 2004. The structure-specific endonuclease Ercc1–Xpf Is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol 24: 5776–5787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ 2004. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell 15: 607–620 [DOI] [PubMed] [Google Scholar]
- Nijman SMB, Huang TT, Dirac AMG, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, Bernards R 2005. The Deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell 17: 331–339 [DOI] [PubMed] [Google Scholar]
- Novatchkova M, Bachmair A, Eisenhaber B, Eisenhaber F 2005. Proteins with two SUMO-like domains in chromatin-associated complexes: The RENi (Rad60–Esc2–NIP45) family. BMC Bioinformatics 6: 22 doi: 10.1186/1471-2105-6-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestergaard VH, Langevin F, Kuiken HJ, Pace P, Niedzwiedz W, Simpson LJ, Ohzeki M, Takata M, Sale JE, Patel KJ 2007. Deubiquitination of FANCD2 Is required for DNA crosslink repair. Mol Cell 28: 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogi T, Kannouche P, Lehmann AR 2005. Localisation of human Y-family DNA polymerase κ: Relationship to PCNA foci. J Cell Sci 118: 129–136 [DOI] [PubMed] [Google Scholar]
- Okada T, Sonoda E, Yoshimura M, Kawano Y, Saya H, Kohzaki M, Takeda S 2005. Multiple roles of vertebrate REV genes in DNA repair and recombination. Mol Cell Biol 25: 6103–6111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orelli B, McClendon TB, Tsodikov OV, Ellenberger T, Niedernhofer LJ, Scharer OD 2010. The XPA-binding domain of ERCC1 is required for nucleotide excision repair but not other DNA repair pathways. J Biol Chem 285: 3705–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski RZ, Kuhn DJ 2008. Proteasome inhibitors in cancer therapy: Lessons from the first decade. Clin Cancer Res 14: 1649–1657 [DOI] [PubMed] [Google Scholar]
- Otsuka C, Kunitomi N, Iwai S, Loakes D, Negishi K 2005. Roles of the polymerase and BRCT domains of Rev1 protein in translesion DNA synthesis in yeast in vivo. Mutat Res 578: 79–87 [DOI] [PubMed] [Google Scholar]
- Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ 2010. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science 329: 219–223 [DOI] [PubMed] [Google Scholar]
- Palle K, Vaziri C 2011. Rad18 E3 ubiquitin ligase activity mediates Fanconi anemia pathway activation and cell survival following DNA Topoisomerase 1 inhibition. Cell Cycle 10: 1625–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulo D, Guillouf C, Mohrenweiser H, Moustacchi E 1990a. Hypomutability in Fanconi anemia cells is associated with increased deletion frequency at the HPRT locus. Proc Natl Acad Sci 87: 8383–8387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulo D, Porfirio B, Moustacchi E 1990b. Mutagenic response of Fanconi's anemia cells from a defined complementation group after treatment with photoactivated bifunctional psoralens. Cancer Res 50: 3289–3294 [PubMed] [Google Scholar]
- Park HK, Wang H, Zhang J, Datta S, Fei P 2010. Convergence of Rad6/Rad18 and Fanconi anemia tumor suppressor pathways upon DNA damage. PLoS ONE 5: e13313 doi: 10.1371/journal.pone.0013313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DJ, Lesueur F, Nguyen-Dumont T, Pertesi M, Odefrey F, Hammet F, Neuhausen SL, John EM, Andrulis IL, Terry MB, et al. 2012. Rare mutations in XRCC2 increase the risk of breast cancer. Am J Hum Genet 90: 734–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JL, Bielen AB, Dikic I, Ulrich HD 2007. Contributions of ubiquitin- and PCNA-binding domains to the activity of Polymerase η in Saccharomyces cerevisiae. Nucleic Acids Res 35: 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar K, D'Andrea AD, Niedernhofer LJ 2009. Mouse models of Fanconi anemia. Mutat Res 668: 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433 [DOI] [PubMed] [Google Scholar]
- Plosky BS, Vidal AE, de Henestrosa ARF, McLenigan MP, McDonald JP, Mead S, Woodgate R 2006. Controlling the subcellular localization of DNA polymerases ι and η via interactions with ubiquitin. EMBO J 25: 2847–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC 2008. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134: 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado IV, Langevin F, Crossan GP, Takata M, Patel KJ 2011. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nat Struct Mol Biol 18: 1432–1434 [DOI] [PubMed] [Google Scholar]
