The transcription factors Nanog and Gata6 are critical to specify the epiblast versus primitive endoderm (PrE) lineages. However, little is known about the mechanisms that regulate the protein stability and activity of these factors in the developing embryo. In this study by Azuara and colleagues, a novel function for the Polycomb group member Bmi1 during early development is discovered. The authors show that Bmi1 stabilizes Gata6 protein by physically interacting with Gata6 in PrE-derived cells and inhibiting Gata6 ubiquitination and proteasome-dependent degradation. In addition, loss-of-function experiments in chimeric embryoid bodies demonstrate a direct role for Bmi1 in PrE lineage formation. Thus, this study provides insight into novel mechanisms regulating protein stability and activity directly during early mammalian development.
Keywords: Bmi1, Nanog, Gata6, cell fate, early mouse embryo, stem cells
Abstract
The transcription factors Nanog and Gata6 are critical to specify the epiblast versus primitive endoderm (PrE) lineages. However, little is known about the mechanisms that regulate the protein stability and activity of these factors in the developing embryo. Here we uncover an early developmental function for the Polycomb group member Bmi1 in supporting PrE lineage formation through Gata6 protein stabilization. We show that Bmi1 is enriched in the extraembryonic (endoderm [XEN] and trophectodermal stem [TS]) compartment and repressed by Nanog in pluripotent embryonic stem (ES) cells. In vivo, Bmi1 overlaps with the nascent Gata6 and Nanog protein from the eight-cell stage onward before it preferentially cosegregates with Gata6 in PrE progenitors. Mechanistically, we demonstrate that Bmi1 interacts with Gata6 in a Ring finger-dependent manner to confer protection against Gata6 ubiquitination and proteasomal degradation. A direct role for Bmi1 in cell fate allocation is established by loss-of-function experiments in chimeric embryoid bodies. We thus propose a novel regulatory pathway by which Bmi1 action on Gata6 stability could alter the balance between Gata6 and Nanog protein levels to introduce a bias toward a PrE identity in a cell-autonomous manner.
During early mouse development, the transition from morula to blastocyst around embryonic day 3.5 (E3.5) marks the onset of differentiation into the inner cell mass (ICM) and trophectoderm (TE) (Jedrusik et al. 2008; Rossant 2008). At this stage, the ICM is heterogeneous and composed of pluripotent epiblast and extraembryonic primitive endoderm (PrE) progenitors, as revealed by a “salt-and-pepper” distribution of the key epiblast (Nanog) and PrE (Gata6) markers at E3.75 (Koutsourakis et al. 1999; Chambers et al. 2003, 2007; Mitsui et al. 2003; Chazaud et al. 2006; Plusa et al. 2008; Silva et al. 2009). Compartmentalization of two distinct expression domains for Nanog and Gata6 is then achieved by cell sorting and apoptosis and strictly delineates the newly formed epiblast and PrE lineages (E4.5) (Plusa et al. 2008; Meilhac et al. 2009). These confined expression patterns are stably maintained in blastocyst-derived embryonic (ES) and extraembryonic endoderm (XEN) stem cells—two cell populations that retain the properties of the epiblast and PrE, respectively (Evans and Kaufman 1981; Martin 1981; Kunath et al. 2005; Rossant 2008). Prior to blastocyst formation, however, Nanog and Gata6 are seen to overlap in most cells from the eight-cell up to the morula stage. This early expression pattern is also characterized by highly dynamic and variable protein levels among blastomeres (Dietrich and Hiiragi 2007; Plusa et al. 2008). How Nanog and Gata6 segregation is triggered and how their expression is stabilized in the epiblast and PrE progenitors remain largely unknown.
Epigenetic factors have emerged as key regulators of cell fate decisions during early development (Torres-Padilla et al. 2007). Among them, the Polycomb-repressive complexes PRC1 (Ring1A, Ring1B, Bmi1, and Mel18) and PRC2 (Ezh2, Suz12, and Eed) are known to maintain the early-determined gene expression patterns of key developmental regulators such as homeobox genes (Satijn and Otte 1999; van Lohuizen 1999). In the early embryo, PRCs are involved in specifying epigenetic asymmetry between parental genomes (Arney et al. 2001; Santos et al. 2005; Puschendorf et al. 2008). Loss-of-function studies also demonstrated a crucial role for these complexes in maintaining the integrity of ES cells in culture (Azuara et al. 2006; Boyer et al. 2006; Jorgensen et al. 2006; Leeb and Wutz 2007; Chamberlain et al. 2008; Endoh et al. 2008; van der Stoop et al. 2008). Although Polycomb group members are dynamically expressed throughout preimplantation development (O'Carroll et al. 2001; Puschendorf et al. 2008), their specific function in blastocyst lineage formation remains elusive. In this study, we identified a novel regulatory pathway that underlies cell fate allocation during early development. This process mechanistically links Bmi1 to the lineage-specific transcription factors Nanog and Gata6.
We show that Bmi1 is repressed by Nanog in ES cells and highly expressed in extraembryonic endoderm ([XEN] and trophectodermal [TS]) stem cells where Nanog is not present. In vivo investigation of expression patterns by immunostaining and single-cell PCR analysis established that Bmi1 first overlaps with Nanog and Gata6 to then preferentially segregate alongside Gata6 in PrE progenitors. In the absence of Bmi1, PrE formation is severely impaired in a cell-autonomous manner, as demonstrated in vitro in chimeric embryoid bodies (EBs). Critically, we demonstrate that Bmi1 physically interacts with Gata6 in PrE-derived XEN cells and controls its protein stability and resultant activity by inhibiting Gata6 ubiquitination and proteasome-mediated degradation. Collectively, these findings provide novel evidence to suggest how Bmi1 action on Gata6 stability could impact on cell fate decisions between epiblast and PrE lineages, most likely by altering the balance between Nanog and Gata6 protein levels in individual cells. Interestingly, Bmi1 also interacts with and maintains high Gata3 protein levels in TE-derived TS cells (Tanaka et al. 1998), suggesting a broader function for Bmi1 in extraembryonic lineage formation and/or maintenance.
Results
Bmi1 is a direct target of Nanog in pluripotent stem cells
To investigate a possible transcriptional link between Nanog and PRC members, we took advantage of genetically modified ES cell lines with distinct Nanog expression levels (Chambers et al. 2003, 2007). Quantitative RT–PCR (qRT–PCR) analysis was performed in control RCN(t), Nanog−/− RCNβH(t), and Nanog-overexpressing EF4 ES cells maintained in self-renewing conditions (Fig. 1A). Among the PRC members analyzed, Bmi1 was uniquely identified as being expressed inversely to Nanog (Fig. 1A; data not shown). While detected at low levels in control cells, Bmi1 transcript was markedly up-regulated in RCNβH(t) cells and repressed in EF4 cells, suggesting that Nanog negatively controls Bmi1 expression in ES cells. Eight putative Nanog-binding sites (BS) were identified across the Bmi1 locus based on the Nanog consensus sequence motif (Supplemental Fig. S1A; Mitsui et al. 2003). Nanog occupancy was assessed at these sites by chromatin immunoprecipitation (ChIP)-qPCR in undifferentiated ES cells. Results showed high enrichment levels at BS1 located 4 kb upstream of the Bmi1 transcription start site in Nanog-expressing cells but not in Nanog−/− ES cells, as expected (Fig. 1B; Supplemental Fig. S1B). Gata6, a known Nanog target gene, was used as positive control in these experiments (Singh et al. 2007; Frankenberg et al. 2011). To directly assess Nanog action on Bmi1 expression, a 1.9-kb fragment from the Bmi1 regulatory region spanning BS1 (Bmi1 Reg) was inserted into a pGL3 promoter vector, and the luciferase reporter construct was transfected into HEK293 cells. Cotransfection with Nanog significantly reduced Bmi1 Reg activity (Fig. 1C). This repressive effect was abolished when BS1 was mutated (Bmi1 Reg MUT), demonstrating that Nanog represses Bmi1 expression via the identified binding site.
