Abstract
REST is a master repressor of neuronal genes; however, whether it has any role during nervous system development remains largely unknown. Here, we analyzed systematically the role of REST in embryonic stem cells and multipotent neural stem/progenitor (NS/P) cells, including neurogenic and gliogenic NS/P cells derived from embryonic stem (ES) cells or developing mouse embryos. We showed that REST-null ES cells remained pluripotent and generated teratomas consisting of the three germ layers. By contrast, multipotent NS/P cells lacking REST displayed significantly reduced self-renewal capacity owing to reduced cell cycle kinetics and precocious neuronal differentiation. Importantly, although early-born neurogenic NS/P cells that lack REST were capable of differentiating to neurons and glia, the neuronal and oligodendrocytic pools were significantly enlarged and the astrocytic pool was shrunken. However, gliogenic NS/P cells lacking REST were able to generate a normal astrocytic pool size, suggesting that the shrinkage of the astrocytic pool generated from neurogenic NS/P cells lacking REST probably occurs by default. Microarray profiling of early-born NS/P cells lacking REST showed upregulation of neuronal as well as oligodendrocytic genes, specifically those involved in myelination. Furthermore, chromatin immunoprecipitation analyses showed that some of the upregulated oligodendrocytic genes contain an RE1 motif and are direct REST targets. Together, our data support a central role for REST during neural development in promoting NS/P cell self-renewal while restricting the generation and maturation of neurons and oligodendrocytes.
Keywords: Embryonic stem cells, Neural development, Neural stem/progenitor cells, Neurons, Oligodendrocytes, REST, Mouse
INTRODUCTION
Nervous system development involves complex extrinsic and intrinsic signaling pathways to precisely coordinate the maintenance of the necessary neural stem/progenitor (NS/P) pool and, at the same time, the orderly acquisition of neural cell fate. Neurons and glia, comprising astrocytes and oligodendrocytes, arise from multipotent neural stem cells in a temporally defined order: generation of neurons precedes glia and generation of astrocytes precedes oligodendrocytes (Edlund and Jessell, 1999; Okano and Temple, 2009; Qian et al., 2000). It is well established that the coordinated activation or suppression of transcriptional activators and repressors, which regulate the expression of a network of lineage-specific genes, plays a major role in this process (Cheng et al., 2005; Edlund and Jessell, 1999; Ross et al., 2003). Nevertheless, the mechanism utilized by specific regulators to control the proper progression of neural development remains mostly unknown.
A potential candidate protein, which could play an essential role in this process, is the transcriptional repressor REST (RE1 silencing transcription factor, also called NRSF) (Chong et al., 1995; Schoenherr and Anderson, 1995). REST regulates a large number of neuronal genes encoding fundamental neuronal traits as well as non-coding brain-specific microRNA genes (Bruce et al., 2004; Johnson et al., 2007; Otto et al., 2007). In non-neural cells, REST binds a conserved 23 bp element, RE1 (repressor element 1, also called NRSE) (Chong et al., 1995; Schoenherr and Anderson, 1995), which is present in its target genes, and acts to suppress their expression via two distinct repressor domains (Andres et al., 1999; Ballas et al., 2001; Roopra et al., 2000). Our previous studies showed that, although REST, in conjunction with its co-repressor CoREST (RCOR1 – Mouse Genome Informatics), silences neuronal genes in non-neuronal cells outside the nervous system, the same genes are repressed and poised for expression in pluripotent embryonic stem (ES) cells (Ballas et al., 2005; Ballas and Mandel, 2005). This differential regulation of the same sets of genes by REST is mediated by the differential recruitment of chromatin modifiers by CoREST (Ballas and Mandel, 2005). Importantly, REST itself is differentially regulated throughout neural development; whereas REST is expressed at a high level in ES cells, it is downregulated to a minimal level in NS/P cells. As NS/P cells differentiate, REST remains present in glia (Abrajano et al., 2009; Dewald et al., 2011) but is largely absent in neurons, allowing the restricted expression of its target genes specifically in neurons (Ballas et al., 2005). Whereas the high level of REST expression in ES cells is mediated by NANOG and OCT4 (POU5F1 – Mouse Genome Informatics)) (Boyer et al., 2005; Loh et al., 2006), the low level in NS/P cells is mediated, at least in part, by proteasomal degradation via the E3 ubiquitin ligase β-TRCP (BTRC – Mouse Genome Informatics) (Ballas et al., 2005; Westbrook et al., 2008). However, the function and significance of this differential regulation of REST during nervous system development and whether the normal low level of REST present in NS/P cells has any roles in maintaining NS/P cell self-renewal and/or differentiation along the different neural lineages remains largely unknown.
Here, we analyzed the function of REST at different stages of neural development and show that, although the high level of REST in ES cells is essential for its full functioning as a repressor of neuronal genes, neither REST nor CoREST is required for maintaining ES cell pluripotency. Furthermore, REST is not required for the conversion of ES cells to neurogenic or gliogenic NS/P cells; however, REST is crucial for maintaining NS/P cell self-renewal and proper ratios of neurons:glia during differentiation. Although the loss of REST in neurogenic NS/P cells accelerated neuronal differentiation by depleting the NS/P and astrocytic pools, it had no effect on the pool size of astrocytes generated from gliogenic NS/P cells. Importantly, we show that REST regulates genes encoding not only neuronal but also oligodendrocyte specification and maturation and that the loss of REST in NS/P cells resulted in de-repression of these two gene classes and acceleration of both neuronal and oligodendrocyte differentiation.
MATERIALS AND METHODS
Animal care
All experiments were performed in accordance with research guidelines set by the Institutional Animal Care and Use Committee (IACUC) of Stony Brook University.
ES cells and conversion to NS/P cells
Wild-type (Rest+/+), heterozygous (Rest+/–) and homozygous (Rest–/–) ES cells were described previously (Jorgensen et al., 2009b). ES cells were cultured as described (Ballas et al., 2005). Conversion of ES cells to NS/P cells was performed using the ES cell-derived neurosphere system as described (Okada et al., 2008) except that ultra low attachment dishes were prepared with Glycosan HyStem (BioTime).
Teratoma formation
Teratomas were generated from Rest+/+ and Rest–/– ES cells by subcutaneous injection of 5×106 cells into the left or right side of the spine of B6.Cg-Foxn1nu/J nude mice (The Jackson Laboratory). After 3 weeks, the teratomas were dissected, fixed with 4% paraformaldehyde, embedded, sectioned and stained with Hematoxylin and Eosin using a standard protocol.
Primary cultures of NS/P cells
NS/P cells were isolated from cortices of embryonic day (E) 12.5 mouse embryos as described (Ballas et al., 2005) and cultured in the presence of 20 ng/ml epidermal growth factor (EGF; R&D Systems) and basic fibroblast growth factor (bFGF; Millipore). NS/P cells from cortices of E17.5 mouse embryos were isolated in a similar manner except that the acutely isolated cortical NS/P cells were labeled with CD15 (LeX/SSEA1)-FITC (BD Biosciences) and sorted by fluorescence-activated cell sorting (FACS) for LeX-positive cells as described (Ballas et al., 2005).
Transduction of ES and NS/P cells with lenti virus-carrying shRNA
The different shRNAs were constructed in a lenti viral vector (a gift from Dr Lemischka, Mount Sinai Medical Center) under the human H1 promoter as described (Ivanova et al., 2006). The vector also contained gfp cDNA driven by the ubiquitin C promoter and served as a marker for the transduced cells. Sequences used for shRNA design: Rest shRNA#1, 5′-GCCGAATCTGAAGAGCAGT-3′; Rest shRNA#2, 5′-GTGTAATCTACAATACCA-3′; CoRest shRNA#1, 5′-AGTCTTATTTGAGCAAGCC-3′; CoRest shRNA#2, 5′-GTCTTATTTGAGCAAGCCT-3′; Scrambled shRNA, 5′-ATCATTCCGGATACTGCGA-3′. Mouse embryonic fibroblast (MEF)-free ES cells were transduced in suspension for 12 hours at 37°C in the presence of polybrene (100 μg/ml), washed, cultured on MEFs until 50% confluent, FACS sorted for GFP+ cells, and cultured on MEFs. E12.5 or E17.5 NS/P cells were dissociated at their first passage, plated on coverslips or 60-mm dishes coated with poly-d-lysine/laminin (BD Biosciences) and transduced with lenti virus after one day in culture. Cells were washed, cultured in NS/P media supplemented with EGF and FGF for 6 days, FACS sorted for GFP and cultured as above. Unless otherwise specified, shRNA#2 and shRNA#1 were used to knock down REST and CoREST, respectively.
Competition assay
NS/P cells were transduced with lenti virus carrying scrambled or Rest shRNA and gfp (H1PshRNA-UbiqC-EGFP) or with lenti virus carrying dsRed (H1P-UbiqC-dsRed). GFP- or dsRed-positive cells were FACS sorted after 6 days in culture and replated at a 3:1 GFP:DsRed ratio for 7 days with fresh EGF and FGF. The cells were dissociated and FACS sorted. The number of GFP and dsRed cells per sample was recorded and the mixed cells were replated as above. This procedure was repeated for three passages.
