The Ras/mitogen-activated protein kinase (MAPK) pathway plays a critical role in transducing mitogenic signals from receptor tyrosine kinases. Loss-of-function mutations in one feedback regulator of Ras/MAPK signaling, SPRED1, causes Legius syndrome, an autosomal dominant human disorder that resembles Neurofibromatosis type 1 (NF1). Spred1 functions as a negative regulator of the Ras/MAPK pathway; however, the underlying molecular mechanism is poorly understood. In this study, McCormick and colleagues demonstrate that Spred1 binding to neurofibromin induces the plasma membrane localization of NF1, which subsequently down-regulates Ras-GTP levels through its RasGAP activity. Thus, this study delineates a novel mechanism for the regulation of neurofibromin.
Keywords: Legius syndrome, NF1, signal transduction, Ras/MAPK, Sprouty
Abstract
The Ras/mitogen-activated protein kinase (MAPK) pathway plays a critical role in transducing mitogenic signals from receptor tyrosine kinases. Loss-of-function mutations in one feedback regulator of Ras/MAPK signaling, SPRED1 (Sprouty-related protein with an EVH1 domain), cause Legius syndrome, an autosomal dominant human disorder that resembles Neurofibromatosis-1 (NF1). Spred1 functions as a negative regulator of the Ras/MAPK pathway; however, the underlying molecular mechanism is poorly understood. Here we show that neurofibromin, the NF1 gene product, is a Spred1-interacting protein that is necessary for Spred1's inhibitory function. We show that Spred1 binding induces the plasma membrane localization of NF1, which subsequently down-regulates Ras-GTP levels. This novel mechanism for the regulation of neurofibromin provides a molecular bridge for understanding the overlapping pathophysiology of NF1 and Legius syndrome.
Spred1 (Sprouty-related protein with an EVH1 domain) functions as a negative regulator of growth factor, cytokine, and chemokine-induced ERK activation by specifically inhibiting the Ras/Raf/MEK/ERK pathway (Wakioka et al. 2001; Miyoshi et al. 2004; Nonami et al. 2004). Spred1 is comprised of an N-terminal Ena/Vasp homology 1 (EVH1) domain, a central c-Kit-binding domain (KBD), and a cysteine rich C-terminal Sprouty (SPR) domain. Mammals contain three homologs, Spred 1, Spred 2, and Spred 3, each of which can negatively regulate the Ras/Raf/mitogen-activated protein kinase (MAPK) pathway (Kato et al. 2003; King et al. 2006). However, the precise mechanism by which the Spreds act remains unclear, and it has been reported to function both upstream of and downstream from Ras (Wakioka et al. 2001; King et al. 2005). Overexpression of Spred1 can increase Raf's recruitment to the plasma membrane, where it associates with Ras without stimulating Raf activation (Wakioka et al. 2001). Yet, this mechanism has never been fully refined and fails to explain how Spred1 prevents Raf activation. A separate report suggests that Spred1 may prevent Ras activation, as evidenced by decreased Ras-GTP levels when Spred1 is overexpressed (King et al. 2005).
Heterozygous germline loss-of-function mutations have been identified in SPRED1 in Legius syndrome, a developmental disorder that shares a number of phenotypes with Neurofibromatosis-1 (NF1) (Brems et al. 2007). Both syndromes are characterized as rasopathies, congenital developmental syndromes caused by germline mutations that affect the Ras/MAPK pathway. Legius syndrome has been characterized as a milder form of NF1, with individuals displaying multiple café-au-lait spots, axillary freckling, and macrocephaly, but lacking other common NF1 manifestations such as Lisch nodules, neurofibromas, osseous lesions, or optic pathway gliomas. Initial experiments revealed that Spred1 mutations were loss-of-function mutations, incapable of inhibiting the Ras/MAPK pathway (Brems et al. 2007). SPRED1 mutations account for at least 2% of the pathogenic mutations associated with patients clinically diagnosed with NF1 (Brems et al. 2007; Messiaen et al. 2009; Pasmant et al. 2009; Spurlock et al. 2009; Muram-Zborovski et al. 2010; Denayer et al. 2011; Laycock-van Spyk et al. 2011; Spencer et al. 2011).
NF1, an autosomal dominant, multisystem disorder that affects approximately one in 3500 individuals, was the first disorder found to originate from a component of the Ras/MAPK pathway. The NF1 gene product neurofibromin negatively regulates Ras signaling by functioning as a Ras GTPase-activating protein (RasGAP) to accelerate the hydrolysis of active Ras-GTP to inactive Ras-GDP (Martin et al. 1990; Xu et al. 1990). Loss of neurofibromin leads to hyperactive Ras signaling, as observed by elevated Ras pathway activity in cells (Basu et al. 1992; DeClue et al. 1992; Bollag et al. 1996). Despite the identification of neurofibromin as a RasGAP 20 years ago, the regulation of neurofibromin activity remains poorly understood. Neurofibromin has been reported to be positively and negatively regulated by various phosphorylation events, but the context of this regulation is largely unknown (Feng et al. 2004; Mangoura et al. 2006; Leondaritis et al. 2009). In addition, neurofibromin protein levels are negatively regulated by the ubiquitin proteasome system following growth factor stimulation (Cichowski et al. 2003; McGillicuddy et al. 2009). However, the exact regulatory mechanism that couples receptor growth factor activation to neurofibromin's suppression of Ras activation remains unclear.
Here we report that Spred1 down-regulates the Ras/MAPK pathway through an interaction with the NF1 protein neurofibromin. Importantly, this interaction functions to recruit neurofibromin to the plasma membrane. Furthermore, loss-of-function Spred1 mutants observed in Legius syndrome are either unable to bind neurofibromin or incapable of recruiting it to the membrane. Our data provide evidence for a molecular link between the phenotypically overlapping developmental disorders Legius syndrome and NF1.