Figure 1.
Bmi1 is repressed by Nanog in ES cells and constitutes an early hallmark of extraembryonic differentiation. (A) Relative transcript levels for Nanog, Oct3/4, Bmi1, and Gata6 as assessed by qRT–PCR in control RCN(t), Nanog−/− RCNβH(t), and Nanog-overexpressing EF4 ES cells maintained in self-renewing conditions. Data were normalized to S17 and L19 and expressed relative to RCN(t) cells. Error bars represent the SD of three biological replicates. P-values were calculated using the Student's t-test. (B) Schematic representation of the Bmi1 locus (University of California at Santa Cruz Genome Browser NCBI37/mm9) showing Nanog putative binding sites (BS; top panel) and abundance of Nanog at the Bmi1 BS1 and Gata6 promoter in RCN(t), RCNβH(t), and EF4 ES cells (bottom panel). The arrow indicates the position of the Bmi1 transcription start site. The red line highlights a 1.9-kb fragment from the Bmi1 regulatory region cloned into a pGL3 promoter vector for luciferase reporter assays as shown in C. Nanog enrichment levels were assessed by ChIP and qPCR and expressed relative to input. Unspecific immunoprecipitation was monitored by control IgG antibodies; background levels are denoted by dotted lines. Error bars represent the SD of three biological replicates. P-values were calculated using the Student's t-test. (C) Luciferase assay for Bmi1 regulatory region. HEK293T cells were transfected with the Bmi1 regulatory region containing Bmi1 BS1 (Bmi1 Reg; AATCCATTATG) or its mutated version (Bmi1 Reg MUT; AATCAGGCCTG) reporter constructs with or without Nanog. Luciferase activity was normalized to the control pGL3 promoter and Renilla. Error bars represent the SD of three biological replicates. P-values were calculated using the Student's t-test. (D) Example of Bmi1 and GFP/Nanog coimmunostaining in GFP/Nanog knock-in TNG ES cell colonies grown in self-renewing conditions. Bars, 10 μm. (E) GFP intensity profile of TNG ES cells as assessed by FACS analysis. Gates used for sorting are indicated. (F) Expression levels of GFP/Nanog, Oct3/4, Bmi1, and Gata6 as assessed in sorted GFPLow and GFPHigh TNG ES cells by qRT–PCR. Error bars represent the SD of three biological replicates. P-values were calculated using the Student's t-test. (G) Immunofluorescence analysis showing the colocalization of Bmi1 and Gata6 in a subset of Nanog−/− RCNβH(t) ES cells. Bars, 10 μm.
Bmi1 expression is mosaic among undifferentiated ES cells
Nanog is heterogeneously expressed within Oct3/4-positive ES cell cultures (Chambers et al. 2007; Singh et al. 2007). A knock-in GFP/Nanog reporter line (TNG) revealed that ES cells oscillate between Nanog-low and Nanog-high states, with Nanog-low cells being more prone to differentiate (Chambers et al. 2007). Using the same TNG reporter line, we checked whether Bmi1 was predominantly present in primed, Nanog-low ES cells. Immunostaining revealed a mosaic expression pattern for Bmi1 within ES cell colonies. As illustrated in Figure 1D, low levels of Bmi1 protein were detected in a manner mutually exclusive to GFP/Nanog signals. This was confirmed at the mRNA level in FACS-sorted GFP/Nanog-low and GFP/Nanog-high ES cell populations (Fig. 1E). In contrast to Nanog, Oct3/4 and Sox2 expression was equally high in both cell populations, highlighting the undifferentiated state of sorted cells (Fig. 1F; data not shown). Bmi1 transcript was consistently enriched in GFP/Nanog-low ES cells, with relatively lower levels being detected in the Nanog-high state (Fig. 1F). These results indicate that Nanog dynamically regulates Bmi1 expression in pluripotent cells and further suggest that Bmi1 might be an early hallmark of differentiation.
Bmi1 is an early marker of extraembryonic endoderm cell commitment
Remarkably, however, Bmi1 was not up-regulated in all Nanog−/− RCNβH(t) ES cells (n = 61/265) (data not shown), but instead was selectively detected in a subset of cells that coimmunostained for Gata6 (n = 58/61, P < 0.01, Wilcoxon test) (Fig. 1G). This confined expression pattern was confirmed in tamoxifen-inducible Nanog−/− RCNβHB ES cells, where Bmi1 and Gata6 were promptly and simultaneously induced upon Nanog depletion, followed by Gata4 and Dab2—two late markers of the PrE lineage (Supplemental Fig. S2A; Yang et al. 2002; Capo-Chichi et al. 2005). Colocalization of Bmi1, Gata6, and Gata4 protein in Nanog-depleted cells was verified by immunostaining (Supplemental Fig. S2B), further pointing to a close association between Bmi1 up-regulation and the acquisition of an extraembryonic cell identity. Consistently, we found that Bmi1 was highly expressed in XEN cells as well as in TS cells—two stem cell populations derived from the PrE and TE lineages that lack Nanog, in contrast to ES cells (Supplemental Fig. S3). Taken together, these data demonstrate that Bmi1 is rapidly up-regulated in PrE-like cells upon Nanog depletion and suggest a role for Bmi1 in extraembryonic lineages.
Bmi1 is dynamically expressed alongside Nanog and Gata6 in vivo
To explore this function, we investigated Bmi1 expression profile alongside Nanog and Gata6 in the early developing embryo. Bmi1 is a maternally inherited factor that is highly expressed in cleavage stage embryos (Puschendorf et al. 2008). Consistently, Bmi1 protein was homogeneously detected in all blastomeres of four-cell stage embryos (Fig. 2A). From eight-cell up to the early morula stage (20 cells, E3.0), Bmi1 overlapped in most cells with the nascent Nanog and Gata6 protein (Fig. 2B; data not shown). This pattern was dynamically altered around cavitation (E3.25), when cell heterogeneity arose among blastomeres. In particular, we observed the emergence of a subpopulation of cells (14.6%) that coexpress Bmi1 and Gata6 but not Nanog (29- to 43-cell embryos; P < 0.0001, Wilcoxon test) (Fig. 2B,C). Bmi1 protein staining became noticeably weaker in the developing blastocyst (E3.5–E4.5) (data not shown) despite Bmi1 transcript being detected throughout (see below), possibly reflecting a change in Bmi1 post-translational modifications (Voncken et al. 2005). These data reveal a dynamic protein expression pattern for Bmi1 and confirm its close association with Gata6 in vivo.
Figure 2.
Bmi1 protein expression profile in the early mouse embryo. (A) Bmi1 staining in a four-cell stage embryo as assessed by immunofluorescence. (B) Examples of Bmi1 staining alongside Nanog and Gata6 in eight-cell, 29-cell, and 40-cell stage embryos. The white arrow highlights an example of blastomeres coexpressing Bmi1 and Gata6 but not Nanog. (C) Counting of blastomeres expressing Bmi1, Gata6, and/or Nanog in 29-cell up to 43-cell stage embryos (n = 26 embryos, 493 cells). P-values were calculated using the Wilcoxon test.
