This study focuses on the regulation of cohesin during DNA damage repair. Wu et al. establish that the Smc5/6 complex influences cohesin function at DNA double-strand break sites by sumoylating the cohesin subunit Scc1. This modification is found to be critical for sister chromatid recombination, and the work further delineates the mechanism of sumoylated Scc1 action via antagonizing the negative regulator Wapl.
Keywords: cohesin, DNA repair, Smc5, Smc6, SUMO
Abstract
DNA double-strand breaks (DSBs) fuel cancer-driving chromosome translocations. Two related structural maintenance of chromosomes (Smc) complexes, cohesin and Smc5/6, promote DSB repair through sister chromatid homologous recombination (SCR). Here we show that the Smc5/6 subunit Mms21 sumoylates multiple lysines of the cohesin subunit Scc1. Mms21 promotes cohesin-dependent small ubiquitin-like modifier (SUMO) accumulation at laser-induced DNA damage sites in S/G2 human cells. Cells expressing the nonsumoylatable Scc1 mutant (15KR) maintain sister chromatid cohesion during mitosis but are defective in SCR and sensitive to ionizing radiation (IR). Scc1 15KR is recruited to DNA damage sites. Depletion of Wapl, a negative cohesin regulator, rescues SCR defects of Mms21-deficient or Scc1 15KR-expressing cells. Expression of the acetylation-mimicking Smc3 mutant does not bypass the requirement for Mms21 in SCR. We propose that Scc1 sumoylation by Mms21 promotes SCR by antagonizing Wapl at a step after cohesin loading at DSBs and in a way not solely dependent on Smc3 acetylation.
A DNA double-strand break (DSB) is the most deleterious form of DNA damage and can cause chromosome translocations, a major class of cancer-driving mutations (Khanna and Jackson 2001; West 2003). Homologous recombination (HR) is a key DSB repair pathway and requires an undamaged DNA template: sister or homologous chromosomes. Because HR between homologous chromosomes can result in loss of heterozygosity (LOH), DSB repair with HR between sister chromatids (SCR) is preferable during the mitotic cell cycle. Two related structural maintenance of chromosomes (Smc) complexes, cohesin and the Smc5/6 complex, play critical roles in SCR-dependent repair of DSBs (Watrin and Peters 2006; De Piccoli et al. 2009; Potts 2009).
The primary function of cohesin is to establish sister chromatid cohesion in S phase and maintain this cohesion until the metaphase–anaphase transition (Onn et al. 2008; Peters et al. 2008; Nasmyth and Haering 2009). Although cohesin is loaded onto chromosomes during telophase or G1, it only becomes cohesive during S phase. Cohesion establishment in S phase requires Smc3 acetylation by the acetyltransferase Eco1 at K112 and K113 in yeast (Rolef Ben-Shahar et al. 2008; Unal et al. 2008; Rowland et al. 2009) or by the Eco1 homologs Esco1/2 at K105 and K106 in human cells (Hou and Zou 2005; Zhang et al. 2008). In human cells, Smc3 acetylation enables the chromatin binding of sororin through Pds5, which protects cohesin from the cohesin inhibitor Wapl, thus establishing functional cohesion (Rankin et al. 2005; Kueng et al. 2006; Nishiyama et al. 2010).
The Smc5/6 complex has multiple functions in DNA damage response (De Piccoli et al. 2009; Potts 2009). It is recruited to DSBs and is critical for their repair through SCR in yeast and humans (De Piccoli et al. 2006; Lindroos et al. 2006; Potts et al. 2006). The Smc5/6 complex component Mms21 is a ligase for small ubiquitin-like modifier (SUMO) (Potts and Yu 2005; Zhao and Blobel 2005). Although it is clear that the SUMO ligase activity of Mms21 is required for DNA repair (Potts and Yu 2005; Zhao and Blobel 2005), the critical targets of Mms21 in DSB repair have not been established.
In addition to its role in global sister chromatid cohesion and chromosome segregation in mitosis, cohesin is loaded locally at DSBs to facilitate SCR in organisms from yeast to humans. In yeast, cohesin is loaded de novo at DSBs in a process that requires the cohesin-loading complex Scc2/4 (Strom et al. 2004; Unal et al. 2004). DNA damage then induces ATR/Chk1-dependent phosphorylation of the cohesin subunit Scc1 at S83, which in turn enables its acetylation at K84 and K210 by Eco1 (Heidinger-Pauli et al. 2008, 2009). Scc1 acetylation antagonizes Wapl to establish DNA damage-induced (DI) cohesion (Strom et al. 2007; Unal et al. 2007; Heidinger-Pauli et al. 2009). Thus, Eco1 has distinct targets during replicative and DI cohesion establishment in yeast. The Smc5/6 complex is loaded at DSBs (De Piccoli et al. 2006; Lindroos et al. 2006). It is not required for the initial loading of cohesin at DSBs but is required for DI cohesion establishment (Strom et al. 2007). The mechanism by which Smc5/6 contributes to DI cohesion establishment is unknown.
In human cells, both cohesin and the Smc5/6 complex are loaded at endonuclease-induced DSBs and collaborate in the same pathway to promote SCR (Potts et al. 2006). In contrast to yeast, however, it is unclear whether DNA damage triggers local DI cohesion in human cells, and if so, how this cohesion is established. The Chk1 phosphorylation site (S83) and the Eco1 acetylation sites (K84 and K210) in yeast Scc1 are not conserved in human Scc1 (see Supplemental Fig. S5), suggesting that if human cells establish DI cohesion, they might use a different mechanism to do so.
In this study, we show that SUMO accumulates at laser-induced DNA damage sites in S/G2 human cells and this accumulation requires both cohesin and Mms21. We further identify Scc1 as an Mms21-dependent SUMO substrate. Mms21 sumoylates multiple lysines in the central region of Scc1. We created an Scc1 mutant named 15KR, which cannot be sumoylated by Mms21. Cells expressing Scc1 15KR in place of the endogenous Scc1 maintain proper sister chromatid cohesion during mitosis but are defective in SCR and sensitive to ionizing radiation (IR). Scc1 15KR is still recruited to laser-induced DNA damage sites. Importantly, depletion of Wapl rescues the SCR defect and IR sensitivity of Mms21-deficient or Scc1 15KR-expressing cells. Thus, Scc1 sumoylation by Mms21 promotes SCR through antagonizing Wapl at a step after cohesin recruitment to DSBs. Expression of the acetylation-mimicking Smc3 mutant fails to bypass the requirement for Mms21 in SCR. Smc3 acetylation, therefore, is unlikely to be the sole critical event downstream from Scc1 sumoylation in SCR. Our study reveals both conserved principles and organism-specific features in the function and regulation of cohesin in DNA repair.
Results
Mms21 sumoylates Scc1 at multiple sites
We first verified that cohesin was required for SCR and DSB repair in human cells. For this purpose, we performed colony survival and sister chromatid exchange (SCE) assays on HeLa Tet-On cells depleted of cohesin subunits by RNAi. Depletion of Smc1, Smc3, Scc1, or SA1/2 expectedly rendered these cells sensitive to IR (Supplemental Fig. S1A). SCE is one form of SCR that involves the resolution of double Holliday junctions in a crossover-producing pathway (West 2003). In the SCE assay, cells were incubated with the bulky nucleotide analog BrdU through two rounds of DNA replication and division. In the end, both DNA strands of one sister chromatid incorporated BrdU, while only one strand of the other sister contained BrdU. The sister with two BrdU-containing strands was more weakly stained with Acridine Orange, a DNA-intercalating dye. SCE was induced by the topoisomerase I inhibitor camptothecin and then scored in metaphase spreads by counting the number of crossover events between sister chromatids. Both Scc1-RNAi and SA1/2-RNAi cells showed a decrease in SCE frequency (Supplemental Fig. S1B,C). Depletion of each cohesin subunit was confirmed by Western blots (Supplemental Fig. S1D). Depletion of a given cohesin subunit often reduced the protein levels of other subunits, suggesting that cohesin existed as a single functional entity. Therefore, cohesin is required for SCR and DSB repair in human cells. We note that cohesin depletion also caused sister chromatid separation in mitosis in a fraction of the metaphase spreads. We were only able to count SCE events in the cells that retained cohesion and presumably had incomplete cohesin knockdown. Thus, the decrease in SCE seen in cohesin RNAi cells was likely an underestimation.
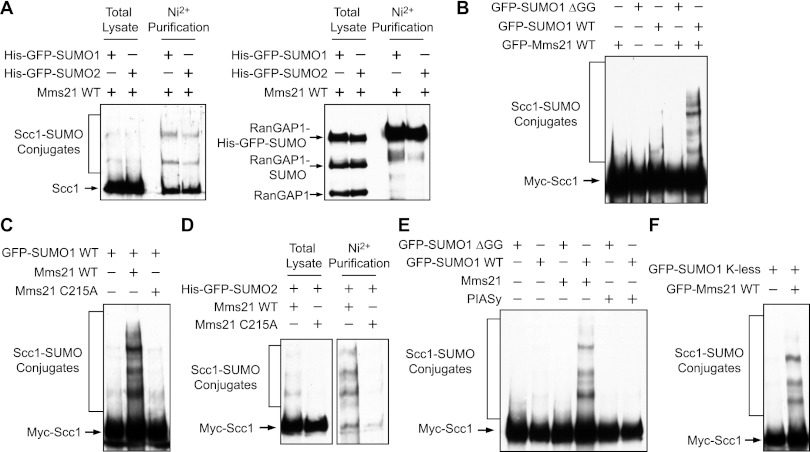
We showed previously that the SUMO ligase activity of human Mms21 was required for DNA repair (Potts and Yu 2005). Mms21 could sumoylate Scc1 and SA2, two cohesin subunits (Potts et al. 2006). In yeast, Mms21 had been shown to sumoylate the cohesin subunits Smc1 and Smc3 (Takahashi et al. 2008). To determine whether Mms21-dependent sumoylation of cohesin was functionally important, we decided to characterize the sumoylation of Scc1 by Mms21. We thus transfected HeLa cells with plasmids encoding Mms21 and His6-GFP-SUMO1/2, lysed the cells with a protein-denaturing buffer, performed Ni2+ bead pull-down from the lysates, and blotted the lysates and proteins bound to beads with anti-Scc1. As a positive control, we blotted the lysates and beads with anti-RanGAP1, a known SUMO-conjugated protein (Matunis et al. 1998). RanGAP1 that was conjugated to His6-GFP-SUMO1/2 was highly enriched by Ni2+ bead pull-down, validating the assay (Fig. 1A). Several Scc1 bands were also enriched by Ni2+ bead pull-down, indicating that they were Scc1–SUMO conjugates (Fig. 1A). Thus, the endogenous cohesin indeed underwent Mms21-dependent sumoylation. This sumoylation was not further enhanced by IR treatment (data not shown). Despite repeated attempts, we could not detect sumoylation of endogenous cohesin by immunoblotting in G2 human cells with or without IR in the absence of Mms21 or SUMO overexpression (data not shown), presumably because only a small pool of cohesin was sumoylated.