Results and Discussion
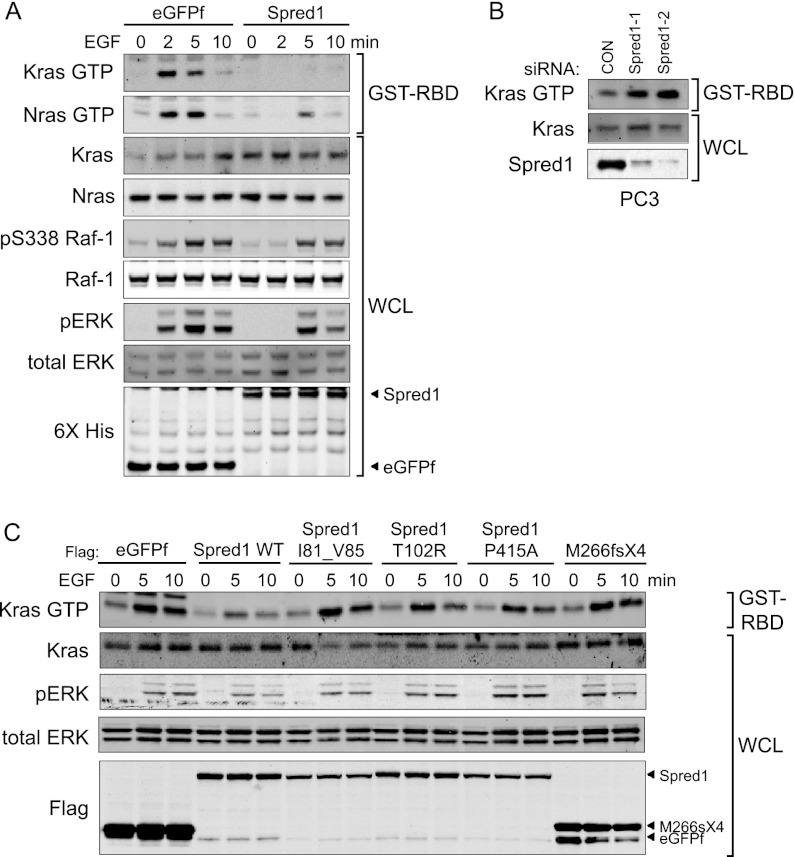
Ectopic expression of Spred1 results in suppression of ERK activation following acute agonist stimulation by various growth factors, including EGF and FGF (Supplemental Fig. S1; Wakioka et al. 2001). Spred1 has been described as functioning both upstream of and downstream from Ras (Wakioka et al. 2001; King et al. 2005). To address these discrepancies, we tested the hypothesis that Spred1 acts upstream of Ras by examining whether Spred1 could affect cellular Ras-GTP levels following growth factor stimulation (Fig. 1A). Inducible expression of Spred1 in T-REx-293 cells significantly reduced Ras-GTP levels following EGF stimulation, suggesting that Spred1 might interfere with Ras regulation. Furthermore, ectopic expression of Spred1 suppressed downstream MAPK signaling, as evidenced by decreases in phospho-Raf and phospho-ERK. To further investigate the hypothesis that Spred1 affects Ras regulation, we depleted Spred1 from a cancer cell line with reportedly high Spred1 protein levels (Fig. 1B; Li et al. 2010). Depletion of Spred1 from the PC3 prostate cell line resulted in elevated levels of Ras-GTP following EGF stimulation. Taken together, these data support the model that Spred1 can function upstream of Ras to negatively regulate its activation, although we cannot exclude the possibility that Spred1 may also independently function downstream from Ras activation as well.
Figure 1.
Spred1 suppresses Ras activity. (A) TET-inducible 6X-HIS eGFPf or Spred1-expressing T-REx-293 cells were stimulated with EGF (10 ng/mL) for the indicated times, and whole-cell lysates were affinity-purified using GST-Raf1 RBD to precipitate active Ras. Immunoblot analysis was performed with the antibodies indicated on the left. (B) PC3 cells were transfected with the indicated siRNA oligos, serum-starved for 12 h, and stimulated with EGF for 2 min. Cell lysates were subjected to GST-Raf1 RBD assays to detect Ras-GTP levels. (C) T-REx-293 cells stably expressing Flag-tagged eGFPf, Spred1, or the indicated Spred1 mutants were stimulated with 10 ng/mL EGF for the indicated times.
We next asked whether SPRED1 loss-of-function mutations found in Legius syndrome (Supplemental Table S1) might similarly affect Ras activity (Fig. 1C). Interestingly, we found that ectopic expression of Spred1-bearing pathogenic missense mutations in the N-terminal EVH1 domain and C-terminal SPR domain are defective in suppressing Ras-GTP levels and inhibiting ERK activation following growth factor treatment. The truncation mutant M266fsX4, which retains the N-terminal EVH1 domain but lacks the SPR and internal Kit-binding domains, was also defective in regulating Ras-GTP and phospho-ERK levels.