Bmi1 preferentially cosegregates with Gata6 in PrE progenitors in the developing blastocyst
Single-cell PCR analysis was exploited to dissect RNA segregation events and further examine the relationship between Bmi1 and Gata6 expression during epiblast/PrE lineage specification. ICMs were isolated from blastocysts by immunosurgery and dissociated into single blastomeres. Embryos analyzed in these experiments were staged based on the average cell number scored among littermates. Bmi1 expression was then examined by qRT–PCR in each individual blastomere, alongside Gapdh, Gata6, Gata4, Nanog, and Ring1B (Fig. 3). In the early blastocyst (49- to 50-cell stage; E3.25), Gata6 and Nanog were expressed in most, if not all, ICM cells, with little variability between blastomeres (Fig. 3A, top panel). Mutually exclusive expression of Gata6 and Nanog emerged at the 75- to 91-cell stage (E3.5) (Fig. 3A, middle panel) and became more prominent at the 163- to 227-cell stage (E4.5) (Fig. 3A, bottom panel) (P < 0.05 at E3.5 and P < 0.01 at E4.5; Spearman test) (Fig. 3B), as previously reported (Kurimoto et al. 2006; Guo et al. 2010). At these developmental stages, Gata6 expression was correlated with Gata4 (P < 0.05 at E3.25 and P < 0.01 at E3.5 and E4.5, Spearman test), denoting PrE lineage emergence and establishment within the ICM. Bmi1 expression was similarly detected in almost all ICM cells of the early blastocyst (E3.25), and its expression was gradually restricted to Gata6-positive/Nanog-negative, presumptive PrE cells (Fig. 3A). Remarkably, at E3.5 and E4.5, Bmi1 expression exhibited a significant correlation with Gata6 (P < 0.01, Spearman test) (Fig. 3B). In contrast, the expression of another PRC1 component, Ring1B, did not correlate with Gata6 in the late blastocyst (E4.5). These results establish that Bmi1 preferentially cosegregates with Gata6 at the transcript level in nascent PrE progenitors during blastocyst development.
Figure 3.
Bmi1 RNA expression profile during epiblast/PrE lineage specification. (A) Gene expression analysis by qRT–PCR for Gapdh, Gata6, Gata4, Bmi1, Nanog, and Ring1B in individual blastomeres derived from the ICMs of developing blastocysts; 49- to 50-cell stage (E3.25) (top panel), 75- to 91-cell stage (E3.5) (middle panel), and 163- to 227-cell stage (E4.5) (bottom panel). Graphs represent the estimated copy number of gene transcripts, with each colored bar corresponding to a single blastomere. Data were sorted based on Gata6 transcript level. (B) Gene expression correlations between Gata6, Gata4, and Bmi1 in 49- to 50-cell stage (E3.25) (top panel), 75- to 91-cell stage (E3.5) (middle panel), and 163- to 227-cell stage (E4.5) (bottom panel) embryos. Scatter plots represent the estimated copy number of gene transcripts. Detection thresholds (Ct values >35) are denoted by red lines. P-values as indicated in the text were calculated using the Spearman test.
Bmi1 is physically associated with Gata6 in extraembryonic XEN cells
The observed association between Bmi1 and Gata6 prompted us to investigate a possible transcriptional cross-regulation between the two factors. Ectopically expressing Bmi1 in ES cells did not, however, impact on Gata6 expression. Conversely, Gata6 overexpression, carried out as previously described (Fujikura et al. 2002; Shimosato et al. 2007), only led to a slight increase in Bmi1 mRNA levels (data not shown), suggesting that no direct transcriptional cross-regulation operates between Bmi1 and Gata6. To test whether Bmi1 and Gata6 could be part of a same protein complex, Cos-7 cells were cotransfected with Gata6 and Bmi1, and cell lysates were subjected to anti-Gata6 immunoprecipitation. Bmi1 was strongly coimmunoprecipitated with Gata6, as revealed by anti-Bmi1 immunoblotting, thus demonstrating that Bmi1 and Gata6 are indeed physically associated (Supplemental Fig. S4A). Importantly, we confirmed that endogenous Bmi1 and Gata6 protein can also be successfully coimmunoprecipitated with anti-Bmi1 (Fig. 4A, top panel) or anti-Gata6 (Fig. 4A,bottom panel) antibodies in XEN cells and furthermore found this association to be DNA-independent, as indicated by Benzonase treatment of the protein extracts (data not shown). In contrast, the Bmi1 paralog Mel18 failed to interact with Gata6 in parallel experiments, highlighting the specificity of Bmi1/Gata6 association (Supplemental Fig. S4B). As expected, Bmi1 could readily interact with other core PRC1 members expressed in XEN cells, including Ring1B and Cbx8 (Supplemental Fig. S4C, left panels). However, Gata6 was not detected in the same complex (Supplemental Fig. S4C, right panels), further suggesting that Bmi1/Gata6 interaction might not take place in a canonical PRC1 complex.
Figure 4.
Bmi1 interacts with Gata6 and regulates its stability and activity in XEN cells. (A) Coimmunoprecipitation of Gata6 and Bmi1 proteins in XEN cells. Protein extracts were immunoprecipitated (IP) with control anti-IgG, anti-Bmi1 (top panel), or anti-Gata6 (bottom panel) antibodies and subjected to immunoblotting (IB) with anti-Gata6 and anti-Bmi1 antibodies, respectively. Three independent experiments were performed with similar results. (B) Gata6 and Gata4 stability assay in control (XENControl) and Bmi1 knockdown (XENBmi1KD) XEN cells. Bmi1, Gata6, and Gata4 protein levels were assessed by immunoblotting in XENControl and XENBmi1KD cells cultured with CHX for the indicated times. The amount of Gata6 and Gata4 protein was quantified using ImageJ software and normalized to Actin as shown in graphs. Error bars represent the SD of four biological replicates. Similar results were obtained using two independent shRNA vectors targeting Bmi1 in XEN cells (data not shown). (C,D) Same experiment as in B in the presence of the proteasome inhibitor MG132 (1 μM) and E1 ubiquitin ligase inhibitor PYR41 (1 μM), respectively. (E) Gata6 and Gata4 ubiquitination levels in XENControl and XENBmi1KD cells. XENControl and XENBmi1KD cells were cultured for 7 h in the presence of MG132. Protein extracts were subjected to immunoprecipitation with anti-Gata6 (left panel) or anti-Gata4 (right panel) antibodies, and the levels of multiubiquitination (Ub) were revealed by immunoblotting with anti-ubiquitin antibodies. Anti-Gata6 and anti-Gata4 immunoblots confirm the uniform recovery of Gata6 or Gata4 protein by immunoprecipitation across cell samples. (F) Gata6 reporter assay in XENControl and XENBmi1KD cells. Both cell populations were transiently transfected with Gata6-dependent Hnf4 reporter, and luciferase activity was assessed 48 h post-transfection. Data were normalized to Renilla. Error bars represent the SD of three biological replicates. The P-value was calculated using the Student's t-test.
Bmi1 stabilizes Gata6 protein levels and enhances its transcriptional activity
Critically, we next demonstrated that Bmi1/Gata6 association directly impacts on Gata6 stability and degradation in PrE derivatives. In this analysis, XEN cells were stably transfected with two shRNA vectors targeting Bmi1 (Supplemental Fig. S4D,E). Gata6 protein levels were assessed in control (XENControl) and Bmi1 knockdown (XENBmi1KD) XEN cells cultured for 0, 1, 2, and 4 h in the presence of cycloheximide (CHX). Without protein synthesis, Gata6 protein levels gradually decreased in control cells, with little or no effect on Gata6 transcription (Fig. 4B; data not shown). This trend was dramatically accelerated in the absence of Bmi1, suggesting that the Bmi1/Gata6 interaction protects Gata6 from degradation. In contrast, no such difference was detected in Gata4 protein decay (Fig. 4B), a factor that was not found to interact with Bmi1 by coimmunoprecipitation in XEN cells (data not shown). The addition of the proteasome inhibitor MG132 alongside CHX confirmed that Gata factor degradation was proteasome-dependent (Fig. 4C). Moreover, blocking ubiquitination with the E1 ubiquitin ligase inhibitor PYR41 also abolished Gata protein degradation (Fig. 4D), with Gata6 becoming detected in both the nucleus and cytosol of treated cells (Supplemental Fig. S4F). A role for Bmi1 in regulating this process was then directly validated by comparing the levels of Gata6 multiubiquitination (Ub) in XENControl and XENBmi1KD cells following proteasome inhibition. The absence of Bmi1 resulted in an increased accumulation of Gata6 ubiquitinated forms (Fig. 4E), which strictly mirrors its decreased protein stability (Fig. 4B). Gata4's ubiquitination status remained unchanged, as expected (Fig. 4E). Interestingly, Bmi1-mediated Gata6 stabilization also enhanced the transcriptional activity of Gata6, as assessed by luciferase assays using a Gata6-dependent Hnf4 promoter reporter (Morrisey et al. 1998) in XEN cells in the presence or absence of Bmi1 (Fig. 4F). Collectively, these results demonstrate that Bmi1/Gata6 association regulates Gata6 protein stability and enhances its transcriptional activity through the inhibition of Gata6 ubiquitination and proteasome-mediated degradation.