Figure 1.
Mms21 stimulates cohesin sumoylation in human cells. (A) HeLa Tet-On cells transfected with Mms21 and His6-GFP-SUMO1/2 plasmids were lysed with a protein-denaturing buffer and subjected to Ni2+ bead pull-down. The total lysates and pull-down were blotted with α-Scc1 (left panel) or α-RanGAP1 (right panel). (B) Mms21 enhances Scc1 sumoylation in human cells. Myc-Scc1 was coexpressed with GFP-SUMO1 wild type (WT) or ΔGG in the presence or absence of GFP-Mms21 in HeLa cells for 24 h. The cell lysates were blotted with α-Myc. The positions of unmodified and sumoylated Scc1 are labeled. (C) The ligase activity of Mms21 is required for Scc1 sumoylation. Lysates of HeLa cells transfected with the indicated plasmids were blotted with α-Myc. The C215A mutation abolished the ligase activity of Mms21. (D) HeLa Tet-On cells transfected with Myc-Scc1, Mms21 wild type/C215A, and His6-GFP-SUMO2 plasmids were lysed with a protein-denaturing buffer and subjected to Ni2+ bead pull-down. The total lysates (left panel) and pull-down (right panel) were blotted with α-Myc. (E) Lysates of HeLa Tet-On cells transfected with the indicated plasmids were blotted with α-Myc. The positions of sumoylated Scc1 are labeled. (F) Mms21 sumoylates multiple lysines of Scc1. HeLa cells were transfected with a plasmid encoding GFP-SUMO1 K-less (the lysine-less SUMO1 mutant that cannot form chains) in the presence or absence of the GFP-Mms21 plasmid. Lysates were blotted with α-Myc.
We next performed sumoylation assays on ectopically expressed Scc1. We transfected HeLa cells with different combinations of plasmids encoding GFP-SUMO1, GFP-SUMO1 ΔGG (a SUMO mutant lacking the two C-terminal glycines required for conjugation), GFP-Mms21, and Myc-Scc1 and blotted the cell lysates with anti-Myc (Fig. 1B). Myc-Scc1 formed high-molecular-mass species in the presence of GFP-SUMO1 but not GFP-SUMO1 ΔGG. Mms21 greatly increased the intensity of these Myc-Scc1 species. In contrast, the ligase-dead mutant of Mms21 (C215A) did not stimulate the production of the modified forms of Myc-Scc1 (Fig. 1C). Moreover, when His6-GFP SUMO2 was cotransfected with Myc-Scc1 and Mms21 wild type or C215A, these modified forms of Myc-Scc1 were absent in the Mms21 C215A sample and highly enriched by Ni2+ bead pull-down, indicating that these species were Scc1–SUMO conjugates (Fig. 1D). Finally, another SUMO ligase, PIASy, failed to stimulate the sumoylation of Myc-Scc1 under the same conditions (Fig. 1E). Thus, these results confirmed that Scc1 was sumoylated by Mms21 in human cells.
The Scc1–SUMO conjugates formed several discernable bands. To determine whether these conjugates contained SUMO chains or represented Scc1 sumoylation at multiple sites, we performed the sumoylation assay with GFP-SUMO1 K-less, a SUMO1 mutant with all lysines mutated to arginine (Gocke et al. 2005). A similar multiband sumoylation pattern of Myc-Scc1 was observed with SUMO1 K-less (Fig. 1F). Because SUMO1 K-less could not form chains, this result indicated that Mms21 sumoylated multiple residues of Scc1. Likewise, both SUMO2 wild type and K11R (a SUMO2 mutant that cannot efficiently form chains) produced Scc1 sumoylation patterns similar to SUMO1 (data not shown), indicating that Mms21 also catalyzed conjugation of SUMO2 to multiple sites in Scc1.
Construction of a nonsumoylatable Scc1 mutant
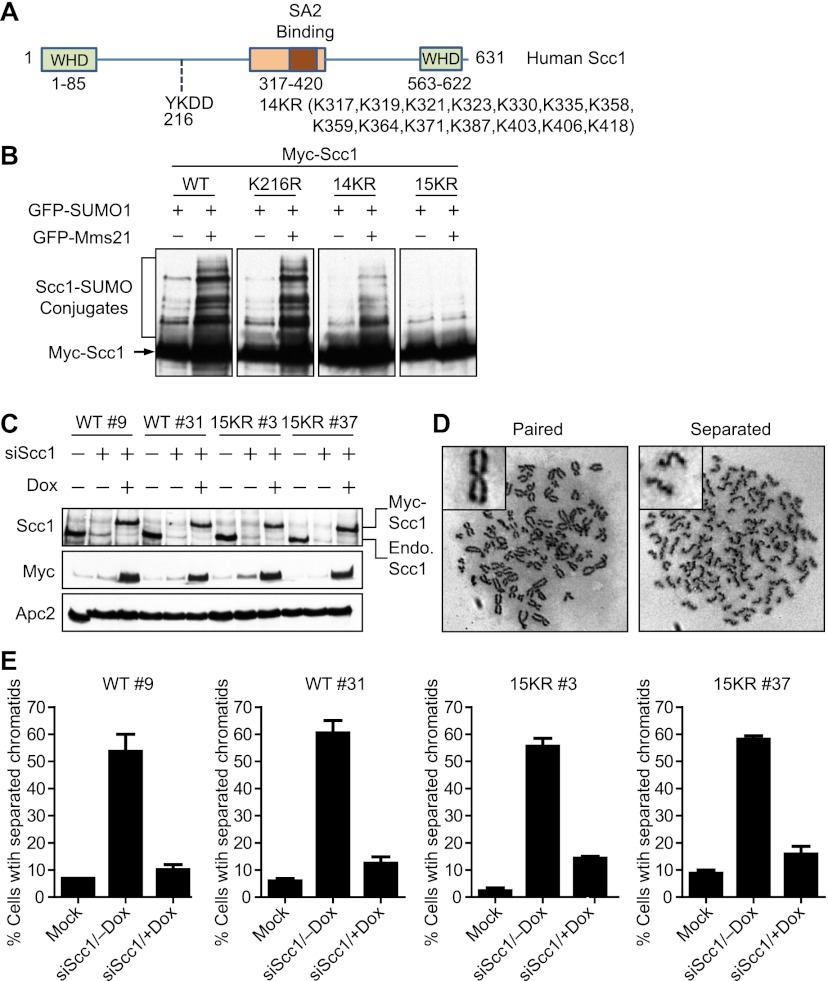
To elucidate the potential roles of Mms21-dependent sumoylation of Scc1 in DSB repair, we needed to create an Scc1 mutant that could no longer be sumoylated by Mms21. Sumoylation often occurs on lysines within ΨKXD/E (where Ψ is a large hydrophobic residue, and X is any residue) consensus motifs, which directly interact with Ubc9 (Gareau and Lima 2010). Scc1 contained one such motif at K216 (Fig. 2A). However, Scc1 K216R was still efficiently sumoylated by Mms21 (Fig. 2B), consistent with our finding that Mms21 sumoylated Scc1 at multiple sites.
Figure 2.
The nonsumoylatable Scc1 15KR mutant is functional in mitotic cohesion. (A) Schematic drawing of the domains and motifs of human Scc1. The positions of two winged helix domains (WHD) required for Smc1/3 binding, the central conserved domain that binds to SA, and a consensus sumoylation motif at K216 are indicated. The Scc1 15KR mutations contain the K216R mutation and mutations of all 14 lysines in the central region (residues 317–420) to arginines (14KR). (B) Myc-Scc1 wild type (WT) and mutants were expressed together with GFP-SUMO1 in the presence or absence of GFP-Mms21 in HeLa cells. The cell lysates were blotted with α-Myc. (C) Construction of stable HeLa Tet-On cell lines that inducibly expressed Myc-Scc1 wild type or 15KR. Two clones of each cell line cultured with or without doxycycline (Dox) were either mock-transfected or transfected with siScc1. Cell lysates were blotted with the indicated antibodies. The positions of Myc-Scc1 or endogenous (Endo.) Scc1 are labeled. (D) Representative metaphase spreads of cells in C with paired (left panel) or separated (right panel) sister chromatids. (E) Quantification of the percentage of mitotic cells in C with separated sister chromatids. More than 200 cells in each sample were counted. The mean and standard deviation of data in two independent experiments are shown.
To identify the Mms21 sumoylation sites of Scc1, we generated a series of truncation mutants of Scc1 (Supplemental Fig. S2) and tested each fragment in the in vivo sumoylation assay. Scc1I (residues 317–420) containing the SA-binding domain was the smallest fragment that retained Mms21-dependent sumoylation (Supplemental Fig. S2). A SUMO ligase can catalyze sumoylation in two ways: It can bring the Ubc9–SUMO thioester and substrate into a complex or stimulate Ubc9 to discharge SUMO to substrate (Gareau and Lima 2010). When coexpressed in HeLa cells, Myc-Scc1 indeed bound to HA-Mms21 (data not shown). Scc1I was the smallest Scc1 fragment that retained both Mms21 binding and Mms21-dependent sumoylation (Supplemental Fig. S2). These results suggest that Mms21 (possibly as a part of the intact Smc5/6 complex) simultaneously binds to Scc1 and Ubc9–SUMO and brings them into close proximity, thereby facilitating Scc1 sumoylation.
