Abstract
During central nervous system development, neural progenitors are patterned to form discrete neurogenic and non-neurogenic zones. In the zebrafish hindbrain, neurogenesis is organised by Fgf20a emanating from neurons located at each segment centre that inhibits neuronal differentiation in adjacent progenitors. Here, we have identified a molecular mechanism that clusters fgf20a-expressing neurons in segment centres and uncovered a requirement for this positioning in the regulation of neurogenesis. Disruption of hindbrain boundary cell formation alters the organisation of fgf20a-expressing neurons, consistent with a role of chemorepulsion from boundaries. The semaphorins Sema3fb and Sema3gb, which are expressed by boundary cells, and their receptor Nrp2a are required for clustering of fgf20a-expressing neurons at segment centres. The dispersal of fgf20a-expressing neurons that occurs following the disruption of boundaries or of Sema3fb/Sema3gb signalling leads to reduced FGF target gene expression in progenitors and an increased number of differentiating neurons. Sema3 signalling from boundaries thus links hindbrain segmentation to the positioning of fgf20a-expressing neurons that regulates neurogenesis.
Keywords: Boundaries, Neurogenesis, Segmentation, FGF signalling, Semaphorin
INTRODUCTION
The complex and stereotyped organisation of neurons in the central nervous system is established during development by the differentiation of progenitor cells in the neural epithelium, their migration to the correct location in the mantle zone, and the extension of axons to connect to appropriate targets. The initial steps require the strict regulation of cell differentiation such that the correct number of each neural cell type is formed while retaining sufficient progenitors to generate later-forming cell types. In part this is achieved through the action of inhibitory factors that limit the amount of differentiation in all progenitors (Bylund et al., 2003; Graham et al., 2003) and through Notch-mediated lateral inhibition within the neural epithelium, in which forming neurons inhibit the differentiation of their neighbours (Louvi and Artavanis-Tsakonas, 2006; Kageyama et al., 2007). In some regions of the neural epithelium, there is also a large-scale patterning of neurogenesis to form discrete neurogenic and non-neurogenic zones (Bally-Cuif and Hammerschmidt, 2003; Diez del Corral et al., 2003). For example, three neurogenic zones form along the dorsoventral axis of the spinal cord during primary neurogenesis due to inhibition of differentiation in the intervening regions by specific Hes/Her and Zic genes (Brewster et al., 1998; Bae et al., 2005). Similarly, a non-neurogenic zone forms at the midbrain-hindbrain boundary due to the inhibitory action of Hes/Her family members (Geling et al., 2003; Geling et al., 2004). In these examples, the expression of transcription factors that inhibit neuronal differentiation appears to be linked to dorsoventral and anteroposterior patterning within the neural epithelium that underlies cell type and regional specification.
Another striking example of spatially restricted neurogenesis occurs within segments in the zebrafish hindbrain, where neuronal differentiation is absent at the boundaries and centre of each segment (rhombomere) and thus becomes confined to zones flanking each segment boundary (Cheng et al., 2004). Furthermore, specific neuronal and glial cell types are organised in a segmentally repeated pattern; for example, primary reticulospinal neurons are located at each segment centre (Hanneman et al., 1988) and the fibres of radial glial cells form a curtain that flanks segment boundary cells (Trevarrow et al., 1990). These observations raise the question of how the patterning of neurogenesis within segments is established. In view of the mirror-image symmetric organisation of neurogenesis around hindbrain boundaries, one possibility is that each boundary serves as a signalling centre that regulates cell differentiation, analogous to the roles of boundaries in the Drosophila wing disc (Irvine and Rauskolb, 2001; Tamagnone and Comoglio, 2004; Cavodeassi and Houart, 2012). Indeed, specialised boundary cells form at the interface of hindbrain segments that have distinct molecular and cellular properties (Guthrie and Lumsden, 1991; Heyman et al., 1995; Xu et al., 1995). The absence of neurogenesis at hindbrain boundaries might be due to Notch activation promoted by Rfng, which is expressed by boundary cells (Cheng et al., 2004; Qiu et al., 2004; Qiu et al., 2009). Apparent support for the role of a boundary signal in organising neurogenesis came from the effects of knockdown of Wnt genes expressed in hindbrain boundaries (Riley et al., 2004; Amoyel et al., 2005); however, later work revealed that the altered neurogenesis was due to non-specific activation of the Bcl-caspase pathway, which has a non-apoptotic role in hindbrain boundary cell formation (Gerety and Wilkinson, 2011).
A different hypothesis for how boundaries could underlie the patterning of hindbrain neurogenesis is suggested by studies that reveal a key role of fgf20a-expressing neurons, which are located in the mantle region at each segment centre and are required for formation of the non-neurogenic zone in the adjacent neural epithelium (Gonzalez-Quevedo et al., 2010). In fgf20a mutant embryos, ectopic neurogenesis occurs throughout segment centres, probably owing to a lack of inhibition of neuronal differentiation rather than to altered proliferation of a progenitor population, as FGF signalling does not affect hindbrain cell proliferation at these stages (Gonzalez-Quevedo et al., 2010). Concurrent with the inhibition of neurogenesis, FGF signalling may promote gliogenesis in segment centres (Esain et al., 2010) and thus underlie a switch in cell fate. These findings suggest that it is the localisation of fgf20a expression to form a signalling centre in the mantle zone that underlies the stereotyped patterning of neurogenesis. We therefore set out to elucidate how fgf20a expression is restricted and, in particular, to address the possibility that it involves signalling from hindbrain boundaries.
By analysing the effects of disrupting hindbrain boundaries, we find that boundary cells are required to maintain a tight cluster of fgf20a-expressing neurons in each segment centre. We show that two semaphorin family members expressed at hindbrain boundaries, Sema3fb and Sema3gb, act through Nrp2a to position fgf20a-expressing neurons in segment centres. The dispersal of these neurons that occurs upon disruption of semaphorin function leads to an abnormal organisation of neurogenesis and to an increase in the number of differentiating neurons. Hence, there is an interdependence between the stereotyped positioning of fgf20a-expressing neurons and the regulation of neurogenesis.
MATERIALS AND METHODS
Fish maintenance and transgenic and mutant zebrafish lines
Zebrafish embryos were obtained by natural spawning and raised at 28.5°C as described (Westerfield, 1993). The tp53 mutant line (Berghmans et al., 2005) was acquired from the Zebrafish International Resource Center (University of Oregon, Eugene, OR, USA).
