In this study, Ruvkun and colleagues identify a novel cytological domain of RNAi, which they term the “Mutator organelle.” In observing the entire set of Mutator proteins, the authors observe their localization to punctate foci at the germline nuclei periphery, which are seeded by MUT-16. This Mutator organelle appears to constitute an siRNA amplification compartment that is key for robust RNA silencing.
Keywords: RNAi, C. elegans, mutator, MUT-16, siRNA, P granule
Abstract
RNA silencing can be initiated by endogenous or exogenously delivered siRNAs. In Caenorhabditis elegans, RNA silencing guided by primary siRNAs is inefficient and therefore requires an siRNA amplification step involving RNA-dependent RNA polymerases (RdRPs). Many factors involved in RNA silencing localize to protein- and RNA-rich nuclear pore-associated P granules in the germline, where they are thought to surveil mRNAs as they exit the nucleus. Mutator class genes are required for siRNA-mediated RNA silencing in both germline and somatic cells, but their specific roles and relationship to other siRNA factors are unclear. Here we show that each of the six mutator proteins localizes to punctate foci at the periphery of germline nuclei. The Mutator foci are adjacent to P granules but are not dependent on core P-granule components or other RNAi pathway factors for their formation or stability. The glutamine/asparagine (Q/N)-rich protein MUT-16 is specifically required for the formation of a protein complex containing the mutator proteins, and in its absence, Mutator foci fail to form at the nuclear periphery. The RdRP RRF-1 colocalizes with MUT-16 at Mutator foci, suggesting a role for Mutator foci in siRNA amplification. Furthermore, we demonstrate that genes that yield high levels of siRNAs, indicative of multiple rounds of siRNA amplification, are disproportionally affected in mut-16 mutants compared with genes that yield low levels of siRNAs. We propose that the mutator proteins and RRF-1 constitute an RNA processing compartment required for siRNA amplification and RNA silencing.
Small RNA pathways protect the genome against foreign elements, such as viruses and transposons, and have important roles in development, chromosome segregation, and gamete production. There are three major classes of small RNAs: microRNAs (miRNAs), piwi-interacting RNAs (piRNAs), and siRNAs. siRNAs can be grouped into either exogenous siRNAs (exo-siRNAs), derived from dsRNA taken up from the environment, or endogenous siRNAs (endo-siRNAs), which are derived from coding genes, transposons, and aberrant transcripts. One mechanism of siRNA biogenesis involves cleavage of a longer dsRNA by the RNase III enzyme Dicer (Bernstein et al. 2001; Ketting et al. 2001). siRNAs are loaded into an effector complex containing an Argonaute protein and accessory factors, where they guide silencing of complementary RNAs by transcriptional and post-transcriptional gene repression (Hutvagner and Simard 2008; Guang et al. 2010). In some organisms, such as Caenorhabditis elegans, plants, and many fungi, RNA silencing is enhanced or maintained through the activity of RNA-dependent RNA polymerases (RdRPs), which synthesize antisense RNAs that are processed into additional siRNAs (Gu et al. 2009; Gent et al. 2010; Vasale et al. 2010).
The endo-siRNA pathways in C. elegans involve several expanded gene families, including RdRPs and Argonautes, which mediate an elaborate siRNA amplification and gene silencing circuit. Deep sequencing of small RNAs has revealed distinct types of siRNAs that can be broadly classified as either 26G (26 nucleotides [nt] long, 5′ monophosphorylated G) or 22G (22 nt long, 5′ triphosphorylated G) siRNAs. They can be further classified according to the Argonaute they associate with: ERGO-1 and ALG-3/4 class 26G siRNAs and WAGO and CSR-1 class 22G siRNAs (Ruby et al. 2006; Claycomb et al. 2009; Gu et al. 2009; Han et al. 2009; Conine et al. 2010; Vasale et al. 2010). 26G siRNAs are primary siRNAs that are Dicer dependent and require enhancer of RNAi (eri) class genes for their production (Han et al. 2009; Conine et al. 2010; Gent et al. 2010; Vasale et al. 2010; Fischer et al. 2011). They predominantly target spermatogenesis-enriched genes (ALG-3/4 class) and duplicated gene families (ERGO-1 class) (Conine et al. 2010; Vasale et al. 2010; Fischer et al. 2011). 26G siRNAs trigger formation of WAGO class 22G siRNAs via the RdRP RRF-1 (Gent et al. 2010). The majority of WAGO class 22G siRNAs, however, do not require a 26G siRNA trigger and target thousands of genes, including protein-coding genes, transposons, and pseudogenes (Gu et al. 2009; Zhang et al. 2011). The CSR-1 class 22G siRNA pathway targets a large proportion of coding genes; however, rather than directing RNA silencing, low levels of CSR-1 class 22G siRNAs are required for centromere formation and chromosome segregation (Claycomb et al. 2009). The mechanism by which genes are routed into each of these siRNA pathways remains a mystery.
A conserved function of endogenous small RNAs is to silence transposons in the germline. The most well-characterized DNA transposon family in C. elegans is Tc1, of which there are ∼32 intact copies present in the genome (Fischer et al. 2003). Mutations that cause activation of Tc1 in the germline were identified from genetic screens for germline mobilization of transposons and are referred to as mutator (mut) class genes (Ketting et al. 1999). A screen for mutations that cause defects in RNAi identified a largely overlapping panel of genes, demonstrating that the silencing of transposons is an endogenous function of the RNAi pathway (Tabara et al. 1999). Many of the mutator class genes have been identified as components of endogenous and exogenous small RNA-mediated gene silencing pathways and act in the same pathway as the better-known Dicer and Argonaute proteins, but at unknown steps in the trajectory of siRNA production and target mRNA encounter. The mutator proteins include the nucleotidyl transferase MUT-2/RDE-3, the 3′–5′ exonuclease MUT-7, the DEAD-box RNA helicase MUT-14, the glutamine/asparagine (Q/N) motif-rich protein MUT-16/RDE-6, and two proteins of unknown function, RDE-2/MUT-8 and MUT-15/RDE-5 (Ketting et al. 1999; Tijsterman et al. 2002; Vastenhouw et al. 2003; Chen et al. 2005; Tops et al. 2005). C. elegans with mutations in any of these genes have active transposons, defects in exogenous RNAi, temperature-sensitve sterility, and elevated male production indicative of chromosome segregation defects. mut-2, mut-7, and mut-16 mutants have been analyzed by deep sequencing and show defects in WAGO class 22G siRNA production or stability (Gu et al. 2009; Zhang et al. 2011). Additionally, several of the mutator genes are required for ERGO-1 class 26G siRNA formation or stability (Zhang et al. 2011). Other components of the WAGO class 22G siRNA pathway form a complex containing the Tudor domain protein EKL-1, the DEAD-box RNA helicase DRH-3, and one of two partially redundant RdRPs, EGO-1 and RRF-1 (Gu et al. 2009; Thivierge et al. 2011). Unlike the mutator genes and rrf-1, ekl-1, drh-3, and ego-1 are also required for the CSR-1 class 22G siRNA pathway (Claycomb et al. 2009).
In many organisms, including insects and mammals, components of the transposon silencing pathway are localized to perinuclear germline granules (Lim and Kai 2007; Aravin et al. 2009; Lim et al. 2009; Olivieri et al. 2010). In C. elegans, germ granules are referred to as P granules and contain many proteins associated with RNA metabolism. Functionally, P granules are similar to P bodies and stress granules (Gallo et al. 2008). P granules are found on the cytoplasmic surface of the nuclear envelope and associate with clusters of nuclear pores (Pitt et al. 2000). In fact, ∼75% of all nuclear pores are found in these clusters, and most, if not all, mRNAs likely pass from the nucleus through P granules to the cytoplasm (Schisa et al. 2001; Sheth et al. 2010). Several endo-siRNA pathway components localize to P granules, including DRH-3, EGO-1, and the Argonautes ALG-3, CSR-1, and WAGO-1 (Claycomb et al. 2009; Gu et al. 2009; Conine et al. 2010; Vasale et al. 2010). However, the subcellular localization of most endo-siRNA and exo-siRNA pathway components is unknown. Here we show that mutator proteins localize, along with the RdRP RRF-1, to perinuclear germline foci adjacent to P granules, but are not dependent on P granule components for their stability. We also show that MUT-16 is uniquely required for formation and proper localization of the core protein complex that constitutes Mutator foci. Small RNA profiling in mut-16 mutants suggests that the mutator complex is required for siRNA amplification. Thus, we propose that Mutator foci are RNA processing compartments where siRNA amplification and RNA silencing occurs.