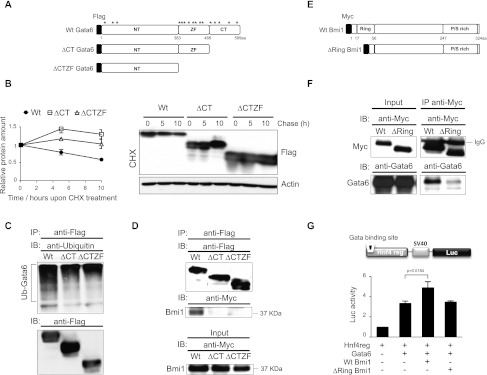
The C-terminal domain of Gata6 mediates its interaction with Bmi1 and ubiquitin-dependent degradation
In an attempt to establish which domain of Gata6 was critical for ubiquitin-dependent proteasome degradation, we next generated truncated Gata6 mutants lacking the C-terminal domain alone (ΔCT) or including its zinc finger region (ΔCTZF), where putative lysine ubiquitination sites are preferentially mapped (highlighted by asterisks in Fig. 5A). Flag-tagged wild-type, ΔCT, or ΔCTZF Gata6 constructs were transfected into Cos-7 cells, and Gata6 protein decay and ubiquitination status were assessed as previously performed (Fig. 4). Both deletions resulted in an increased Gata6 protein stability (Fig. 5B) and a reduced sensitivity to ubiquitination (Fig. 5C), identifying the C-terminal domain as being critical for Gata6 degradation via ubiquitination. Moreover, this domain was found to be equally important for Gata6 interaction with Bmi1, as demonstrated by coimmunoprecipitation assays (Fig. 5D). Conversely, using a similar mutagenesis approach for Bmi1, we validated that Bmi1/Gata6 interaction is mediated via the Bmi1 Ring finger domain (Fig. 5E,F; Hosokawa et al. 2006) and furthermore demonstrated that an intact Bmi1 is required for enhancing Gata6 transcriptional activity (Fig. 5G). Taken together, these results reiterate the functional importance of Bmi1/Gata6 interaction and further suggest how Bmi1 binding can confer protection against Gata6 ubiquitination and degradation, most likely by masking lysine residues in the Gata6 C-terminal domain from ubiquitin-conjugating enzymes.
Figure 5.
The C-terminal domain of Gata6 is critical to trigger its ubiquitin-dependent degradation and interacts with the Bmi1 Ring domain. (A) Scheme depicting Flag-tagged Gata6 wild-type and truncated forms. Wild-type Gata6 (Wt) mouse cDNA was flagged, and mutants lacking either the C-terminal domain (ΔCT) alone or including its zinc finger region (ΔCTZF) were generated by PCR-based mutagenesis. Asterisks highlight the location of putative ubiquitination lysine sites in Gata6 protein. (B) Comparative protein stability assay using wild-type (Wt) and mutant Gata6 forms. Cos-7 cells were transiently transfected with wild-type, ΔCT, or ΔCTZF Gata6. Flagged Gata6 protein levels were assessed by immunoblotting following CHX treatment for the indicated times. The amount of Gata6 protein was quantified using ImageJ software and normalized to Actin as shown in graphs. Error bars represent the SD of three biological replicates. (C) Ubiquitination status of Gata6 mutant forms. Cos-7 cells were transiently transfected with Flagged wild-type, ΔCT, or ΔCTZF Gata6 and cultured for 7 h in the presence of MG132. (Top panel) Protein extracts were subjected to immunoprecipitation with anti-Flag antibodies, and the levels of Gata6 multiubiquitination (Ub) were revealed by immunoblotting with anti-ubiquitin antibodies. Anti-Flag immunoblots confirmed the efficient recovery of Gata6 protein following immunoprecipitation across cell samples. (D) Coimmunoprecipitation of Bmi1 and Gata6 mutant forms. (Bottom panel) Cos-7 cells were transiently cotransfected with Flag-tagged wild-type, ΔCT, or ΔCTZF Gata6 and Myc-tagged Bmi1, and protein extracts were immunoprecipitated (IP) with anti-Flag antibodies and subsequently subjected to immunoblotting (IB) with anti-Flag (recovery control) or anti-Myc antibodies to detect Bmi1 protein. (E) Scheme depicting Myc-tagged wild-type and truncated Bmi1 forms. Bmi1 mouse cDNA was Myc-tagged (wild-type [Wt]), and a mutant lacking the Ring domain (ΔRing) was generated. (F) Coimmunoprecipitation of Gata6 and wild-type or ΔRing Bmi1. Cos-7 cells were transiently cotransfected with Gata6 and Myc-tagged wild-type or ΔRing Bmi1, and protein extracts were subjected to immunoprecipitation (IP) with anti-Myc antibodies (top panel) and immunoblotting (IB) with anti-Gata6 antibodies (bottom panel). Inputs and anti-Myc immunoblots confirmed homogeneous levels of different transfected forms. Three independent experiments were performed with similar results. (G) Gata6 reporter assay using wild-type and ΔRing Bmi1 forms. HEK293 cells were transiently cotransfected with Gata6-dependent Hnf4 reporter and wild-type versus ΔRing Bmi1 constructs, and luciferase activity was assessed 48 h post-transfection. Data were normalized to Renilla. Error bars represent the SD of three biological replicates. The P-value was calculated using the Student's t-test.
Bmi1 promotes the emergence of PrE-like cells upon EB formation
The experiments described thus far show that Bmi1 cosegregates with Gata6 in PrE derivatives, where it interacts with and stabilizes Gata6 protein levels. This suggests an early developmental function for Bmi1 in regulating extraembryonic endoderm lineage formation that we investigated in ES-derived EBs. In this system, ES cells are induced to form aggregates in hanging drops, and differentiation is allowed to proceed over 5 d. During this time window, PrE- and epiblast-like cells first emerge in a salt-and-pepper manner (Rula et al. 2007). They then segregate with the formation of an organized, outer PrE-like layer that costains for Bmi1, Gata6, and Gata4, as visualized by immunofluorescence on day 5 EBs (Fig. 6A). As previously reported, Nanog-overexpressing EBs were not capable of forming a proper outer layer (Chambers et al. 2003; Niakan et al. 2010), and this phenotype was associated with a loss of Bmi1 induction alongside Gata6 and Gata4 (data not shown).
Figure 6.
Bmi1 promotes PrE emergence in a cell-autonomous manner. (A) Coimmunostainings for Bmi1, Gata6, and Gata4 were performed on agarose-embedded and microsectioned EBs cultured for 5 d. (B) Relative transcript levels for Bmi1, Oct3/4, Nanog, Gata6, Gata4, and Sox17 as assessed by qRT–PCR in control (ESControl) and Bmi1 knockdown (ESBmi1KD) ES cells upon EB formation for 5 d. Data were normalized to S17 and L19 and expressed relative to undifferentiated ESControl cells. Error bars represent the SD of two biological replicates. (C) Coimmunostainings for Gata6 and Gata4 performed on EBs cultured for 5 d in the presence (ESControl; top panel) or absence (ESBmi1KD; bottom panel) of Bmi1. Bars, 20 μM. (D) Schematic of chimeric EB formation. GFP-labeled ESControl or ESBmi1KD cells were mixed with unlabeled ESControl cells (ratio 1/1–1/3) and allowed to differentiate for 5 d upon EB formation. (E) Coimmunostainings for GFP and Gata6 performed on chimeric EBs formed as described in D. The outer layer of the EB structure is denoted by dotted lines. Bars, 20 μM.