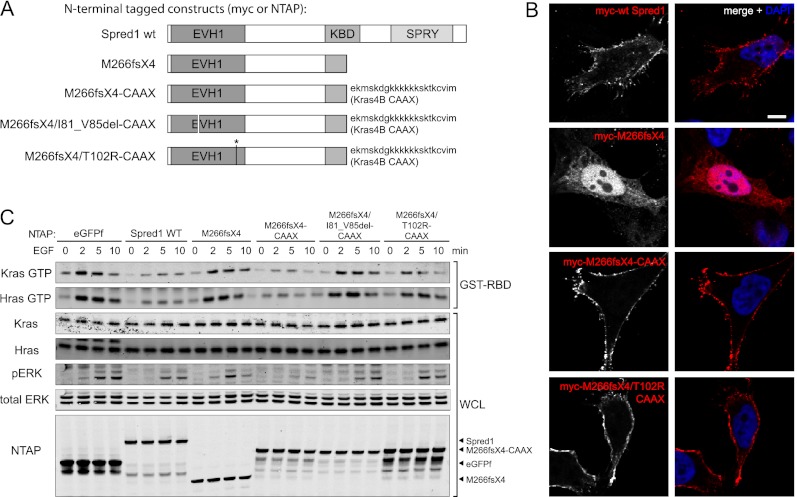
The majority of reported Spred1 mutations produce a premature stop codon, resulting in C-terminally truncated protein products (Brems et al. 2007; Messiaen et al. 2009; Pasmant et al. 2009; Spurlock et al. 2009; Muram-Zborovski et al. 2010; Denayer et al. 2011; Laycock-van Spyk et al. 2011; Spencer et al. 2011). As the SPR domain has previously been reported to target Spred1 to the plasma membrane via interactions with phospholipids and caveolin-1 (Lim et al. 2002; Nonami et al. 2005), we surmised that the truncated mutants were unable to function as a result of mislocalization. To test this hypothesis, we artificially localized the Spred1 C-terminal truncation mutant (M266fsX4) to the membrane by fusing it to the membrane targeting CAAX motif of Kras4B (Fig. 2A). Immunofluorescence microscopy revealed that the mutant Spred1 protein localized primarily to the cytoplasm, whereas the mutant CAAX fusion protein was localized to the plasma membrane (Fig. 2B). The mutant CAAX protein also showed a restored ability to reduce EGF-induced Ras activation and inhibit ERK activation (Fig. 2C), indicating that the inhibitory activity of Spred1 resides at the N terminus. Intriguingly, this also suggests that defective membrane targeting of Spred1 is responsible for loss-of-function of Spred1 in Legius syndrome-associated C-terminal truncation mutants.
Figure 2.
Spred1's inhibitory activity resides in the EVH1 domain. (A) Domain structure of wild-type Spred1 and the Spred1 mutants used in this study. (B) HeLa cells were transfected with myc-Spred1, myc-M266fsX4, myc-M266fsX4-CAAX, or myc-M266fsX4/T102R-CAAX. Cells were serum-starved for 16 h, fixed, and immunostained with antibodies against myc (red) and DNA (DAPI). Bars, 5 μm. (C) T-REx-293 cells stably expressing TET-inducible NTAP-tagged eGFPf, Spred1, or the indicated Spred1 mutants were stimulated with 10 ng/mL EGF for the indicated times. NTAP-Spred1 mutants with a C-terminal CAAX motif rescued suppression of ERK phosphorylation. Introducing known pathogenic missense mutations into NTAP-M266fsX4-CAAX abolished its ability to rescue.
Thus far, ∼12 missense mutations in SPRED1 have been characterized as pathogenic in Legius syndrome (11 point mutations and one four-amino-acid in-frame deletion), five of which are used in this study (Supplemental Table S1; Brems et al. 2007; Messiaen et al. 2009; Pasmant et al. 2009; Spurlock et al. 2009; Denayer et al. 2011; Spencer et al. 2011). Of these 12 missense mutations, nine localize to the EVH1 domain. When we introduced two of these mutations (T102R and I81_V85del) into the truncated CAAX fusion protein, both prevented the ability of the CAAX motif to rescue Spred1 function (Fig. 2C). Together, these results underscore the importance of the EVH1 domain of Spred1 in inhibiting ERK activation, and the importance of the SPR domain in directing the plasma membrane localization of Spred1.
EVH1 domains are protein–protein interaction modules that recognize and bind proline-rich sequences (Gertler et al. 1996). The crystal structure of the Xenopus tropicalis Spred1 EVH1 domain suggests a distinct peptide-binding mechanism compared with other identified EVH1 domains (Harmer et al. 2005). Although an interacting protein for the Spred2 EVH1 domain has recently been reported (NBR1) (Mardakheh et al. 2009), proteins that specifically interact with the Spred1 EVH1 domain have yet to be determined. Since the EVH1 domain appears to be essential for Spred's function, we hypothesized that an unidentified binding partner of the EVH1 domain might be required for Spred1's inhibitory activity. To identify novel Spred1 EVH1-interacting proteins, we performed tandem affinity purification (TAP) (Burckstummer et al. 2006) using the truncation mutant-CAAX fusion protein (Fig. 2A) as bait. Following mass spectrometry analysis, we identified the NF1 gene product neurofibromin as a novel Spred1 EVH1-binding partner (Supplemental Fig. S2). Neurofibromin is a tumor suppressor and a RasGAP (Martin et al. 1990; Xu et al. 1990) that regulates Ras GTPase activity by catalyzing the hydrolysis of active GTP-bound Ras to inactive GDP-bound Ras. Loss of neurofibromin function removes this regulation and leads to uncontrolled cell growth and proliferation. The identification of this RasGAP as a binding partner of Spred1 was consistent with our biochemical results that Spred1 suppresses Ras-GTP levels and was particularly intriguing given the overlapping phenotypes observed in Legius syndrome and NF1.