Results
Mutator proteins localize to perinuclear germline foci
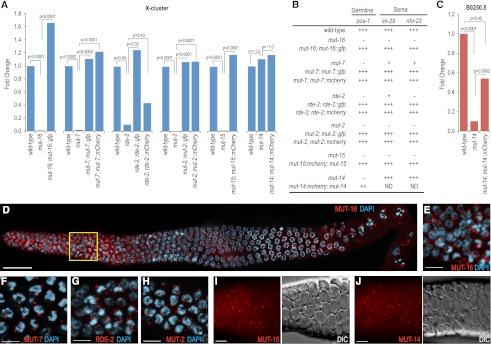
Mutator genes are essential factors in RNA silencing, yet their specific roles in small RNA pathways are poorly understood. We generated C-terminal GFP or mCherry fusions to each mutator gene so that we could characterize their roles in small RNA formation and activity. The genomic region surrounding each mutator gene, including 5′ and 3′ regulatory sequences, was PCR-amplified and fused to GFP (mut-16, mut-7, rde-2, and mut-2) and/or mCherry (mut-7, rde-2, mut-2, mut-15, and mut-14). Mutations in mutator genes for which the global levels of siRNAs have been determined (mut-2, mut-7, and mut-16) have reduced levels of most WAGO class 22G siRNAs, including those targeting the well-characterized X-cluster locus (Gu et al. 2009; Zhang et al. 2011). Thus, we tested each transgene for its ability to rescue small RNA defects in the corresponding mutant strain. We used TaqMan quantitative RT–PCR to examine levels of the most abundant X-cluster siRNA, 22G siR-1 (Montgomery et al. 2012), in each of the mutator mutants in the presence and absence of the putative rescuing transgene. mut-2(ne298), mut-7(pk720), rde-2(pk1657), mut-15(tm1358), and mut-16(pk710) mutants each displayed substantially reduced levels of 22G siR-1 relative to wild type (P < 0.05) (Fig. 1A). Introduction of the respective GFP or mCherry fusion construct to each mutant strain rescued 22G siR-1 to near wild-type levels (P < 0.02 for all constructs except rde-2::mCherry) (Fig. 1A). In addition to deficiencies in endogenous siRNA production, mut-2, mut-7, rde-2, mut-15, and mut-16 mutants are defective in exogenous RNAi (Fig. 1B; Tabara et al. 1999; Zhang et al. 2011). Introduction of the respective GFP or mCherry mutator fusion construct into each of these mutants restored the normal silencing response to dsRNAs targeting the germline gene pos-1, which causes embryonic lethality, and the somatic genes nhr-23 and lin-29, which cause adult lethality in wild-type animals (Fig. 1B).
Figure 1.
Mutator proteins localize to perinuclear foci in the germline. (A) Fluorescently tagged mutator proteins rescue mutations in their respective genes as assayed by TaqMan quantitative RT–PCR to detect the X-Cluster siRNA 22G siR-1. The mean is calculated from two biological replicates for each strain. (B) Susceptibility of mutator mutant worms to germline and somatic RNAi in the presence and absence of rescuing transgenes. pos-1 RNAi scored as complete embryonic lethality (that is, normal response to pos-1 RNAi) (+++), complete embryonic viability (that is, an RNAi-defective response) (−), or ∼50% embryonic viability (++). lin-29 RNAi scored as 100% vulval bursting (+++), 100% viable adults (−), or adults with morphological defects (i.e., protruding vulva) (+). nhr-23 RNAi scored as 100% larval arrest (+++), 100% viable adults (−), or adults with morphological defects (+). (C) mCherry-tagged MUT-14 rescues the mut-14(pk738) mutant as assayed by TaqMan quantitative RT–PCR of the germline-expressed B0250.8 siRNA. The mean is calculated from two biological replicates for each strain. (D) MUT-16 (red) localizes throughout the germline, but is brightest in the mitotic proliferation and transition zone regions as well as in the diplotene/diakinesis stages of meiosis. The yellow box is magnified in E. Image depicts entire dissected gonad stained with DAPI (blue) and anti-GFP (recognizing MUT-16::GFP). Image is an assembly of four three-dimensional (3D) data stacks following deconvolution. Bars, 20 μm. (E–J) Mutator proteins localize to foci in the germlines of adult hermaphrodites. MUT-16::GFP (E), MUT-7::GFP (F), RDE-2::GFP (G), and MUT-2::GFP (H) associate with the nuclear periphery as visualized by DAPI staining (blue). MUT-15::mCherry (I) and MUT-14::mCherry (J) mCherry fluorescence images are displayed next to the corresponding DIC images. All animals were dissected prior to imaging. Bars, 5 μm.
mut-14(pk738) is the only mutator gene that did not affect 22G siR-1 levels (Fig. 1A). X-cluster siRNAs are enriched approximately fivefold in worms without a germline, suggesting that these siRNAs are predominantly expressed in somatic tissue (Vasale et al. 2010). It is possible that mut-14 is specifically required for endogenous siRNA produced in the germline. Thus, we performed TaqMan quantitative RT–PCR of an abundant siRNA targeting a germline gene (B0250.8). The B0250.8 siRNA is reduced by >90% in mut-14(pk738) (P < 0.0001) (Fig. 1C). Introduction of the mut-14::mCherry transgene significantly, although not completely, restored B0250.8 siRNA levels in mut-14(pk738) (P = 0.0002) (Fig. 1C). mut-14 mutants are defective in RNAi targeting germline genes (Tijsterman et al. 2002). RNAi against the germline gene pos-1 caused embryonic lethality in wild-type animals but not in mut-14 mutants (Fig. 1C). Introduction of the mut-14::mCherry transgene into mut-14(pk738) partially restored lethality caused by pos-1 RNAi (Fig. 1B), indicating that mut-14::mCherry can substantially rescue the mut-14(pk738) mutant phenotype.
To determine where each of the mutator proteins function, we examined the localization of each of the mutator transgenes by immunofluorescence and live imaging. MUT-16::GFP formed punctate foci throughout the germline, as determined by immunofluorescence. The brightest and most concentrated foci were in the mitotic region and the transition zone (leptotene/zygotene) regions of the germline (Fig. 1D,E). The MUT-16::GFP foci were predominantly perinuclear from the mitotic region through pachytene, but by the diakinesis stage of meiosis, some of the foci had detached from the nuclear periphery and become cytoplasmically localized. MUT-2, MUT-7, and RDE-2 GFP fusion proteins and MUT-14 and MUT-15 mCherry fusion proteins displayed punctate foci similar to what was observed for MUT-16::GFP (Fig. 1F–J). The Mutator foci were present during larval stages through adult development and were visible in both hermaphrodite and male germlines (Supplemental Fig. S1).