Primers
Primers used were (5′-3′): Sema3fbFw(ClaI), ATAATCGATATGCTCTTGGACAGTCTATGGCCAGTC; Sema3fbRe(BsiWI), ATACGTACGTCATGTCTCCTCCTCGTCCCCG; Sema3gbFw(ClaI), ATAATCGATATGTCATCTCTGCTTTTCGTCTTG; Sema3gbRe(BsiWI), ATACGTACGTCACTGCTCCTCCTCTGGTTCTCC; efnb3bEx1Fw, TTCCGAGTCCAGAGATCTCCA; efnb3bEx2Re, ATGATGAAGTAGTCGTGGTTGG; and efnb3bIn1Re, TAATGTGGCGGCTACTGTCG.
Morpholino oligonucleotides
Morpholino oligonucleotides (MOs) were purchased from Gene Tools. One- to four-cell embryos were microinjected with 1.8 nl MO diluted to 5-10 ng/nl in water. All experiments were performed with at least two independent replicates, and injections with control MO were carried out on the same clutch as experimental injections. tp53–/– embryos were used for MO injections to avoid off-target gene expression caused by toxicity (Gerety and Wilkinson, 2011). The following MOs (5′-3′) have been validated previously: epha4a (AACACAAGCGCAGCCATTGGTGTC) (Cooke et al., 2005); rfng (TGGAGGCGACATGGGATAAGTGCAT) (Cheng et al., 2004); sema3gb (ATCGCAACATTTCTCACCTTTGTAT) (Yu and Moens, 2005); and sema3fb (CATAGACTGTCCAAGAGCATGGTGC), nrp2a (CTTGGTGTGATATCCAGAAATCCAT) and nrp2b (CGCGTAGAGGAAAAAGCTGAAGTTC) (Tanaka et al., 2007).
Splice-blocking efnb3b MO (TTGCGGCTCTTACCTTTTGTTCAAG, efnb3b-SB MO) was used at 6 ng/nl. A scheme of the effect on transcript production is shown in supplementary material Fig. S1D. The efficacy of this MO was determined by RT-PCR analysis, which showed that efnb3b transcript splicing is blocked by efnb3b-SB MO but not control MO (supplementary material Fig. S1F). The loss of rfng expression in efnb3b-SB MO-injected embryos (supplementary material Fig. S1B) is similar to that observed in embryos injected with translation-blocking efnb3b MO (ACTCCCATCAAAGCCGTGTGCGGGA; efnb3b-TB) (supplementary material Fig. S1C).
In situ hybridisation and immunohistochemistry
Embryos were grown at 28.5°C to the desired stage, fixed in 4% paraformaldehyde in PBS overnight, then stored in 100% methanol, or processed immediately for in situ hybridisation or immunohistochemistry. The in situ hybridisation probes for neurog1, rfng (Amoyel et al., 2005), fgf20a and etv5b (Gonzalez-Quevedo et al., 2010) are as previously described. Based on published sequence information, PCR products were cloned as probe templates into pGemT-Easy (Promega, A1360) using the primers described above for sema3gb (AY766121), sema3fb (AY766119.1), nrp2a (NM_212965.1) and nrp2b (NM_212966.1). Digoxigenin-UTP-labelled riboprobes were synthesized according to the manufacturer’s instructions (Roche) and in situ hybridisation and colour development with NTB/BCIP were performed as described (Xu and Wilkinson, 1998). Fluorescent in situ hybridisations combined with NTB/BCIP were performed as described (Hauptmann and Gerster, 1994) with minor modifications. Colour detection was performed using Fast Red tablets (Roche, 11496549001).
For antibody staining after fluorescent in situ hybridisation, embryos were washed with PBS containing 0.1% Tween 20, then blocked in PBS containing 0.1% Tween 20 and 5% goat serum. Antibodies were diluted in this blocking solution and incubated overnight at 4°C. Rabbit anti-EphA4 (1:450) (Irving et al., 1996), rabbit anti-GFP (1:200; Torrey Pines, TP 401) and anti-HuC/D (1:200; Invitrogen, A21271) were used as primary antibodies and detected with Alexa Fluor 488, 594 or 647 goat anti-rabbit IgG (1:450; Invitrogen). Fluorescent images were captured using a Leica TCS SP2 confocal microscope.
Quantitations
Stacks of confocal images were collected of fgf20a-expressing neurons or HuC/D-expressing neurons, each combined with Epha4a immunocytochemistry to detect the location of segment interfaces. Images were processed with FIJI (ImageJ) and Adobe Photoshop. To quantitate the distribution of fgf20a-expressing neurons, images of r3, r4 and r5 were processed to an identical size along the anteroposterior (AP) axis from one boundary to the next. A scheme of the procedure and examples of clusters in the different MO conditions are shown in supplementary material Fig. S4. The images were used for two quantitations: (1) the AP distance from the rhombomere centre to the furthest edge of the neuron cluster compared with the centre-to-boundary distance (α; 100 arbitrary units) and (2) the AP distance between the two edges of the cluster compared with the total rhombomere length (β; 100 arbitrary units).
To quantitate the fusion of r5 and r6 neurons in efnb3b morphants, stacks of confocal images were used to measure the AP distance from the posterior edge of the r5 fgf20a-expressing neuronal cluster to the anterior edge of the r6 cluster. Stacks of confocal images were used to count the number of Hu-expressing neurons or fgf20a-expressing neurons in r3-r5, and to measure the AP length of r3, r4 and r5 in the midline region. All data were processed with Microsoft Excel and the significance of results assessed using the two-tailed Student’s t-test.
RESULTS
The stereotyped organisation of neurogenesis within hindbrain segments in zebrafish can potentially be explained by a role of hindbrain boundary cells in restricting fgf20a expression to neurons (for brevity referred to hereafter as fgf20a neurons) located in the centre of segments. Such a role could be mediated through one of two alternative mechanisms: (1) that boundary cells are a source of signals that position neurons which intrinsically express fgf20a; or (2) that a diffusible signal emanating from boundary cells extrinsically represses fgf20a expression in the vicinity. These models can be assessed by analysing the effects of disrupting the formation of hindbrain boundary cells.