To directly assess the effect of Bmi1 depletion on this process, Bmi1 knockdown ES cells were established by stable transfection with different Bmi1 shRNA vectors. These cells showed no increased incidence of differentiation when grown in self-renewing conditions (data not shown). Importantly, Bmi1 knockdown was efficiently maintained upon EB formation, as assessed at the mRNA and protein levels (Fig. 6B; data not shown). Here, we observed a pronounced defect on PrE-like cell differentiation in the absence of Bmi1. While Oct3/4 and Nanog were down-regulated in both control and Bmi1 knockdown EBs, the induction of the PrE markers Gata6 and Sox17 was impaired (Fig. 6B), with no proper outer layer organization (Fig. 6C). This phenotype most closely resembles that of Gata6−/− EBs but differs from that of Sox17−/− EBs, in accord with Bmi1 action on Gata6 stability (Fig. 4; Koutsourakis et al. 1999; Fujikura et al. 2002; Niakan et al. 2010; Artus et al. 2011). Moreover, and as shown in Figure 6C, only very few PrE-like, Bmi1-depleted cells emerged, and these cells expressed Gata4 protein alongside Gata6, further highlighting that the emergence of PrE-like progenitors, rather than their maturation, might be directly affected by Bmi1 depletion.
Bmi1 biases cell fate toward a PrE identity in a cell-autonomous manner
To assess whether the observed defect was cell-autonomous, we repeated these experiments and mixed control (ESControl) cells with Bmi1 knockdown (ESBmi1KD) ES cells to form chimeric EBs. Cells were first labeled by stable Gfp transfection followed by FACS sorting, and the GFP-labeled ESControl or ESBmi1KD cells aggregated with unlabeled ESControl cells upon EB formation (Fig. 6D). The fate of labeled cells was assessed based on their position within the EB structure (inner/outer), and the emergence of PrE-like cells was monitored by looking at Gata6 expression (Fig. 6E; Supplemental Fig. S5). Remarkably, GFP-ESBmi1KD cells were preferentially located within the inner part of EBs (P < 0.05, Student's t-test) (Supplemental Fig. S5A), in contrast to GFP-ESControl cells, which appeared to be evenly distributed. This observation was consistent with a lower frequency of Gata6-positive, GFP-ESBmi1KD cells detected in day 5 EBs as compared with controls (P < 0.005, Student's t-test) (Supplemental Fig. S5B). Taken together, these results demonstrate a direct role for Bmi1 in cell allocation between a PrE- and an epiblast-like fate upon EB formation.
Discussion
In this study, we identify a novel role for the Polycomb group member Bmi1 in regulating cell fate choice between extraembryonic endoderm and pluripotent lineages (Fig. 7). We show that Bmi1 is readily detected in vivo in all blastomeres of cleavage stage embryos and overlaps with Nanog and Gata6 from the eight-cell stage onward. This pattern dynamically changes upon blastocyst formation, when Bmi1 becomes mosaic among ICM cells, preferentially cosegregating with Gata6 in nascent PrE progenitors. Critically, we demonstrate that Bmi1 controls Gata6 protein stability and its resultant activity by conferring protection against ubiquitination and proteasome-dependent degradation, as confirmed by Bmi1 knockdown in XEN cells. This effect is thought to be mediated through Bmi1/Gata6 interaction via the Bmi1 Ring domain, which could, in turn, alter Gata6 protein conformation and/or mask lysine residues in the Gata6 C-terminal domain from ubitiquin-conjugating enzymes (Clurman et al. 1996). Importantly, we establish that Bmi1 plays a cell-autonomous role in promoting the induction of the PrE lineage, as assessed in vitro in chimeric EBs. In the context of the early embryo, Gata6 and Nanog expression is first stochastic (Dietrich and Hiiragi 2007), and cell fate is thought to remain flexible (Yamanaka et al. 2010). We propose here that Bmi1 action on Gata6 stability could directly alter the balance between Gata6 and Nanog protein levels in individual blastomeres and thus impact on cell fate decisions by introducing a bias toward a PrE identity. Interestingly, Gata4 does not interact with Bmi1, and this together with the late onset of Gata4 protein expression in vivo further suggests that Gata4 and Sox17 could, in turn, reinforce Gata6 function in a Bmi1-independent manner.
Figure 7.
Model of Bmi1 function in extraembryonic endoderm lineage emergence. Nanog transcription factor protects the pluripotent state by repressing Bmi1 and Gata6 PrE-associated genes (depicted as black solid lines) and activating miR-200c, a microRNA also shown to modulate Bmi1 protein expression in ES cells (depicted as a black dotted line, indicating no evidence that this regulation is direct). (BS) Location of the Nanog-binding site upstream of the Bmi1 and miR-200c transcription start sites (arrows). Bmi1 physically interacts with and stabilizes Gata6 protein levels by protecting it from ubiquitination and proteasomal degradation (depicted as red triangles and dashed box). Formation of this complex enhances Gata6 transcriptional activity on target genes and promotes PrE emergence. Gata6 lies upstream of Gata4 and Sox17 in differentiation cascade (depicted as red dotted lines, indicating no evidence that these regulations are direct). Bmi1 expression also leads to a reduction in Nanog levels by an unknown mechanism (depicted as a black dotted line).
Interestingly, we provide novel evidence that Nanog represses Bmi1 in ES cells, as for Gata6 (Fig. 7; Singh et al. 2007), further supporting the view that Nanog actively suppresses the PrE identity in pluripotent cells while being required for proper PrE differentiation in a non-cell-autonomous manner (Silva et al. 2009; Messerschmidt and Kemler 2010; Frankenberg et al. 2011). We demonstrate that Bmi1 expression mirrors Nanog fluctuations within ES cell cultures and constitutes an early hallmark of extraembryonic differentiation upon Nanog depletion. Interestingly, while Bmi1 protein expression is mutually exclusive with Nanog, the Bmi1 transcript remained detected, although at variable levels, in Nanog-high and Nanog-low ES cell populations (Fig. 1), suggesting that Bmi1 itself may be post-transcriptionally regulated in this context. Several microRNAs were previously shown to target the Bmi1 3′ untranslated region (UTR) in various cancer cell lines (Shimono et al. 2009). Here we identified miR-200c as being uniquely expressed in ES cells, as opposed to XEN and TS cells (Supplemental Fig. S6A), and positively regulated by Nanog (Supplemental Fig. S6A,B). Moreover, inhibiting miR-200c was found sufficient to release Bmi1 protein expression in ES cells (Supplemental Fig. S6C,D), adding another level of Bmi1 expression control mediated by Nanog (Fig. 7). Conversely, we found that ectopic expression of Bmi1 in ES cells reduces Nanog mRNA levels without altering Oct4, Sall4, or Klf4, yet is not capable of driving PrE differentiation alone (data not shown). Similar cross-regulatory events between Bmi1 and Nanog might also take place in vivo as Bmi1/Gata6 and Nanog transcripts segregate upon epiblast/PrE lineage specification, a pattern that is stably established in the late blastocyst by E4.5. By comparison, Bmi1 and Gata6 protein were seen to cosegregate in a subset of blastomeres in the early blastocyst around E3.25, suggesting that the Bmi1/Gata6 protein–protein interaction could be an early event in PrE emergence, while Nanog-mediated transcriptional repression of Bmi1, Gata6, and other factors would “lock in” or reinforce the epiblast/pluripotent identity both in vivo and in vitro. Additional mechanisms, such as the timing of cell internalization and signaling cascades, have also been shown to act in concert to dictate or consolidate PrE lineage specification (Nichols et al. 2009; Morris et al. 2010; Yamanaka et al. 2010).