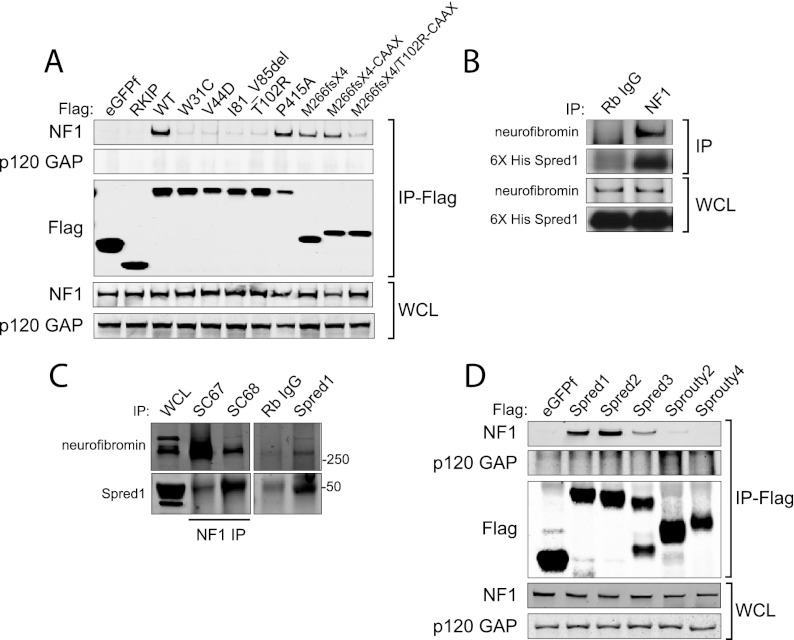
To confirm this interaction, we performed coimmunoprecipitation experiments in mammalian cells. Ectopically expressed wild-type Spred1 coprecipitated with endogenous neurofibromin in human embryonic kidney (HEK) 293T cells but not with p120 GAP, another RasGAP (Fig. 3A). In a reciprocal experiment, immunoprecipitated endogenous neurofibromin could coprecipitate stably expressed full length Spred1 (Fig. 3B). In order to detect endogenous Spred1–NF1 binding, we used postnatal day 0 (P0) mouse brain lysates for endogenous interaction assays, since Spred1 is expressed predominantly in the brain (Bundschu et al. 2006). Endogenous neurofibromin could be immunoprecipitated with endogenous Spred1, and inversely, Spred1 could be immunoprecipitated with neurofibromin, supporting a physiologically relevant interaction between Spred1 and neurofibromin (Fig. 3C). Furthermore, we were able to detect endogenous Spred1–NF1 binding in a high-expressing Spred1 cell line, DLD-1, a colon cancer cell line (Supplemental Fig. S3).
Figure 3.
Neurofibromin is a novel Spred1-binding partner. (A) Flag-tagged eGFPf, RKIP, wild-type Spred1, or the indicated Spred1 mutants were transfected into HEK-293T cells, serum-starved for 16 h, and lysed 24 h post-transfection. Anti-Flag immunoprecipitates were blotted for endogenous neurofibromin and p120 GAP. (B) Endogenous neurofibromin was immunoprecipitated from T-REx-293 cell lysates stably expressing TET-inducible 6X-HIS Spred1. (C) Endogenous neurofibromin coimmunoprecipitated with endogenous Spred1 from P0 whole mouse brain lysates using an anti-Spred1 antibody. Endogenous Spred1 also coimmunoprecipitated with endogenous neurofibromin with two different NF1 antibodies. (D) Ectopic Spred2 and Spred3 also coprecipitated endogenous neurofibromin in T-REx-293 cells, whereas Sprouty2 and Sprouty4 were unable to do so.
We also found that the two other Spred1 family members, Spred2 and Spred3, could associate with neurofibromin (Fig. 3D). However, Sprouty proteins, which lack the N-terminal EVH1 domain, did not coprecipitate neurofibromin. Moreover, pathogenic point mutations in the EVH1 domain of Spred1 prevented association with neurofibromin, whereas constructs encoding the wild-type EVH1 domain either alone or fused to a CAAX motif were able to interact with neurofibromin (Fig. 3A). Finally, a pathogenic point mutation in the C-terminal SPR domain P415A did not prevent Spred1 from binding neurofibromin. These results indicated that the ability of Spred1 to associate with neurofibromin is dependent on its EVH1 domain. These data support the model that missense EVH1 mutations are pathogenic due to loss of an interaction with neurofibromin.
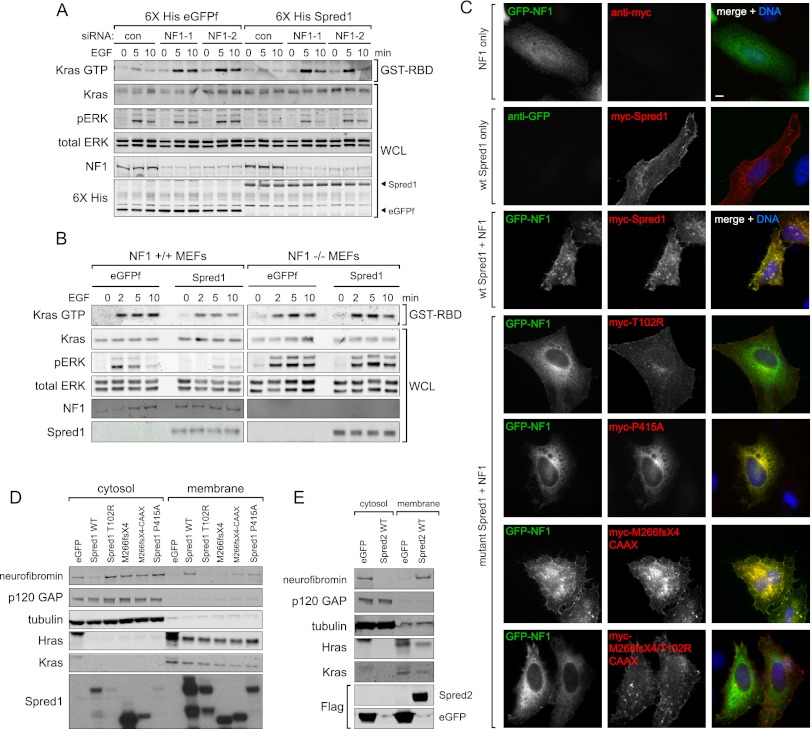
To determine whether neurofibromin is necessary for Spred1 to inhibit Ras activation, we depleted neurofibromin with siRNA in T-REx-293 cells stably expressing Spred1 (Fig. 4A). Reducing neurofibromin expression with two different siRNA oligos suppressed Spred1's ability to reduce Ras-GTP levels and decrease ERK phosphorylation, indicating that neurofibromin is necessary for Spred1 function. We next examined Spred1's activity in NF1−/− mouse embryonic fibroblasts (MEFs) (Fig. 4B). Whereas ectopic Spred1 was able to reduce Ras-GTP levels when introduced into wild-type NF1+/+ MEFs, Spred1 overexpression in NF1−/− MEFs failed to reduce Ras activity. Taken together, these results show that neurofibromin is necessary for Spred1's inhibitory activity on the Ras pathway. We next tested whether neurofibromin requires Spred to negatively regulate Ras. In PC3 or U87 glioblastoma cells, NF1 expression slightly dampened Ras-GTP levels in control and Spred1 siRNA-depleted cells. However, this effect was abrogated in cells depleted of both Spred1 and Spred2 (Supplemental Fig S4). These results suggest that the Spreds are necessary for neurofibromin's inhibitory activity.