Loss of a subset of segment boundaries leads to repositioning of fgf20a neurons
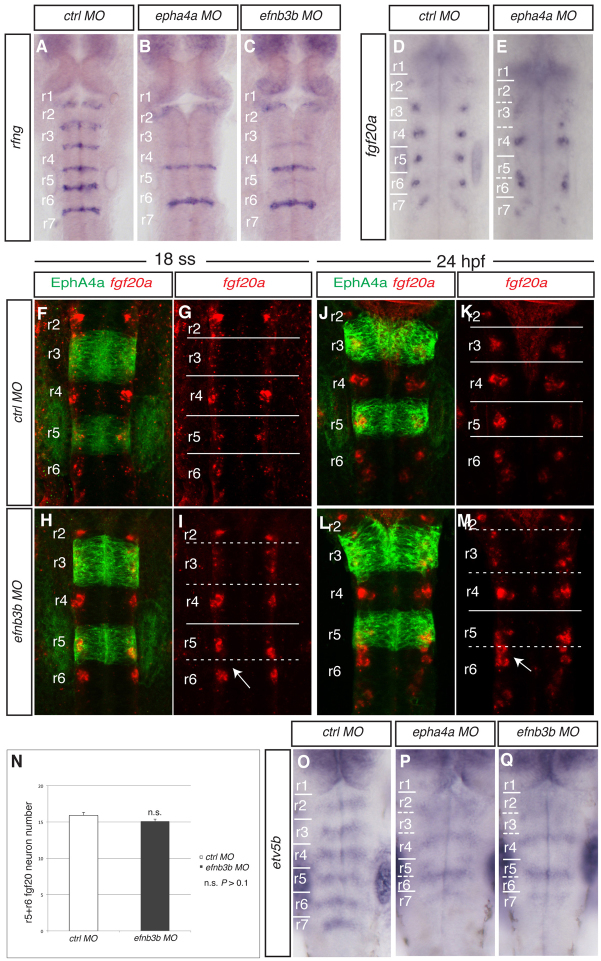
As a first approach toward addressing the potential role of hindbrain boundary cells, we took advantage of the findings that Eph receptor and ephrin signalling not only underlies the formation of sharp segment interfaces, but also is required for the upregulation of boundary cell markers (Xu et al., 1995; Cooke et al., 2005). Knockdown of epha4a leads to decreased expression of the boundary cell marker sema3gb at some segment interfaces, but not others (Cooke et al., 2005), which is likely to reflect the overlapping expression and function of other Eph receptors. We verified this finding using rfng as a boundary cell marker and found loss or substantial downregulation of its expression at the interface between segments 2/3, 3/4 and 5/6, whereas expression was maintained at the 4/5 and 6/7 boundaries (Fig. 1A,B). We found that epha4a knockdown leads to an altered organisation of fgf20a-expressing cells at 24 hours postfertilisation (hpf): whereas fgf20a expression in rhombomere (r) 4 seemed unaffected, the cluster of fgf20a neurons in r3 was more dispersed, and in r5 and r6 the clusters were relocated and became fused near to the r5/r6 interface (Fig. 1D,E). As the altered organisation is not accompanied by an increase in the number of fgf20a neurons (Fig. 1N), these observations argue against boundary cells acting to repress fgf20a expression and are more consistent with a role in positioning of fgf20a neurons.
Fig. 1.
Disruption of a subset of rhombomere boundaries affects the position of fgf20a neurons. Brightfield and confocal images show dorsal views of the zebrafish hindbrain, anterior to the top, following in situ hybridisation (blue or red) and staining for EphA4 protein (green). (A-C) rfng expression in 21 ss embryos after injection of control MO (A), epha4a MO (B) or efnb3b-SB MO (C). There is decreased boundary marker expression selectively at the r2/r3, r3/r4 and r5/r6 borders following epha4a (90%, n=32) or efnb3b (82%, n=23) knockdown. (D,E) fgf20a expression at 24 hpf after injection of control MO (D) or epha4a MO (E). Knockdown of epha4a leads to altered localisation of fgf20a neurons (88%, n=26). (F-M) fgf20a expression (red) and EphA4 antibody staining (green) in embryos injected with control MO (F,G,J,K) or efnb3b-SB MO (H,I,L,M) at 18 ss (F-I) and 24 hpf (J-M). Arrow points to r5 and r6 fgf20a neuronal clusters that approach and fuse near to the r5/r6 interface. efnb3b morphants do not have the r5-r6 fusion of fgf20a neurons at 18 ss (95%, n=20), but do at 24 hpf (83%, n=18). (N) Quantitation of the number of fgf20a neurons in r5 plus r6. There is no significant difference between control versus efnb3b knockdown embryos (15.9±0.4 versus 15.1±0.3 neurons, average ± s.e.m., n=11 each; P=0.19). (O-Q) etv5b expression at 30 hpf in embryos injected with control MO (O), epha4a MO (P) and efnb3b-SB MO (Q). Knockdown of epha4a or efnb3b leads to an altered pattern with fewer stripes of etv5b expression (94%, n=35; 88%, n=24, respectively). Dashed white lines indicate the position of missing boundary marker expression, and continuous white lines indicate the position of remaining boundary cells. r, rhombomere.
An alternative interpretation is that knockdown disrupts the organisation of fgf20a neurons in r3 and r5, where epha4a is normally expressed, due to a role of this Eph receptor within r3/r5 rather than in boundary cell formation. To address this, we knocked down ephrin B3b (efnb3b), a ligand of EphA4 (Gale et al., 1996), which is expressed in r2/r4/r6 (Chan et al., 2001), complementary to epha4a. We found that efnb3b knockdown leads to a similar disruption of boundary marker expression as epha4a knockdown (Fig. 1C; supplementary material Fig. S1), consistent with these factors acting as a receptor-ligand pair. By detecting Epha4a protein as a marker of r3/r5 following knockdown of efnb3b, we visualised the relationship between fgf20a neurons and segment interfaces (Fig. 1F-M). We found that efnb3b knockdown leads to a similar alteration in the positioning of fgf20a neurons at 24 hpf as epha4a knockdown, with a dispersal of fgf20a neurons in r3 and an ectopic cluster at the r5/r6 interface (Fig. 1L,M). The r5/r6 clusters are separate at the 18-somite stage (ss) (Fig. 1H,I) but are fused by 24 hpf (Fig. 1L,M), suggesting that they undergo a progressive displacement from their original position at the centre of r5 and r6. The similar effect of knockdown of epha4a and efnb3b – which in the hindbrain interact only at segment borders – suggests that the altered organisation of fgf20a neurons is a consequence of the depletion of boundary cells rather than reflecting an autonomous role in the segments within which they are expressed. The finding that the organisation of fgf20a neurons in efnb3b morphants appears normal at early stages suggests that boundary cells are not required for the initial positioning of these neurons.
To determine whether the altered position of fgf20a neurons affects the pattern of FGF receptor activation, we analysed the expression of etv5b (erm), a target of Fgf20a signalling at this stage of development (Gonzalez-Quevedo et al., 2010). In control embryos, etv5b is restricted to the centre of each of the six rhombomeres (Fig. 1O). Knockdown of either epha4a (Fig. 1P) or efnb3b (Fig. 1Q) leads to a similar change, in which there are fewer stripes of etv5b expression, and these correlate with the altered position of fgf20a neurons in these knockdown conditions: there is strong etv5b expression in r4 and at the r5/r6 border where clustering of fgf20a neurons occurs, but expression is weaker in the other segments.