Neurosphere formation assay
For primary neurospheres, NS/P cells transduced with lenti virus carrying Rest or scrambled shRNA and FACS sorted for GFP were plated at a density of 50,000 cells/well in a 12-well plate (BD Biosciences) with EGF and FGF for 6 days. For generation of secondary neurospheres, the primary neurospheres were dissociated and replated at similar density as described above.
Alkaline phosphatase assay
ES cells were cultured on MEFs in a 12-well plate and fixed with 2% paraformaldehyde. Vector Red substrate working solution (Vector Laboratories) was applied for 30 minutes followed by washes with 100 mM Tris-HCl buffer (pH 8.0).
Immunocytochemistry
Immunostaining of cells on coverslips was performed as described (Ballas et al., 2005). ES cell-derived neurospheres were fixed in 4% paraformaldehyde, cryoprotected in 30% sucrose in PBS overnight, washed in PBS, frozen in Neg 50 (Richard-Allan Scientific), sectioned at 10 μm on a cryostat and mounted onto gelatin-coated slides. Primary antibodies used were: chicken anti-GFP (1:500, AbCam); rabbit anti-GFAP (1:500, Dako); mouse anti-TUJ1 (1:500, Covance); mouse anti-CNPase (1:500, Sigma); mouse anti-nestin (1:500, Millipore); mouse anti-Ki67 (1:50, BD Biosciences); mouse anti-phosphohistone H3 (1:50, Cell Signaling); mouse anti-SSEA1 (CD15) (1:50, BD Biosciences); rabbit anti-OCT4 (1:100, AbCam). Secondary antibodies were conjugated to Alexa Fluor (Invitrogen). Images were collected on a Zeiss confocal laser scanning LSM 510 microscope.
Western blot analysis
Whole cell extracts were prepared using a modified Dignam method as described (Ballas et al., 2005). Primary antibodies used were: rabbit anti-REST-C (1:1000) and rabbit anti-CoREST (1:800) as described (Ballas et al., 2005); rabbit anti-β-actin (1:5000, Sigma); rabbit anti-Sin3A (1:1000, Santa Cruz).
Quantitative real-time RT-PCR analysis
Total RNA was isolated using Trizol (Invitrogen). Quantitative real-time PCR was performed following reverse transcription using a StepOnePlus Realtime PCR System (Applied Biosystems). The relative amount of the specific mRNAs was normalized to the level of Gapdh mRNA. The sequences of primers used are listed in supplementary material Table S1.
Microarray analysis of NS/P cells
Microarray analysis was performed at the Bionomics Research and Technology Center (BRTC), University of Medicine and Dentistry of New Jersey. E12.5 NS/P cells were transduced with lenti virus carrying Rest or scrambled shRNA, FACS sorted after 6 days in culture, and replated in the presence of EGF and FGF for 3 days. Total RNA was isolated from three independent experiments and analyzed individually. Microarray data have been deposited in the Gene Expression Omnibus database under accession number GSE38538.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed on E12.5 NS/P cells cultured with EGF and FGF as previously described (Ballas et al., 2005). The sheared chromatin was immunoprecipitated using rabbit anti-REST antibodies generated against the N terminus (anti-REST p73) (Chong et al., 1995) or the C terminus (anti-REST-C) (Ballas et al., 2005). Normal rabbit IgG (Santa Cruz) was used as a control. Primer sets designed to amplify the RE1-containing regions of the specific neuronal and oligodendrocytic genes were as follows: Syt4, forward 5′-GCGTCAATTCGGGTTTCAGCCACGTC-3′, reverse 5′-CAAGTTCAGCAACTTGCTCACCGAAT-3′; Snap25, forward 5′-GCTACAGGCGAGCATGTGCTGCAAAC-3′, reverse 5′-GTGAGGAAGAGAGCAGGCAATTGTC-3′; Mal, forward 5′-GTCTACTGAAAGAACGGGAACC-3′, reverse 5′-CTTTGGCAAGAGTACTGGGAAC-3′; Myt1, forward 5′-CAAGCAGGAGCTGTTTGAGTTGGT-3′, reverse 5′-TGACAGTGTTGGAGAACAAAGCGG-3′; Myt1L, forward 5′-CCCTCCCAAAGTTAATCACCCATC-3′, reverse 5′-CAGTGAACCTGTAGGAAGTCTCTG-3′.
Statistical analysis
Data were analyzed for statistical significance using Student’s t-test. Comparisons were interpreted as significant when associated with P<0.05. For the microarray, data were analyzed for statistical significance using paired t-test.
RESULTS
REST functions as a repressor of neuronal genes in ES cells but is not required for maintaining ES cell pluripotency or for their conversion to NS/P cells
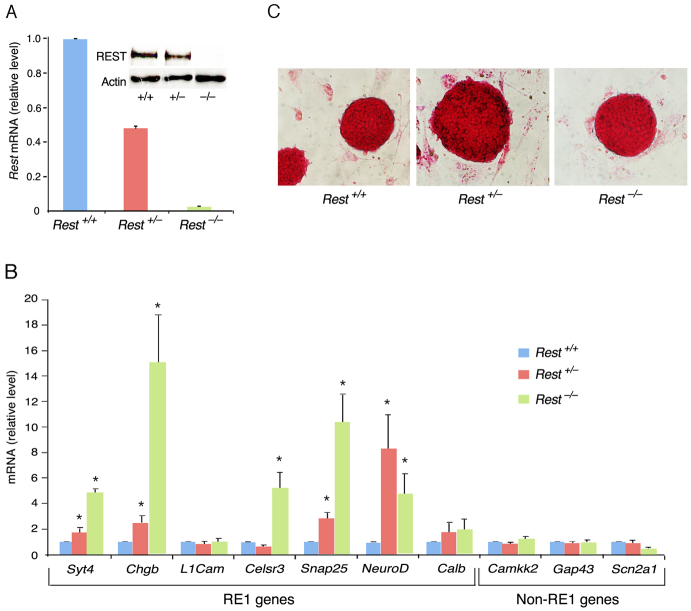
Because REST is a key regulator of neuronal genes and is controlled by pluripotent transcription factors, it could potentially be involved in maintaining ES cell pluripotency. Recent studies attempting to analyze whether REST is involved in maintaining the pluripotency state of ES cells remain controversial (Buckley et al., 2009; Jorgensen et al., 2009a; Jorgensen and Fisher, 2010; Singh et al., 2008). Thus, we sought to analyze systematically the state of ES cells lacking REST. We compared homozygous Rest knockout (Rest–/–) ES cells, isolated from a previously generated Rest-null mouse model (Chen et al., 1998), as well as heterozygous (Rest+/–) ES cells containing half of the normal amount of REST, to wild-type (Rest+/+) ES cells (Fig. 1A). We examined first the repressor activity of REST and found that most of the tested REST target genes, containing the RE1 motif, were derepressed in the Rest–/–, and to a lesser extent in Rest+/– ES cells (Fig. 1B), indicating that REST is a functional and indispensable repressor of neuronal genes in ES cells and that its high level is essential for its full repressor activity. Of the three non-RE1-containing neuronal genes analyzed, none was derepressed in the absence of REST, suggesting that the derepression of REST target genes is unable to drive broad neuronal gene expression.
Fig. 1.
The lack or reduced level of REST in ES cells abrogates neuronal gene repression but does not compromise ES cell pluripotency. (A) Quantitative RT-PCR showing that Rest mRNA is reduced by 50% in Rest+/– ES cells and is absent in Rest–/– ES cells. Inset shows reduced level of REST protein in Rest+/– ES cells and its absence in Rest–/– ES cells. (B) Quantitative RT-PCR of neuronal gene mRNAs indicating that most of the RE-1-containing genes were derepressed in Rest–/– ES cells and Rest+/– ES cells. (C) Alkaline phosphatase (AP) staining is similar in Rest+/+, Rest+/– and Rest–/– ES cells cultured on mouse embryonic fibroblasts (MEFs). All error bars are mean ± s.e.m. based on three independent experiments. *P<0.05.