Figure 4.
NF1 is necessary for Spred1's inhibitory function. (A) Neurofibromin was siRNA-depleted from TET-inducible 6X-HIS eGFPf and Spred1 cell lines, serum-starved, and stimulated with EGF for the times indicated. Active Ras was precipitated from whole-cell lysates using GST-Raf RBD beads. (B) MEFs were transduced with eGFPf- or Spred1-expressing retrovirus. Following serum starvation, cells were treated with EGF for the indicated times, lysed, and analyzed by immunoblot. (C) HeLa cells were transfected with either GFP-NF1 alone, myc-Spred1 alone, GFP-NF1 and myc-Spred1, or GFP-NF1 and the indicated myc-Spred1 mutant and serum-starved and for 16 h. Fixed cells were immunostained for myc (red), GFP (green), and DNA (DAPI). (D,E) HEK-293T cells ectopically expressing either wild-type Spred1 or the indicated Spred1 mutant (D) or Spred2 (E) were subjected to biochemical fractionation, and the resulting fractions were resolved by immunoblot analysis.
GAPs can be recruited to the plasma membrane via adapter proteins; for example, p120 translocation to the plasma membrane is dependent on receptor tyrosine kinase (RTK) binding (Cleghon et al. 1998). Because Spred1 has previously been reported to interact with Ras directly (Wakioka et al. 2001), and localizing the Spred1 EVH1 domain to the plasma membrane rescues Spred1's inhibitory function (Fig. 2), we reasoned that Spred1 might recruit neurofibromin to active signaling complexes at the plasma membrane. To test this hypothesis, we monitored neurofibromin localization in the presence or absence of Spred1 by immunofluorescence microscopy. Expressed alone, full-length neurofibromin localized primarily to the cytoplasm, whereas Spred1, when expressed alone, localized to the plasma membrane (Fig. 4C). When coexpressed, however, Spred1 and neurofibromin colocalized at the membrane, suggesting that Spred1 can facilitate neurofibromin translocation to the plasma membrane. We also examined the effect of the Spred1 point mutations on neurofibromin localization and observed that the T102R EVH1 domain mutant was still able to localize to the plasma membrane but failed to recruit neurofibromin to the membrane (Fig. 4C). In contrast, the P415A Spry domain mutant colocalized with neurofibromin at the cytoplasm, rather than at the plasma membrane. These data were consistent with our observations that the T102R mutant failed to interact with neurofibromin by coimmunoprecipitation, whereas the pathogenic P415A mutant could still bind neurofibromin.
To confirm that neurofibromin recruitment was dependent on the EVH1 domain, we also examined neurofibromin localization after coexpression of the M266fsX4-CAAX mutant (Fig. 4C; Supplemental Fig. S5). Consistent with our biochemical interaction data, we observed that neurofibromin translocated to the membrane in the presence of the membrane targeted (CAAX) Spred1 mutant. However, when the T102R point mutation was introduced into the M266fsX4-CAAX construct, mutant Spred1 still localized to the membrane but failed to recruit neurofibromin, which remained cytoplasmic (Fig. 4C, bottom panel).
To biochemically confirm that Spred1 was capable of localizing neurofibromin to the plasma membrane, Spred1-expressing 293 cells were subjected to biochemical fractionation (Fig. 4D). Endogenous neurofibromin was distributed primarily to the cytoplasmic fraction. However, following exogenous expression of Spred1, there was a robust change in subcellular distribution of neurofibromin in that cytoplasmic neurofibromin significantly decreased and there was a large increase in neurofibromin in the membrane fraction. Consistent with our immunofluorescence data, the Spred1 EVH1 T102R mutant was found in the membrane fraction but did not alter neurofibromin's cytoplasmic localization. The ectopic expression of Spred2 also led to a significant redistribution of neurofibromin from the cytoplasm to the membrane fraction (Fig. 4E).
Additionally, the siRNA knockdown of both Spred1 and Spred2 in PC3 cells led to a significant reduction of endogenous neurofibromin in the membrane fraction (Supplemental Fig. S6). In summary, Spred's inhibitory activity on the Ras/MAPK pathway depends on the recruitment of neurofibromin to the plasma membrane.