Loss of all segment boundaries leads to dispersal of fgf20a neurons
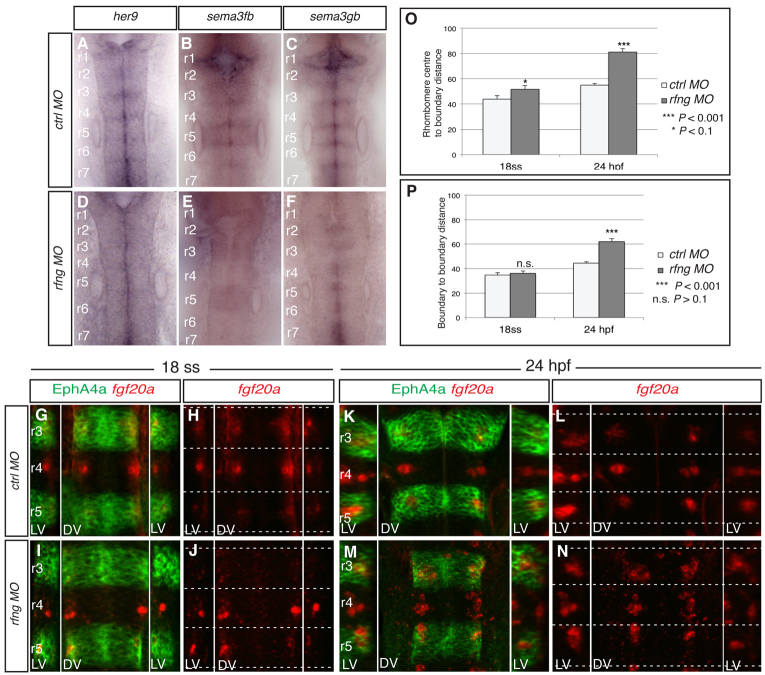
To further test the role of hindbrain boundary cells, we sought to disrupt their formation or maintenance independently of loss of Eph/ephrin signalling. Previous studies have shown that a deficiency of Notch activation leads to progressive depletion of hindbrain boundary cells, probably because Notch limits the differentiation of neural progenitors (Cheng et al., 2004; Qiu et al., 2009). rfng is a good candidate to be involved in boundary cell maintenance because its expression is restricted to rhombomere boundaries (Cheng et al., 2004; Qiu et al., 2004) and, by glycosylating Notch, Rfng promotes Notch activation in other contexts (Irvine and Wieschaus, 1994). We knocked down rfng and found that this leads to decreased boundary expression of her9, sema3gb and sema3fb (Fig. 2A-F), whereas borders of epha4a expression remain sharp (Fig. 2G,I). Unlike knockdown of epha4a or efnb3b, rfng knockdown disrupts boundary cell marker expression at all segment borders.
Fig. 2.
Disruption of all rhombomere boundaries leads to spreading of fgf20a neurons. (A-F) Expression of her9 (A,D), sema3fb (B,E) and sema3gb (C,F) in 24-hpf zebrafish embryos following injection of control MO (A-C) or rfng MO (D-F). rfng knockdown leads to loss of these markers at hindbrain boundaries (100%, n=14; 94%, n=16; 93%, n=15, respectively). (G-N) Expression of fgf20a mRNA (red) and EphA4a protein (green) at 18 ss (G-J) and 24 hpf (K-N) in control MO embryos (G,H,K,N) and rfng MO embryos (I,J,M,N). rfng knockdown has little effect on the organisation of fgf20a neurons at 18 ss (100%, n=8), but a strong effect at 24 hpf (90%, n=18). Dashed white lines indicate the position of segment borders. Left and right reconstructed lateral views (LV) and dorsal views (DV) are shown. (O) The average distance from fgf20 neuronal cluster edge to boundary. y-axis represents the distance from the rhombomere centre (0) to the boundary (100 arbitrary units, A.U.). For control MO the average distance from centre to cluster edge at 18 ss is 43.8±2.8 A.U. (n=15) and at 24 hpf is 54.9±1.5 A.U. (n=48). For rfng MO the distance at 18 ss is 51.8±2.8 A.U. (n=30; P=0.09) and at 24 hpf is 81.2±2.6 A.U. (n=48; P=9.4×10–16). On average, clusters in rfng morphants are 18% closer to boundaries than in controls at 18 ss, which increases significantly to a difference of 48% by 24 hpf. (P) Average anteroposterior (AP) length of fgf20 neuronal clusters. y-axis represents the distance from one rhombomere boundary (0) to another boundary (100 A.U.). For control MO at 18 ss the distance is 34.8±1.9 A.U. (n=15) and at 24 hpf is 44.6±0.9 A.U. (n=48). For rfng MO embryos the average length at 18 ss is 36.1±1.9 A.U. (n=30; P=0.67) and at 24 hpf is 62.1±2.4 A.U. (n=29; P=4.4×10–7). At 18 ss the difference is not significant, whereas at 24 hpf there is a significant 39% increase in length compared with control embryos. Values are average ± s.e.m. The orientation of embryos is as in Fig. 1.
We found that rfng knockdown leads to alterations in the position of fgf20a neurons at 24 hpf (Fig. 2K-N) and in the pattern of etv5b expression, which becomes less localised to segment centres and/or decreases in level (supplementary material Fig. S2C). Whereas in control embryos the clusters of fgf20a neurons are at rhombomere centres (Fig. 2K,L), following rfng knockdown they are either closer to one of the boundaries or more spread out along the anteroposterior (AP) axis of the rhombomere (Fig. 2M,N). Quantitation of the number of fgf20a neurons revealed no significant change in rfng morphants (supplementary material Fig. S3A) and thus the altered organisation appears to be due to mispositioning rather than excess production. Unlike the situation in efnb3b morphants, we did not detect a systematic anterior or posterior movement of specific clusters. Since the organisation of fgf20a neurons in rfng morphants appeared normal at 18 ss (Fig. 2G-J), we conclude that boundaries are required to maintain rather than establish their positioning.