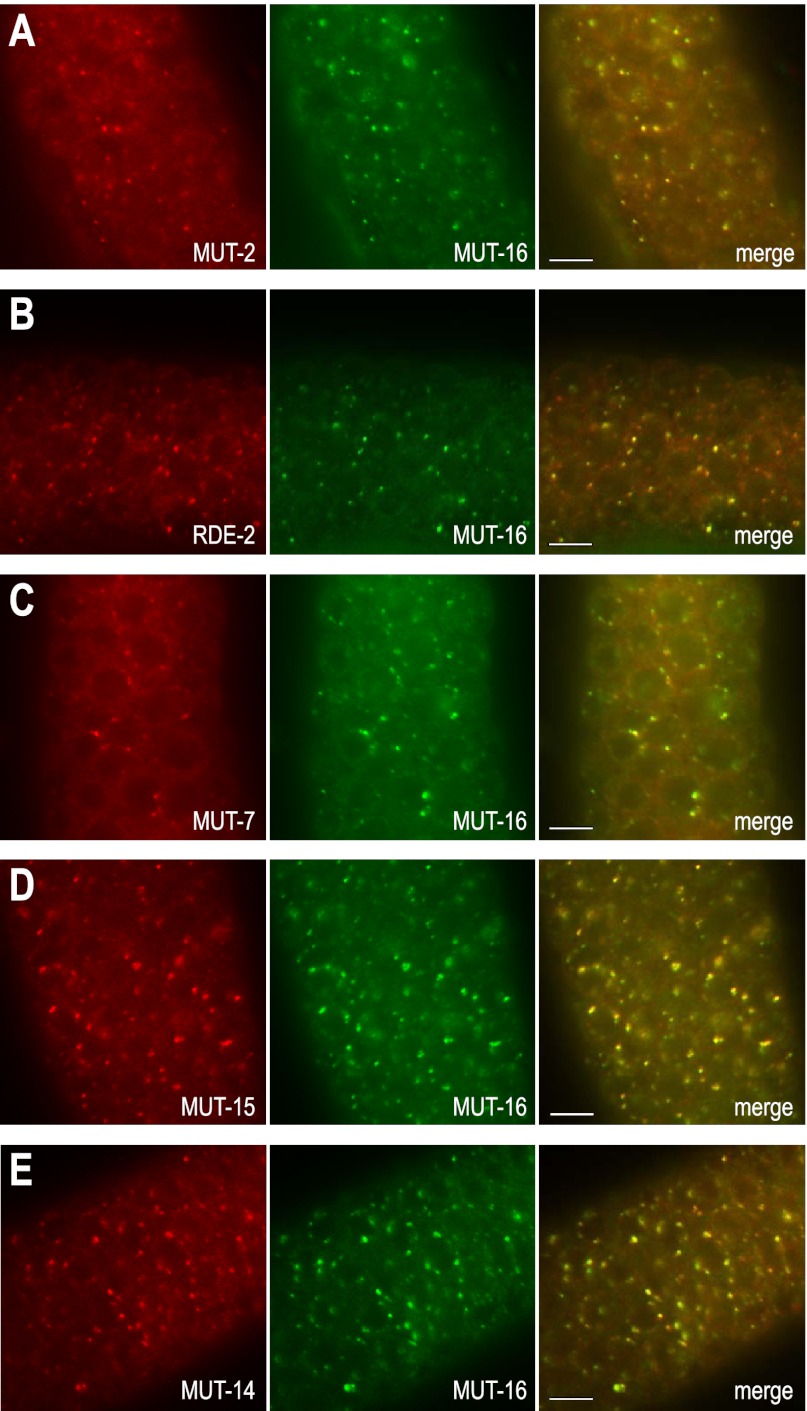
To determine whether the mutator proteins colocalize to the same germline foci, we introduced the mut-16::GFP transgene into individual C. elegans strains containing each of the other mutator genes fused to mCherry. MUT-2, MUT-7, RDE-2, MUT-14, and MUT-15 mCherry fusion proteins each colocalized with MUT-16::GFP (Fig. 2), suggesting that the mutator proteins share a common perinuclear space in which they direct RNA silencing.
Figure 2.
MUT-16 colocalizes with other mutator proteins at germline foci. MUT-2::mCherry (A), RDE-2::mCherry (B), MUT-7::mCherry (C), MUT-15::mCherry (D), and MUT-14::mCherry (E) colocalize with MUT-16::GFP. All animals were dissected prior to imaging. Bars, 5 μm.
Mutator foci are distinct from P granules
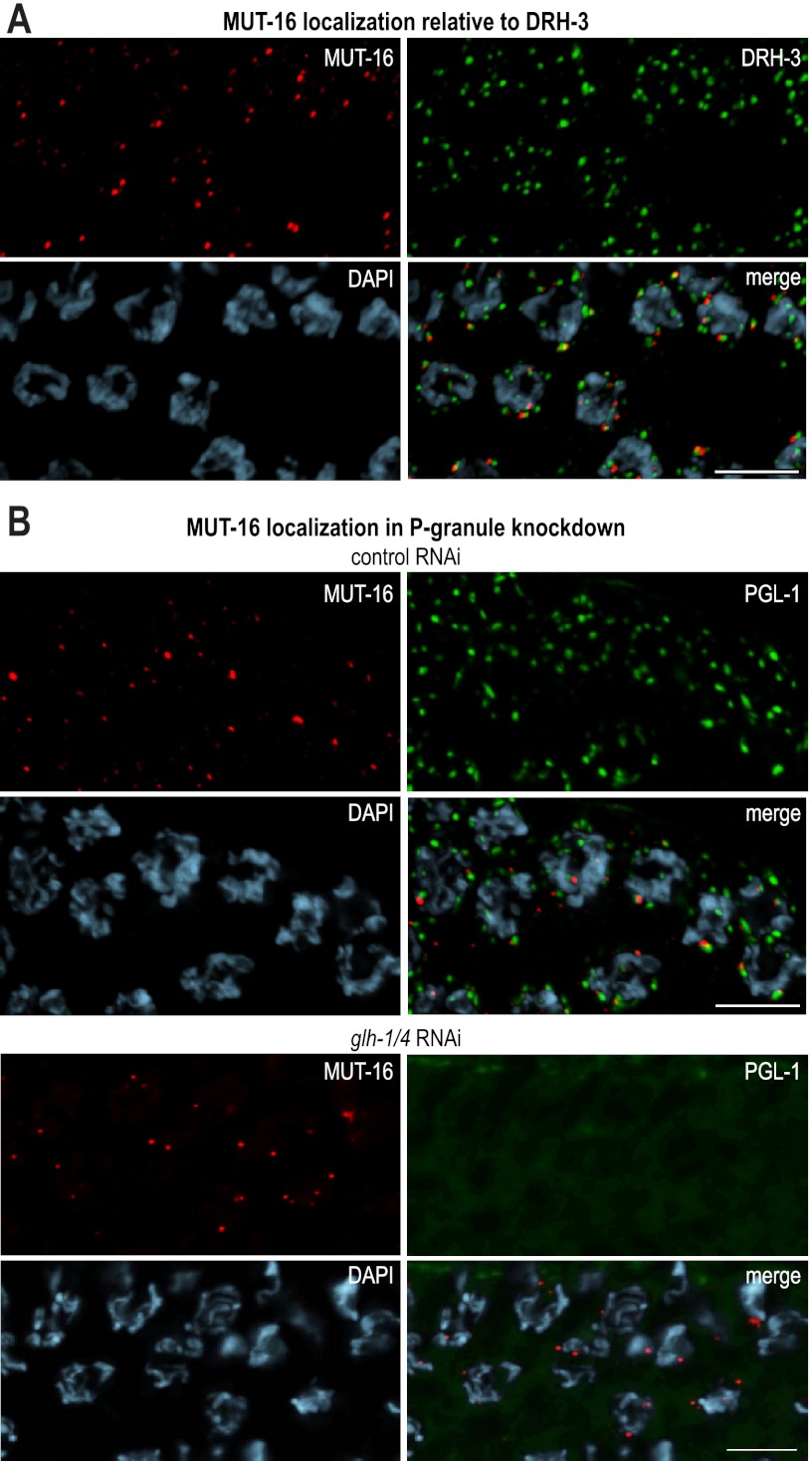
Several components of endo-siRNA pathways associate with P granules, including ALG-3, CSR-1, WAGO-1, DRH-3, and EGO-1 (Claycomb et al. 2009; Gu et al. 2009; Conine et al. 2010). Given the perinuclear pattern of Mutator foci and the requirement for mutator proteins in RNAi, it is possible that they also associate with P granules, although we observed that Mutator foci tended to be smaller and more punctate than most P granules. Similar to P-granule components, the mutator proteins localize to the nuclear periphery near nuclear pores (Supplemental Fig. S2A). To determine whether mutator proteins colocalize with known endo-siRNA pathway proteins at P granules, we immunostained the MUT-16::GFP strain with an antibody that recognizes the helicase DRH-3 (Gu et al. 2009). MUT-16 foci were nearly always adjacent to DRH-3 foci, but were rarely completely overlapping (Fig. 3A). Additionally, when introduced into a strain carrying the fluorescent P-granule marker PGL-1::RFP (Gu et al. 2009) or immunostained with an anti-PGL-1 antibody, MUT-16::GFP was nearly always adjacent to but only partially overlapping with PGL-1 foci (Fig. 3B; Supplemental Fig. S2B). To test whether Mutator foci depend on P-granule components for proper localization to the nuclear periphery, C. elegans containing the mut-16::GFP and pgl-1::RFP transgenes were treated with RNAi targeting the well-characterized P-granule components pgl-1, glh-1, and glh-4 (Gruidl et al. 1996; Kawasaki et al. 1998; Kuznicki et al. 2000; Spike et al. 2008). These RNAi treatments disrupted P granules—pgl-1 RNAi silenced PGL-1::RFP expression, while glh-1 and glh-1/glh-4 RNAi caused PGL-1::RFP expression to become diffuse throughout the cytoplasm (Fig. 3B; Supplemental Fig. S2C). In contrast, pgl-1, glh-1, or glh-1/glh-4 RNAi failed to disrupt MUT-16::GFP localization (Fig. 3B; Supplemental Fig. S2C).