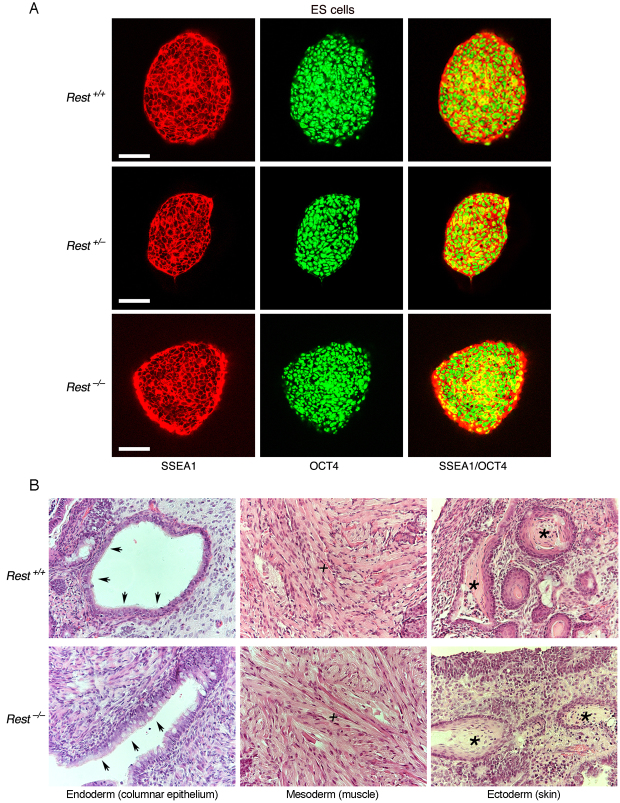
Next, we analyzed the different ES cell types for the presence of alkaline phosphatase (AP) activity, a marker for pluripotency, and found normal morphology, typical of pluripotent ES cell colonies, and no difference in AP staining (Fig. 1C). Importantly, high-resolution images for immunostaining showed that the pluripotency markers SSEA1 (stage-specific embryonic antigen 1) and OCT4 were uniformly present in the colonies of the different ES cell types (Fig. 2A). Likewise, knockdown of REST or CoREST by shRNAs had no effect on the morphology of ES cell colonies or AP staining (supplementary material Fig. S1), nor did the cells lose the pluripotency markers SSEA1 and OCT4 (supplementary material Fig. S2). However, although the presence of pluripotency factors serve as an indicator of the state of ES cells, they do not reflect actual pluripotency. Therefore, we analyzed the pluripotency state of REST-null ES cells in vivo using a teratoma formation assay, a reliable indicator for pluripotency (Lensch et al., 2007). We found that, like the wild-type ES cells (Rest+/+), ES cells lacking REST (Rest–/–) were able to generate teratomas and differentiate into the three germ layers: endoderm, mesoderm and ectoderm (Fig. 2B). Together, these data suggest that REST is not required for maintaining ES cell pluripotency.
Fig. 2.
The lack of REST in ES cells does not compromise ES cell pluripotency. (A) Immunostaining of Rest+/+, Rest+/– and Rest–/– ES cells, indicating that the pluripotency markers SSEA1 and OCT4 are uniformly present in the ES colonies. Scale bars: 50 μm. (B) Teratomas generated from Rest+/+ or Rest–/– ES cells differentiated into the three germ layers. Arrows indicate ciliated columnar epithelial, plus sign indicates striated muscle, asterisk indicates epidermal structures.
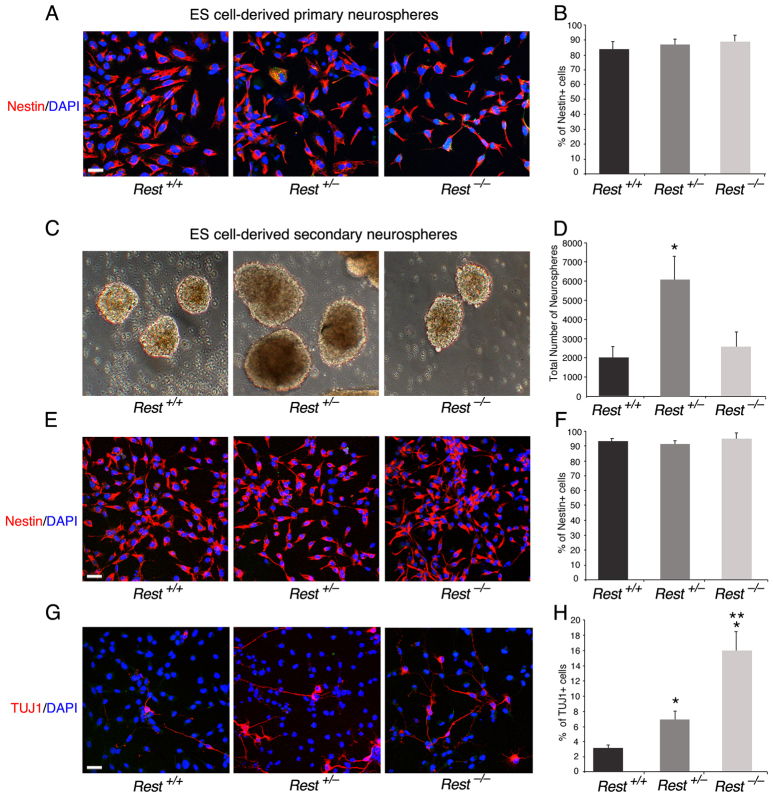
We then analyzed whether the presence of REST in ES cells has any role in their subsequent conversion to NS/P cells. We employed the ES cell-derived neurosphere system, which recapitulates the temporal progression of CNS development (Okada et al., 2008). In this system, the capacity of the ES cell-derived neurospheres switches with time from neurogenic to gliogenic, and, whereas the primary neurospheres give rise mainly to neurons, secondary and tertiary neurospheres give rise to neurons and glia with the tertiary neurospheres being more gliogenic than neurogenic (Okada et al., 2008; Okano and Temple, 2009). We compared the ability of the three different ES cell types, Rest+/+, Rest+/– and Rest–/–, to generate primary, secondary and tertiary neurospheres. Primary neurospheres generated from Rest–/– ES cells showed a lack of uniformity compared with those generated from Rest+/+ or Rest+/– ES cells (supplementary material Fig. S3A); however, >85% of the cells in the cultured neurospheres were nestin positive, similar to cultured primary neurospheres derived from Rest+/– and Rest+/+ ES cells (Fig. 3A,B). Potentially, the dissociation and culturing of the neurospheres could select for specific cell type, e.g. neural progenitors. To rule out this possibility, we analyzed sectioned neurospheres and found that all three types of primary neurospheres consisted mostly of nestin-positive cells, which were uniformly distributed throughout the spheres (supplementary material Fig. S4A). Furthermore, the Rest–/– primary neurospheres were also able to generate secondary neurospheres (Fig. 3C). The heterozygous primary neurospheres generated two- to threefold more secondary neurospheres than did wild-type or homozygous primary neurospheres, which were also larger and more uniform in structure (Fig. 3C,D; supplementary material Fig. S3B). Similar results were obtained when tertiary neurospheres were generated from secondary neurospheres (supplementary material Fig. S3C,D). Analysis of the different types of secondary neurospheres for the presence of cell division markers showed no significant differences in the fraction of Ki67-positive cells; however, heterozygous neurospheres had a higher mitotic index than the wild-type or homozygous neurospheres as indicated by the higher fraction of phosphohistone H3-positive cells (supplementary material Fig. S5A-D). This suggests that the precise low level of REST in NS/P cells is likely to be crucial for efficient neurosphere generation and proliferation.
Fig. 3.
ES cells that lack REST are able to generate nestin-positive neurospheres albeit with some precocious neuronal differentiation. (A) Primary neurospheres generated from ES cells comprise largely nestin-positive NS/P cells. Immunostaining of dissociated neurospheres is shown. Nestin, NS/P marker; DAPI, nuclear marker. (B) Bar graph showing the fraction of nestin-positive NS/P cells in the different neurospheres. (C) Rest+/– and Rest–/– ES cells are able to generate secondary neurospheres similar to Rest+/+ ES cells. (D) Bar graphs indicating equal number of neurospheres generated from wild-type and Rest–/– ES cells but significantly higher number generated from Rest+/– ES cells (*P<0.05). (E) Secondary neurospheres generated from Rest+/– or Rest–/– ES cells comprise largely nestin-positive NS/P cells. Immunostaining of dissociated neurospheres is shown. Nestin, NS/P marker; DAPI, nuclear marker. (F) Bar graphs demonstrating no significant difference in the number of nestin-positive cells between the different secondary neurospheres. (G) Secondary neurospheres generated from Rest+/– and Rest–/– ES cells show some precocious neuronal differentiation. Immunostaining of dissociated neurospheres is shown. TUJ1, neuronal marker; DAPI, nuclear marker. (H) Bar graph indicating low but significant increase in the number of TUJ1-positive cells in secondary neurospheres generated from Rest+/– and Rest–/– ES cells (*P<0.05). There is also a significant increase in the number of TUJ1-positive cells in Rest–/– relative to Rest+/– secondary neurospheres (**P<0.05). All error bars are mean ± s.e.m. based on three independent experiments. Scale bars: 50 μm.
Analysis of the secondary neurospheres for the presence of the neural precursor marker nestin and the neuronal marker TUJ1 (TUBB3 – Mouse Genome Informatics) showed that, like the primary neurospheres, sectioned and cultured secondary neurospheres consisted mostly of nestin-positive cells (Fig. 3E,F; supplementary material Fig. S4B). However, although relatively low, there was increased precocious neuronal differentiation, as shown by the presence and morphology of TUJ1-positive cells, in Rest+/– (up to 7%) and Rest–/– (up to 16%) compared with Rest+/+ (up to 3%) cultured secondary neurospheres (Fig. 3G,H). Together, these data suggest that, although some precocious neuronal differentiation was evident in the cultured neurospheres, the lack of REST had essentially no effect on the ability of ES cells to generate neurospheres that consist mostly of nestin-positive NS/P cells.