The altered distribution of fgf20a neurons varies between rfng morphants (supplementary material Fig. S4C) and even between the left and right sides of the same rhombomere (Fig. 2N). We quantitated and assessed the statistical significance of the changes (supplementary material Fig. 4A) by measuring the position of the anterior and posterior edges of the fgf20a neuron population in relation to the boundary-to-boundary length of each segment (which is not significantly altered in rfng morphants; supplementary material Fig. S3B). We used this to calculate the distance from the segment centre to the boundary occupied by fgf20a neurons. Whereas in control 24-hpf embryos fgf20a neurons occupy 55% of the centre-to-boundary distance, in rfng knockdown embryos they occupy 81% (Fig. 2O). We also measured the average length of the clusters along the AP axis and found that this increased from 45% in control embryos to 62% in rfng knockdown embryos (Fig. 2P). In contrast to this significant spreading at 24 hpf, there was little difference between control and rfng morphant embryos at 18 ss (Fig. 2O,P).
Semaphorins expressed at hindbrain boundaries maintain the positioning of fgf20a neurons
The migration of fgf20a neurons to the r5/r6 border following loss of epha4a or efnb3b function might be because boundary cells are depleted only at specific segment interfaces, as this phenotype is not seen following rfng knockdown, in which all boundaries are disrupted. If boundary cells are a source of a repellent cue, signalling from residual boundaries would act to relocalise fgf20a neurons, whereas depletion of all boundaries would decrease chemorepulsion and lead to a spreading of these neurons. Specifically, this model can explain why in epha4a and efnb3 knockdown embryos the fgf20a neurons in r5 and r6 move to the r5/r6 border, equidistant from the remaining boundary cells, although it does not account for the unaltered distribution of r4 neurons. This raises the question of whether hindbrain boundary cells express any chemorepellents. Semaphorins are good candidates to fit this role of a repellent because, in addition to roles in axon guidance, they have been implicated in directing the migration of neuronal cell bodies (Tamagnone and Comoglio, 2004; Ayala et al., 2007; Marín et al., 2010). Two members of this ligand family, sema3gb and sema3fb, are specifically expressed in rhombomere boundaries (Fig. 2B,C) (Cooke et al., 2005). Furthermore, both sema3gb and sema3fb are downregulated, as with all boundary markers tested, following knockdown of epha4a (Cooke et al., 2005) or rfng (Fig. 2E,F). sema3fb is also segmentally expressed in r3 and r5, and this aspect of its expression is not altered following disruption of boundary cell formation (Fig. 2B,E).
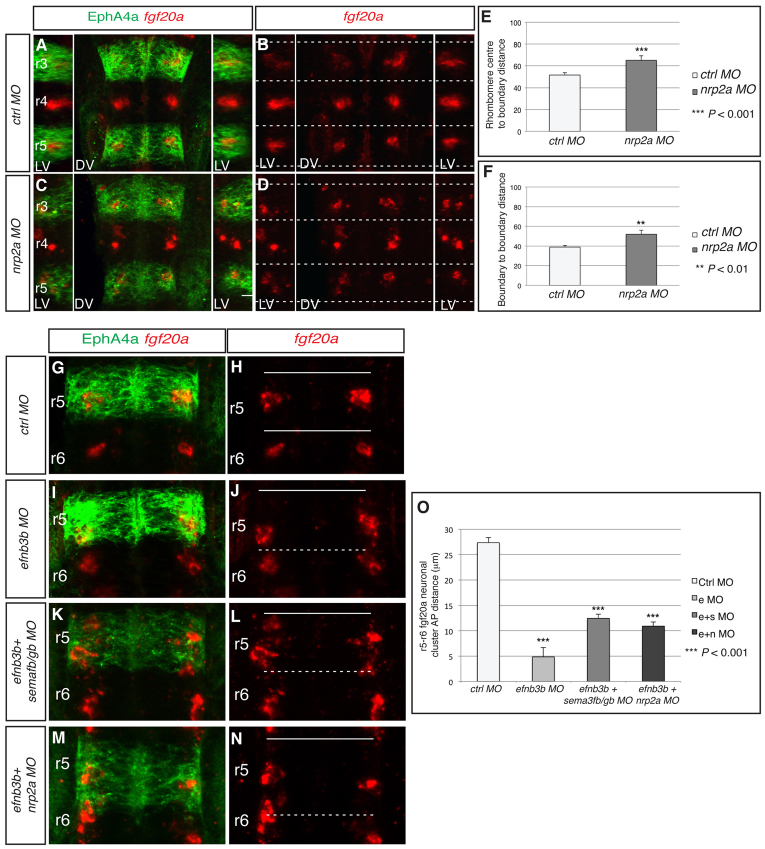
We carried out knockdowns to determine whether sema3gb or sema3fb regulates the positioning of fgf20a neurons. We found that single knockdowns of sema3gb or sema3fb do not alter the organisation of fgf20a neurons, which, because these semaphorins act through the same receptor (Pellet-Many et al., 2008), could be due to overlapping functions. By contrast, knockdown of sema3gb plus sema3fb led to spreading or mispositioning of fgf20a neurons (Fig. 3A-D; supplementary material Fig. S4D) and decreased or mislocalised etv5b expression (supplementary material Fig. S2E,F), without any significant change in the number of these neurons (supplementary material Fig. S3A). Quantitation of the distribution of fgf20a neurons in sema3fb+sema3gb morphants reveals a significant increase in the distance that they occupy from segment centre to boundary and in the AP length of the cluster (Fig. 3E,F). The similar effect of knockdown of sema3fb+sema3gb and rfng, in which the clusters of fgf20a neurons are mislocalised or spread out towards boundaries, is consistent with these semaphorins acting as the proposed repellent cues. The remaining segmental expression of sema3fb in r3 and r5 might explain why the fgf20a neurons in r4 are not mislocalised in epha4a knockdown embryos despite the depletion of anterior boundary cells of this segment.
Fig. 3.