Figure 3.
Mutator proteins localize independently of P-granule components. (A) MUT-16 (red) and DRH-3 (green) form distinct foci adjacent to germline nuclei. Staining was performed using antibodies against GFP (MUT-16 in red), DRH-3 (green), and DAPI (blue). (B) MUT-16 and PGL-1 foci partially overlap in adult C. elegans feeding on Escherichia coli expressing control (empty vector) dsRNA. Upon treatment with glh-1/glh-4 dsRNA, PGL-1 becomes diffuse, but MUT-16 is unchanged. Proteins were visualized using anti-GFP (MUT-16 in red) and anti-dsRed (PGL-1 in green). DNA was stained by DAPI (blue). Bars, 5 μm.
To determine whether the mutator proteins are required for localization of other P-granule components, we performed PGL-1 immunostaining in the C. elegans mutant for either mut-16, rde-2, mut-2, mut-7, mut-14, or mut-15. In each mutant, PGL-1 localization was indistinguishable from wild type (Supplemental Fig. S2D), indicating that the mutator proteins are not required for the localization of P granules. These results suggest that the mutator proteins form RNA silencing bodies that are at least partially distinct from P granules, as well as other endo-siRNA factors that associate with P granules.
MUT-16 is uniquely required for formation of Mutator foci
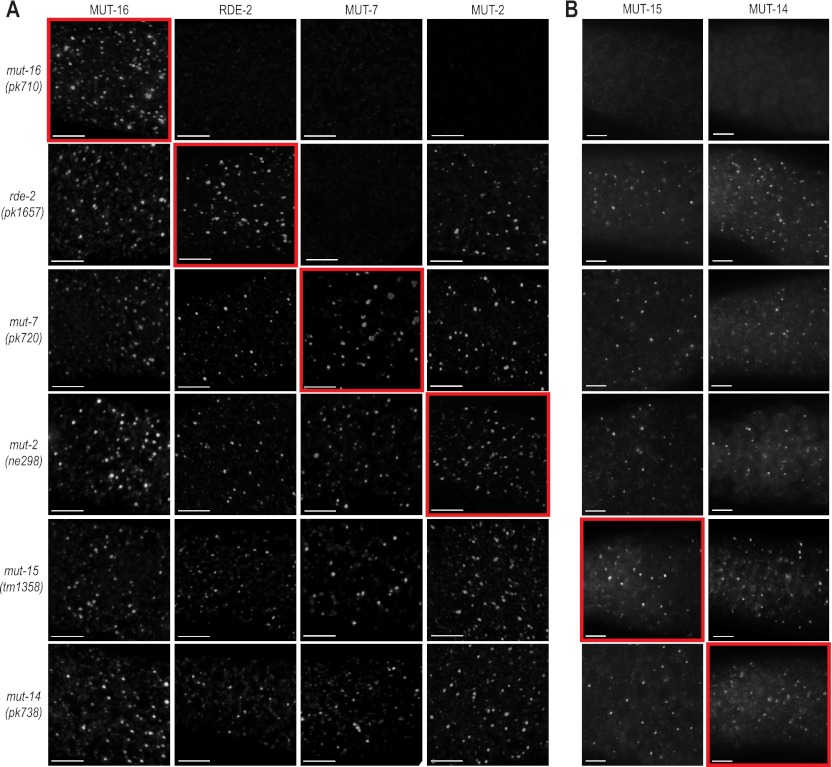
To determine the genetic requirements for proper localization of the mutator proteins, we introduced each of the GFP and mCherry mutator fusion transgenes into strains carrying mutations in each of the other mutator genes. Each of the mutator proteins localized independently of one another, with two exceptions: rde-2 was required for the localization of MUT-7::GFP, and mut-16 was required for the localization of each of the other mutator proteins to the perinuclear foci (Fig. 4). RDE-2 and MUT-7 interact in yeast two-hybrid and coimmunoprecipitation assays (Tops et al. 2005); thus, RDE-2, which has no known domains, may simply be required to recruit MUT-7 to Mutator foci. These data suggest that MUT-16 is the only mutator protein essential for the formation of Mutator foci and is therefore likely the primary component of Mutator foci.
Figure 4.
Genetic requirements for mutator protein localization. (A) MUT-16::GFP, RDE-2::GFP, MUT-7::GFP, and MUT-2::GFP expression in each of the mutator mutants. Images highlighted by red boxes display expression from a transgene in the corresponding mutant. All animals were dissected and stained with anti-GFP. Bars, 5 μm. (B) MUT-15::mCherry or MUT-14::mCherry were introduced into each of the six mutator mutants. Images highlighted by red boxes display expression from a transgene in the corresponding mutant. Bars, 5 μm.
ego-1, drh-3, and ekl-1 are required for the production of both CSR-1 and WAGO class 22G siRNAs and localize to P granules (Claycomb et al. 2009; Gu et al. 2009). We examined MUT-16::GFP localization in drh-3, ekl-1, and the RdRP double mutant ego-1 rrf-1. We also examined MUT-7::GFP localization in the ego-1 rrf-1 mutant. MUT-16 and MUT-7 were still localized primarily to the nuclear periphery in the drh-3, ekl-1, and rrf-1 ego-1 mutants (Supplemental Fig. S3). These results suggest that drh-3, ekl-1, ego-1, and rrf-1 are not directly involved in assembly of the Mutator foci.
To determine whether other small RNA factors are required for the formation or stability of Mutator foci, we screened, by RNAi, a panel of genes implicated in siRNA pathways for disruption of MUT-7::GFP localization (including ergo-1, dcr-1, rde-4, csr-1, cde-1, rde-1, drh-1, rsd-2, ppw-2, and sago-1). The panel included factors required for WAGO class 22G, CSR-1 class 22G, and ERGO-1 and ALG-3/4 class 26G endo-siRNA pathways, as well as factors involved in exo-RNAi pathways (for review, see Fischer 2010; Ketting 2011). Of all of the factors tested, only mut-16 and mut-7 RNAi disrupted MUT-7::GFP localization (Supplemental Table S1). These results suggest that Mutator foci form independent of many, if not all, other factors involved in siRNA formation or activity.