E12.5 NS/P cells that lost REST have significantly reduced ability to self-renew
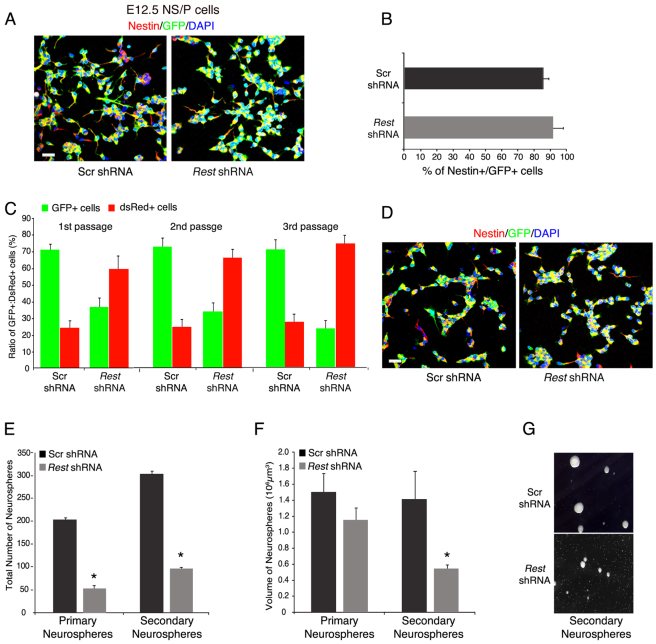
We analyzed in further detail the effect of loss of REST on early-born, neurogenic NS/P cells isolated from embryos at E12.5. First, we investigated whether the loss of REST in neurogenic NS/P cells could affect their identity or capacity to self-renew. For this, we knocked down REST using a lenti virus carrying Rest-specific shRNA and GFP (supplementary material Fig. S6). Analysis of the NS/P cells lacking REST showed that, like control NS/P cells expressing scrambled shRNA, >85% were nestin positive (Fig. 4A,B). We then analyzed the NS/P cells lacking REST for self-renewal in a competition-based assay. In this assay, GFP-positive NS/P cells, expressing either Rest shRNA or scrambled shRNA, were mixed with NS/P cells expressing DsRed, in a 3:1 ratio, respectively, and cultured for three passages; in each passage, GFP- and DsRed-positive cells were counted by FACS sorting before replating. Our data show that, although the ratio of the mixed NS/P cells expressing scrambled shRNA (GFP+) and NS/P cells expressing DsRed (DsRed+) remained 3:1 throughout the three passages, the ratio of the mixed NS/P cells expressing Rest shRNA (GFP+) and DsRed+ was inverted with passages from the initial 3:1 ratio to 1:3 (Fig. 4C). However, when we analyzed the GFP-positive cells after the third passage, we found that the majority of the cells remained nestin positive, irrespective of whether they expressed control shRNA or Rest shRNA (Fig. 4D). We examined the NS/P cells lacking REST for their ability to generate neurospheres and found that the number and volume of the primary and secondary neurospheres were significantly reduced relative to control NS/P cells (Fig. 4E-G). Together, these data suggest that E12.5 NS/P cells lacking REST have significantly reduced capacity to self-renew, although the majority of the NS/P cells did not lose their identity.
Fig. 4.
E12.5 N/SP cells that lack REST have reduced capacity to proliferate or generate neurospheres. (A) Immunostaining of E12.5 NS/P cells expressing Rest shRNA indicating that they remained largely nestin positive. GFP represents transduced cells. Scrambled (Scr) shRNA served as a control. (B) Bar graph indicating no significant differences in the number of nestin-positive NS/P cells, whether expressing scrambled or Rest shRNA. (C) NS/P cells lacking REST show reduced proliferation rate in a competition-based assay. Bar graph shows that mixed NS/P cells initially cultured at a ratio of 3:1 GFP/Rest shRNA-expressing NS/P cells:dsRed-expressing NS/P cells, respectively, ended up at a 1:3 ratio after three passages. (D) Immunostaining after the third passage indicates that GFP+ cells, expressing Rest shRNA, remained largely Nestin-positive NS/P cells. (E,F) The number (E) and volume (F) of neurospheres generated from NS/P lacking REST (Rest shRNA) are significantly reduced. *P<0.05. (G) Representative images showing that secondary neurospheres generated from Rest shRNA-expressing NS/P cells are smaller than scrambled shRNA-expressing NS/P cells. Scale bars: 50 μm. All error bars are mean ± s.e.m. based on three independent experiments.
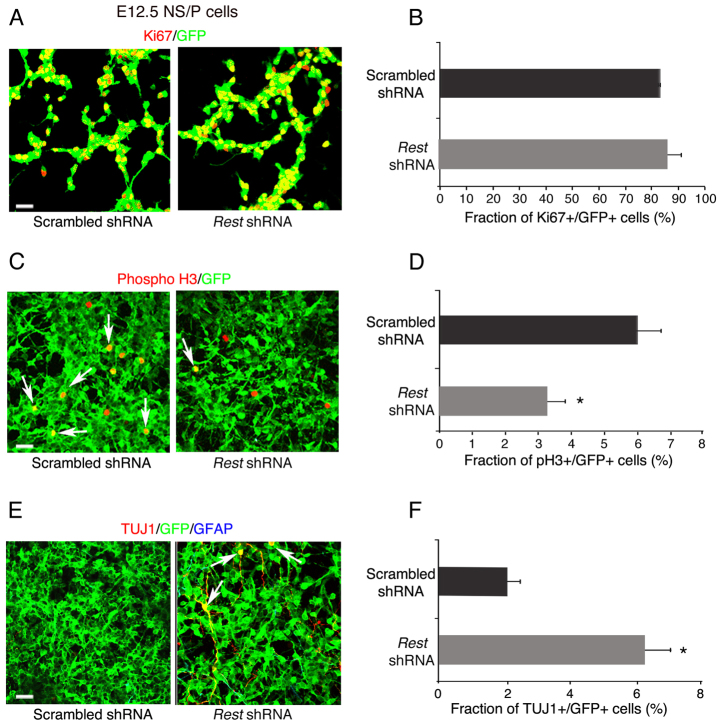
Next, we used cell division and neuronal-specific markers to examine the underlying cause of reduced self-renewal of E12.5 NS/P cells lacking REST. Our results show that, similar to control NS/P cells, 85% of the NS/P cells lacking REST expressed Ki67 (Fig. 5A,B); however, their mitotic index (phosphohistone H3+/GFP+ cells), measured by the end of the first and second passages, was reduced by 45-50% compared with control NS/P cells (Fig. 5C,D; supplementary material Fig. S7A,B). Immunostaining for the presence of the neuronal marker TUJ1 revealed higher precocious neuronal differentiation (8% and 3% in first and second passages, respectively) than in control NS/P cells (0.5% and 2% in first and second passages, respectively) (Fig. 5E,F; supplementary material Fig. S7C,D). No glial precocious differentiation was observed based on the absence of GFAP (Fig. 5E,F; supplementary material Fig. S7C,D). These data suggest that the reduced capacity of E12.5 NS/P cells lacking REST to self-renew is mainly due to reduced cell cycle kinetics and, to a certain extent, due to precocious neuronal differentiation. Supporting this view is our finding that at least three genes involved in cell cycle progression, Cdk6, cyclin B2 and Cdk1c, were downregulated in E12.5 NS/P cells lacking REST (supplementary material Fig. S7E).
Fig. 5.
E12.5 NS/P cells that lack REST do not exit the cell cycle but have significantly reduced mitotic activity. (A) Immunostaining showing that most of the NS/P cells expressing Rest shRNA express the cell division marker Ki67. GFP represents cells expressing shRNA. (B) Bar graph indicating no significant difference in the number of Ki67+ cells in NS/P cells expressing Rest or scrambled shRNA. (C) Immunostaining showing fewer NS/P cells expressing the mitotic marker phosphohistone H3 (phospho H3) when Rest shRNA is expressed. Arrows point to GFP+ cells expressing phospho H3. (D) The mitotic index (fraction of phosphoH3+/GFP+ cells) of NS/P cells expressing Rest shRNA is significantly reduced. (E) Immunostaining showing some precocious neuronal differentiation as indicated by the presence of TUJ1+ cells in NS/P cells expressing Rest shRNA (arrows). (F) Bar graph indicating low but significant number of Rest shRNA-expressing NS/P cells that are TUJ1+. Scale bars: 50 μm. All error bars are mean ± s.e.m. based on three independent experiments. *P<0.05.