We could not detect a physical interaction between the endogenous Scc1 and Mms21 in human cells with or without IR. We thus fractionated lysates of HeLa cells arrested in G2 and treated with or without IR on a gel filtration column (Supplemental Fig. S3). All cohesin subunits eluted in the same fractions. All tested Smc5/6 components also cofractioned with one another. IR treatment did not significantly alter the fractionation profiles of cohesin, Smc5/6, and the cohesin regulators Pds5A/B and Wapl. Therefore, our data suggested that the bulk of Mms21 existed as a component of the Smc5/6 complex in human cells with or without DNA damage. It might sumoylate cohesin through a transient interaction between the two intact complexes. We cannot exclude the possibility that a small pool of Mms21 shuttles between Smc5/6 and cohesin complexes.
On the other hand, Mms21 stimulated the sumoylation of an Scc1 fragment (Scc1F) that could not bind to Smc1 or Smc3 (Supplemental Fig. S4A), suggesting that sumoylation of Scc1 by Mms21 did not require the intact cohesin. To test whether free Mms21 stimulated sumoylation of Scc1, we expressed and purified from bacteria the full-length human Mms21. Recombinant Mms21 stimulated the sumoylation of in vitro translated full-length Myc-Scc1 or Scc1F in the presence of SUMO1, Aos1/Uba2 (E1), and Ubc9 (E2) (Supplemental Fig. S4B,C). Therefore, Mms21 is sufficient to sumoylate Scc1 in vitro, although cohesin sumoylation in vivo might be mediated through a transient interaction between the intact Smc5/6 and cohesin complexes.
Scc1I contained 14 lysines, some of which were conserved in metazoans (Supplemental Fig. S5). Mutation of all 14 lysines to arginines (14KR) in the context of the full-length Scc1 or deleting residues 317–420 greatly reduced Scc1 sumoylation (Fig. 2B; Supplemental Fig. S4D). Mutations of different subsets of these 14 lysines did not appreciably diminish Scc1 sumoylation (data not shown), suggesting that sumoylation of these lysines might be distributive, with many of them being potential SUMO acceptors. Finally, we created the Scc1 15KR mutant, which contained the K216R and 14KR mutations. Scc1 15KR was only weakly sumoylated in the presence of SUMO1 (Fig. 2B). This weak sumoylation was not stimulated by Mms21. Thus, Scc1 15KR has lost its ability to be sumoylated by Mms21.
Scc1 15KR is functional in sister chromatid cohesion during mitosis
To study the functions of Scc1 sumoylation, we generated stable HeLa Tet-On cell lines that expressed Myc-Scc1 wild-type or 15KR transgenes driven by a tetracycline-inducible promoter. Both Scc1 transgenes contained silent mutations to make them resistant to RNAi-mediated depletion. For subsequent experiments, we chose two clones for each cell line: clones 9 and 31 for Myc-Scc1 wild type, and clones 3 and 37 for Myc-Scc1 15KR. In the presence of doxycycline, these clones expressed Myc-Scc1 wild type or 15KR at levels comparable with that of the endogenous Scc1 (Fig. 2C).
We first tested whether Scc1 15KR supported replicative cohesion establishment and proper chromosome segregation in mitosis. In the metaphase spread of a cell with normal cohesion, sister chromatids were paired at their centromeres and had an X shape, while sister chromatids had lost their connection at centromeres and were not paired in cohesion-defective cells (Fig. 2D). In the absence of doxycycline, depletion of the endogenous Scc1 caused premature sister chromatid separation in >50% of mitotic cells in all four clones (Fig. 2E). In the presence of doxycycline, expression of Myc-Scc1 wild type or 15KR largely rescued the cohesion defect caused by Scc1 RNAi. Thus, Scc1 15KR supports proper sister chromatid cohesion in mitosis and, by inference, is capable of establishing replicative cohesion in normal S phase. Consistently, Scc1 15KR could still bind to Smc1, Smc3, and SA2 (data not shown).
Scc1 15KR is defective in DSB repair and SCR
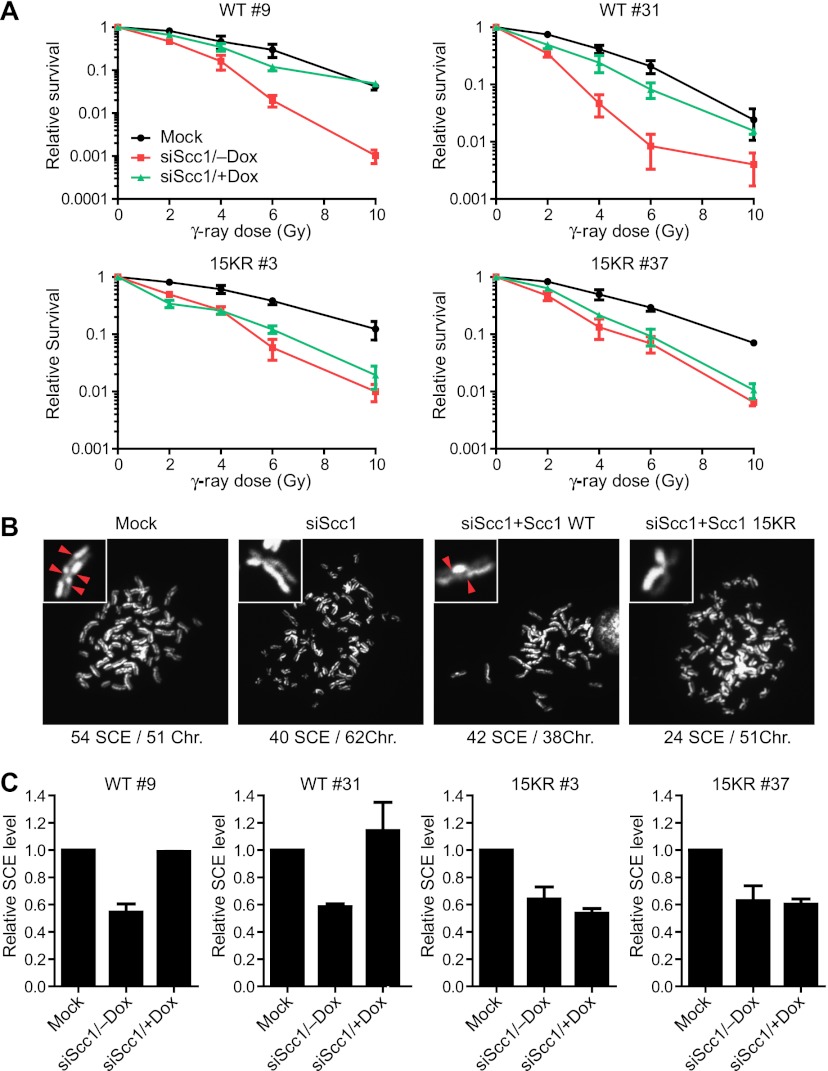
Next, we examined whether Scc1 15KR supported the DNA repair function of cohesin. In the absence of doxycycline, depletion of Scc1 by RNAi rendered cells expressing Myc-Scc1 wild type or 15KR sensitive to IR (Fig. 3A). Doxycycline-induced expression of Myc-Scc1 wild type, but not 15KR, rescued the IR sensitivity caused by Scc1 depletion. Depletion of Scc1 also reduced the frequency of SCE in these cells in the absence of doxycycline (Fig. 3B,C). Induced expression of Myc-Scc1 wild type restored the SCE frequency to that of the mock-depleted cells. In contrast, expression of Myc-Scc1 15KR did not rescue the SCE defect caused by Scc1 knockdown. These results indicate that Scc1 15KR is defective in DSB repair and SCR and further suggest that Mms21-dependent sumoylation of Scc1 might be critical for DNA repair.
Figure 3.
Cells expressing Scc1 15KR are sensitive to IR and defective in SCR. (A) Two clones each of HeLa Tet-On cell lines that inducibly expressed Myc-Scc1 wild type (WT) or 15KR were cultured with or without doxycycline (Dox), mock-transfected or transfected with siScc1, and irradiated with different doses of IR. The relative colony survival numbers are plotted against IR doses. Each data point represents the mean and standard deviation of values in two independent experiments, with duplicate samples in each experiment. The differences in the IR sensitivity seen in different clones of the Myc-Scc1-expressing lines after Scc1 RNAi are likely due to clonal variation. (B) Representative images of the SCE assays on cells in A. The numbers of SCE events and chromosomes in each image are shown below. A pair of sister chromatids for each image is magnified and shown in the inset, with the SCE events marked by red arrowheads. (C) Quantification of the relative SCE levels of cells described in A. The mean and standard deviation of data from two experiments are shown. About 30 cells were counted in each experiment.
Scc1 sumoylation is not required for cohesin recruitment to DNA damage sites
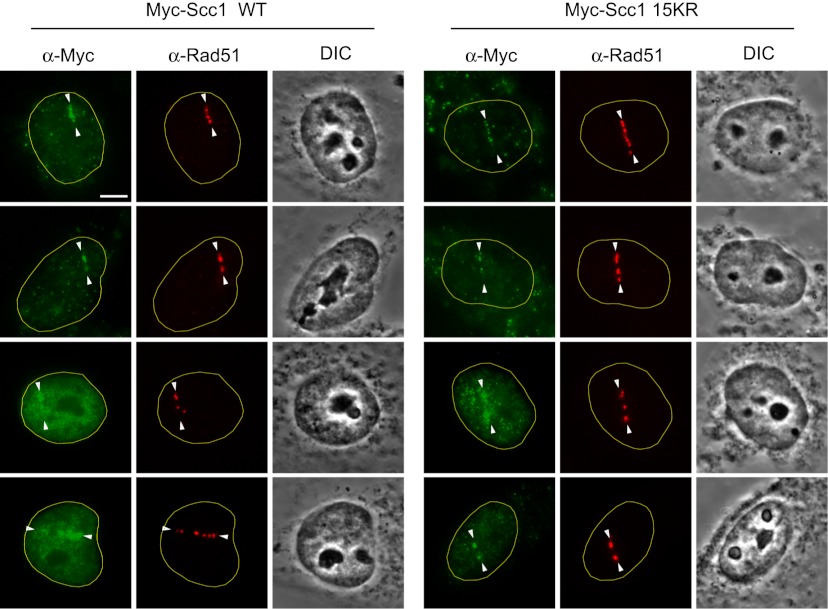
Because the nonsumoylatable Scc1 15KR mutant was defective in DSB repair and SCR, we wished to examine whether Scc1 15KR was recruited to DNA damage sites. We thus examined the recruitment of Myc-Scc1 wild type and 15KR to laser-induced DNA damage sites in fixed cells with immunofluorescence. HeLa Tet-On cells stably expressing Myc-Scc1 wild type or 15KR were synchronized at S/G2 by a thymidine arrest release protocol. DSBs were generated along a straight line using nanosecond green laser microirradiation. This system was used previously to demonstrate S/G2-specific and Mre11–Rad50-dependent cohesin recruitment to damage sites in human cells (Kim et al. 2002). This Mre11 dependence in cohesin recruitment to DSBs was corroborated by chromatin immunoprecipitation (ChIP) analysis in yeast (Strom et al. 2004; Unal et al. 2004), supporting the physiological relevance of our laser system. Cells were fixed at 1 h post-damage and stained with antibodies against Myc and the key HR protein, Rad51. Both Scc1 wild type and 15KR were recruited to the laser-induced DNA damage foci, along with Rad51 (Fig. 4). It is well established that cohesin loading to chromatin per se is insufficient to establish functional cohesion (Rolef Ben-Shahar et al. 2008; Unal et al. 2008; Rowland et al. 2009). Our result suggests that sumoylation of Scc1 does not affect cohesin recruitment to DSBs but might affect its function at a later step during SCR.