- Ross AL, Simpson LJ, Sale JE 2005. Vertebrate DNA damage tolerance requires the C-terminus but not BRCT or transferase domains of REV1. Nucleic Acids Res 33: 1280–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale JE, Lehmann AR, Woodgate R 2012. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol 13: 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Davies AA, Ulrich HD, McHugh PJ 2006. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase ζ. EMBO J 25: 1285–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab RA, Blackford AN, Niedzwiedz W 2010. ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J 29: 806–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Wood RD 2008. DNA polymerase κ (POLQ) can extend from mismatches and from bases opposite a (6-4) photoproduct. DNA Repair (Amst) 7: 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamseldin HE, Elfaki M, Alkuraya FS 2012. Exome sequencing reveals a novel Fanconi group defined by XRCC2 mutation. J Med Genet 49: 184–186 [DOI] [PubMed] [Google Scholar]
- Shima N, Munroe RJ, Schimenti JC 2004. The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol Cell Biol 24: 10381–10389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson LJ, Sale JE 2003. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J 22: 1654–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Saro D, Ali AM, Zheng X-F, Du C-h, Killen MW, Sachpatzidis A, Wahengbam K, Pierce AJ, Xiong Y, et al. 2010. MHF1–MHF2, a Histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell 37: 879–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, Clark AB, Kunkel TA, Harper JW, Colaiácovo MP, et al. 2010. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell 39: 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommers JA, Rawtani N, Gupta R, Bugreev DV, Mazin AV, Cantor SB, Brosh RM 2009. FANCJ uses its motor ATPase to destabilize protein–DNA complexes, unwind triplexes, and inhibit RAD51 strand exchange. J Biol Chem 284: 7505–7517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y 2004. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci 101: 14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song IY, Palle K, Gurkar A, Tateishi S, Kupfer GM, Vaziri C 2010. Rad18-mediated translesion synthesis of bulky DNA adducts is coupled to activation of the Fanconi anemia DNA repair pathway. J Biol Chem 285: 31525–31536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E, Okada T, Zhao GY, Tateishi S, Araki K, Yamaizumi M, Yagi T, Verkaik NS, van Gent DC, Takata M, et al. 2003. Multiple roles of Rev3, the catalytic subunit of polζ in maintaining genome stability in vertebrates. EMBO J 22: 3188–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoepker C, Hain K, Schuster B, Hilhorst-Hofstee Y, Rooimans MA, Steltenpool J, Oostra AB, Eirich K, Korthof ET, Nieuwint AWM, et al. 2011. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet 43: 138–141 [DOI] [PubMed] [Google Scholar]
- Suhasini AN, Rawtani NA, Wu Y, Sommers JA, Sharma S, Mosedale G, North PS, Cantor SB, Hickson ID, Brosh RM 2011. Interaction between the helicases genetically linked to Fanconi anemia group J and Bloom's syndrome. EMBO J 30: 692–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen JM, Harper JW 2010. GEN1/Yen1 and the SLX4 complex: Solutions to the problem of Holliday junction resolution. Genes Dev 24: 521–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O'Connell BC, Gygi SP, Elledge SJ, Harper JW 2009. Mammalian BTBD12/SLX4 assembles a holliday junction resolvase and is required for DNA repair. Cell 138: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy M-C, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT 2008. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol 10: 538–546 [DOI] [PubMed] [Google Scholar]
- Ulrich HD 2009. Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair (Amst) 8: 461–469 [DOI] [PubMed] [Google Scholar]
- Unk I, Hajdu I, Fatyol K, Szakal B, Blastyak A, Bermudez V, Hurwitz J, Prakash L, Prakash S, Haracska L 2006. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc Natl Acad Sci 103: 18107–18112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unk I, Hajdu I, Fatyol K, Hurwitz J, Yoon J-H, Prakash L, Prakash S, Haracska L 2008. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc Natl Acad Sci 105: 3768–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, Neveling K, Endt D, Kesterton I, Autore F, et al. 2010. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet 42: 406–409 [DOI] [PubMed] [Google Scholar]