The lack of REST or CoREST in neurogenic NS/P cells enhances neuronal differentiation and maturation
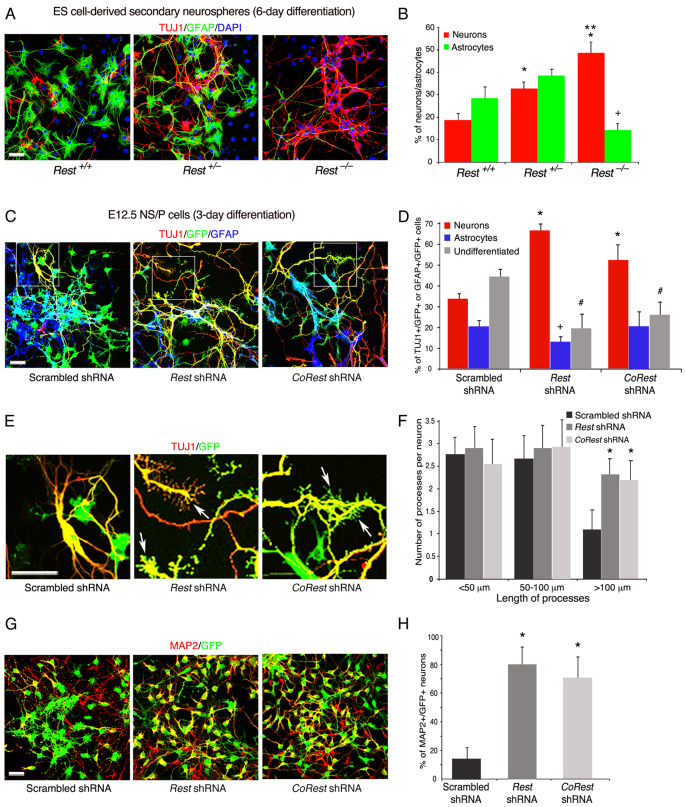
We examined further the nestin-positive NS/P cells lacking REST for their capacity to differentiate to neurons and glia. We first analyzed the ability of Rest+/+, Rest+/– and Rest–/– ES cell-derived secondary neurospheres (Fig. 3) to differentiate into neurons and astrocytes upon withdrawal of EGF and FGF. Our data show that Rest–/– and Rest+/– secondary neurospheres produced significantly more neurons than Rest+/+neurospheres (50:30:20%, respectively) (Fig. 6A,B). However, whereas the heterozygous and wild-type neurospheres produced comparable numbers of astrocytes, the homozygous produced fewer astrocytes than did wild-type or heterozygous secondary neurospheres (15% versus 30-40%) (Fig. 6A,B). These data show that although ES cells that lack REST generate neurospheres composed largely of nestin-positive NS/P cells, their neuronal differentiation was enhanced, resulting in altered neurons:glia ratios.
Fig. 6.
The lack of REST or CoREST in neurogenic E12.5 NS/P cells enhances neuronal differentiation and maturation. (A) Immunostaining of Rest+/+, Rest+/– and Rest–/– secondary neurospheres (SNS) after 6-day differentiation. TUJ1, GFAP and DAPI, represent neuronal, astrocytic and nuclear markers, respectively. (B) Bar graph showing the fraction of neurons and astrocytes in the different differentiated SNS. Rest+/– and Rest–/– SNS generated significantly more neurons (TUJ1+ cells) than did Rest+/+SNS (*P<0.05), and Rest–/– SNS generated significantly more neurons than Rest+/– SNS (**P<0.05). Rest–/– SNS generated significantly fewer astrocytes (GFAP+ cells) than Rest+/+ or Rest+/– SNS (+P<0.05). (C) Immunostaining of the indicated E12.5 NS/P cells after 3-day differentiation upon withdrawal of growth factors. TUJ1 and GFAP represent neuronal and astrocytic markers, respectively. GFP represents shRNA-expressing cells. Note the long processes extended from TUJ1+ cells expressing Rest shRNA or CoRest shRNA. Boxes indicate the areas shown in E. (D) Bar graph showing the fraction of neurons, astrocytes and undifferentiated NS/P cells after 3-day differentiation of NS/P cells expressing the indicated shRNAs. NS/P cells expressing Rest shRNA or CoRest shRNA generated significantly more neurons (TUJ1+ cells). (*P<0.05). NS/P cells expressing Rest shRNA generated significantly fewer astrocytes (GFAP+ cells) (+P<0.05). The number of undifferentiated cells in NS/P cells expressing Rest or CoRest shRNA was significantly reduced (#P<0.05). (E) Neurons differentiated from NS/P cells expressing Rest shRNA or CoRest shRNA have a growth cone-like structure. Enlargement of the framed areas in C. TUJ1, pan neuronal marker; GFP, indicating cells expressing shRNA. Arrows point to the areas with many short protrusions extended from axonal tips. (F) Bar graph showing significant increase in the number of long processes (>100 μm) in neurons (TUJ1+) differentiated for 3 days from NS/P cells expressing Rest shRNA or CoRest shRNA (*P<0.05). (G) Immunostaining for MAP2 of the indicated E12.5 NS/P cells after 3-day differentiation. MAP2, dendritic marker; GFP represents cells expressing shRNA. (H) Bar graph showing significantly higher number of MAP2+ neurons in the absence of REST (Rest shRNA) or CoRest shRNA (*P<0.05). Scale bars: 50 μm. All error bars are mean ± s.e.m. based on three independent experiments.
Similar to the Rest–/– ES cell-derived NS/P cells (Fig. 6A,B), E12.5 NS/P cells lacking REST (Rest shRNA) generated twice as many neurons and 40% fewer astrocytes compared with control NS/P cells, after 3 days differentiation in the absence of growth factors (Fig. 6C,D). Knockdown of CoREST in E12.5 NS/P cells (supplementary material Fig. S6) resulted in up to 60% more neurons, but a similar number of astrocytes, compared with control differentiated NS/P cells (Fig. 6C,D). Importantly, we found that both NS/P cells lacking REST or CoREST had significantly fewer undifferentiated, nestin-positive NS/P cells than did the control NS/P cells (20:25:45%, respectively) (Fig. 6D), suggesting that loss of REST and CoREST enhances the production of neurons while depleting the NS/P pool. Interestingly, neurons generated from NS/P cells lacking REST or CoREST often showed growth cone-like structures with abundant short neurite extensions reminiscent of filopodia, which were lacking in the control NS/P-differentiated neurons (Fig. 6E). These neurons also had significantly more processes, which were >100 μm in length compared with control NS/P-differentiated neurons (Fig. 6C,F). Finally, immunostaining for the dendritic marker MAP2 (MTAP2 – Mouse Genome Informatics) showed that there was a dramatic increase in the number of MAP2-positive neurons in differentiated cultures of NS/P cells lacking REST or CoREST, compared with control NS/P-differentiated neurons (80:70:15%, respectively) (Fig. 6G,H). Together, these data suggest that the lack of REST or CoREST in neurogenic NS/P cells enhances not only the production of neurons but also their maturation.
Loss of REST in gliogenic NS/P cells enhances the generation of neurons and oligodendrocytes but does not affect the generation of astrocytes
The intrinsic capacity of early-born NS/P cells switches with time from neurogenic to more gliogenic (Okano and Temple, 2009; Qian et al., 2000). Therefore, we analyzed the effect of loss of REST or CoREST on the generation of neurons and glia from: (1) E12.5 NS/P cells when maintained in culture long-term (10-day differentiation); (2) E17.5 NS/P cells, which are generally considered to be more gliogenic than E12.5 NS/P cells (supplementary material Fig. S8); (3) tertiary neurospheres derived from ES cells.
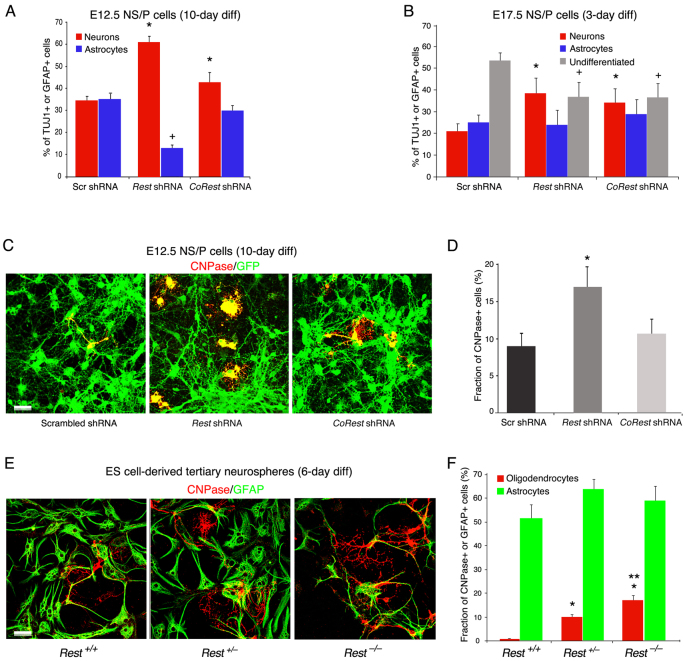
Normal E12.5 NS/P cells produced more neurons and fewer astrocytes after short-term differentiation (3 days) (Fig. 6D, scrambled shRNA; supplementary material Fig. S8) but after long-term differentiation (10 days), they generated an equal number of neurons and astrocytes (Fig. 7A, scrambled shRNA), similar to E17.5 NS/P cells when differentiated for 3 days (Fig. 7B, scrambled shRNA; supplementary material Fig. S8). However, like E12.5 NS/P cells lacking REST and differentiated for 3 days (Fig. 6D, Rest shRNA), when differentiated for 10 days, E12.5 NS/P cells still produced significantly more neurons and fewer astrocytes than the control NS/P cells (Fig. 7A). This suggests that the reduced number of astrocytes generated in the absence of REST is not due to a delay in their production. Unlike REST, lack of CoREST, although accelerating the production of neurons to a certain extent, did not compromise the production of astrocytes during short- or long-term NS/P cell differentiation (Fig. 6D, Fig. 7A). When E17.5 NS/P cells lacking REST or CoREST were differentiated for 3 days, there was also a significant increase in the number of neurons and a decrease in the number of undifferentiated NS/P cells, but to a lesser extent than was observed in E12.5 NS/P cells (compare Fig. 6D and Fig. 7B). Notably, unlike E12.5 NS/P cells, the fraction of astrocytes generated from E17.5 NS/P cells lacking REST was not significantly different from control E17.5 NS/P cells (Fig. 7B), suggesting that loss of REST in NS/P cells enhances the production of neurons primarily from the neurogenic pool.