Figure 4.
Scc1 15KR is recruited to laser-induced DNA damage sites. HeLa Tet-On cells expressing Myc-Scc1 wild type (WT) or 15KR were cultured in the presence of doxycycline, synchronized at S/G2 by thymidine arrest and release, and subjected to laser irradiation along a straight line. At 1 h after laser cutting, cells were fixed and stained with α-Myc (green) and α-Rad51 (red). (DIC) Differential interference contrast. We irradiated 20 Myc-Scc1 wild-type-expressing and 20 Myc-Scc1 15KR-expressing cells. Among them, 16 Myc-Scc1 wild-type and 15 Myc-Scc1 15KR cells were Myc-positive. All Myc-positive cells in both groups exhibited enrichment of Myc signals at DNA damage sites, with 12 of 16 in the wild-type group and 11 of 15 in the 15KR group showing strong recruitment of Myc-Scc1 proteins to damage sites. Four representative cells of each cell line are shown. Bar, 10 μm.
Smc5/6 is dispensable for cohesin recruitment to DNA damage sites in human cells
We showed previously that the Smc5/6 complex was recruited to I-SceI endonuclease-induced DSBs by ChIP analysis and that depletion of Mms21 and Smc5 abolished cohesin recruitment to these sites in human cells (Potts et al. 2006), implicating Smc5/6 in cohesin recruitment. The fact that Scc1 15KR was still recruited to DNA damage sites was inconsistent with these previous results. Furthermore, in yeast, although the Smc5/6 complex is required for DI cohesion establishment, it is not required for cohesin recruitment to DSBs (Strom et al. 2007). Finally, the Mms21 siRNA used in our previous work (siMms21-7) and certain Smc5 siRNAs (siSmc5-5) were recently shown by others to cause premature sister chromatid separation (Behlke-Steinert et al. 2009), prompting the investigators to suggest that Smc5 and Mms21 might have roles in replicative cohesion establishment or maintenance. We decided to reinvestigate the role of Smc5/6 in cohesin recruitment to laser- and endonuclease-induced DNA damage sites in human cells.
First, we tested whether Smc5/6 was indeed required for proper chromosome segregation in human cells. We depleted Mms21 from HeLa cells using seven siRNAs. Although six Mms21 siRNAs depleted Mms21 efficiently, only siMms21-3, siMms21-5, and siMms21-7 caused premature sister chromatid separation (Supplemental Fig. S6). Moreover, ectopic expression of Mms21 resistant to siMms21-7 failed to rescue the chromosome segregation defects in cells transfected with siMms21-7 (data not shown). Similarly, only one of the six Smc5 siRNAs and none of the Smc6 siRNAs caused premature sister chromatid separation (Supplemental Fig. S6). Note that depletion of Smc5 also reduced the protein levels of Smc6, and vice versa. These results indicate that Smc5/6 and Mms21 are not required for replicative cohesion establishment and proper chromosome segregation during mitosis. siMms21-7 and siSmc5-5 may have depleted cohesin regulators through off-target effects.
Consistent with our previous ChIP analysis of endonuclease-induced DSB sites (Potts et al. 2006), Smc5 and Mms21 were recruited to laser-induced damage sites (Fig. 5A). The localization of Mms21 to these sites was dependent on Smc5, suggesting that it was recruited as a part of the Smc5/6 complex. Scc1 depletion had no effect on Mms21 localization (Fig. 5B), indicating that the Smc5/6 complex localized to damage sites independently of cohesin, consistent with the ChIP analysis (Potts et al. 2006). Transfection of siMms21-7 and siSmc5-7 diminished cohesin recruitment to laser-induced DNA damage sites (Supplemental Fig. S6F). In contrast, depletion of Smc5 and Mms21 with the siRNAs (siSmc5-2 and siMms21-6) that did not cause premature sister chromatid separation did not affect cohesin recruitment to DNA damage sites (Fig. 5C; Supplemental Fig. S6F). Depletion of Smc6 also did not prevent cohesin recruitment to damage sites (data not shown). Therefore, Smc5/6 and Mms21 are not required for the initial cohesin loading at laser-induced DNA damage sites.
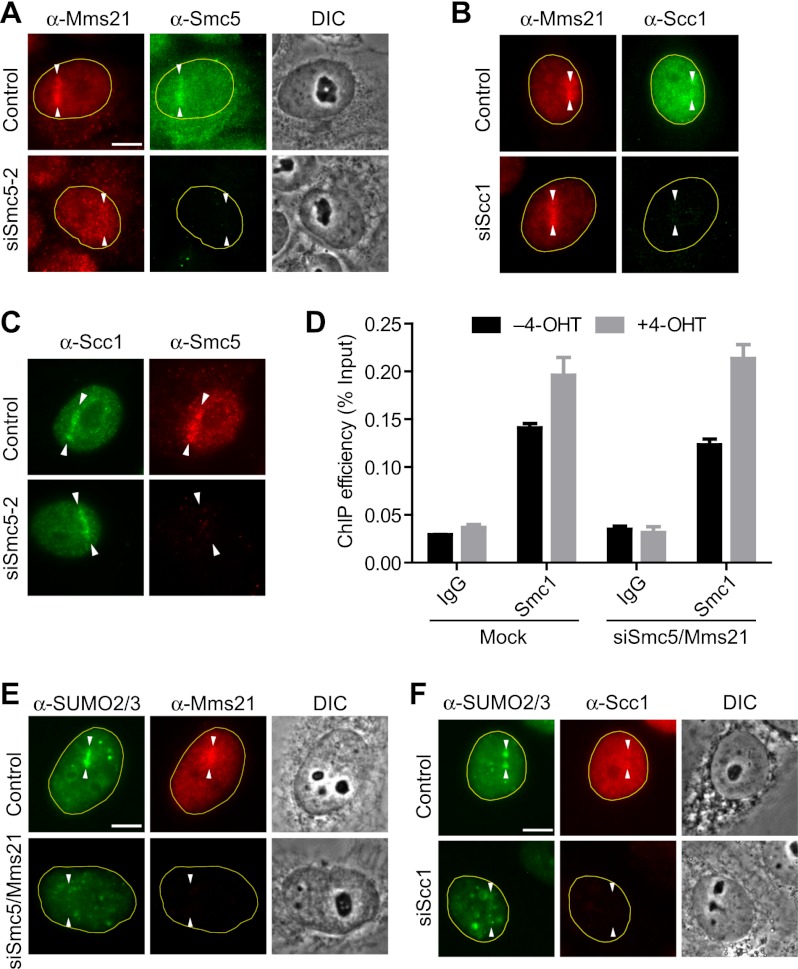
Figure 5.
The Smc5/6 complex is dispensable for cohesin loading at DNA damage sites. (A) HeLa cells transfected with siControl or siSmc5-2 were synchronized at S/G2 by thymidine arrest and release and subjected to laser irradiation along a straight line. At 1 h after laser cutting, cells were fixed and stained with α-Smc5 (green) and α-Mms21 (red). (DIC) Differential interference contrast. Bar, 10 μm. (B) HeLa cells transfected with siControl or siScc1 were synchronized at S/G2 and subjected to laser irradiation along a straight line. At 1 h after laser cutting, cells were fixed and stained with α-Scc1 (green) and α-Mms21 (red). (C) HeLa cells transfected with siControl or siSmc5-2 were synchronized at S/G2 by thymidine arrest and release and subjected to laser irradiation along a straight line. At 1 h after laser cutting, cells were fixed and stained with α-Scc1 (green) and α-Smc5 (red). (D) HeLa Tet-On cells were transfected with siControl or siSmc5-2/siMms21-6 and then transfected with the ER-I-PpoI plasmid. Cells were then treated with or without 4-OHT for 12 h and subjected to ChIP by IgG and α-Smc1 followed by qPCR analysis. The mean and standard deviation of two independent experiments are shown. (E) HeLa cells transfected with siControl or siSmc5-2/siMms21-6 were synchronized at S/G2 and subjected to laser irradiation along a straight line. At 1 h after laser cutting, cells were fixed and stained with α-SUMO2/3 (green) and α-Mms21 (red). (DIC) Differential interference contrast. Bar, 10 μm. (F) HeLa cells transfected with siControl or siScc1 were synchronized at S/G2 and subjected to laser irradiation along a straight line. At 1 h after laser cutting, cells were fixed and stained with α-SUMO2/3 (green) and α-Scc1 (red). (DIC) Differential interference contrast. Bar, 10 μm. Note that the Scc1 accumulation was not as prominent as in C because a different Scc1 antibody was used for this experiment to allow the costaining of SUMO2/3 and Scc1.