Bmi1 is a pleiotropic factor with roles linked to cell cycle regulation and cancer (Bruggeman et al. 2007; Grinstein and Mahotka 2009) as well as to the homeostasis of adult stem cells (van der Lugt et al. 1994; Molofsky et al. 2003; Park et al. 2003; Bruggeman et al. 2007). Our study unveils a previously unrecognized developmental function for Bmi1 (Puschendorf et al. 2008), acting as a key post-transcriptional regulator of Gata6, a factor essential for extraembryonic endoderm development both in vitro and in vivo (Koutsourakis et al. 1999; Lim et al. 2008). Of interest, Bmi1 was also found to interact with Gata3 in TE-derived TS cells (Supplemental Fig. S7A; Home et al. 2009; Ralston et al. 2010). Bmi1 depletion in TS cells notably led to a loss of stem cell identity accompanied by a rapid and drastic reduction in Gata3 protein levels (Supplemental Fig. S7B–E). This indicates that Bmi1 might regulate Gata3 protein expression in TS cells and suggests a broader role for Bmi1 in the formation and/or maintenance of extraembryonic lineages during early mouse development.
Materials and methods
Cell culture
Mouse ES, TS, and XEN cell lines were grown as previously described (Tanaka et al. 1998; Kunath et al. 2005; Alder et al. 2010). For Nanog depletion, the RCNβHB cell line was treated with 1 μg/mL 4′-OH-tamoxifen (Chambers et al. 2007). Transfections were carried out using Lipofectamine 2000 (Invitrogen, 11668) following the manufacturer's recommendations. For stable clone derivation, cells were treated 24 h post-transfection with puromycin (Sigma, p8833) at 1 μg/mL for 8–10 d. Clones were then pooled or picked individually, depending on the experimental design. EB formation was induced in hanging drops as previously described (Lavial et al. 2007).
Antibodies
Anti-Bmi1 (Millipore ,F6), anti-Gata6 (R&D Systems, AF1700), anti-Gata4 (Santa Cruz Biotechnology, sc-9053), anti-Gata3 (Santa Cruz Biotechnology, sc-268), anti-Cdx2 (Biogenex, MU392A-UC), anti-Nanog (Cosmobio, RCA B000 2P-F), anti-ubiquitin (Biomol, FK2), anti-Ring1B (Active Motif, 39663), anti-Cbx8 (Bethyl Laboratories, A300-882A), anti-Mel18 (Abcam, ab5267), anti-GFP (Abcam, AB290), anti-Myc (Santa Cruz Biotechnology, sc-40), anti-Flag (Sigma, M2), and anti-Actin (Abcam, AB8227) were used. For immunoprecipitation experiments, anti-Gata6 (Santa Cruz Biotechnology, sc-9055) was used. For immunofluorescence, Alexa secondary antibodies were used (Invitrogen). For immunoblotting and coimmunoprecipitation experiments, mouse (Santa Cruz Biotechnology), rabbit (Santa Cruz Biotechnology), and goat (Dako) secondary antibodies were used.
RNA expression analysis
Total RNA was isolated using the Qiagen RNeasy minikit and DNase I-treated. Samples were oligo(dT) reverse-transcribed using Invitrogen SuperScript III or M-MLV following the manufacturer's recommendations and analyzed by qRT–PCR using Sigma Jumpstart SYBR Green. Primer sequences are available on request.
Vector construction
pLKO.1 vectors containing hairpins directed against Bmi1 cDNA were purchased from Sigma: shRNA vector 1 (CCGGCCAGCAAGTATTGTCCTATTTCTCGAGAAATAGGACAATACTTGCTGGTTTTT) and shRNA vector 2 (CCGGCCTGAACATAAGGTCAGATAACTCGAGTTATCTGACCTTATGTTCAGGTTTTT). The Bmi1 2.4-kb promoter was PCR-amplified on mouse ES cell genomic DNA using long expand Taq (Roche Biomedicals, 11681834001) and Bmi1prom-F (5′-TCCCTGCCAGACTGTTTCTT-3′) and Bmi1prom-R (5′-CGTAAATGACCACGGGGATT-3′) primers. Taq polymerase (Invitrogen 10342-020) was used to add adenines and clone the fragment into pGEMTeasy (Promega, TM042). The Bmi1 1.9-kb promoter fragment was then subcloned into the pGL3 promoter (Promega, E1761) using MluI and BglII restriction enzymes (New England Biolabs). Mutations in the Nanog-binding site BS1 were inserted by using PfuTurbo polymerase (Stratagene 600250), DpnI restriction enzyme (New England Biolabs), and Bmi1mut-F (5′-TAAAATGTCTGGTCGCAGACTGCAATTGTCAGGCCTGATTAAGAGCGTACTTTAAGACAAATCACTT-3′) and Bmi1mut-R (5′-AAGTGATTTGTCTTAAAGTACGCTCTTAATCAGGCCTGACAATTGCAGTCTGCGACCAGACATTTTA-3′) primers. Gata6 cDNA was subcloned in the BglII site of pSG5-Flag (Stratagene) using primers G6-F (5′-ATAGATCTAGCCTTGACTGACGGCGGC-3′) and G6-R (5′-ATAGATCTATCAGGCCAGGGCCAGAGC-3′). Gata6 truncated forms were generated with primers ΔCTZF-F (5′-CCTGTCGGAGAGCCGCTGATAGATCTGGTACC-3′), ΔCTZF-R (5′-GGTACCAGATCTATCAGCGGCTCTCCGACAGG-3′), ΔCT-F (5′-GGAATTCAAACCAGGAAACGAAAATGATAGATCTGGTACCACTA-3′), and ΔCT-R (5′-TAGTGGTACCAGATCTATCATTTTCGTTTCCTGGTTTGAATTCC-3′). Recombinant lentiviruses were generated using a three-plasmid system in 293T cells as previously described (Kutner et al. 2009). Virus-containing culture supernatants were collected 24 and 48 h after transfection, pooled, concentrated, and used for infection. Control and Bmi1KD-infected TS cells were collected 4 d post-infection as previously described (Alder et al. 2010). The Gata6-dependent Hnf4 promoter was cloned into the pGL3 promoter as previously described (Morrisey et al. 1998). miR-200c inhibitor was purchased from Exiqon and transfected following the manufacturer's recommendations.
Luciferase reporter assay
Luciferase assays were carried out using 2.5 × 105 HEK293T cells in 96-well plates. Transfections with pGL3 promoter vectors and control peGFPC1 (Clontech) or pRenilla-Tk (Promega) were performed using Lipofectamine 2000 (Invitrogen, 11668-019). Luciferase activity was assessed 48 h post-transfection using a Steadylite kit (PerkinElmer, 6016756) following the manufacturer's recommendations. Transfection efficiency was corrected using GFP or Renilla levels.
Immunoblotting analysis
Cells lysis was carried out using RIPA buffer (50 mM Tris at pH 8, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0,1% sodium deoxycholate, 140 mM NaCl) supplemented with a protease inhibitor tablet (Roche Diagnostics, 11836153001). Protein fractionation was performed using NE-PER kit (Thermo Fisher Scientific, 78833). Protein concentrations of whole-cell extracts were measured using a Bradford assay (Thermo Fisher Scientific, 23225). Thirty-microgram samples were loaded onto 10% acrylamide gels and blotted onto methanol-activated polyvinylidene fluoride membranes (Millipore, IPFL00010) using a semidry or wet transfer system. Membranes were treated with enhanced chemiluminescent substrate (Thermo Fisher Scientific, 32106).