- Wang W 2007. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet 8: 735–748 [DOI] [PubMed] [Google Scholar]
- Wang AT, Sengerova B, Cattell E, Inagawa T, Hartley JM, Kiakos K, Burgess-Brown NA, Swift LP, Enzlin JH, Schofield CJ, et al. 2011. Human SNM1A and XPF–ERCC1 collaborate to initiate DNA interstrand cross-link repair. Genes Dev 25: 1859–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington MT, Johnson RE, Prakash S, Prakash L 1999. Fidelity and processivity of Saccharomyces cerevisiae DNA Polymerase η. J Biol Chem 274: 36835–36838 [DOI] [PubMed] [Google Scholar]
- Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC 2009. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev 73: 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SA, Longerich S, Sung P, Vaziri C, Kupfer GM 2011a. The E3 ubiquitin ligase RAD18 regulates ubiquitylation and chromatin loading of FANCD2 and FANCI. Blood 117: 5078–5087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SA, Wilson JB, Clark AP, Mitson-Salazar A, Tomashevski A, Ananth S, Glazer PM, Semmes OJ, Bale AE, Jones NJ, et al. 2011b. Functional and physical interaction between the mismatch repair and FA-BRCA pathways. Hum Mol Genet 20: 4395–4410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltrout ME, Walker GC 2011. The DNA polymerase activity of Saccharomyces cerevisiae Rev1 is biologically significant. Genetics 187: 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben JP, Reshmi SC, Gollin SM, Wood RD 2006. Loss of DNA polymerase ζ causes chromosomal instability in mammalian cells. Cancer Res 66: 134–142 [DOI] [PubMed] [Google Scholar]
- Wu HI, Brown JA, Dorie MJ, Lazzeroni L, Brown JM 2004. Genome-wide identification of genes conferring resistance to the anticancer agents cisplatin, oxaliplatin, and mitomycin C. Cancer Res 64: 3940–3948 [DOI] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM 2006. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell 22: 719–729 [DOI] [PubMed] [Google Scholar]
- Xie K, Doles J, Hemann MT, Walker GC 2010. Error-prone translesion synthesis mediates acquired chemoresistance. Proc Natl Acad Sci 107: 20792–20797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Zhang P, Liu L, Lee MYWT 2001. A novel PCNA-binding motif identified by the panning of a random peptide display library. Biochemistry 40: 4512–4520 [DOI] [PubMed] [Google Scholar]
- Yamamoto KN, Kobayashi S, Tsuda M, Kurumizaka H, Takata M, Kono K, Jiricny J, Takeda S, Hirota K 2011. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc Natl Acad Sci 108: 6492–6496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Delannoy M, Ling C, Daee D, Osman F, Muniandy PA, Shen X, Oostra AB, Du H, Steltenpool J, et al. 2010. A Histone-fold complex and FANCM Form a conserved DNA-remodeling complex to maintain genome stability. Mol Cell 37: 865–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Moldovan GL, D'Andrea AD 2010. RAD18-dependent recruitment of SNM1A to DNA repair complexes by a ubiquitin-binding zinc finger. J Biol Chem 285: 19085–19091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Moldovan GL, Vinciguerra P, Murai J, Takeda S, D'Andrea AD 2011. Regulation of the Fanconi anemia pathway by a SUMO-like delivery network. Genes Dev 25: 1847–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz O, Majumder S, Kramer B, Sekelsky JJ 2002. Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol Cell 10: 1503–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikiyo K, Kratz K, Hirota K, Nishihara K, Takata M, Kurumizaka H, Horimoto S, Takeda S, Jiricny J 2010. KIAA1018/FAN1 nuclease protects cells against genomic instability induced by interstrand cross-linking agents. Proc Natl Acad Sci 107: 21553–21557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Liu X, Li L, Legerski R 2007. Double-strand breaks induce homologous recombinational repair of interstrand cross-links via cooperation of MSH2, ERCC1–XPF, REV3, and the Fanconi anemia pathway. DNA Repair (Amst) 6: 1670–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, Yu X 2009. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol 19: 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietlow L, Smith LA, Bessho M, Bessho T 2009. Evidence for the involvement of human DNA polymerase N in the repair of DNA interstrand cross-links. Biochemistry 48: 11817–11824 [DOI] [PMC free article] [PubMed] [Google Scholar]