We next examined cohesin recruitment to endonuclease-induced DSBs by ChIP in human cells (Berkovich et al. 2007). We transfected HeLa Tet-On cells with a plasmid encoding I-PpoI fused to estrogen receptor (ER-I-PpoI) and added 4-hydroxytamoxifen (4-OHT) to induce the nuclear translocation of ER-I-PpoI and DNA cleavage. Addition of 4-OHT indeed induced DNA damage in a time-dependent manner, as indicated by the increase in γH2AX and phospho-Chk1 signals, which peaked at 12 h after 4-OHT addition (Supplemental Fig. S7). I-PpoI has been shown to induce DSBs in the native rDNA locus (Berkovich et al. 2007). We then performed ChIP experiments on cells treated with or without 12-h 4-OHT treatment and measured the amounts of rDNA in the immunoprecipitations using quantitative PCR (qPCR) (Fig. 5D). There was basal-level cohesin binding to the rDNA locus in the absence of DNA damage. I-PpoI-mediated DSB formation enhanced cohesin binding at this locus. Depletion of Smc5 and Mms21 with siSmc5-2 and siMms21-6 did not affect either the basal cohesin binding or the DSB-induced cohesin enrichment at the native rDNA locus. Thus, the Smc5/6 complex is dispensable for cohesin loading to endonuclease-induced DSBs in human cells. Taken together, our previous finding that depletion of Mms21 and Smc5 prevented cohesin loading at DSBs was due to off-target effects of the particular siRNAs used, and Mms21 and cohesin are recruited to DNA damage sites independently of each other.
Because cohesin was sumoylated by Mms21 in vitro and in human cells, we examined whether this occurred at damage sites. Since cohesin was recruited to damage sites only in the S/G2 phase of the cell cycle (Kim et al. 2002), we examined the SUMO signal at damage sites in S/G2 cells. A prominent SUMO2/3 signal was observed at damage sites, which was diminished by Smc5 and Mms21 depletion at this cell cycle stage (Fig. 5E). Furthermore, cohesin depletion greatly diminished the SUMO2/3 signal at damage sites (Fig. 5F). These results suggest that cohesin is the major target of Mms21-dependent sumoylation at damage sites. Taken together, our data suggest that cohesin sumoylation is not required for the initial cohesin targeting to damage sites but is important for a later function of cohesin in SCR.
Wapl depletion bypasses the requirement of Mms21 in SCR
Our data thus far support a role of Mms21-dependent sumoylation of Scc1 in promoting SCR at a step after the initial cohesin recruitment. Because Wapl is a negative regulator of cohesin throughout the cell cycle, we hypothesized that Mms21 promoted cohesin's function at DSBs through counteracting Wapl. We tested this hypothesis using three types of assays.
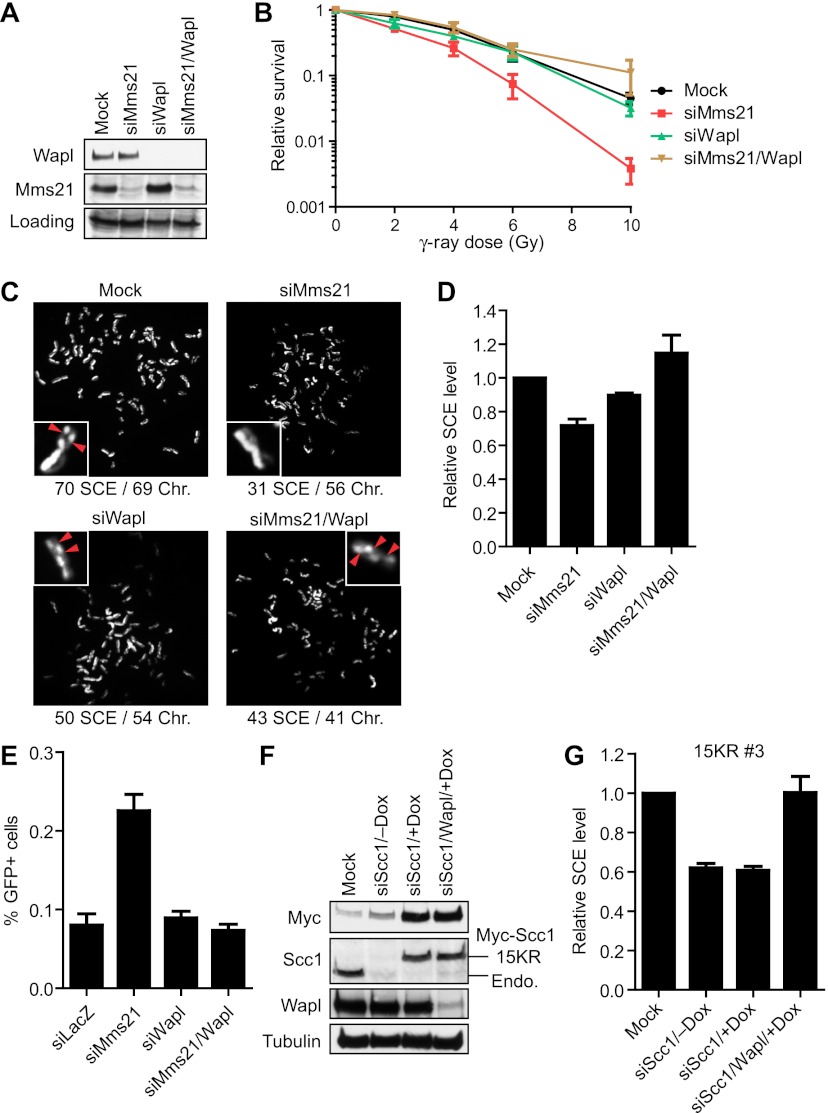
First, in the colony survival assay, Mms21 depletion rendered HeLa cells sensitive to IR, while Wapl depletion had no effect (Fig. 6A,B). Strikingly, codepletion of Wapl along with Mms21 rescued the IR sensitivity of Mms21-RNAi cells. Second, in the SCE assay, codepletion of Mms21 and Wapl restored the SCE frequency to that of the mock-transfected cells (Fig. 6C,D). Consistent with previous reports (Kueng et al. 2006), sister chromatids were less resolved in cells depleted of Wapl alone and cells depleted of both Wapl and Mms21 (Fig. 6C).
Figure 6.
Wapl depletion rescues IR sensitivity and SCR defects of Mms21-RNAi or Scc1 15KR-expressing cells. (A) Lysates of HeLa cells transfected with the indicated siRNAs were blotted with α-Wapl and α-Mms21. A cross-reacting band in the α-Mms21 blot served as the loading control. (B) IR colony survival assay of cells described in A. Each data point represents the mean and standard deviation of values in three independent experiments, with duplicate samples in each experiment. (C) Representative images from the SCE assay on cells in A. The numbers of SCE events and chromosomes in each image are shown below. A pair of sister chromatids for each image is magnified and shown in the inset, with the SCE events marked by red arrowheads. (D) Quantification of the relative SCE levels of cells in A. The mean and standard deviation of data from three experiments are shown. More than 30 cells were counted in each experiment. (E) Quantification of the normalized percentage of GFP-positive cells in the I-SceI-based gene targeting assay of 293/A658 cells transfected with the indicated siRNAs. The cells were cotransfected with siRNAs and the repair plasmid. Three days later, cells were harvested and analyzed by FACS. (F) HeLa Tet-On cells expressing Myc-Scc1 15 KR (clone 3) cultured in the absence or presence of doxycycline (Dox) were transfected with the indicated siRNAs. Lysates were blotted with the indicated antibodies. The positions of Myc-Scc1 and the endogenous (Endo.) Scc1 are labeled. (G) Quantification of the relative SCE levels of cells described in F. The mean and standard deviation of data from two experiments are shown. About 30 cells were counted in each experiment.
Finally, we used a GFP-based gene targeting assay to confirm the antagonism between Mms21 and Wapl. In this assay, an artificial gene target (containing a mutated GFP gene with in-frame stop codons and an I-SceI recognition site inserted) was stably integrated at a single genomic locus in 293 cells (Potts et al. 2006). These cells were then transfected with a repair plasmid that contained the I-SceI gene and a truncated GFP gene. Neither the mutated GFP gene integrated in the genome nor the truncated GFP gene on the repair plasmid could express functional GFP. Expression of I-SceI introduced a DSB within the integrated GFP locus. HR between the mutated GFP gene in the genome and the truncated GFP gene on the plasmid reconstituted a functional GFP gene. The percentage of GFP-positive cells as determined by flow cytometry then provided a quantitative measure of the frequency of gene targeting. Consistent with a previous study (Potts et al. 2006), depletion of Mms21 increased the percentage of GFP-positive cells in this assay because inactivation of Mms21 decreased SCR, an HR pathway that was not expected to produce GFP-positive cells (Fig. 6E). While Wapl depletion alone did not alter the gene targeting efficiency, codepletion of Wapl nullified the increase in the gene targeting rate caused by Mms21 depletion. Taken together, our results from all three assays are consistent with the notion that Mms21 promotes SCR by counteracting Wapl. Our results further implicate cohesin as a major downstream target of Mms21 in DSB repair in human cells.
If Scc1 is a key downstream target of Mms21 in this process, one would expect that depletion of Wapl should also rescue the SCR defect of cells expressing Scc1 15KR. Consistent with the results in Figure 3C, doxycycline-induced expression of Myc-Scc1 15KR did not rescue the SCE defect caused by Scc1 depletion (Fig. 6F,G). Depletion of Wapl in these cells, however, restored SCE to normal levels. This result suggests that Scc1 sumoylation by Mms21 functionally opposes Wapl to promote SCR.
Expression of the Smc3 acetylation-mimicking mutant fails to bypass the requirement for Mms21 in SCR
We next explored the mechanism by which Scc1 sumoylation counteracted Wapl. Mms21-dependent sumoylation occurs in the central region of Scc1. A similar region has been implicated in Wapl binding (Shintomi and Hirano 2009). One possibility is that Scc1 sumoylation directly blocks Wapl binding to cohesin. On the other hand, the recombinant purified N-terminal domain of Wapl could still bind to sumoylated Scc1 in vitro (data not shown), disfavoring the possibility of direct inhibition.