ChIP and coimmunoprecipitation
ChIP was carried out as previously described (Alder et al. 2010). For coimmunoprecipitation experiments, 800 μg of cell protein extracts was precleared with protein A Sepharose beads (Sigma, P3391) for 2 h at 4°C and then incubated overnight at 4°C with the indicated antibodies. Protein A beads were then added for 5 h, washed with RIPA and TSE buffer (2 mM EDTA, 20 mM Tris at pH 8, 150 mM NaCl) four times, and loaded on either 7% or 14% acrylamide gels. Protein extracts were treated for 2 h at 4°C with 50 U of Benzonase (Merck, 71205) where indicated.
Protein stability assay
Cells were split, and 3 × 106 cells were plated back into 10-cm plates. On the following day, cells were treated with 100 μM CHX (Sigma), CHX plus 1 μM MG132 (Calbiochem, 474790), or CHX plus 1 μM PYR41 (Calbiochem, 662105) for the indicated times. Protein amounts were quantified using ImageJ software and normalized to Actin levels.
Ubiquitination assay
Cells were treated for 7 h with 1–5 μM MG132, lysed in the presence of deubiquitination inhibitor NEM (Sigma, E3876), sonicated, and subjected to overnight immunoprecipitation with control anti-IgG, anti-Gata6, anti-Gata4, or anti-Flag antibodies. Protein G beads (GE Healthcare, 17-0618-01) were added for 4 h at 4°C, and the levels of ubiquitination were subsequently revealed by immunoblotting with anti-ubiquitin antibody.
Immunofluorescence analysis
Cells were seeded on gelatinized glass coverslips and fixed in PBS with 4% paraformaldehyde. Samples were permeabilized and blocked at room temperature before incubation with the indicated antibodies. Coverslips were mounted on VectaShield with DAPI (Vector Laboratories, H-1200) and examined using a Leica SP5 confocal microscope (40× or 63× lens). Embryo immunostainings were performed as previously described (Chazaud et al. 2006). EBs were fixed overnight in formalin at 4°C and embedded in agarose and wax. Five micromolar sections were used for stainings and observations on a Leica SP5 confocal microscope.
Embryo collection and staging for single-cell PCR analysis
BL/6xC3H F1 mice were bred naturally, and the embryos were recovered at E3.25, E3.5, or E4.5 by flushing either the oviduct or uterus. ICMs were isolated from blastocysts by immunosurgery and further dissociated into single blastomeres by pipetting in a solution of 1 mM EDTA dissolved in HBS after treatment with 1% trypsin (Sigma, T-4549) and 1 mM EDTA in HBS. Staging of embryos subjected to single-cell PCR analysis was defined as follows. Upon recovery, average-size embryos were selected for subsequent analysis, and the remaining littermates were fixed in PBS with 4% paraformaldehyde (Electron Microscopy Sciences, 19208) and stained in PBS with 10 μM DAPI (Molecular Probes, D3571) and 5 U/mL Alexa Fluor 633 or Alexa Fluor 564 phalloidin (Molecular Probes, A22284 or A22283, respectively). Images were acquired on a Zeiss LSM 510 META or 710 microscope and analyzed using IMARIS software (Bitplane). The total cell number of each embryo was counted, and an average cell number of littermates (excluding those with maximum and minimum cell numbers) was used to define the developmental stage of each embryo processed for single-cell PCR analysis. Experiments were performed in accordance with European Union guidelines for the care and use of laboratory animals.
Single-cell cDNA amplification
Single-cell cDNA amplification from each blastomere was performed as previously reported (Kurimoto et al. 2006). Briefly, single blastomeres were lysed in individual tubes without purification, and first strand cDNAs were synthesized using a modified poly(dT)-tailed primer. The unincorporated primer was specifically digested by exonuclease, and the second strands were generated with a second poly(dT)-tailed primer after poly(dA) tailing of the first strand cDNAs. The cDNAs were amplified by PCR first with poly(dT)-tailed primers and subsequently with primers bearing the T7 promoter sequence. The resultant cDNA products were used for further real-time PCR analysis. Primer sequences are available on request. Note that “spike” RNAs that consist of poly(A)-tailed RNAs artificially designed from Bacillus subtilis genes were added to each sample as amplification control to estimate the copy number of gene transcripts analyzed. A mixture of four distinct “spike” RNAs—Lys, Thr, Phe, and Dap (American Type Culture Collection 87482, 87483, 87484, and 87486)—were prepared so that each tube contained 1000, 100, 20, and five copies of each spike RNA, respectively.
Statistics analysis
Statistical analyses were performed using GraphPad Prism 5. For single-cell PCR analysis, the Spearman's rank correlation coefficient test was performed using R software to evaluate gene expression correlations with Gata6.
Acknowledgments
We thank N. Brockdorff for the B1-TS cell line, J. Rossant for the IM8A1 XEN cell line, R. Guyot for the pSG5-Flag plasmid, and P. Ovando Roche for his technical assistance. H. Acloque, H. Jorgensen, M. Parker, T. Rodriguez, and all the members of the Epigenetics and Development group are acknowledged for discussion and/or critical reading of the manuscript. This work was supported by the BBSRC, Genesis Research Trust, FCT, Imperial College London, ARC, LNCC, ERC, DFG, and ministry of Education, Culture, Sports, Science, and Technology in Japan.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.188193.112.
References
- Alder O, Lavial F, Helness A, Brookes E, Pinho S, Chandrashekran A, Arnaud P, Pombo A, O'Neill L, Azuara V 2010. Ring1B and Suv39h1 delineate distinct chromatin states at bivalent genes during early mouse lineage commitment. Development 137: 2483–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arney KL, Erhardt S, Drewell RA, Surani MA 2001. Epigenetic reprogramming of the genome–from the germ line to the embryo and back again. Int J Dev Biol 45: 533–540 [PubMed] [Google Scholar]
- Artus J, Piliszek A, Hadjantonakis AK 2011. The primitive endoderm lineage of the mouse blastocyst: Sequential transcription factor activation and regulation of differentiation by Sox17. Dev Biol 350: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. 2006. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Bruggeman SW, Hulsman D, Tanger E, Buckle T, Blom M, Zevenhoven J, van Tellingen O, van Lohuizen M 2007. Bmi1 controls tumor development in an Ink4a/Arf-independent manner in a mouse model for glioma. Cancer Cell 12: 328–341 [DOI] [PubMed] [Google Scholar]
- Capo-Chichi CD, Rula ME, Smedberg JL, Vanderveer L, Parmacek MS, Morrisey EE, Godwin AK, Xu XX 2005. Perception of differentiation cues by GATA factors in primitive endoderm lineage determination of mouse embryonic stem cells. Dev Biol 286: 574–586 [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T 2008. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells 26: 1496–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113: 643–655 [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A 2007. Nanog safeguards pluripotency and mediates germline development. Nature 450: 1230–1234 [DOI] [PubMed] [Google Scholar]
- Chazaud C, Yamanaka Y, Pawson T, Rossant J 2006. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2–MAPK pathway. Dev Cell 10: 615–624 [DOI] [PubMed] [Google Scholar]
- Clurman BE, Sheaff RJ, Thress K, Groudine M, Roberts JM 1996. Turnover of cyclin E by the ubiquitin–proteasome pathway is regulated by cdk2 binding and cyclin phosphorylation. Genes Dev 10: 1979–1990 [DOI] [PubMed] [Google Scholar]
- Dietrich JE, Hiiragi T 2007. Stochastic patterning in the mouse pre-implantation embryo. Development 134: 4219–4231 [DOI] [PubMed] [Google Scholar]
- Endoh M, Endo TA, Endoh T, Fujimura Y, Ohara O, Toyoda T, Otte AP, Okano M, Brockdorff N, Vidal M, et al. 2008. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development 135: 1513–1524 [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292: 154–156 [DOI] [PubMed] [Google Scholar]
- Frankenberg S, Gerbe F, Bessonnard S, Belville C, Pouchin P, Bardot O, Chazaud C 2011. Primitive endoderm differentiates via a three-step mechanism involving Nanog and RTK signaling. Dev Cell 21: 1005–1013 [DOI] [PubMed] [Google Scholar]