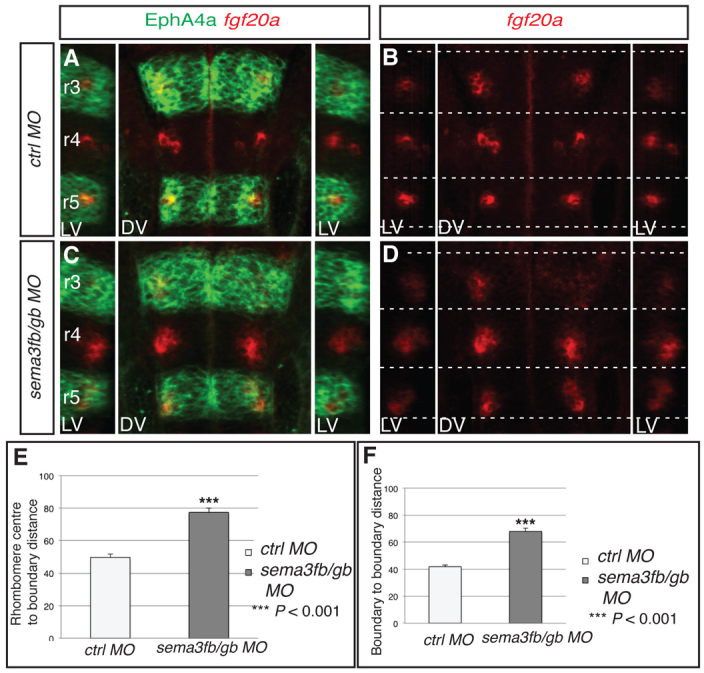
Sema3fb and Sema3gb position fgf20a neurons. (A-D) fgf20a expression (red) combined with EphA4 antibody staining (green) at 24 hpf in zebrafish embryos injected with control MO (A,B) or sema3fb+sema3gb MO (C,D). Dashed white lines indicate the position of boundary marker expression. Left and right reconstructed lateral views (LV) plus dorsal views (DV) are shown. Knockdown of sema3fb+sema3gb disrupts the organisation of fgf20a neurons (79%, n=24). (E,F) The average distance from fgf20a neuronal clusters to boundary (E) and average AP length of clusters (F); see Fig. 2 legend for methodology. In control MO embryos, the average distance to the boundary is 49.7±2.1 A.U. (n=24); in sema3fb+sema3gb MO embryos this increases to 77.4±2.8 A.U. (n=24; P=2.2×10–8), resulting in sema3fb+sema3gb clusters being on average half the distance from boundaries than control clusters. In control MO embryos the average cluster length is 41.8±1.4. A.U. (n=24), which increases in sema3fb+sema3gb MO embryos to 68.0±2.3 A.U. (n=24; P=3.0×10–9), a 62% increase in length. Values are average ± s.e.m. The orientation of embryos is as in Fig. 1.
To further address the role of sema3fb and sema3gb in positioning fgf20a neurons, we analysed the expression and role of their receptors, nrp2a and nrp2b. We found that nrp2a expression at segment centres (Yu et al., 2004; Yu and Moens, 2005) is coincident with fgf20a expression, whereas nrp2b is not expressed (supplementary material Fig. S5). Following nrp2a knockdown (but not nrp2b knockdown), fgf20a neurons are more spread out and closer to the boundary (Fig. 4A-D, quantitation in 4E,F; supplementary material Fig. S4E), but do not significantly change in number (supplementary material Fig. S3A).
Fig. 4.
Nrp2a is required to position fgf20a neuronal clusters. (A-D) fgf20a expression (red) combined with EphA4 antibody staining (green) at 24 hpf in zebrafish embryos injected with (A,B) control MO or (C,D) nrp2a MO. Dashed white lines indicate the position of segment borders. Left and right reconstructed lateral views (LV) plus dorsal views (DV). nrp2a knockdown disrupts the organisation of fgf20a neurons (70%, n=17). (E,F) The average distance from fgf20 neuronal clusters to boundary (E) and average AP length of clusters (F). In control MO embryos, the average distance to the boundary is 51.6±2.2 A.U. (n=24), which in nrp2a MO embryos increases to 65.1±4.1 A.U. (n=24; P=5×10–4), i.e. 22% closer to boundaries. In control MO embryos the average cluster length is 38.8±1.6 A.U. and this increases in nrp2a morphants to 52.0±4.0 A.U. (n=24; P=4×10–3), a 34% increase in length. (G-N) fgf20a mRNA expression (red) and EphA4 antibody staining (green) of embryos injected with control MO (G,H), efnb3b MO (I,J), efnb3b+sema3fb+sema3gb MOs (K,L) or efnb3b+nrp2a MOs (M,N). Dashed white lines indicate the position of depleted boundary cells; continuous white lines indicate the position of the remaining boundaries. The migration of fgf20a neurons towards the r5/r6 interface following efnb3b knockdown was partly blocked when combined with sema3fb+sema3gb knockdown (76%, n=17) or nrp2a knockdown (70%, n=23). (O) Quantitation of the distance between r5 and r6 fgf20a neuronal clusters in the different knockdown experiments. There is a significant decrease in the distance between both clusters in efnB3b morphants (e MO), as compared with control MO, that is significantly rescued by co-injection of sema3fb+sema3gb MOs (e+s MO) or nrp2a MO (e+n MO): control MO 27.3±1.0 μm (n=10) versus efnb3b MO 4.9±1.6 μm (n=10), P=3.1×10–9; e+s MO 12.4±0.8 μm (n=10), P=9.5×10–10; e+n MO 10.9±0.8 μm (n=10), P=2.0×10–10. Values are average ± s.e.m.
sema3fb+sema3gb or nrp2a knockdown alleviates fgf20a neuron migration induced by disrupting a subset of boundaries
The finding that, following epha4a or efnb3b knockdown, fgf20a neurons in r5 and r6 become relocalised to the r5/r6 border might be explained by chemorepulsion from the boundary cells, which remain at the r4/r5 and r6/r7 interfaces. This model predicts that knockdown of sema3fb+sema3gb will rescue the mislocalisation of these cells following efnb3b knockdown. We carried out knockdown of efnb3b and visualised segment borders by detection of EphA4, and, as shown above, found that the fgf20a-expressing clusters in r5 and r6 are now located closer to the r5/r6 border at 24 hpf (Fig. 4I,J) than in controls (Fig. 4G,H). However, when efnb3b knockdown is combined with sema3fb+sema3gb knockdown, there is less movement of fgf20a neurons towards the r5/r6 border and the r5 and r6 clusters do not collide (Fig. 4K,L, quantitated in 4O). Instead, the fgf20a neurons are more spread out both anteriorly and posteriorly, a characteristic of knockdown of sema3fb+sema3gb. Similarly, nrp2a knockdown decreased the movement of fgf20a neurons towards the r5/r6 border in efnb3b morphants (Fig. 4M-O). These results confirm that Sema3fb and Sema3gb act through their common receptor Nrp2a as repellent cues that position fgf20a neuronal clusters at segment centres.