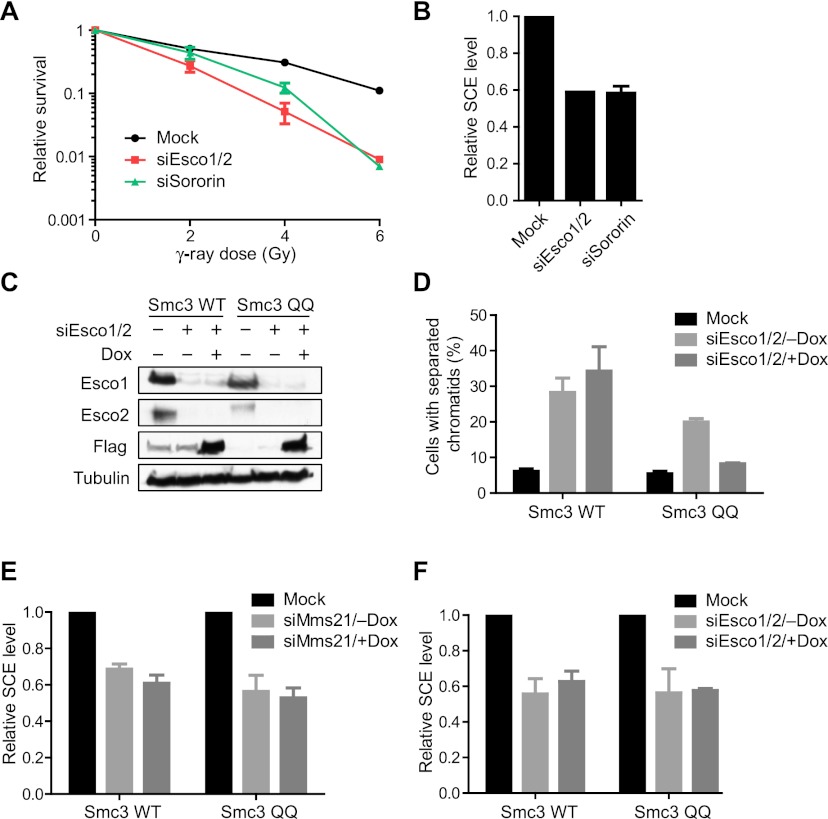
In yeast, Eco1 acetylates Scc1 and is required for DNA damage-induced cohesion generation (Heidinger-Pauli et al. 2009). In human cells, replicative cohesion establishment during S phase requires Esco1/2-dependent acetylation of Smc3 at K105 and K106 (Zhang et al. 2008). Smc3 acetylation enables sororin binding to Pds5A/B, which disrupts a binding interface between Wapl and Pds5A/B and inhibits Wapl's ability to release cohesin from chromatids (Nishiyama et al. 2010). Sororin has also been implicated in DNA repair (Schmitz et al. 2007). Because our results described here implicated Wapl as a downstream effector of Scc1 sumoylation by Mms21 in SCR, we next sought to examine the potential interplays among Scc1 sumoylation, Smc3 acetylation, and sororin in this process. Depletion of Esco1/2 or sororin by RNAi caused IR sensitivity and SCE defect in HeLa cells, confirming their involvement in DNA repair (Fig. 7A,B; Schmitz et al. 2007).
Figure 7.
Expression of the Smc3 acetylation-mimicking mutant does not bypass the requirement for Mms21 in SCR. (A) IR colony survival assay of HeLa cells transfected with the indicated siRNAs. Each data point represents the mean and standard deviation of values in two independent experiments, with duplicate samples in each experiment. (B) Quantification of the relative SCE levels of cells in A. The mean and standard deviation of data from two experiments are shown. About 30 cells were counted in each experiment. (C) 293T cell lines inducibly expressing Flag-Smc3 wild type (WT) or K105Q/K106Q (QQ) were cultured in the absence or presence of doxycycline (Dox) and then either mock-transfected or transfected with siEsco1/2. Lysates were blotted with the indicated antibodies. (D) Quantification of the percentage of mitotic cells in C with separated sister chromatids. The mean and standard deviation of three independent experiments are shown. (E) 293T cell lines inducibly expressing Flag-Smc3 wild type or K105Q/K106Q (QQ) were cultured in the absence or presence of doxycycline (Dox) and then either mock-transfected or transfected with siMms21. The relative SCE levels of these cells were quantified. The mean and standard deviation of data from two experiments are shown. About 30 cells were counted in each experiment. (F) Quantification of the relative SCE levels of cells in C. The mean and standard deviation of data from two experiments are shown. About 30 cells were counted in each experiment.
We next examined the relationship between Smc3 acetylation and Scc1 sumoylation. Consistent with a previous report (Zhang et al. 2008), we showed that doxycycline-induced ectopic expression of Smc3 K105Q/K106Q (QQ; an acetylation-mimicking Smc3 mutant with K105 and K106 mutated to glutamine), but not Smc3 wild type, rescued the sister chromatid cohesion defect of 293T cells depleted of Esco1/2 by RNAi (Fig. 7C,D). This result confirmed that Smc3 QQ indeed functionally mimicked acetylation at these two sites and that Smc3 K105 and K106 are the major targets of Esco1/2 in replicative sister chromatid cohesion. However, expression of Smc3 QQ did not rescue the SCE defect of Mms21-RNAi cells (Fig. 7E). Thus, Smc3 acetylation might not be the sole critical downstream event of Mms21-dependent Scc1 sumoylation in SCR. Furthermore, Smc3 QQ expression did not rescue the SCE defect of Esco1/2 RNAi cells (Fig. 7F), suggesting that Esco1/2 had target sites in addition to (or instead of) Smc3 K105 and K106 in DNA repair in human cells, similar to yeast Eco1.
Scc1 is the critical target of Eco1 in DNA repair (Heidinger-Pauli et al. 2009). Although the acetylation sites of yeast Scc1 are not conserved in human Scc1, and previous mass spectrometry analysis failed to uncover acetylation sites in human Scc1 (Kim et al. 2010), it remained formally possible that in addition to abolishing Mms21-dependent sumoylation, Scc1 15KR also eliminated yet unidentified acetylation sites on Scc1. We immunoprecipitated Myc-Scc1 wild type and 15KR from lysates of HeLa Tet-On cells with or without IR treatment and blotted the immunoprecipitations with the antibody against several pan-specific acetyl-lysine antibodies. Two pan-acetyl-lysine antibodies detected bands that corresponded to the size of Myc-Scc1, suggesting that Scc1 was acetylated (Supplemental Fig. S8). However, Scc1 acetylation was not stimulated by IR. Scc1 15KR was as efficiently acetylated as Scc1 wild type. Thus, there was no evidence to indicate that the 15KR mutation adversely affected Scc1 acetylation. A pan-acetyl-lysine antibody also detected another cohesin-associated protein, whose identity remained to be determined.
Discussion
In this study, we provide evidence to suggest that Mms21-dependent Scc1 sumoylation is necessary for SCR in human cells. It functionally opposes the negative cohesin regulator Wapl at a step after cohesin recruitment to DSBs. We further show that Smc3 acetylation is not the sole critical molecular event downstream from Scc1 sumoylation in DNA repair. Our study thus establishes a general framework for the function and regulation of cohesin in SCR in human cells.
Role of Scc1 sumoylation by Mms21 in SCR
We showed convincingly that Mms21 is sufficient to promote Scc1 sumoylation in vitro and in human cells. We also provided strong evidence to suggest that a small pool of endogenous cohesin is sumoylated at laser-induced DNA damage sites in an Smc5/6-dependent manner. On the other hand, we could not biochemically detect the sumoylation of endogenous cohesin without overexpression of Mms21 or SUMO under normal or DNA damage conditions. The difficulty of detecting sumoylation of endogenous proteins is well documented. Only a handful of proteins are sumoylated at appreciable steady-state levels in human cells. The underlying reasons for the low steady-state levels of sumoylation are not understood, but have been attributed in part to the highly dynamic nature of this modification. Furthermore, our fractionation (Supplemental Fig. S3) experiments show that DNA damage does not induce global changes in the composition of cohesin and its molecular interactions with known regulators. Therefore, our results suggest that only a small pool of cohesin at DSBs is sumoylated, providing a possible explanation for our failure to detect sumoylation of endogenous cohesin in cells with native levels of Mms21 and SUMO.
Scc1 sumoylation by overexpressed Mms21 and SUMO is not further stimulated by DNA damage (data not shown). This result suggested that the SUMO ligase activity of the bulk of Mms21 was not directly regulated by DNA damage. Cohesin sumoylation might be triggered by a transient interaction between cohesin and the Smc5/6 complex when both are independently recruited to DNA damage sites. On the other hand, we cannot exclude trivial explanations for the apparent lack of regulation of Mms21-dependent Scc1 sumoylation by DNA damage. For example, overexpressed Mms21 might not behave the same as the endogenous Mms21 in this regard. A definitive answer to this question awaits the development of a method that can selectively isolate DSB-bound cohesin from human cells.
Although our results show that Scc1 15KR is defective in DSB repair and SCR, we cannot completely rule out the possibility that in addition to abolishing Mms21-dependent sumoylation, the 15KR mutation affects other functions of Scc1. Scc1 15KR is still functional in maintaining mitotic sister chromatid cohesion, indicating that it does not have gross structural defects. On the other hand, yeast strains expressing Scc1 at 30% of wild-type levels are deficient in DNA repair (Heidinger-Pauli et al. 2010). In contrast, sister chromatid cohesion and chromosome segregation remain normal even when Scc1 is reduced to 13% of wild-type levels in yeast cells. Similarly, certain siRNAs against Scc1 that depleted cohesin poorly caused IR sensitivity but produced small defects in sister chromatid cohesion in human cells (data not shown). Thus, partial inactivation of cohesin reveals its role in DNA repair, while a more complete inactivation of cohesin is needed to reveal its function in sister chromatid cohesion. A simple quantitative difference between the activities of Scc1 wild type and 15KR could explain why Scc1 15KR is capable of supporting sister chromatid cohesion but fails to support proper DNA repair. Finally, because human Esco1/2 acetylate unknown substrates in DNA repair, and because Scc1 acetylation is critical for DI cohesion in yeast, the 15KR mutation could conceivably eliminate a yet unidentified acetylation site. We do not have evidence for such a scenario because Scc1 15KR does not alter Scc1 acetylation as detected by pan-acetyl-lysine antibodies.
How does Scc1 sumoylation by Mms21 contribute to SCR? We show that the Smc5/6 complex and Scc1 sumoylation are dispensable for cohesin recruitment to DSBs. Depletion of Wapl, a negative regulator of cohesin, rescues the IR sensitivity and SCE defect of Mms21-deficient or Scc1 15KR-expressing cells. Our results support a two-step model for cohesin regulation in SCR (Supplemental Fig. S9). In the first step, both cohesin and the Smc5/6 complex are independently recruited to DNA damage sites. The loaded cohesin is not yet cohesive and may be removed by Wapl. In the second step, a transient interaction between the loaded Smc5/6 and cohesin complexes enables Mms21-dependent sumoylation of Scc1 locally at DSBs. This local cohesin sumoylation counteracts Wapl, stabilizing cohesin around DSBs and facilitating SCR.