MUT-16 is a Q/N-rich protein essential for mutator complex formation
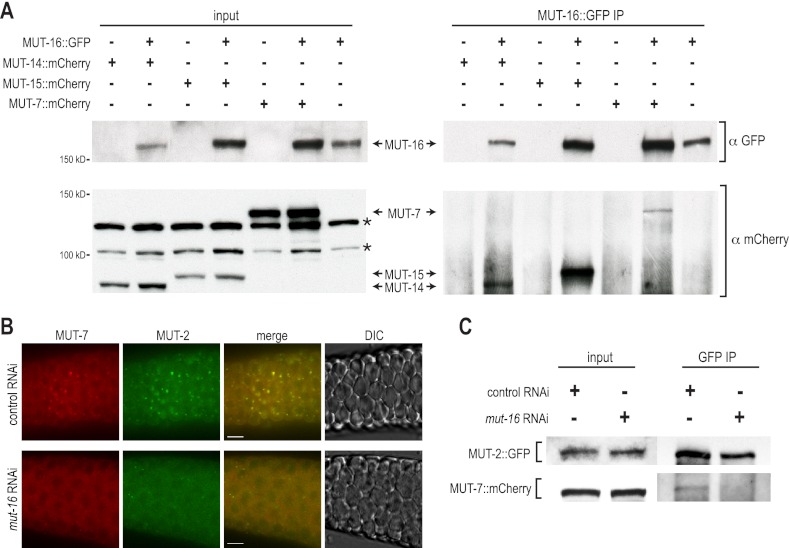
To determine whether the mutator proteins form a complex with one another, we tested whether MUT-16::GFP interacts with MUT-14::mCherry, MUT-15::mCherry, and MUT-7::mCherry by MUT-16::GFP coimmunoprecipitation assays. Each of the mutator proteins tested coimmunoprecipitated with MUT-16::GFP but were undetectable in immunoprecipitation assays lacking MUT-16::GFP (Fig. 5A). Because MUT-16 is required for the formation of Mutator foci, it is possible that it is specifically required to promote formation of the mutator complex. To test this possibility, we examined interactions between MUT-7::mCherry and MUT-2::GFP using immunofluorescence and coimmunoprecipitation assays following control or mut-16 RNAi. MUT-7::mCherry and MUT-2::GFP colocalized at germline foci when treated with control RNAi but failed to form foci when treated with mut-16 RNAi (Fig. 5B). Furthermore, MUT-7::mCherry and MUT-2::GFP, which coimmunoprecipitated when treated with control RNAi, failed to coimmunoprecipitate when treated with RNAi targeting mut-16 (Fig. 5C).
Figure 5.
MUT-16 is essential for mutator complex formation. (A) GFP and mCherry proteins from total lysate (input, left panels) and GFP-IP (right panels) from the indicated transgenic strains as assayed by Western blot. MUT-14::mCherry (93.1 kDa), MUT-15::mCherry (96.1 kDa), and MUT-7::mCherry (139.5 kDa) coimmunoprecipitate with MUT-16::GFP (154.4 kDa). Asterisks mark bands resulting from cross-reactivity of anti-mCherry with non-mutator proteins. (B) MUT-7::mCherry (red) and MUT-2::GFP (green) expression in C. elegans treated with control or mut-16 RNAi. (C) GFP and mCherry proteins from total lysate (input, left panels) and GFP-IP (right panels) from the transgenic strain containing MUT-2::GFP (86.6 kDa) and MUT-7::mCherry (139.5 kDa) treated with control or mut-16 RNAi and assayed by Western blot.
MUT-16 is a Q/N-rich protein—Q/N domains have been implicated in protein–protein interactions (Michelitsch and Weissman 2000), suggesting that MUT-16 may promote the assembly of the mutator complex. To determine whether the Q/N-rich nature of MUT-16 is conserved, we identified MUT-16 orthologs in several closely related nematode species, including Caenorhabditis briggsae (Cbr-mut-16/CBG03869), Caenorhabditis remanei (Cre-mut-16/CRE08100), Caenorhabditis brenneri (CBN32703), and Caenorhabditis japonica (Cjp-mut-16/CJA22296) (Supplemental Fig. S4). Conservation between MUT-16 orthologs is limited to the N-terminal half of the proteins; thus, we performed BLAST alignments with either the N-terminal region of C. elegans MUT-16 (amino acids 1–530) or the C-terminal region (amino acids 531–1050). BLAST with the N-terminal region of the protein identified each of the orthologs with 38%–42% identity, whereas BLAST with the C-terminal region of the protein failed to identify significant similarity to any other proteins (Supplemental Table S2). Importantly, while amino acid conservation was greatest in the N-terminal region of MUT-16, each ortholog had enrichment of glutamine and asparagine residues in the C-terminal region. In C. elegans MUT-16, 28.5% of amino acids between positions 707 and 959 are glutamine or asparagine, and an additional 17% are proline. Similarly, in C. briggsae, C. remanei, and C. japonica, 45%–50% of the amino acids across the same region are glutamine, asparagine, or proline (Supplemental Table S2). These results suggest that although there are likely conserved functional domains within the N-terminal region of MUT-16, the Q/N-rich nature of the C-terminal region is also important for its function. We conclude that MUT-16 is the core subunit of the mutator complex, and our data suggest that it is involved in tethering the complex together at Mutator foci by mediating interactions between the mutator proteins.
Mutator foci are sites of siRNA amplification
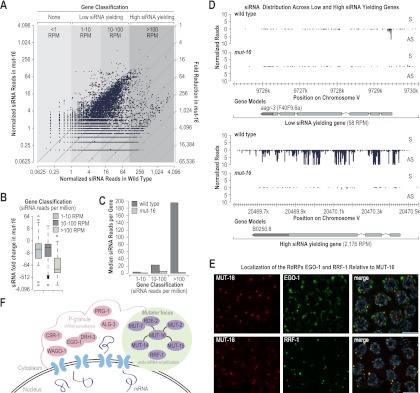
To determine the role of the mutator complex in RNA silencing, we analyzed small RNA defects in a presumably null allele of mut-16 (pk710) using small RNA high-throughput sequencing data (Zhang et al. 2011). From a wild-type small RNA library (Zhang et al. 2011), we classified genes as either low siRNA yielding (either 1–10 reads per million total reads [RPM] or 10–100 RPM) or high siRNA yielding (>100 RPM) and asked whether genes that produce high levels of siRNAs had a greater tendency to be depleted of siRNAs in mut-16. When plotted as a function of the number of siRNA reads in wild type versus mut-16 mutants, siRNAs from high siRNA yielding genes were almost entirely mut-16-dependent (Fig. 6A). Of the 546 genes classified as high siRNA yielding, ∼90% were depleted of siRNA by more than threefold in mut-16 mutants (Supplemental Table S3). In contrast, of the 9625 low siRNA yielding genes, only ∼55% were depleted of siRNAs by more than threefold in mut-16 (Supplemental Table S3). The proportion of genes that were depleted of siRNAs in mut-16 was similar between the 5158 that yield 1–10 RPM and the 4467 that yield 10–100 RPM (∼58% and ∼52%, respectively) (Supplemental Table S3). The median number of siRNA reads from high siRNA yielding genes was ∼70-fold lower in mut-16 relative to wild type, whereas the median number of siRNA reads from low siRNA yielding genes (1–10 RPM and 10–100 RPM) was only approximately fourfold lower in mut-16 (Fig. 6B). Thus, high siRNA yielding genes were depleted of siRNAs by ∼18-fold more than low siRNA yielding genes in mut-16 mutants. Mutations in other mutator class genes, including mut-2 and mut-7, also tend to have a greater effect on high siRNA yielding genes compared with low siRNA yielding genes (Gu et al. 2009). The median number of residual siRNA reads per gene was similar between low and high siRNA yielding genes in mut-16 mutants (∼0.7, ∼7, and ∼3 RPM for 1–10, 10–100, and >100, respectively), indicating that regardless of the total number of siRNAs produced from mut-16 targeted genes, they tend to be depleted to a similar level in mut-16 mutants (Fig. 6C). A major role of the mutator proteins is to silence transposons, and thus they tend to be hypersusceptible to mut-16-dependent silencing (Zhang et al. 2011). Of the 49 transposons that yield >100 RPM, all but three are depleted of siRNAs by >98% (Supplemental Table S4).
Figure 6.
MUT-16 is required for siRNA amplification. (A) Scatter plots display small RNA RPM on a log2 scale for each annotated coding gene in wild-type (bottom axis) and mut-16 mutants (left axis). The fold reduction of siRNA reads in mut-16 mutants relative to wild type is indicated by the diagonal lines on the right axis. (B) Box plots display ratio of siRNA reads on a log2 scale in mut-16 relative to wild type for low siRNA yielding genes (1–10 RPM or 10–100 RPM) and high siRNA yielding genes (>100 RPM). (C) Median siRNA reads per gene for low siRNA yielding genes (1–10 RPM or 10–100 RPM) or high siRNA yielding genes (>100 RPM) in wild-type and mut-16 mutants. (D) Small RNA distribution across the low siRNA yielding gene aagr-3 and the high siRNA yielding gene B0250.8 in wild-type and mut-16 mutants. (E) Localization of HA::EGO-1 and Flag::RRF-1 relative to MUT-16::GFP in dissected germlines immunostained with anti-GFP and either anti-HA or anti-Flag antibodies. Bars, 5 μm. (F) Model depicting the composition and localization of Mutator foci and P granules adjacent to nuclear pores.