This work identifies a new biochemical link between Legius syndrome and NF1 and provides the first evidence for the molecular mechanism of Spred1 function, invoking a novel and critical regulatory mechanism for neurofibromin. On the basis of the data described here, we propose a new model for Spred1 inhibition of the Ras/MAPK pathway. We show that the N-terminal EVH1 domain of Spred1 interacts with neurofibromin and mediates its translocation to the plasma membrane, where neurofibromin can perform its function as a RasGAP. This model is consistent with the ability of Spred1 to reduce cellular Ras-GTP levels in the absence of any known enzymatic activity. Previously, Spred1 and Spred2 have been reported to translocate to the plasma membrane following growth factor stimulation (Wakioka et al. 2001) and inhibit mitogenic signaling induced by a range of growth factors and cytokines (Nonami et al. 2004; Bundschu et al. 2006); future studies should aim to identify specific receptors that may engage Spred1 to recruit neurofibromin and modulate Ras-GTP signaling.
Given the overlapping clinical phenotypes of Legius syndrome and NF1, our findings provide a molecular basis for these similarities and support a role for Spred1 as a novel regulator of neurofibromin function. Furthermore, since Spred2 and Spred3 can also interact with neurofibromin, these two isoforms may compensate for loss of Spred1 and thus may help explain the milder phenotype associated with Legius syndrome in comparison with NF1. Thus, these results not only identify a novel regulatory module for neurofibromin and Ras/MAPK signaling, but importantly, also provide the first molecular link between NF1 and Legius syndrome.
Materials and methods
Plasmid constructs
N-terminal-tagged full-length Spred1 and M266fsX4 were made by Gateway cloning (Invitrogen) according to the manufacturer's instructions. Spred1 truncation mutants were made by PCR cloning. SPRED1 missense mutants were made by PCR-directed mutagenesis and verified by sequencing. Full-length NF1 was synthesized by GeneArt and transferred to various Gateway Destination vectors (Invitrogen). The N terminus the GS TAP cassette was obtained from the Euroscarf collection (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/index.html).
Immunochemical analysis
Cell culture, transfections, and cell lysis were performed as described (Rodriguez-Viciana et al. 2006). T-REx-293 was used to generate stable HEK-293 cell lines with tetracycline-inducible expression of His-tagged or NTAP-tagged eGFPf, Spred1, or Spred1 mutants. NF1+/+ and NF1−/− MEFs were a kind gift from J. Nakamura (University of California at San Francisco). The following antibodies were used for immunoblotting: Kras (F234), Nras (F155), Hras (C20), Myc (A-14), neurofibromin (D), GFP (B-2), SBP (SB19-C4), and GST (B-14) were from Santa Cruz Biotechnology; total ERK, phospho-ERK (Thr 202/Tyr 204), and phospho-S338 Raf were from Cell Signaling; Spred1 (rabbit) was from Abcam; Spred1 (sheep) was from R&D Systems; Flag M2 was from Sigma; 6X-His was from Clontech; and Raf-1 (610151) was from BD Transduction Laboratories.
siRNA transfections
Transfection of siRNA oligonucleotides into cells was performed with RNAiMAX (Invitrogen) according to the manufacturer's protocol. siRNA transfections were performed twice. Forty-eight hours after the first transfection, cells were serum-starved. Seventy-two hours after the initial transfection, the appropriate growth factor was added, and cells were harvested for further analysis.
The following target sequences (Qiagen) were used: Spred1-1, TTCACGTATCATTCTGCTAAA; Spred1-2, TAGGGTCCCTTTGAAATCAAT; Spred2-1, AAGGACTTGGTCTACACCAAA; NF1-1, CAGGTGGCTTGGGATCAATAA; NF1-2, TACAGTAATAGCACTAACCAA.
Nonsilencing control siRNAs were from Dharmacon.
TAP and mass spectrometry (MS)
T-REx-293 cells (Invitrogen) stably expressing tetracycline-inducible TAP-tagged wild-type Spred1 or M266fsX4-CAAX were plated in five 15-cm dishes. Twenty-four hours following induction with doxycycline (1 μg/mL), cells were stimulated with EGF (10 ng/mL). Pooled cell lysates were purified and analyzed as described (Burckstummer et al. 2006; Rodriguez-Viciana et al. 2006) with the following differences for the MS analysis: Digests were analyzed using a QSTAR Elite mass spectrometer (Applied Biosystems/MDS Sciex). Data were searched against the UniProtKB.2010.08.10 as of August 10, 2010, using in-house ProteinProspector version 5.2.2 (a public version is available online).
Immunofluorescence
Following transfection, HeLa cells were grown in serum-free medium overnight, washed with PBS, and fixed in 4% paraformaldehyde or stimulated with EGF (10 ng/mL) prior to fixation. Cells were washed with PBS, followed by extraction and blocking with PBS containing 3% BSA (Sigma-Aldrich), 0.1% Triton X-100, and 0.02% sodium azide (PBS-BT). Coverslips were incubated sequentially with primary antibodies diluted in PBS-BT for 1 h at room temperature or overnight at 4°C. Alexa Fluor dye-conjugated secondary antibodies (Invitrogen) were diluted in PBS-BT 1:250 and incubated sequentially for 1 h at room temperature. Coverslips were mounted using anti-fade mounting medium containing PBS, glycerol, and P-phenylenediamine. For standard immunofluorescence, images for Figure 2B were acquired with a Leica SP6; for Figure 4C and Supplemental Figure S5, images were acquired with Openlab 4.0.4 (PerkinElmer) using an Axiovert 200M (Carl Zeiss, Inc). Images were processed using Photoshop (Adobe Systems, Inc).