Antagonism between Mms21 and Wapl
We do not have evidence that Scc1 sumoylation directly prevents Wapl binding to cohesin in vitro and in cells. Furthermore, Wapl does not completely dissociate from functional cohesin that has Smc3 acetylation and is bound to Pds5 and sororin. How Wapl promotes cohesin release from chromatin and how sororin antagonizes Wapl remain poorly understood. Without a mechanistic understanding of how Wapl works, biochemical studies on how Scc1 sumoylation affects Wapl function are premature. Our data thus only establish Wapl as a downstream effector of the Mms21 pathway but do not prove a causative mechanistic link between the two.
During the unperturbed S phase, Esco1/2 acetylates Smc3 at K105 and K106 in a process that is coupled to DNA replication (Zhang et al. 2008). Expression of the Smc3 acetylation-mimicking mutant rescues the sister chromatid cohesion defect of Esco1/2-RNAi cells but fails to bypass the requirement for Mms21 in SCR. Therefore, in the context of SCR, Smc3 acetylation is unlikely to be the sole downstream event that is regulated by Scc1 sumoylation. As discussed above, we cannot rule out the possibility that Smc3 QQ is not a perfect mimic of acetylation. It supports sister chromatid cohesion but does not support DNA repair because the latter process requires a larger amount of functional cohesin. On the other hand, because sororin and Esco1/2 are required for DBS repair and SCR (Schmitz et al. 2007), it is conceivable that Scc1 sumoylation acts indirectly through Esco1/2 and sororin to oppose Wapl function.
Similarities and differences of cohesin regulation in DNA repair in yeast and humans
In yeast, it has been conclusively shown that DNA damage induces functional cohesion locally at DSBs and globally throughout the genome (Strom et al. 2007; Unal et al. 2007). Because we do not have a strategy to inactivate the existing functional cohesin specifically in G2 human cells, we cannot definitively test whether DNA damage actually establishes functional cohesion at damage sites in human cells. However, our results do reveal common themes shared by yeast and human cells in terms of cohesin regulation in DNA repair. First, the involvement of cohesin in DNA repair is a multistep process in both organisms. The mere loading of cohesin to damage sites is insufficient. Post-translational modifications (Scc1 acetylation in yeast and sumoylation in humans) are necessary to make the loaded cohesin functional. Second, in both organisms, Wapl is the critical downstream effector. Post-translational modifications of cohesin antagonize Wapl to promote SCR. Third, replicative cohesion establishment and the DNA repair function of cohesin require different modifications of cohesin and its regulators. In yeast, Eco1 targets Smc3 in replicative cohesion establishment but targets Scc1 to establish DI cohesion. Likewise, Esco1/2 in humans also have distinct targets in replicative cohesion establishment and in DNA repair, although the target of Esco1/2 in the latter process remains to be identified.
There are also important differences in the mechanisms by which cohesin promotes DNA repair in yeast and human cells. The acetylation and phosphorylation sites in yeast Scc1 that are critical for DNA repair and DI cohesion are not conserved in human Scc1. Furthermore, sororin is required for DSB repair and SCR in human cells. Functional sororin homologs have not been identified in yeast.
In conclusion, our results establish a new post-translational regulatory mechanism of cohesin during DNA repair and reveal both conserved principles and organism-specific features in cohesin regulation during sister chromatid recombination.
Materials and methods
Cell culture, transfection, and synchronization
HeLa Tet-On, 293/A658, and 293T cells were grown in DMEM (Invitrogen) supplemented with 10% FBS, 100 μg/mL penicillin and streptomycin, and 2 mM L-glutamine. Site-directed mutagenesis was performed using the QuikChange kit (Qiagen). For plasmid transfections, cells were transfected at 50% confluency with the Effectene reagent (Qiagen) according to the manufacturer's protocols. For siRNA transfections, cells were transfected at 20% confluency with Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's protocols. For plasmid and siRNA double transfections, cells were transfected at 80% confluency with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocols.
To establish stable cell lines, HeLa Tet-On cells were transfected with pTRE2-Myc-based plasmids encoding siRNA-resistant Scc1 wild type or 15KR with a C-terminal Myc6 tag. Clones were selected in the presence of 200 μg/mL hygromycin B. Inducible expression was screened in the absence or presence of 1 μg/mL doxycycline (Invitrogen). 293T cell lines expressing Smc3 wild type and Smc3 QQ were kindly provided by Dr. Jun Qin (Baylor College of Medicine). For the functional rescue experiments, 1 μg/mL doxycycline was added into the medium to induce protein expression at 8 h before siRNA transfection. To arrest cells in S/G2, cells were treated with 2 mM thymidine for 18 h and released into fresh medium for 5–6 h. To arrest cells in G2, cells were treated with the Cdk1 inhibitor RO3306 (EMD) at 10 μM for 20 h.
The siRNAs used in this study (Supplemental Table S1) were chemically synthesized by or purchased from Dharmacon or Qiagen.
Antibodies, immunoblotting, and immunoprecipitation
To generate antibodies against Scc1 and Wapl, fragments of Scc1 (residues 211–316) and Wapl (residues 601–1190) were produced in bacteria as His6-tagged fusion proteins and purified. The proteins were used to immunize rabbits at Yenzym Antibodies. Production of the α-Mms21 antibody was described previously (Potts and Yu 2005). Antibodies against sororin, Esco1/2, and Smc3 K105Ac were gifts from Susannah Rankin (Oklahoma Medical Research Foundation), Hui Zou (University of Texas Southwestern), and Jun Qin (Baylor College of Medicine), respectively. The commercial antibodies used in this study were α-Myc (Roche, 11667203001), α-HA (Roche, 11666606001), α-Scc1 (Bethyl Laboratories, A300-080A), mouse α-Scc1 (Upstate Biotechnologies, #05-908), α-Flag (Stratagene, 200472), α-Smc1 (Bethyl Laboratories, A300-055A), α-phospho-Smc1 S966 (Bethyl Laboratories, A300-050A), α-Smc3 (Bethyl Laboratories, A300-060A), α-Smc5 (Bethyl Laboratories, A300-236A), goat α-Smc5 (Santa Cruz Biotechnology, sc-47627), α-Smc6 (Bethyl Laboratories, A300-237A), α-Smc6 (Santa Cruz Biotechnology, sc-365742), α-SA2 (Santa Cruz Biotechnology, sc-81852), α-Pds5A (Bethyl Laboratories, A300-089A), α-Pds5B (Bethyl Laboratories, A300-537A), α-Pds5B (Bethyl Laboratories, A300-538A), α-Chk1 phospho-S317 (Cell Signaling, #2344), γH2AX (Millipore, 05-636), α-SUMO3 (ABGENT, AM1201a), α-RanGAP1 (Santa Cruz Biotechnology, sc-25630), α-Rad51 (Santa Cruz Biotechnology, sc-6862), α-acetylated-lysine Ab1 (Immunechem, ICP0380), and α-acetylated-lysine Ab2 (Cell Signaling, #9441).
For immunoblotting, cells were lysed in SDS sample buffer, sonicated, boiled, separated by SDS-PAGE, and blotted with the desired antibodies. Horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse IgGs (Amersham Biosciences) were used as secondary antibodies. Immunoblots were developed using the ECL reagent (Amersham Biosciences) according to the manufacturer's protocols. For immunoprecipitation, lysate was prepared in the lysis buffer containing 25 mM Tris-HCl (pH 7.7), 50 mM NaCl, 0.1% (v/v) NP-40, 2 mM MgCl2, 10% (v/v) glycerol, 5 mM NaF, 0.3 mM NaVO4, 10 mM β-glycerophosphate, 1 mM DTT, and 1× protease inhibitor cocktail (Roche). Turbo Nuclease was also added. Cells were broken by passing through a small-gauge needle 10 times. After incubating for 1 h on ice and 10 min at room temperature, all samples were centrifuged at 14,000 rpm for 20 min at 4°C. Protein A beads (Bio-Rad) and the desired antibody at a concentration of 1 μg/mL were incubated with the supernatant for 3 h at 4°C. The beads were then washed four times with the lysis buffer. The proteins bound to the beads were dissolved in SDS sample buffer, separated by SDS-PAGE, and blotted with the appropriate antibodies.
Sumoylation assay
Scc1 substrates were in vitro transcribed and translated (IVT) from pCS2-Myc plasmids in reticulocyte lysate (Promega) in the presence of cold or 35S-methionine. One microliter of IVT protein was then mixed with 1 μg of Aos1–Uba2, 0.1 μg of Ubc9, 10 μg of SUMO1, and 1 μL of energy mix (150 mM phosphocreatine, 20 mM ATP, 2 mM EGTA, 20 mM MgCl2 at pH 7.7) in the absence or presence of 0.5 μg of His6-Mms21 for the indicated time at 30°C. Reaction mixtures were adjusted to 10 μL with XB buffer (10 mM HEPES at pH 7.7, 1 mM MgCl2, 0.1 mM CaCl2, 100 mM KCl, 50 mM sucrose). SDS sample buffer was added to stop the reactions. The samples were boiled and subjected to SDS-PAGE followed by immunoblotting or autoradiography.
For the denaturing Ni2+ bead pull-down from HeLa cell lysates to enrich SUMO conjugates, cells were incubated with the lysis buffer containing 6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris-HCl (pH 8.0), 5 mM imidazole, and 10 mM β-ME; sonicated; and mixed with Ni2+-NTA beads for 2 h at room temperature. The beads were then washed with the following buffers: buffer I (6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris-HCl at pH 8.0, 10 mM β-ME), buffer II (8 M urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris-HCl at pH 8.0, 10 mM β-ME), buffer III (8 M urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris-HCl at pH 6.3, 10 mM β-ME, 0.2% [v/v] Triton X-100), buffer IV (8 M urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris-HCl at pH 6.3, 10 mM β-ME), and buffer V (8 M urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris-HCl at pH 6.3, 10 mM β-ME, 0.1% [v/v] Triton X-100). The proteins bound to beads were dissolved in SDS sample buffer, separated by SDS-PAGE, and blotted with the appropriate antibodies.