Individual genes that fall into either the low or high siRNA classes displayed a similar trend in siRNA depletion: Low-abundance siRNAs were only modestly affected, while high-abundance siRNAs were uniformly depleted, although the high siRNA yielding genes tended to be more heavily depleted of siRNAs (Fig. 6D). Many if not all genes in C. elegans are targeted by the low-abundance CSR-1 class 22G siRNAs (Claycomb et al. 2009), which are not mut-16-dependent (Zhang et al. 2011). Therefore, it is likely that the low-level residual siRNAs produced from mut-16 targets belong to one or more distinct classes of small RNAs. These results suggest that high-abundance siRNAs—those that are produced through successive rounds of amplification—are entirely mut-16-dependent, and we propose that Mutator foci are the sites at which siRNA amplification occurs.
RRF-1 localizes to Mutator foci
If Mutator foci are siRNA amplification compartments, then presumably they contain an RdRP. To determine whether either of the partially redundant RdRPs—EGO-1 and RRF-1—associates with Mutator foci, we generated a construct containing the ego-1 rrf-1 operon with an HA epitope sequence fused to ego-1 and a Flag epitope sequence fused to rrf-1. To demonstrate the functionality of this construct, we introduced it into the ego-1 rrf-1 double mutant. Mutations in ego-1 cause sterility, while mutations in rrf-1 result in defects in somatic RNAi. The HA::ego-1 Flag::rrf-1 construct rescued both the fertility and somatic RNAi defects caused by the ego-1 and rrf-1 mutations (Supplemental Fig. S5A). We introduced mut-16::GFP into the HA::ego-1 Flag::rrf-1 strain and examined the localization of HA::EGO-1 and Flag::RRF-1 with respect to MUT-16::GFP and the Mutator foci. Consistent with previous reports of EGO-1 localization (Claycomb et al. 2009), HA::EGO-1 localized primarily to P granules, although we also observed occasional partial overlap with MUT-16::GFP (Fig. 6E; Supplemental S5B, top panels). In contrast, Flag::RRF-1 did not associate with P granules; rather, its germline localization completely overlapped with that of MUT-16::GFP (Fig. 6E; Supplemental S5B, bottom panels). These data suggest that RRF-1 is the primary RdRP at Mutator foci and functions in siRNA amplification and that EGO-1 is the primary RdRP at P granules, where it functions in the formation of the low-abundance CSR-1 class siRNAs.
Discussion
We developed fluorescently tagged transgene constructs for each of the mutator class genes. Using these constructs, we showed that the mutator proteins and the RdRP RRF-1 associate with germline-specific perinuclear puncta we termed Mutator foci. Formation of Mutator foci was dependent on the Q/N-rich protein MUT-16. In the absence of mut-16, each of the other mutator proteins was mislocalized, and protein interactions within the mutator complex were disrupted. We also demonstrated that high-abundance siRNAs, which are produced through multiple rounds of siRNA amplification, are mut-16-dependent, suggesting a role for Mutator foci in siRNA amplification.
The perinuclear structures to which MUT-16 and the other mutator proteins localize are highly reminiscent of P granules, and yet in the absence of many P granule components, Mutator foci are still present. It is important to note that while knockdown of GLH-1 causes the dispersal of several P-granule components, including PGL-1 and PGL-3, as well as loss of P-granule-associated mRNAs, small abnormal electron-dense structures still associate with nuclear pores at sites normally occupied by P granules (Schisa et al. 2001; Sheth et al. 2010). These structures lack the granular matrix seen in wild-type P granules, but it is possible that they retain certain P-granule components required to recruit mutator proteins. Alternatively, the Mutator foci may interact directly with components of the nuclear pore and could be the electron-dense structures still present in the glh-1 mutant animals. In support of this idea, several genome-wide screens have identified members of the nuclear pore complex as being required for RNA silencing (Vastenhouw et al. 2003; Kim et al. 2005; Zhou et al. 2008).
In P granules, mRNAs are surveilled as they exit the nuclear pore and enter the cytoplasm. It has been proposed that P granules act as hydrophobic barriers to slow diffusion of nascent mRNAs, extending the period in which appropriate regulatory molecules can find their targets (Sheth et al. 2010). The small RNA factors residing in P granules are mainly associated with primary and nonprocessive modes of siRNA production, including the 26G siRNA-associated Argonaute ALG-3 and the piRNA (21U)-associated Argonaute PRG-1 (Batista et al. 2008; Wang and Reinke 2008; Conine et al. 2010). Primary siRNAs are generally produced at levels too low to efficiently silence their targets; thus, RNA silencing requires siRNA amplification (Yigit et al. 2006; Pak and Fire 2007; Sijen et al. 2007). Both the piRNA and 26G siRNA pathways engage secondary 22G siRNAs to direct RNA silencing (Das et al. 2008; Conine et al. 2010). The CSR-1 siRNA pathway components CSR-1, EGO-1, and DRH-3 are also found in P granules. CSR-1 class siRNAs are produced at low levels and are not involved in RNA silencing (Claycomb et al. 2009).
We propose that mRNAs marked for siRNA-mediated RNA silencing are routed from P granules into Mutator foci, where, in conjunction with the RdRP RRF-1, the mutator complex acts as an amplification module to churn out sufficiently high levels of siRNAs to affect gene silencing (Fig. 6F). Although our results indicate that RRF-1 localizes primarily to Mutator foci, while EGO-1 associates primarily with P granules, EGO-1 is redundant with RRF-1 in the production of mutator-dependent germline 22G siRNAs (Gu et al. 2009), suggesting that it may substitute for RRF-1 in the mutator complex when RRF-1 is absent. Neither of the two other C. elegans RdRP proteins—RRF-3, required for 26G siRNAs, or RRF-2, which has no clearly described function—can substitute for RRF-1 or EGO-1 in 22G siRNA production (Gu et al. 2009; Gent et al. 2010; Vasale et al. 2010).
It is surprising that we do not see analogous perinuclear localization of the mutator proteins in somatic cells, despite a clear role for most of these proteins in somatic endo-siRNA and exo-siRNA pathways. The germline and soma produce different classes of endo-siRNAs, which have distinct genetic requirements (Gu et al. 2009; Han et al. 2009; Gent et al. 2010; Vasale et al. 2010; Fischer et al. 2011; Maniar and Fire 2011). In the germline, siRNA pathways have essential roles in silencing transposons. Thus, mutator proteins may be expressed at higher levels or associate with different complexes in the germline compared with in the cytoplasm. It is also possible that the mutator proteins reside in a similar complex in the soma, but that this complex is diffuse in the cytoplasm, rather than localized at the nuclear periphery, making it difficult to distinguish localization from background autofluorescence or nonspecific antibody staining. rrf-1 is also essential for 22G siRNA formation in somatic tissues (Gent et al. 2010; Vasale et al. 2010), suggesting that RRF-1 functions with the mutator complex in siRNA formation throughout development.
The Q/N- and proline-rich domains of MUT-16 may serve as a protein–protein interaction domain for multimerization. Q/N-rich motifs are common among eukaryotic organisms (107–472 per proteome) and are associated with prions in yeast and aggregation of proteins involved in neurodegenerative diseases. These proteins are thought to have a propensity toward self-aggregation and protein–protein interaction (Michelitsch and Weissman 2000). This region of MUT-16 is also enriched for prolines, which also play a role in protein–protein interaction (Williamson 1994). Q/N-rich motifs have also been found in several P-body components—these proteins are prone to aggregation, and this tendency to aggregate may aid in efficient accumulation of these proteins in P bodies (Reijns et al. 2008).