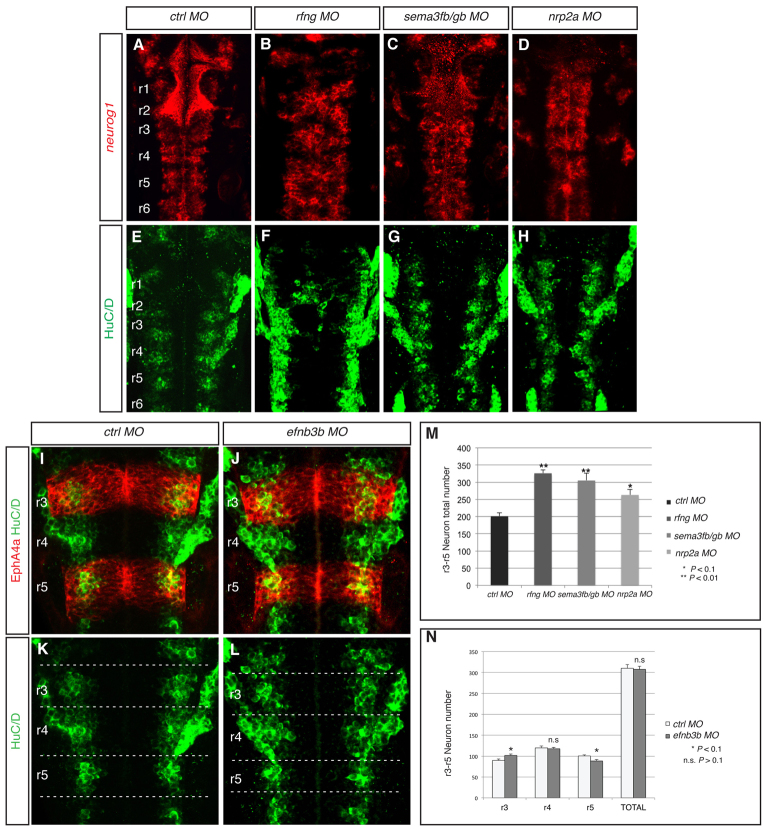
Dispersal of fgf20a neurons leads to increased neurogenesis
Previous work showing that fgf20a suppresses neurogenesis at segment centres (Gonzalez-Quevedo et al., 2010) implies that the appropriate positioning of fgf20a neurons is required to achieve the correct patterning of neurogenesis. Indeed, we find that knockdown of rfng, sema3fb+sema3gb, or nrp2a each leads to disorganisation of the non-neurogenic zone, as marked by a lack of neurog1 expression (Fig. 5A-D). To examine whether the dispersal of fgf20a neurons following loss of hindbrain boundary cells affects the overall amount of neurogenesis, we detected HuC/D (Elavl3/4) protein, which marks all differentiating neurons in the mantle zone, and quantitated the number of expressing cells in confocal stacks throughout the dorsoventral axis. We found that knockdown of rfng, sema3fb+sema3gb, or nrp2a led to a 30-60% increase in the number of neurons (Fig. 5E-H, quantitated in 5M). The pattern of HuC/D staining in these morphants retains a segmented distribution, with a gap around hindbrain boundaries; this is likely to reflect persistence of the curtains of radial glial fibres that flank boundaries and physically exclude neurons (Trevarrow et al., 1990).
Fig. 5.
Mispositioning of fgf20a neurons affects the patterning and amount of neurogenesis. (A-H,M) Expression at 30 hpf of neurog1, which marks differentiating neurons in the neural epithelium (red), and HuC/D, which marks neurons in the mantle zone (green), in embryos injected with control MO (A,E), rfng MO (B,F), sema3fb+sema3gb MOs (C,G) or nrp2a MO (D,H). Knockdown of rfng (85%, n=14), sema3fb+sema3gb (65%, n=23) or nrp2a (59%, n=17) leads to disorganisation of the neurogenic zones. (M) Quantitation of the number of HuC/D-expressing cells in r3 to r5 in control MO embryos (201±9.7 neurons, n=4) reveals that these increase following knockdown of rfng (326±16.2 neurons, n=4; P=0.008), sema3fb+sema3gb (306±9.4 neurons, n=4; P=0.001) or nrp2a (263±9.4 neurons, n=4; P=0.06). (I-L,N) HuC/D-expressing neurons (green) combined with EphA4 antibody staining (red) at 30 hpf in control (L,K) and efnb3b (J,L) knockdown embryos. (N) Quantitation of neurons in both conditions in r3, r4 and r5 separately plus total number. There is a significant increase in the number of neurons in r3 (control MO 90±2.7, n=5, versus efnb3b MO 101±3.1; P=0.09), a decrease in r5 (control MO 100±2.7, n=5, versus efnb3b MO 88±3.5; P=0.06), but no significant change in r4 (control MO 120±4.6, n=5, versus efnb3b 118±3.3; P=0.74). The total number of neurons is not significantly different (control MO 310±9.1, n=5, versus efnb3b 307±7.2; P=0.85). The orientation of embryos is as in Fig. 1.
A potential explanation for the increased neurogenesis following the dispersal of fgf20a neurons is suggested by the observation that, in epha4a and efnb3b knockdown embryos, expression of the FGF target gene etv5b occurs at high levels in r4 and at the r5/r6 border, but at lower levels in all other hindbrain segments (Fig. 1P,Q). This pattern of etv5b expression correlates with the clustering of fgf20a neurons in r4 and the r5/r6 border, and suggests that the dispersal of these neurons in the other segments leads to a lower level of FGF pathway activation. We quantified HuC/D-expressing neurons within hindbrain segments following efnb3b knockdown and found a significant increase in r3 (13%) and decrease in r5 (13%), whereas r4 was unaffected (Fig. 5I-L, quantitated in 5N). The amount of neurogenesis thus correlates with the organisation of fgf20a neurons, which in efnb3b morphants are dispersed in r3, clustered normally in r4, and form a fused cluster of r5 and r6 neurons at the r5/r6 border.
DISCUSSION
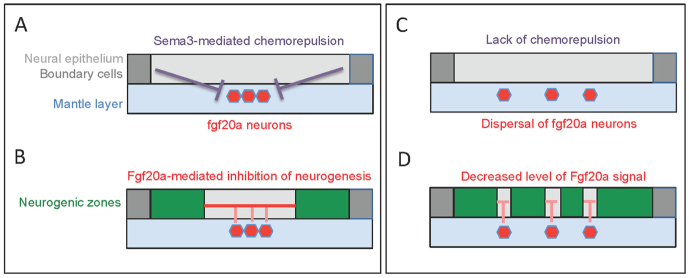
Previous studies have shown that neurogenesis is spatially organised within hindbrain segments in zebrafish through the FGF-mediated inhibition of neurogenesis (Gonzalez-Quevedo et al., 2010). This inhibition is due to signalling from the mantle zone by fgf20a neurons located at each segment centre that underlies the formation of a non-neurogenic zone in the adjacent progenitor cells. These findings suggested that the stereotyped location of fgf20a neurons at segment centres might be important for the patterning of neurogenesis, and raise the question of how this positioning is achieved. Here, we have uncovered a mechanism in which signalling from hindbrain boundaries maintains the tight clustering and positioning of fgf20a neurons at segment centres. We show that selective loss of specific boundaries due to epha4a or efnb3b knockdown leads to an altered organisation of fgf20a neurons, including an abnormal migration of these neurons in r5 and r6 away from the remaining boundaries and towards the deficient r5/r6 border. This mispositioning of fgf20a neurons is matched by an altered pattern of FGF-dependent etv5b gene expression in the adjacent progenitor cells. Furthermore, loss of hindbrain boundary cells at all segment borders following rfng knockdown leads to a spreading and mispositioning of fgf20a neurons. The finding that the organisation of fgf20a neurons in efnb3b and rfng morphants appears normal at early stages argues that boundaries serve to maintain rather than establish the position of these neuronal clusters at segment centres. These observations also argue against a model in which boundaries regulate the organisation of fgf20a neurons by establishing a prepattern of their precursors in the neural epithelium.