Laser microirradiation and fluorescent image analysis
Laser damage induction and image analysis were performed essentially as described (Kim et al. 2002; Kong et al. 2009). Briefly, 532 nm of the second harmonic of a pulsed Nd:YAG laser beam (∼2–3 μJ per pulse energy after objective; ∼4–6 nsec pulse duration; 7.5 Hz, Quantronix-Continuum Lasers) was focused through a 100× oil objective (NA 1.3; Olympus) on a microscope (model IX81; Olympus). The sample stage was repeatedly scanned for 2 min at a scanning rate of ∼10 μm/sec to create a line pattern of microirradiation inside the nucleus. The cells were incubated for 1 h at 37°C and then fixed in 4% formaldehyde (10 min at room temperature). Cells were permeabilized with 0.5% Triton X-100 for 5 min on ice and then immunostained with the appropriate antibodies. Five to seven cells were damaged in one plate, and each experiment was repeated at least three times.
Metaphase spread
Cells were collected by trypsinization and pelleted. One milliliter of medium was left in the tube. Two milliliters of tap water was then added to resuspend the cells. After a 5-min incubation, the fixation solution (1:3 [v/v] glacial acetic acid:methanol) was added. After centrifugation, cells were resuspended in the fixation solution, incubated for 10 min at room temperature, and washed twice with the fixation solution. Cells were resuspended and stored at −20°C. To prepare the slides for Giemsa staining, three drops of fixed cells were dropped onto each slide (pretreated with methanol) and air-dried for 5 min. Five percent Giemsa (EMD) in the Giemsa staining buffer (0.01 M PBS at pH 6.8) was used to stain the slides. The slides were washed twice with running water, dried for 20 min at room temperature, mounted in Entellan mounting medium (Merck), and analyzed by microscopy.
Colony survival assay
HeLa Tet-On cells were transfected with the indicated siRNAs for 24 h and then replated into six-well plates with 500, 2000, 10,000, and 40,000 cells per well. After 1 d, cells were exposed to a Cs-137 sealed irradiator to receive different doses of γ-rays (0, 2, 4, 6, 10 Gy) and then put back into culture for ∼10 d to form colonies. At the day of staining, medium was removed, and crystal violet colony staining solution was added to fix and stain colonies for 20 min at room temperature. After staining, dishes were washed under running water and air-dried for 2 d. Colonies with >50 cells were manually counted. The numbers of colonies were normalized to the unirradiated sample.
SCE assay
HeLa Tet-On cells were transfected with the indicated siRNAs for 24 h and then replated and incubated in the presence of 100 μM BrdU and 2.5 nM camptothecin for 42 h (about two cell divisions). Colcemid (150 ng/mL) was added during the final 2 h to enrich for mitotic cells. Cells were collected by trypsinization and washed with PBS. Cells were swelled in 75 mM KCl for 12 min at 37°C, followed by centrifugation. Cell pellets were resuspended in the fixation solution for 20 min at 4°C. Cells were washed twice with the fixation solution, resuspended, and dropped onto cold slides. After 2–3 d, slides were stained with 0.05 mg/mL Acridine Orange (Molecular Probes) for 5 min, washed under running water for 4 min, and mounted in the Sorenson buffer (0.1 M Na2HPO4, 0.1 M NaH2PO4 at pH 6.8) for 1 min. Slides were immediately viewed under the microscope. For each experiment, the numbers of crossover events and chromosomes were counted in ∼30 mitotic cells. The numbers of SCEs per 100 chromosomes were calculated. All data were normalized to the value of the mock-transfected sample.
ChIP assay
HeLa Tet-On cells were mock-transfected or transfected with siRNAs against Smc5 and Mms21. At 24 h after transfection, cells were transfected with the ER-I-PpoI expression plasmid for 24 h. One micromolar 4-OHT was then added into the medium for 12 h to induce DNA damage. ChIP was performed as described previously (Berkovich et al. 2008). Quantitative real-time PCR was performed with primers toward the rDNA locus containing multiple I-PpoI sites (5′-TGGAGCAGAAGGGCAAAAGC-3′ and 5′-TAGGAAGAGCCGACATCGAAGG-3′).
Acknowledgments
We thank Drs. Jun Qin, Susannah Rankin, Hui Zou, and Mike Kastan for providing key reagents. This work is supported by grants from the Cancer Prevention Research Institute of Texas (to H.Y.), the Welch Foundation (I-1441 to H.Y.), the National Institutes of Health (CA100710 to K.Y.), and the University of California Cancer Research Coordinating Committee (CRCC-46081 to K.Y.). H.Y. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.193615.112.
References
- Behlke-Steinert S, Touat-Todeschini L, Skoufias DA, Margolis RL 2009. SMC5 and MMS21 are required for chromosome cohesion and mitotic progression. Cell Cycle 8: 2211–2218 [DOI] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ Jr, Kastan MB 2007. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol 9: 683–690 [DOI] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ Jr, Kastan MB 2008. Assessment of protein dynamics and DNA repair following generation of DNA double-strand breaks at defined genomic sites. Nat Protoc 3: 915–922 [DOI] [PubMed] [Google Scholar]
- De Piccoli G, Cortes-Ledesma F, Ira G, Torres-Rosell J, Uhle S, Farmer S, Hwang JY, Machin F, Ceschia A, McAleenan A, et al. 2006. Smc5–Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nat Cell Biol 8: 1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Piccoli G, Torres-Rosell J, Aragon L 2009. The unnamed complex: What do we know about Smc5–Smc6? Chromosome Res 17: 251–263 [DOI] [PubMed] [Google Scholar]
- Gareau JR, Lima CD 2010. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 11: 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocke CB, Yu H, Kang J 2005. Systematic identification and analysis of mammalian small ubiquitin-like modifier substrates. J Biol Chem 280: 5004–5012 [DOI] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Unal E, Guacci V, Koshland D 2008. The kleisin subunit of cohesin dictates damage-induced cohesion. Mol Cell 31: 47–56 [DOI] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Unal E, Koshland D 2009. Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol Cell 34: 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Mert O, Davenport C, Guacci V, Koshland D 2010. Systematic reduction of cohesin differentially affects chromosome segregation, condensation, and DNA repair. Curr Biol 20: 957–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F, Zou H 2005. Two human orthologues of Eco1/Ctf7 acetyltransferases are both required for proper sister-chromatid cohesion. Mol Biol Cell 16: 3908–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna KK, Jackson SP 2001. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat Genet 27: 247–254 [DOI] [PubMed] [Google Scholar]
- Kim JS, Krasieva TB, LaMorte V, Taylor AM, Yokomori K 2002. Specific recruitment of human cohesin to laser-induced DNA damage. J Biol Chem 277: 45149–45153 [DOI] [PubMed] [Google Scholar]
- Kim BJ, Li Y, Zhang J, Xi Y, Yang T, Jung SY, Pan X, Chen R, Li W, Wang Y, et al. 2010. Genome-wide reinforcement of cohesin binding at pre-existing cohesin sites in response to ionizing radiation in human cells. J Biol Chem 285: 22784–22792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Mohanty SK, Stephens J, Heale JT, Gomez-Godinez V, Shi LZ, Kim JS, Yokomori K, Berns MW 2009. Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nucleic Acids Res 37: e68 doi: 10.1093/nar/gkp221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM 2006. Wapl controls the dynamic association of cohesin with chromatin. Cell 127: 955–967 [DOI] [PubMed] [Google Scholar]
- Lindroos HB, Strom L, Itoh T, Katou Y, Shirahige K, Sjogren C 2006. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol Cell 22: 755–767 [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Wu J, Blobel G 1998. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol 140: 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH 2009. Cohesin: Its roles and mechanisms. Annu Rev Genet 43: 525–558 [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, Bando M, Shirahige K, Hyman AA, Mechtler K, et al. 2010. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell 143: 737–749 [DOI] [PubMed] [Google Scholar]
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE 2008. Sister chromatid cohesion: A simple concept with a complex reality. Annu Rev Cell Dev Biol 24: 105–129 [DOI] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J 2008. The cohesin complex and its roles in chromosome biology. Genes Dev 22: 3089–3114 [DOI] [PubMed] [Google Scholar]
- Potts PR 2009. The Yin and Yang of the MMS21-SMC5/6 SUMO ligase complex in homologous recombination. DNA Repair (Amst) 8: 499–506 [DOI] [PubMed] [Google Scholar]
- Potts PR, Yu H 2005. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol 25: 7021–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR, Porteus MH, Yu H 2006. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J 25: 3377–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin S, Ayad NG, Kirschner MW 2005. Sororin, a substrate of the anaphase-promoting complex, is required for sister chromatid cohesion in vertebrates. Mol Cell 18: 185–200 [DOI] [PubMed] [Google Scholar]
- Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F 2008. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321: 563–566 [DOI] [PubMed] [Google Scholar]
- Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouet F, Underwood P, Metson J, Imre R, et al. 2009. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell 33: 763–774 [DOI] [PubMed] [Google Scholar]
- Schmitz J, Watrin E, Lenart P, Mechtler K, Peters JM 2007. Sororin is required for stable binding of cohesin to chromatin and for sister chromatid cohesion in interphase. Curr Biol 17: 630–636 [DOI] [PubMed] [Google Scholar]
- Shintomi K, Hirano T 2009. Releasing cohesin from chromosome arms in early mitosis: Opposing actions of Wapl-Pds5 and Sgo1. Genes Dev 23: 2224–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom L, Lindroos HB, Shirahige K, Sjogren C 2004. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell 16: 1003–1015 [DOI] [PubMed] [Google Scholar]
- Strom L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjogren C 2007. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 317: 242–245 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Dulev S, Liu X, Hiller NJ, Zhao X, Strunnikov A 2008. Cooperation of sumoylated chromosomal proteins in rDNA maintenance. PLoS Genet 4: e1000215 doi: 10.1371/journal.pgen.1000215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell 16: 991–1002 [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Koshland D 2007. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science 317: 245–248 [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE 2008. A molecular determinant for the establishment of sister chromatid cohesion. Science 321: 566–569 [DOI] [PubMed] [Google Scholar]
- Watrin E, Peters JM 2006. Cohesin and DNA damage repair. Exp Cell Res 312: 2687–2693 [DOI] [PubMed] [Google Scholar]
- West SC 2003. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol 4: 435–445 [DOI] [PubMed] [Google Scholar]
- Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang Z, Yang T, Fu X, Jung SY, Wang Y, et al. 2008. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell 31: 143–151 [DOI] [PubMed] [Google Scholar]
- Zhao X, Blobel G 2005. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci 102: 4777–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]