The Q/N-rich region of MUT-16 is primarily in the nonconserved, C-terminal region of the protein, whereas the more conserved, N-terminal region, although not bearing any obvious domains, contains numerous candidate phosphorylation motifs as well as other potential modification sites. Perhaps the N-terminal region of the protein provides the specificity for recruitment of specific proteins, whereas the C-terminal region is primarily used for self-interaction. Since MUT-16 is so essential to endo-siRNA and exo-siRNA pathways in C. elegans, it is surprising that it is not conserved outside of nematodes. However, it is plausible that unrelated proteins with Q/N-rich regions or other aggregation-prone domains may act as functional orthologs to generate hubs of RNAi machinery.
Although MUT-16 is not conserved outside of nematodes, other mutator class genes do have clear orthologs known to play roles in small RNA pathways. For example, the DEAD-box RNA helicase MUT-14 is orthologous to Vasa in Drosophila, MVH/DDX4 in mice, and DDX4 in humans, which are required for silencing of retrotransposons by piRNAs and localize to perinuclear germline structures (Liang et al. 1994; Toyooka et al. 2000; Lim and Kai 2007; Kuramochi-Miyagawa et al. 2010). The DEDD family 3′–5′ exonuclease MUT-7 also has clear orthologs—EXD3/mut-7 in humans and Nibbler in Drosophila, which trims one-fourth of all miRNAs by 1–3 nt (Han et al. 2011; Liu et al. 2011). Numerous nucleotidyl transferases, like MUT-2, modify endo-siRNAs, miRNAs, or miRNA precursors to affect their processing or stability (Heo et al. 2009; van Wolfswinkel et al. 2009). The orthologs of other mutator proteins (MUT-15 and RDE-2) are less clear, but like MUT-16, once we can address their role functionally, we may develop a better understanding of their functional orthologs.
Materials and methods
Genetics and generation of transgenic strains
The C. elegans wild-type strain is N2 Bristol. All worms were cultured at 20°C according to standard conditions unless stated otherwise (Brenner 1974). All mutator mutants were outcrossed four times to the wild-type N2 strain. Strains are listed in Supplemental Table S5. The mut-2, mut-7, rde-2, mut-14, mut-15, and mut-16 genes, promoter, and 3′ untranslated regions (UTRs) were amplified from N2 genomic DNA using Phusion polymerase (Finnzymes) and the primers listed in Supplemental Table S6. PCR products were cloned into entry vectors using Gateway BP recombinase (Invitrogen). Destination vectors pCFJ151 (for integration on Ch. II) and pCFJ178 (for integration on Ch. IV) were modified to be compatible with the Invitrogen Multisite Gateway technology, and entry vectors were recombined into these modified vectors using LR recombinase (Invitrogen). All constructs were sequence-verified. Each construct was introduced into C. elegans strain EG4322 (for Ch. II) or EG5003 (for Ch. IV) using Mos1-mediated single-copy insertion (Frøkjaer-Jensen et al. 2008). Because the ego-1 and rrf-1 genes are coexpressed in an operon, the HA::ego-1 Flag::rrf-1 construct was generated to maintain the integrity of the operon, while including the HA and Flag tags. HA::ego-1 and Flag::rrf-1 were initially cloned separately using the Gateway technology into pCFJ151. The region between ego-1 and rrf-1 was included in both constructs. Flag::rrf-1 was introduced into the HA::ego-1 construct using DraIII and SphI restriction sites.
Antibody staining and imaging
C. elegans were dissected in egg buffer containing 0.1% Tween-20 and fixed in 1% formaldehyde in egg buffer as described (Phillips et al. 2009). Samples were immunostained with mouse anti-GFP (Invitrogen, A-11120), rabbit anti-GFP (Invitrogen, A-11122), rabbit anti-dsRed (Clontech, 632496), mouse anti-mAb414 (nuclear pores; Covance, MMS-120P), rabbit anti-DRH-3 (Gu et al. 2009), mouse anti-PGL-1 (K76) (Strome and Wood 1983), rat anti-HA (Roche, 11867423001), or mouse anti-Flag (Sigma, F1804). Alexa-Fluor secondary antibodies were purchased from Invitrogen. For live imaging, animals were dissected in egg buffer and immediately mounted for imaging. All animals were dissected as 1-d-old adults (∼24 h after L4) unless otherwise stated. Imaging was done on a Zeiss Axio Imager Z1 microscope running Axiovision software. When data stacks were collected, deconvolution was performed using Axiovision, and three-dimensional images are presented as maximum intensity projections.
RNA isolation and TaqMan quantitative RT–PCR
RNA was isolated from synchronized 1-d-old adult (66–68 h after L1 arrest) C. elegans using Trizol, followed by chloroform extraction and isopropanol precipitation. RNA samples were normalized to 1.0 μg/μL. TaqMan quantitative RT–PCR assays of small RNA were performed as described (Han et al. 2009) using the following sequences for probe set design: X-Cluster/22G siR-1 (GAATAGATACGCGGTATGAGGT) and B0250.8 (GTTCCAAAATGATTCCAAGGAA). miR-1 (TGGAATGTAAAGAAGTATGTA) was used for normalization. The 2−ΔΔCt method was used for comparing relative levels of each siRNA. P-values were calculated in R using ANOVA and Tukey's HSD tests.
Immunoprecipitation
Synchronized adult C. elegans (66–68 h at 20°C after L1 arrest) were harvested for immunoprecipitation. Approximately 30,000 worms were used per immunoprecipitation (100,000 for MUT-2::GFP; MUT-7::mCherry immunoprecipitations), and all immunoprecipitations were performed in duplicate. The worms were frozen in liquid nitrogen and ground into a powder. After further dilution into lysis buffer (1:10 packed worms:buffer), a sample was taken as “input.” MUT-16::GFP or MUT-2::GFP were immunoprecipitated using Protein A (Bio-Rad, #156-006) and monoclonal mouse anti-GFP (Invitrogen A-11120). Samples were subsequently analyzed by Western blot. For Western blots, proteins were resolved on 5% or 7.5% Tris-HCl polyacrylamide gels (Bio-Rad), transferred to nitrocellulose membranes, and probed with monoclonal rat anti-GFP (Riken, BRC JFP-J5) (Hayashi and Shirao 1999), monoclonal mouse anti-GFP (Roche, #11814460001), or polyclonal rabbit anti-mCherry antibodies (Rizki et al. 2011).
RNAi assays
For RNAi assays, L1 or L2 animals were fed Escherichia coli expressing dsRNA against pos-1, lin-29, or nhr-23. For pos-1, animals were scored ∼4 d later for hatching of the F2 embryos. For lin-29 or nhr-23, animals were scored 2–3 d later for vulval bursting or larval arrest, respectively. For RNAi of P-granule components and small RNA pathway genes, C. elegans were fed E. coli expressing dsRNA against target genes beginning at L1 larval stage. F1 progeny were imaged as ∼1-d-old adults. For RNAi of mut-16, C. elegans were fed E. coli expressing dsRNA against target genes beginning at L1 larval stage. F1 progeny were harvested for eggs as adults, and synchronized F2 L1s were placed again on mut-16 RNAi. F2s were grown for 66 h at 20°C prior to immunoprecipitation or imaging (Kamath et al. 2003; Rual et al. 2004).
Acknowledgments
We thank A. Soukas, C. Mello, the Caenorhabditis Genetics Center (CGC), and Shohei Mitani of the Japanese National Bioresources Project for providing antibodies and strains. This work was supported in part by the National Institutes of Health Grant GM44619 (to G.R.) and the Massachusetts General Hospital Executive Committee of Research Tosteson Post-doctoral Fellowship (to C.M.P). T.A.M. is a Damon Runyon fellow (DRG 2029-09), and C.M.P. is the Marion Abbe fellow of the Damon Runyon Cancer Research Foundation (DRG 1988-08).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.193904.112.