Fig. 7.
The lack of REST in gliogenic NS/P cells significantly enhances oligodendrocytic differentiation whereas astrocytic differentiation was not affected. (A) Bar graph showing that E12.5 NS/P cells expressing Rest shRNA or CoRest shRNA and differentiated for 10 days generated significantly more neurons (TUJ1+) (*P<0.05). Fewer astrocytes (GFAP+) were generated from NS/P cells expressing Rest shRNA (+P<0.05). (B) Bar graph showing that E17.5 NS/P cells expressing Rest shRNA or CoRest shRNA and differentiated for 3 days produced significantly more neurons (TUJ1+) (*P<0.05) whereas the number of undifferentiated NS/P (nestin+) cells was significantly reduced (+P<0.05). The number of astrocytes (GFAP+) generated was not significantly different. (C) Immunostaining for CNPase, an oligodendrocytic marker, in the indicated shRNA-expressing E12.5 NS/P cells (GFP+) after 10-day differentiation. (D) Bar graph showing that the number of oligodendrocytes (CNPase+) generated from E12.5 NS/P cells expressing Rest shRNA after 10-day differentiation is significantly higher than that generated from scrambled or CoRest shRNA-expressing NS/P cells (*P<0.05). (E) Immunostaining for GFAP, an astrocytic marker, and CNPase, an oligodendrocytic marker, in ES cell-derived tertiary neurospheres after 6-day differentiation. (F) Bar graph showing that the number of oligodendrocytes generated from Rest+/– and Rest–/– tertiary neurospheres but not astrocytes is significantly higher than that generated from Rest+/+ tertiary neurospheres (*P<0.05) and the number of oligodendrocytes generated from Rest–/– tertiary neurospheres is significantly higher than that generated from Rest+/– tertiary neurospheres (**P<0.05). Scale bars: 50 μm. All error bars are mean ± s.e.m. based on three independent experiments.
Surprisingly, our analysis of the long-term differentiated culture of E12.5 NS/P cells showed that the production of oligodendrocytes, measured by the presence of CNPase (CNP – Mouse Genome Informatics), was twofold higher in the absence of REST but was not altered in the absence of CoREST (Fig. 7C,D). Enhanced production of oligodendrocytes was also evident in differentiated tertiary neurospheres derived from Rest+/– or Rest–/– ES cells, which generated up to 10- and 20-fold, respectively, more oligodendrocytes than from neurospheres derived from wild-type Rest+/+ ES cells (Fig. 7E,F). Conversely, and like differentiated E17.5 N/P cells (Fig. 7B), the fractions of astrocytes generated from the three different types of tertiary neurospheres were indistinguishable from each other (Fig. 7F). This suggests that in the absence of REST neurogenic NS/P cells are able to acquire gliogenic capacity and that the loss of REST has no effect on the production of astrocytes from NS/P cells, which had already acquired gliogenic fate.
REST is a functional repressor of neuronal and oligodendrocytic lineage genes in NS/P cells
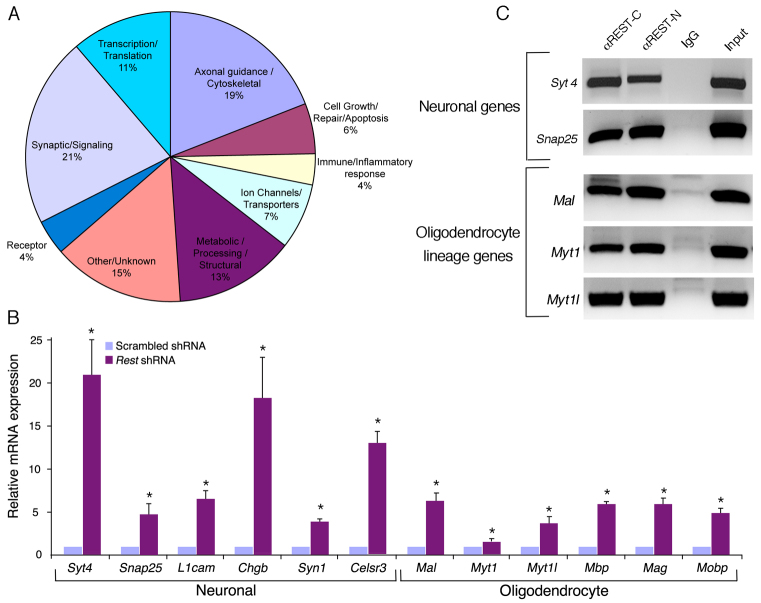
Next, we investigated whether the aberrant pattern of neuronal:glial differentiation is directly related to the type of genes that REST regulates in NS/P cells. To this end, we analyzed the mRNA profile of E12.5 NS/P cells lacking REST and found that a large number of genes (231 in total) were significantly upregulated. The majority of these genes are expressed in the brain, many preferentially, and many are genes involved in synaptic signaling, axonal guidance/cytoskeletal, ion channel transport and transcription/translation (Fig. 8A; supplementary material Table S2. Furthermore, at least 70% of these upregulated genes are RE1-containing genes based on genome-wide analyses of REST occupancy (supplementary material Table S2) or computational analysis based on a previously published algorithm (Otto et al., 2007). Importantly, analysis of the individual genes showed that, although many were neuronal lineage-specific genes, some of the genes were oligodendrocyte-specific genes. We verified their upregulation by quantitative RT-PCR and found that, like the selected neuronal RE1-containing genes, the oligodendrocyte lineage genes were significantly upregulated (Fig. 8B). Remarkably, all the upregulated oligodendrocytic genes are those involved in myelin formation: myelin-associated glycoprotein (Mag), myelin basic protein (Mbp), myelin and lymphocyte protein (Mal), myelin-associated oligodendrocytic basic protein (Mobp), as well as two zinc-finger DNA-binding transcription factors involved in both neuronal and oligodendrocyte lineage specification, myelin transcription factor 1 (Myt1) and myelin transcription factor 1-like (Myt1l) (Fig. 8B). We analyzed these genes for the presence of the RE1 motif using the position-weighted matrix previously generated for the serial analysis of chromatin occupancy of REST (SACO) (Otto et al., 2007) and found that at least three of the upregulated genes, Myt1 and Myt1l and Mal, contained putative RE1 motifs in their regulatory regions. Chromatin immunoprecipitation (ChIP) analysis of E12.5 NS/P cells, using two different REST-specific antibodies, showed that, like in the neuronal RE1-containing genes Syt4 and Snap25, REST is bound to the RE1 motif of the oligodendrocyte genes Mal, Myt1 and Myt1l (Fig. 8C). Together, our results suggest that REST regulates neuronal and oligodendrocyte specific genes, and that the absence of REST in NS/P cells promotes the upregulation of these target genes and, consequently, enhances the generation of neurons and oligodendrocytes.
Fig. 8.
mRNA profiling of E12.5 NS/P cells that lack REST revealed upregulation of many brain-specific RE1-containing genes, including neuronal and oligodendrocyte lineage genes. (A) Microarray analysis. Diagram presenting the function-based distribution of 231 genes that were upregulated >1.5-fold in NS/P cells expressing Rest shRNA versus scrambled shRNA. For a full list of genes, and the presence of the RE1 motif, see supplementary material Table S2. (B) Quantitative RT-PCR of representative neuronal and oligodendrocyte genes that were upregulated >1.5-fold based on the microarray analysis. Error bars are mean ± s.e.m. based on four independent experiments. *P<0.05. (C) Chromatin immunoprecipitation (ChIP) analysis of E12.5 NS/P cells using two different anti-REST antibodies demonstrating that REST is bound to RE1-containing neuronal and oligodendrocytic genes. Rabbit IgG served as negative control. Images represent PCR amplification of the immunoprecipitated DNA around the identified RE1 motif of the specific genes.