- Fujikura J, Yamato E, Yonemura S, Hosoda K, Masui S, Nakao K, Miyazaki Ji J, Niwa H 2002. Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev 16: 784–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinstein E, Mahotka C 2009. Stem cell divisions controlled by the proto-oncogene BMI-1. J Stem Cells 4: 141–146 [PubMed] [Google Scholar]
- Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P 2010. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell 18: 675–685 [DOI] [PubMed] [Google Scholar]
- Home P, Ray S, Dutta D, Bronshteyn I, Larson M, Paul S 2009. GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J Biol Chem 284: 28729–28737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa H, Kimura MY, Shinnakasu R, Suzuki A, Miki T, Koseki H, van Lohuizen M, Yamashita M, Nakayama T 2006. Regulation of Th2 cell development by Polycomb group gene bmi-1 through the stabilization of GATA3. J Immunol 177: 7656–7664 [DOI] [PubMed] [Google Scholar]
- Jedrusik A, Parfitt DE, Guo G, Skamagki M, Grabarek JB, Johnson MH, Robson P, Zernicka-Goetz M 2008. Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes Dev 22: 2692–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen HF, Giadrossi S, Casanova M, Endoh M, Koseki H, Brockdorff N, Fisher AG 2006. Stem cells primed for action: Polycomb repressive complexes restrain the expression of lineage-specific regulators in embryonic stem cells. Cell Cycle 5: 1411–1414 [DOI] [PubMed] [Google Scholar]
- Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F 1999. The transcription factor GATA6 is essential for early extraembryonic development. Development 126: 723–732 [PubMed] [Google Scholar]
- Kunath T, Arnaud D, Uy GD, Okamoto I, Chureau C, Yamanaka Y, Heard E, Gardner RL, Avner P, Rossant J 2005. Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development 132: 1649–1661 [DOI] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Ono Y, Uno KD, Yamada RG, Ueda HR, Saitou M 2006. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res 34: e42 doi: 10.1093/nar/gkl050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutner RH, Zhang XY, Reiser J 2009. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protoc 4: 495–505 [DOI] [PubMed] [Google Scholar]
- Lavial F, Acloque H, Bertocchini F, Macleod DJ, Boast S, Bachelard E, Montillet G, Thenot S, Sang HM, Stern CD, et al. 2007. The Oct4 homologue PouV and Nanog regulate pluripotency in chicken embryonic stem cells. Development 134: 3549–3563 [DOI] [PubMed] [Google Scholar]
- Leeb M, Wutz A 2007. Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J Cell Biol 178: 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CY, Tam WL, Zhang J, Ang HS, Jia H, Lipovich L, Ng HH, Wei CL, Sung WK, Robson P, et al. 2008. Sall4 regulates distinct transcription circuitries in different blastocyst-derived stem cell lineages. Cell Stem Cell 3: 543–554 [DOI] [PubMed] [Google Scholar]
- Martin GR 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci 78: 7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meilhac SM, Adams RJ, Morris SA, Danckaert A, Le Garrec JF, Zernicka-Goetz M 2009. Active cell movements coupled to positional induction are involved in lineage segregation in the mouse blastocyst. Dev Biol 331: 210–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt DM, Kemler R 2010. Nanog is required for primitive endoderm formation through a non-cell autonomous mechanism. Dev Biol 344: 129–137 [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113: 631–642 [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ 2003. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425: 962–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Teo RT, Li H, Robson P, Glover DM, Zernicka-Goetz M 2010. Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc Natl Acad Sci 107: 6364–6369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisey EE, Tang Z, Sigrist K, Lu MM, Jiang F, Ip HS, Parmacek MS 1998. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev 12: 3579–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niakan KK, Ji H, Maehr R, Vokes SA, Rodolfa KT, Sherwood RI, Yamaki M, Dimos JT, Chen AE, Melton DA, et al. 2010. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev 24: 312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Silva J, Roode M, Smith A 2009. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development 136: 3215–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T 2001. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol 21: 4330–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF 2003. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423: 302–305 [DOI] [PubMed] [Google Scholar]
- Plusa B, Piliszek A, Frankenberg S, Artus J, Hadjantonakis AK 2008. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development 135: 3081–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puschendorf M, Terranova R, Boutsma E, Mao X, Isono K, Brykczynska U, Kolb C, Otte AP, Koseki H, Orkin SH, et al. 2008. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet 40: 411–420 [DOI] [PubMed] [Google Scholar]
- Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS, Rossant J 2010. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 137: 395–403 [DOI] [PubMed] [Google Scholar]
- Rossant J 2008. Stem cells and early lineage development. Cell 132: 527–531 [DOI] [PubMed] [Google Scholar]
- Rula ME, Cai KQ, Moore R, Yang DH, Staub CM, Capo-Chichi CD, Jablonski SA, Howe PH, Smith ER, Xu XX 2007. Cell autonomous sorting and surface positioning in the formation of primitive endoderm in embryoid bodies. Genesis 45: 327–338 [DOI] [PubMed] [Google Scholar]
- Santos F, Peters AH, Otte AP, Reik W, Dean W 2005. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol 280: 225–236 [DOI] [PubMed] [Google Scholar]
- Satijn DP, Otte AP 1999. Polycomb group protein complexes: Do different complexes regulate distinct target genes? Biochim Biophys Acta 1447: 1–16 [DOI] [PubMed] [Google Scholar]
- Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al. 2009. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 138: 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimosato D, Shiki M, Niwa H 2007. Extra-embryonic endoderm cells derived from ES cells induced by GATA factors acquire the character of XEN cells. BMC Dev Biol 7: 80 doi: 10.1186/1471-213X-7-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A 2009. Nanog is the gateway to the pluripotent ground state. Cell 138: 722–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AM, Hamazaki T, Hankowski KE, Terada N 2007. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells 25: 2534–2542 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J 1998. Promotion of trophoblast stem cell proliferation by FGF4. Science 282: 2072–2075 [DOI] [PubMed] [Google Scholar]
- Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M 2007. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445: 214–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lugt NM, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H, van der Valk M, Deschamps J, Sofroniew M, van Lohuizen M, et al. 1994. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev 8: 757–769 [DOI] [PubMed] [Google Scholar]
- van der Stoop P, Boutsma EA, Hulsman D, Noback S, Heimerikx M, Kerkhoven RM, Voncken JW, Wessels LF, van Lohuizen M 2008. Ubiquitin E3 ligase Ring1b/Rnf2 of polycomb repressive complex 1 contributes to stable maintenance of mouse embryonic stem cells. PLoS ONE 3: e2235 doi: 10.1371/journal.pone.0002235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lohuizen M 1999. The trithorax-group and polycomb-group chromatin modifiers: Implications for disease. Curr Opin Genet Dev 9: 355–361 [DOI] [PubMed] [Google Scholar]
- Voncken JW, Niessen H, Neufeld B, Rennefahrt U, Dahlmans V, Kubben N, Holzer B, Ludwig S, Rapp UR 2005. MAPKAP kinase 3pK phosphorylates and regulates chromatin association of the Polycomb group protein Bmi1. J Biol Chem 280: 5178–5187 [DOI] [PubMed] [Google Scholar]
- Yamanaka Y, Lanner F, Rossant J 2010. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development 137: 715–724 [DOI] [PubMed] [Google Scholar]
- Yang DH, Smith ER, Roland IH, Sheng Z, He J, Martin WD, Hamilton TC, Lambeth JD, Xu XX 2002. Disabled-2 is essential for endodermal cell positioning and structure formation during mouse embryogenesis. Dev Biol 251: 27–44 [DOI] [PubMed] [Google Scholar]