Biochemical fractionation
Twenty-four hours to 48 h after transfection with GFP or Spred constructs, 293T cells were resuspended in hypotonic lysis buffer (10 mM Tris at pH 7.5, 1 mM EDTA, 1 mM DTT, protease and phosphatase inhibitor cocktails), incubated for 20 min on ice, and then passed through a 25-gauge needle 15 times to lyse. Lysates were replenished with NaCl to a final concentration of 150 mM, spun at 100g for 3 min to pellet nuclei and unbroken cells, and then spun at 10,000g for 15 min to pellet the membrane fraction. The pellet was washed twice with lysis buffer, and membrane proteins were solubilized in TNE buffer (20 mM Tris at pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM DTT, protease and phosphatase inhibitor cocktails); the membrane fraction was then cleared at 15,000g for 10 min. The supernatant following the 10,000g spin was spun at 100,000g for 30 min, and the final supernatant was isolated as the cytoplasmic fraction.
Acknowledgments
We thank A. Young, T. Yuan, and K. Shannon for discussions and critical reading of the manuscript, and A. Balmain and members of the McCormick laboratory for support and discussions. This work was supported by a contribution from Sandra Lloyd and a Young Investigator Award from the Children's Tumor Foundation. The University of California at San Francisco Mass Spectrometry Facility (A. L. Burlingame, director) is supported by the NIH NCRR (grant no. P41RR001614).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.190876.112.
References
- Basu TN, Gutmann DH, Fletcher JA, Glover TW, Collins FS, Downward J 1992. Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature 356: 713–715 [DOI] [PubMed] [Google Scholar]
- Bollag G, Clapp DW, Shih S, Adler F, Zhang YY, Thompson P, Lange BJ, Freedman MH, McCormick F, Jacks T, et al. 1996. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nat Genet 12: 144–148 [DOI] [PubMed] [Google Scholar]
- Brems H, Chmara M, Sahbatou M, Denayer E, Taniguchi K, Kato R, Somers R, Messiaen L, De Schepper S, Fryns JP, et al. 2007. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat Genet 39: 1120–1126 [DOI] [PubMed] [Google Scholar]
- Bundschu K, Walter U, Schuh K 2006. The VASP–Spred–Sprouty domain puzzle. J Biol Chem 281: 36477–36481 [DOI] [PubMed] [Google Scholar]
- Burckstummer T, Bennett KL, Preradovic A, Schutze G, Hantschel O, Superti-Furga G, Bauch A 2006. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods 3: 1013–1019 [DOI] [PubMed] [Google Scholar]
- Cichowski K, Santiago S, Jardim M, Johnson BW, Jacks T 2003. Dynamic regulation of the Ras pathway via proteolysis of the NF1 tumor suppressor. Genes Dev 17: 449–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleghon V, Feldmann P, Ghiglione C, Copeland TD, Perrimon N, Hughes DA, Morrison DK 1998. Opposing actions of CSW and RasGAP modulate the strength of Torso RTK signaling in the Drosophila terminal pathway. Mol Cell 2: 719–727 [DOI] [PubMed] [Google Scholar]
- DeClue JE, Papageorge AG, Fletcher JA, Diehl SR, Ratner N, Vass WC, Lowy DR 1992. Abnormal regulation of mammalian p21ras contributes to malignant tumor growth in von Recklinghausen (type 1) neurofibromatosis. Cell 69: 265–273 [DOI] [PubMed] [Google Scholar]
- Denayer E, Chmara M, Brems H, Kievit AM, van Bever Y, Van den Ouweland AM, Van Minkelen R, de Goede-Bolder A, Oostenbrink R, Lakeman P, et al. 2011. Legius syndrome in fourteen families. Hum Mutat 32: E1985–E1998 doi: 10.1002/humu.21404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Yunoue S, Tokuo H, Ozawa T, Zhang D, Patrakitkomjorn S, Ichimura T, Saya H, Araki N 2004. PKA phosphorylation and 14-3-3 interaction regulate the function of neurofibromatosis type I tumor suppressor, neurofibromin. FEBS Lett 557: 275–282 [DOI] [PubMed] [Google Scholar]
- Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P 1996. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell 87: 227–239 [DOI] [PubMed] [Google Scholar]
- Harmer NJ, Sivak JM, Amaya E, Blundell TL 2005. 1.15 Å crystal structure of the X. tropicalis Spred1 EVH1 domain suggests a fourth distinct peptide-binding mechanism within the EVH1 family. FEBS Lett 579: 1161–1166 [DOI] [PubMed] [Google Scholar]
- Kato R, Nonami A, Taketomi T, Wakioka T, Kuroiwa A, Matsuda Y, Yoshimura A 2003. Molecular cloning of mammalian Spred-3 which suppresses tyrosine kinase-mediated Erk activation. Biochem Biophys Res Commun 302: 767–772 [DOI] [PubMed] [Google Scholar]
- King JAJ, Straffon AFL, D'Abaco GM, Poon CLC, I STT, Smith CM, Buchert M, Corcoran NM, Hall NE, Callus BA, et al. 2005. Distinct requirements for the Sprouty domain for functional activity of Spred proteins. Biochem J 388: 445–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JAJ, Corcoran NM, D'Abaco GM, Straffon AF, Smith CT, Poon CLC, Buchert M, I S, Hall NE, Lock P, et al. 2006. Eve-3: A liver enriched suppressor of Ras/MAPK signaling. J Hepatol 44: 758–767 [DOI] [PubMed] [Google Scholar]