DISCUSSION
Our data show that REST is a functional repressor of neuronal genes in ES cells and that even a 50% reduction in the REST level is sufficient to abrogate neuronal gene repression. This suggests that the high level of REST in ES cells, which is mediated by pluripotent transcription factors, is required for its full repressor activity. These results, which corroborate our previous findings that neuronal genes in ES cells are repressed but their chromatin is poised for activation (Ballas et al., 2005), suggest that this type of repressor mechanism depends mainly on the function of the REST repressor complex. Nevertheless, the derepressed neuronal genes in the absence of REST, including the transcription factor NeuroD, which potentially could drive neuronal differentiation (Cho and Tsai, 2004), were unable to compromise ES cell pluripotency. It is likely that, in ES cells, neuronal genes are also regulated post-transcriptionally such that the elevated neuronal transcripts are not translated into functional proteins. Previous studies addressing the role of REST in maintaining ES cell pluripotency have been controversial. Whereas Singh et al. (Singh et al., 2008) have shown that REST is required for ES cell pluripotency based on reduced levels of pluripotency markers, Jorgensen et al. and Buckley et al. have reported that levels of pluripotency factors remain unchanged in the absence of REST (Buckley et al., 2009; Jorgensen et al., 2009a). It remains unclear whether different experimental conditions used in these studies could have contributed to this discrepancy. However, our analyses using a more direct approach, i.e. teratoma formation, in addition to pluripotency markers, corroborate the studies by Jorgenson et al. and Buckley et al. (Buckley et al., 2009; Jorgensen et al., 2009a) and suggest that REST is not part of the transcription factor network that regulates ES cell pluripotency. That the loss of function of REST in ES cells does not compromise their pluripotency is further supported by earlier work showing that REST knockout mice died at E11 (Chen et al., 1998), well past the ES cell stage (E4-E5).
In contrast to the lack of a role for REST in maintaining ES cell pluripotency, our results point to a central role for REST past the ES cell stage, during neural development. ES cells lacking REST were able to convert to neurospheres with a high percentage of nestin-positive NS/P cells; however, precocious neuronal differentiation was evident, similar to E12.5 NS/P cells lacking REST. The reduced cell cycle kinetics together with precocious neuronal differentiation in E12.5 NS/P cells lacking REST resulted in depletion of the NS/P pool. Recent studies addressing the function of REST in adult hippocampal neurogenesis show that loss of REST also results in precocious neurogenesis and depletion of the adult NS/P pool (Gao et al., 2011), together suggesting a general and crucial role for REST in maintaining both embryonic and adult NS/P cell self-renewal. The number and size of neurospheres generated from Rest–/– ES cells were also significantly lower than those generated from Rest+/– ES cells, but not different from those generated from Rest+/+ ES cells. This might reflect differences between direct conversion of ES cells to NS/P cells via neurospheres in vitro, and NS/P cells isolated from developing embryos. This is because the level of Rest mRNA in E12.5 NS/P (Ballas et al., 2005) is more similar to the level of Rest mRNA in neurospheres derived from Rest+/– ES cells rather than neurospheres derived from Rest+/+ ES cells, which was twofold higher (data not shown). In this scenario, the higher level of REST in neurospheres derived from Rest+/+ relative to those derived from Rest+/– ES cells or E12.5 NS/P cells, probably delayed neurosphere generation. Interestingly, similar to the loss of function of REST in E12.5 NS/P cells, our recent data show that E12.5 NS/P cells overexpressing REST remain mostly nestin positive and do not exit the cell cycle, although reduced cell proliferation was evident (Mandel et al., 2011) (data not shown). These data further suggest that the precisely downregulated level of REST in multipotent stem cells is crucial, and too much or too little of REST negatively affects NS/P cell self-renewal.
Consistent with the precocious neuronal differentiation of NS/P cells lacking REST, their default differentiation resulted in a significantly larger pool of neurons and smaller pools of undifferentiated NS/P cells and astrocytes. This suggests that dysfunction of the REST repressor complex enhances neurogenesis while depleting the NS/P pool (Fig. 9). The shrinkage of the astrocytic pool in the absence of REST could be due to the depletion of neurogenic NS/P cells for the accelerated generation of neurons, which in turn depletes the gliogenic NS/P pool that is normally generated with time from the neurogenic NS/P pool. Alternatively, the absence of REST might drive neuronal differentiation not only from neurogenic, but also from gliogenic, NS/P cells and could thus deplete both the neurogenic and gliogenic NS/P pools, resulting in generation of fewer astrocytes. That the production of neurons from gliogenic NS/P cells lacking REST was accelerated whereas the production of astrocytes was unaffected supports the view that the loss of REST probably enhances the production of neurons only from the neurogenic NS/P pool and does not switch gliogenic NS/P cells to neurogenic. Furthermore, lack of REST also did not affect the transition of neurogenic to gliogenic NS/P cells in the presence of growth factors. Interestingly, our microarray analysis of genes that are up- or downregulated in NS/P cells that lack REST did not identify genes involved in astrocytic lineage specification. Together, these data support the view that depletion of the astrocytic pool during differentiation of neurogenic NS/P cells lacking REST probably occurs indirectly.
Fig. 9.
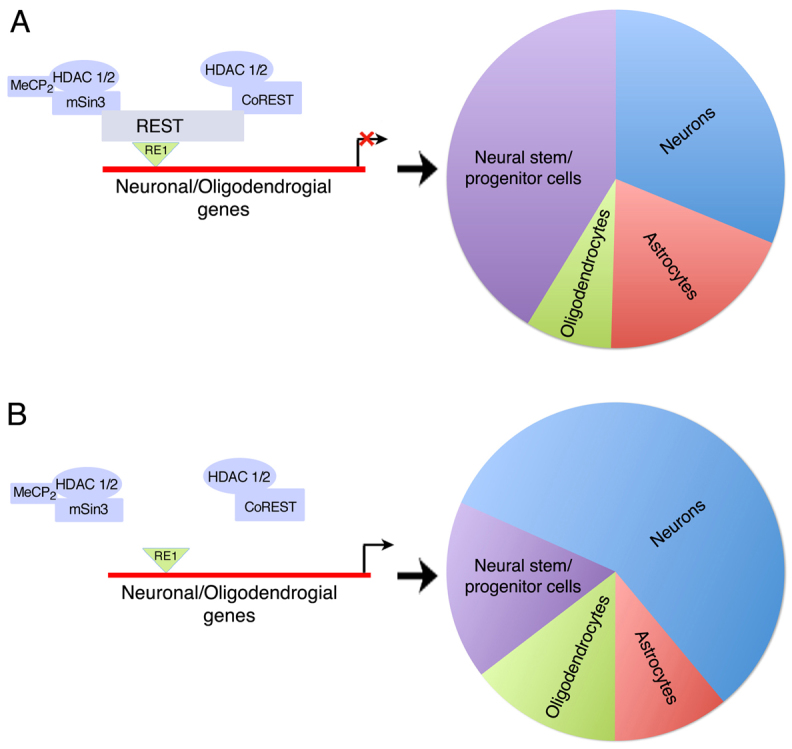
Models for the role of REST in early-born NS/P cells and during their neural differentiation. (A) Left: The REST repressor complex regulates neuronal and oligodendrocyte lineage genes in NS/P cells. Right: Diagram showing the relative pool size of the different neural lineages when REST is present in NS/P cells and during their differentiation. (B) Left: In the absence of REST, the REST co-repressor complex is not recruited to the RE1 sites and neuronal and oligodendrocytic genes are derepressed. Right: Diagram showing that in the absence of REST, the relative pool size of the different lineages is altered, resulting in expansion of the neuronal and oligodendrocytic pools and shrinkage of the NS/P and astrocytic pools.
Loss of CoREST in NS/P cells also accelerated neuronal differentiation and maturation; however, this effect was milder compared with the loss of REST and therefore did not affect the astrocytic pool. Although CoREST could potentially be part of other repressor complexes, the close similarity in the phenotype observed in the absence of REST or CoREST suggests that the predominant function of CoREST during neural development is probably in conjunction with the REST repressor complex.
Our recent data show that gain of function of REST in neurogenic NS/P cells during neocortical development delays, but does not compromise, neuronal differentiation (Mandel et al., 2011). Importantly, gain of function of REST in neurogenic progenitors did not result in generation of glia even though REST remains present in glia and absent in neurons (Mandel et al., 2011). These studies further support the view that loss or gain of function of REST in NS/P cells is incapable of switching NS/P cells from gliogenic to neurogenic or from neurogenic to gliogenic.
Our microarray, ChIP and differentiation analyses of NS/P cells all point to a role for REST not only in neurogenesis but also in oligodendrogenesis (Fig. 9). The absence of REST in NS/P cells resulted in upregulation of neuronal genes as well as oligodendrocyte lineage genes, specifically those involved in myelination. At least three of the six upregulated oligodendrocyte-associated genes (Mal, Myt1 and Myt1l) contain the RE1 motif, which also binds REST. Supporting our data, ChIP-ChIP analysis of REST/CoREST target genes in premature or mature oligodendrocytes identified genes involved in oligodendrocyte specification and maturation (Abrajano et al., 2009).