References
- Aravin AA, van der Heijden GW, Castañeda J, Vagin VV, Hannon GJ, Bortvin A 2009. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet 5: e1000764 doi: 10.1371/journal.pgen.1000764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell 31: 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Brenner S 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-CG, Simard MJ, Tabara H, Brownell DR, McCollough JA, Mello CC 2005. A member of the polymerase β nucleotidyltransferase superfamily is required for RNA interference in C. elegans. Curr Biol 15: 378–383 [DOI] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. 2009. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, Shirayama M, Mello CC 2010. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci 107: 3588–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, et al. 2008. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell 31: 79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer SEJ 2010. Small RNA-mediated gene silencing pathways in C. elegans. Int J Biochem Cell Biol 42: 1306–1315 [DOI] [PubMed] [Google Scholar]
- Fischer SEJ, Wienholds E, Plasterk RHA 2003. Continuous exchange of sequence information between dispersed Tc1 transposons in the Caenorhabditis elegans genome. Genetics 164: 127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer SEJ, Montgomery TA, Zhang C, Fahlgren N, Breen PC, Hwang A, Sullivan CM, Carrington JC, Ruvkun G 2011. The ERI-6/7 helicase acts at the first stage of an siRNA amplification pathway that targets recent gene duplications. PLoS Genet 7: e1002369 doi: 10.1371/journal.pgen.1002369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen S-P, Grunnet M, Jorgensen EM 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet 40: 1375–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo CM, Munro E, Rasoloson D, Merritt C, Seydoux G 2008. Processing bodies and germ granules are distinct RNA granules that interact in embryos. Dev Biol 323: 76–87 [DOI] [PubMed] [Google Scholar]
- Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ 2010. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell 37: 679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruidl ME, Smith PA, Kuznicki KA, McCrone JS, Kirchner J, Roussell DL, Strome S, Bennett KL 1996. Multiple potential germ-line helicases are components of the germ-line-specific P granules of Caenorhabditis elegans. Proc Natl Acad Sci 93: 13837–13842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. 2009. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 36: 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S 2010. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 465: 1097–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry-Mieg D, Thierry-Mieg J, Kim JK 2009. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc Natl Acad Sci 106: 18674–18679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BW, Hung J-H, Weng Z, Zamore PD, Ameres SL 2011. The 3′-to-5′ exoribonuclease Nibbler shapes the 3′ ends of microRNAs bound to Drosophila Argonaute1. Curr Biol 21: 1878–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Shirao T 1999. Change in the shape of dendritic spines caused by overexpression of drebrin in cultured cortical neurons. J Neurosci 19: 3918–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim Y-K, Ha M, Yoon M-J, Cho J, Yeom K-H, Han J, Kim VN 2009. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708 [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ 2008. Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol 9: 22–32 [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237 [DOI] [PubMed] [Google Scholar]
- Kawasaki I, Shim YH, Kirchner J, Kaminker J, Wood WB, Strome S 1998. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94: 635–645 [DOI] [PubMed] [Google Scholar]
- Ketting RF 2011. The many faces of RNAi. Dev Cell 20: 148–161 [DOI] [PubMed] [Google Scholar]
- Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RHA 1999. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99: 133–141 [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RHA 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15: 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli AE, Rual J-F, Kennedy S, Dybbs M, Bertin N, Kaplan JM, et al. 2005. Functional genomic analysis of RNA interference in C. elegans. Science 308: 1164–1167 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, et al. 2010. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev 24: 887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznicki KA, Smith PA, Leung-Chiu WM, Estevez AO, Scott HC, Bennett KL 2000. Combinatorial RNA interference indicates GLH-4 can compensate for GLH-1; these two P granule components are critical for fertility in C. elegans. Development 127: 2907–2916 [DOI] [PubMed] [Google Scholar]
- Liang L, Diehl-Jones W, Lasko P 1994. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development 120: 1201–1211 [DOI] [PubMed] [Google Scholar]
- Lim AK, Kai T 2007. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci 104: 6714–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Tao L, Kai T 2009. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J Cell Biol 186: 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Abe M, Sabin LR, Hendriks G-J, Naqvi AS, Yu Z, Cherry S, Bonini NM 2011. The exoribonuclease nibbler controls 3′ end processing of microRNAs in Drosophila. Curr Biol 21: 1888–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniar JM, Fire AZ 2011. EGO-1, a C. elegans RdRP, modulates gene expression via production of mRNA-templated short antisense RNAs. Curr Biol 21: 449–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelitsch MD, Weissman JS 2000. A census of glutamine/asparagine-rich regions: Implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci 97: 11910–11915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Rim Y-S, Zhang C, Dowen RH, Phillips CM, Fischer SEJ, Ruvkun G 2012. PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet 8: e1002616 doi: 10.1371/journal.pgen.1002616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J 2010. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J 29: 3301–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, Fire A 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315: 241–244 [DOI] [PubMed] [Google Scholar]
- Phillips CM, McDonald KL, Dernburg AF 2009. Cytological analysis of meiosis in Caenorhabditis elegans. Methods Mol Biol 558: 171–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt JN, Schisa JA, Priess JR 2000. P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev Biol 219: 315–333 [DOI] [PubMed] [Google Scholar]
- Reijns MAM, Alexander RD, Spiller MP, Beggs JD 2008. A role for Q/N-rich aggregation-prone regions in P-body localization. J Cell Sci 121: 2463–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizki G, Iwata TN, Li J, Riedel CG, Picard CL, Jan M, Murphy CT, Lee SS 2011. The evolutionarily conserved longevity determinants HCF-1 and SIR-2.1/SIRT1 collaborate to regulate DAF-16/FOXO. PLoS Genet 7: e1002235 doi: 10.1371/journal.pgen.1002235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J-F, Ceron J, Koreth J, Hao T, Nicot A-S, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, et al. 2004. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res 14: 2162–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127: 1193–1207 [DOI] [PubMed] [Google Scholar]
- Schisa JA, Pitt JN, Priess JR 2001. Analysis of RNA associated with P granules in germ cells of C. elegans adults. Development 128: 1287–1298 [DOI] [PubMed] [Google Scholar]
- Sheth U, Pitt J, Dennis S, Priess JR 2010. Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development 137: 1305–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RHA 2007. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315: 244–247 [DOI] [PubMed] [Google Scholar]
- Spike C, Meyer N, Racen E, Orsborn A, Kirchner J, Kuznicki K, Yee C, Bennett K, Strome S 2008. Genetic analysis of the Caenorhabditis elegans GLH family of P-granule proteins. Genetics 178: 1973–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Wood WB 1983. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell 35: 15–25 [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99: 123–132 [DOI] [PubMed] [Google Scholar]
- Thivierge C, Makil N, Flamand M, Vasale JJ, Mello CC, Wohlschlegel JA, Conte D, Duchaine TF 2011. Tudor domain ERI-5 tethers an RNA-dependent RNA polymerase to DCR-1 to potentiate endo-RNAi. Nat Struct Mol Biol 19: 90–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M, Ketting RF, Okihara KL, Sijen T, Plasterk RHA 2002. RNA helicase MUT-14-dependent gene silencing triggered in C. elegans by short antisense RNAs. Science 295: 694–697 [DOI] [PubMed] [Google Scholar]
- Tops BBJ, Tabara H, Sijen T, Simmer F, Mello CC, Plasterk RHA, Ketting RF 2005. RDE-2 interacts with MUT-7 to mediate RNA interference in Caenorhabditis elegans. Nucleic Acids Res 33: 347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y, Tsunekawa N, Takahashi Y, Matsui Y, Satoh M, Noce T 2000. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech Dev 93: 139–149 [DOI] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, Ketting RF 2009. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 139: 135–148 [DOI] [PubMed] [Google Scholar]
- Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte D 2010. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc Natl Acad Sci 107: 3582–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Fischer SEJ, Robert VJ, Thijssen KL, Fraser AG, Kamath RS, Ahringer J, Plasterk RHA 2003. A genome-wide screen identifies 27 genes involved in transposon silencing in C. elegans. Curr Biol 13: 1311–1316 [DOI] [PubMed] [Google Scholar]
- Wang G, Reinke V 2008. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol 18: 861–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson MP 1994. The structure and function of proline-rich regions in proteins. Biochem J 297: 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen C-CG, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC 2006. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127: 747–757 [DOI] [PubMed] [Google Scholar]
- Zhang C, Montgomery TA, Gabel HW, Fischer SEJ, Phillips CM, Fahlgren N, Sullivan CM, Carrington JC, Ruvkun G 2011. mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proc Natl Acad Sci 108: 1201–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Hotta I, Denli AM, Hong P, Perrimon N, Hannon GJ 2008. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol Cell 32: 592–599 [DOI] [PMC free article] [PubMed] [Google Scholar]