- Laycock-van Spyk S, Jim HP, Thomas L, Spurlock G, Fares L, Palmer-Smith S, Kini U, Saggar A, Patton M, Mautner V, et al. 2011. Identification of five novel SPRED1 germline mutations in Legius syndrome. Clin Genet 80: 93–96 [DOI] [PubMed] [Google Scholar]
- Leondaritis G, Petrikkos L, Mangoura D 2009. Regulation of the Ras-GTPase activating protein neurofibromin by C-tail phosphorylation: Implications for protein kinase C/Ras/extracellular signal-regulated kinase 1/2 pathway signaling and neuronal differentiation. J Neurochem 109: 573–583 [DOI] [PubMed] [Google Scholar]
- Li D, Jackson RA, Yusoff P, Guy GR 2010. Direct association of Sprouty-related protein with an EVH1 domain (SPRED) 1 or SPRED2 with DYRK1A modifies substrate/kinase interactions. J Biol Chem 285: 35374–35385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Yusoff P, Wong ES, Chandramouli S, Lao DH, Fong CW, Guy GR 2002. The cysteine-rich sprouty translocation domain targets mitogen-activated protein kinase inhibitory proteins to phosphatidylinositol 4,5-bisphosphate in plasma membranes. Mol Cell Biol 22: 7953–7966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangoura D, Sun Y, Li C, Singh D, Gutmann DH, Flores A, Ahmed M, Vallianatos G 2006. Phosphorylation of neurofibromin by PKC is a possible molecular switch in EGF receptor signaling in neural cells. Oncogene 25: 735–745 [DOI] [PubMed] [Google Scholar]
- Mardakheh FK, Yekezare M, Machesky LM, Heath JK 2009. Spred2 interaction with the late endosomal protein NBR1 down-regulates fibroblast growth factor receptor signaling. J Cell Biol 187: 265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GA, Viskochil D, Bollag G, McCabe PC, Crosier WJ, Haubruck H, Conroy L, Clark R, O'Connell P, Cawthon RM, et al. 1990. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell 63: 843–849 [DOI] [PubMed] [Google Scholar]
- McGillicuddy LT, Fromm JA, Hollstein PE, Kubek S, Beroukhim R, De Raedt T, Johnson BW, Williams SM, Nghiemphu P, Liau LM, et al. 2009. Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer Cell 16: 44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messiaen L, Yao S, Brems H, Callens T, Sathienkijkanchai A, Denayer E, Spencer E, Arn P, Babovic-Vuksanovic D, Bay C, et al. 2009. Clinical and mutational spectrum of neurofibromatosis type 1-like syndrome. JAMA 302: 2111–2118 [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Wakioka T, Nishinakamura H, Kamio M, Yang L, Inoue M, Hasegawa M, Yonemitsu Y, Komiya S, Yoshimura A 2004. The Sprouty-related protein, Spred, inhibits cell motility, metastasis, and Rho-mediated actin reorganization. Oncogene 23: 5567–5576 [DOI] [PubMed] [Google Scholar]
- Muram-Zborovski TM, Stevenson DA, Viskochil DH, Dries DC, Wilson AR, Rong M 2010. SPRED 1 mutations in a neurofibromatosis clinic. J Child Neurol 25: 1203–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonami A, Kato R, Taniguchi K, Yoshiga D, Taketomi T, Fukuyama S, Harada M, Sasaki A, Yoshimura A 2004. Spred-1 negatively regulates interleukin-3-mediated ERK/mitogen-activated protein (MAP) kinase activation in hematopoietic cells. J Biol Chem 279: 52543–52551 [DOI] [PubMed] [Google Scholar]
- Nonami A, Taketomi T, Kimura A, Saeki K, Takaki H, Sanada T, Taniguchi K, Harada M, Kato R, Yoshimura A 2005. The Sprouty-related protein, Spred-1, localizes in a lipid raft/caveola and inhibits ERK activation in collaboration with caveolin-1. Genes Cells 10: 887–895 [DOI] [PubMed] [Google Scholar]
- Pasmant E, Sabbagh A, Hanna N, Masliah-Planchon J, Jolly E, Goussard P, Ballerini P, Cartault F, Barbarot S, Landman-Parker J, et al. 2009. SPRED1 germline mutations caused a neurofibromatosis type 1 overlapping phenotype. J Med Genet 46: 425–430 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Oses-Prieto J, Burlingame A, Fried M, McCormick F 2006. A phosphatase holoenzyme comprised of Shoc2/Sur8 and the catalytic subunit of PP1 functions as an M-Ras effector to modulate Raf activity. Mol Cell 22: 217–230 [DOI] [PubMed] [Google Scholar]
- Spencer E, Davis J, Mikhail F, Fu C, Vijzelaar R, Zackai EH, Feret H, Meyn MS, Shugar A, Bellus G, et al. 2011. Identification of SPRED1 deletions using RT–PCR, multiplex ligation-dependent probe amplification and quantitative PCR. Am J Med Genet A 155A: 1352–1359 [DOI] [PubMed] [Google Scholar]
- Spurlock G, Bennett E, Chuzhanova N, Thomas N, Jim HP, Side L, Davies S, Haan E, Kerr B, Huson SM, et al. 2009. SPRED1 mutations (Legius syndrome): Another clinically useful genotype for dissecting the neurofibromatosis type 1 phenotype. J Med Genet 46: 431–437 [DOI] [PubMed] [Google Scholar]
- Wakioka T, Sasaki A, Kato R, Shouda T, Matsumoto A, Miyoshi K, Tsuneoka M, Komiya S, Baron R, Yoshimura A 2001. Spred is a Sprouty-related suppressor of Ras signalling. Nature 412: 647–651 [DOI] [PubMed] [Google Scholar]
- Xu GF, O'Connell P, Viskochil D, Cawthon R, Robertson M, Culver M, Dunn D, Stevens J, Gesteland R, White R, et al. 1990. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell 62: 599–608 [DOI] [PubMed] [Google Scholar]