Collectively, our data support a central role for the REST repressor complex during neural development in maintaining NS/P cell self-renewal and in regulating the generation of proper pool size of the different neural lineages, including neurons, astrocytes and oligodendrocytes (Fig. 9). By regulating genes involved in neuronal and oligodendrocyte specification and maturation, the REST repressor complex restrains neuronal and oligodendrocyte generation and thus regulates the precise pools of the different neural lineages. Further studies are warranted to understand whether such regulation of neuronal and oligodendrocyte generation and maturation by the REST repressor complex, is dynamically involved in pathological conditions such as acute injury and repair or chronic neurological disorders.
Supplementary Material
Acknowledgments
We thank Dr Sally Temple for valuable discussion about the results and advice; Dr Sean McCorkle for providing his expertise with the bioinformatics; Dr Gail Mandel for providing the anti-REST antibodies; Drs Amanda Fisher, Helle Jorgenson and Zhou-Feng Chen for sharing and providing the different Rest-knockout ES cells; Dr Thomas Zarembinski and BioTime for the generous gift of Glycosan HyStem; the Pathology Translational Research Laboratory for assistance with the teratoma analysis.
Footnotes
Funding
This work was supported by a grant from the National Institutes of Health [NS060797 to N.B.]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.074765/-/DC1
References
- Abrajano J. J., Qureshi I. A., Gokhan S., Zheng D., Bergman A., Mehler M. F. (2009). Differential deployment of REST and CoREST promotes glial subtype specification and oligodendrocyte lineage maturation. PLoS ONE 4, e7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres M. E., Burger C., Peral-Rubio M. J., Battaglioli E., Anderson M. E., Grimes J., Dallman J., Ballas N., Mandel G. (1999). CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. USA 96, 9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N., Mandel G. (2005). The many faces of REST oversee epigenetic programming of neuronal genes. Curr. Opin. Neurobiol. 15, 500–506 [DOI] [PubMed] [Google Scholar]
- Ballas N., Battaglioli E., Atouf F., Andres M. E., Chenoweth J., Anderson M. E., Burger C., Moniwa M., Davie J. R., Bowers W. J., et al. (2001). Regulation of neuronal traits by a novel transcriptional complex. Neuron 31, 353–365 [DOI] [PubMed] [Google Scholar]
- Ballas N., Grunseich C., Lu D. D., Speh J. C., Mandel G. (2005). REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121, 645–657 [DOI] [PubMed] [Google Scholar]
- Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., et al. (2005). Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce A. W., Donaldson I. J., Wood I. C., Yerbury S. A., Sadowski M. I., Chapman M., Gottgens B., Buckley N. J. (2004). Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc. Natl. Acad. Sci. USA 101, 10458–10463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley N. J., Johnson R., Sun Y. M., Stanton L. W. (2009). Is REST a regulator of pluripotency? Nature 457, E5-6; discussion E7 [DOI] [PubMed] [Google Scholar]
- Chen Z. F., Paquette A. J., Anderson D. J. (1998). NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat. Genet. 20, 136–142 [DOI] [PubMed] [Google Scholar]
- Cheng L. C., Tavazoie M., Doetsch F. (2005). Stem cells from epigeneticsto microRNAs. Neuron 46, 363–367 [DOI] [PubMed] [Google Scholar]
- Cho J. H., Tsai M. J. (2004). The role of BETA2/NeuroD1 in the development of the nervous system. Mol. Neurobiol. 30, 35–47 [DOI] [PubMed] [Google Scholar]
- Chong J. A., Tapia-Ramirez J., Kim S., Toledo-Aral J. J., Zheng Y., Boutros M. C., Altshuller Y. M., Frohman M. A., Kraner S. D., Mandel G. (1995). REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80, 949–957 [DOI] [PubMed] [Google Scholar]
- Dewald L. E., Rodriguez J. P., Levine J. M. (2011). The RE1 binding protein REST regulates oligodendrocyte differentiation. J. Neurosci. 31, 3470–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund T., Jessell T. M. (1999). Progression from extrinsic to intrinsic signaling in cell fate specification: a view from the nervous system. Cell 96, 211–224 [DOI] [PubMed] [Google Scholar]
- Gao Z., Ure K., Ding P., Nashaat M., Yuan L., Ma J., Hammer R. E., Hsieh J. (2011). The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J. Neurosci. 31, 9772–9786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I. R. (2006). Dissecting self-renewal in stem cells with RNA interference. Nature 442, 533–538 [DOI] [PubMed] [Google Scholar]
- Johnson D. S., Mortazavi A., Myers R. M., Wold B. (2007). Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497–1502 [DOI] [PubMed] [Google Scholar]
- Johnson R., Teh C. H., Kunarso G., Wong K. Y., Srinivasan G., Cooper M. L., Volta M., Chan S. S., Lipovich L., Pollard S. M., et al. (2008). REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol. 6, e256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen H. F., Fisher A. G. (2010). Can controversies be put to REST? Nature 467, E3-4; discussion E5 [DOI] [PubMed] [Google Scholar]
- Jorgensen H. F., Chen Z. F., Merkenschlager M., Fisher A. G. (2009a). Is REST required for ESC pluripotency? Nature 457, E4-5; discussion E7 [DOI] [PubMed] [Google Scholar]
- Jorgensen H. F., Terry A., Beretta C., Pereira C. F., Leleu M., Chen Z. F., Kelly C., Merkenschlager M., Fisher A. G. (2009b). REST selectively represses a subset of RE1-containing neuronal genes in mouse embryonic stem cells. Development 136, 715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., Haussler D. (2002). The human genome browser at UCSC. Genome Res. 12, 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattin J. E., Schroder K., Su A. I., Walker J. R., Zhang J., Wiltshire T., Saijo K., Glass C. K., Hume D. A., Kellie S., et al. (2008). Expression analysis of G protein-coupled receptors in mouse macrophages. Immunome Res. 4, 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lensch M. W., Schlaeger T. M., Zon L. I., Daley G. Q. (2007). Teratoma formation assays with human embryonic stem cells: a rationale for one type of human-animal chimera. Cell Stem Cell 1, 253–258 [DOI] [PubMed] [Google Scholar]
- Loh Y. H., Wu Q., Chew J. L., Vega V. B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., et al. (2006). The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38, 431–440 [DOI] [PubMed] [Google Scholar]
- Mandel G., Fiondella C. G., Covey M. V., Lu D. D., Loturco J. J., Ballas N. (2011). Repressor element 1 silencing transcription factor (REST) controls radial migration and temporal neuronal specification during neocortical development. Proc. Natl. Acad. Sci. USA 108, 16789–16794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Matsumoto A., Shimazaki T., Enoki R., Koizumi A., Ishii S., Itoyama Y., Sobue G., Okano H. (2008). Spatiotemporal recapitulation of central nervous system development by murine embryonic stem cell-derived neural stem/progenitor cells. Stem Cells 26, 3086–3098 [DOI] [PubMed] [Google Scholar]
- Okano H., Temple S. (2009). Cell types to order: temporal specification of CNS stem cells. Curr. Opin. Neurobiol. 19, 112–119 [DOI] [PubMed] [Google Scholar]
- Otto S. J., McCorkle S. R., Hover J., Conaco C., Han J. J., Impey S., Yochum G. S., Dunn J. J., Goodman R. H., Mandel G. (2007). A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J. Neurosci. 27, 6729–6739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Shen Q., Goderie S. K., He W., Capela A., Davis A. A., Temple S. (2000). Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron 28, 69–80 [DOI] [PubMed] [Google Scholar]
- Roopra A., Sharling L., Wood I. C., Briggs T., Bachfischer U., Paquette A. J., Buckley N. J. (2000). Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3-histone deacetylase complex. Mol. Cell. Biol. 20, 2147–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S. E., Greenberg M. E., Stiles C. D. (2003). Basic helix-loop-helix factors in cortical development. Neuron 39, 13–25 [DOI] [PubMed] [Google Scholar]
- Schoenherr C. J., Anderson D. J. (1995). The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 267, 1360–1363 [DOI] [PubMed] [Google Scholar]
- Singh S. K., Kagalwala M. N., Parker-Thornburg J., Adams H., Majumder S. (2008). REST maintains self-renewal and pluripotency of embryonic stem cells. Nature 453, 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A. I., Wiltshire T., Batalov S., Lapp H., Ching K. A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., et al. (2004). A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 101, 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R. H., Lindblad-Toh K., Birney E., Rogers J., Abril J. F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 [DOI] [PubMed] [Google Scholar]
- Westbrook T. F., Hu G., Ang X. L., Mulligan P., Pavlova N. N., Liang A., Leng Y., Maehr R., Shi Y., Harper J. W., et al. (2008). SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature 452, 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.