The results of loss-of-function experiments implicate Sema3fb and Sema3gb, which are expressed in hindbrain boundaries, in positioning fgf20a neurons by acting through their common receptor Nrp2a. Consistent with this, we find that sema3fb+sema3gb knockdown alleviates the abnormal migration of fgf20a neurons towards the depleted r5/r6 boundary in efnb3b morphants. These findings are consistent with a model in which hindbrain boundary cells are a source of semaphorin signals that chemorepel fgf20a neurons towards segment centres where they are furthest from the flanking borders, leading to inhibition of neurogenesis in the adjacent progenitors (Fig. 6A,B). The tight clustering of fgf20a neurons has an important role in the patterning of neurogenesis: when it is disrupted following knockdown of rfng, sema3fb+sema3gb or nrp2a the organisation of the neurogenic and non-neurogenic zones is altered (Fig. 6C,D).
Fig. 6.
Model of the role of boundaries in neuronal positioning. A single zebrafish hindbrain segment is represented with the neural epithelium in light grey, boundary cells in dark grey, and the mantle layer in blue. (A,B) Normal embryos, in which Sema3fb/Sema3gb-mediated chemorepulsion from boundary cells positions fgf20a neurons (red) in the segment centre. The cluster of fgf20a neurons creates a focussed source of FGF signals (red lines) that inhibit neurogenesis in the adjacent neural epithelium, restricting the neurogenic zones (green) to being adjacent to boundaries. (C,D) Embryos in which boundary cells have been depleted or Sema3fb/Sema3gb signalling disrupted. The consequent dispersal of fgf20a neurons leads to disorganisation of the non-neurogenic zones and diminishes the level of FGF signalling.
Our findings raise the question of how the cluster of fgf20a neurons spreads out and becomes mislocalised in a variable pattern following the disruption of hindbrain boundaries or of sema3fb/sema3gb function. This might be associated with the major expansion of the mantle region as differentiating neurons migrate in from the neural epithelium, as well as with the overall growth of neural tissue. This growth has the potential to drive cell intercalation that separates neighbours, such that repeated rounds of intercalation lead to a variable spreading of cells that are initially clustered.
Roles of boundaries in patterning
One of the important roles of boundary formation during development is to generate specialised boundary cells that are a source of inductive signals that regulate cell differentiation in a concentration-dependent manner (Dahmann and Basler, 1999; Irvine and Rauskolb, 2001; Kiecker and Lumsden, 2005). Our findings reveal a distinctive role of hindbrain boundaries in which, by acting as a source of chemorepellent, they serve to position another signalling source – fgf20a neurons – that patterns cell differentiation. It remains possible that other signals emanating from hindbrain boundaries have an inductive role in which they promote the differentiation of adjacent progenitors, such that the neurogenic zone is positioned by antagonistic inductive and inhibitory signals. However, this seems unlikely because neurogenesis does not decrease – on the contrary, it increases – following depletion of boundary cells. Thus, other boundary signals, such as Wnt family members (Riley et al., 2004; Amoyel et al., 2005), are not essential for neuronal differentiation and might regulate other aspects of hindbrain development.
Neuronal organisation and neurogenesis
The precise positioning of neuronal cell types and their organisation into clusters are ubiquitous aspects of nervous system development. This positioning requires the radial migration of neuronal cell bodies from the neural epithelium, and in some cases tangential migration over long distances, guided by contact-dependent or diffusible cues that include semaphorins (Ayala et al., 2007; Marín et al., 2010). The significance of such migration to position neuronal cell bodies at a particular location is unclear, but is thought to facilitate the formation of functional neuronal circuits (Chen et al., 2006). In the case of reticulospinal neurons, which are located at segment centres in the hindbrain (Hanneman et al., 1988), these are components of an early neuronal circuit that underlies an escape response (Liu and Fetcho, 1999). Our findings suggest another type of role for neuronal clustering.
We find that depletion of hindbrain boundary cells leads to a significant increase in the overall amount of neurogenesis. One potential explanation is that signals from boundary cells inhibit neurogenesis, albeit that this requires other factors to account for the normal formation of neurogenic zones adjacent to boundaries. However, increased neurogenesis also occurs following disruption of Sema3fb/Sema3gb signalling, in which hindbrain boundary cells remain present. These findings argue for a model in which the clustering of fgf20a neurons is required for the correct patterning and amount of neurogenesis. We find that the spreading of fgf20a neurons along the AP axis that occurs following the disruption of boundaries or of Sema3fb/Sema3gb signalling does not lead to a wider non-neurogenic zone, but rather correlates with an increase in the overall amount of neurogenesis. A potential explanation is suggested by the observation that, following epha4 or efnb3b knockdown, expression of the FGF-response gene etv5b remains at a high level in r4 and at the r5/r6 border where clustering of fgf20a neurons still occurs, but is at lower levels in all of the other segments. This suggests that clustering of Fgf20a signalling cells is required to achieve a high level of FGF signal and target gene activation. We therefore propose that neuronal differentiation is inhibited above a threshold level of FGF pathway activation in progenitor cells, and that this requires the tight clustering of fgf20a neurons to provide a focussed source of FGF signals. This is reminiscent of the situation for signalling centres that are induced at boundaries, which require that the border is sharp and straight in order to achieve the appropriate distribution of the graded signal (Dahmann and Basler, 1999). Our findings thus provide a further perspective on the mechanistic importance of establishing a precise organisation of signalling cells and how this can be achieved.
Supplementary Material
Acknowledgments
We thank James Briscoe and Francois Guillemot for helpful discussions and comments on the manuscript and NIMR Biological Services for zebrafish husbandry.
Footnotes
Funding
This work was supported by the UK Medical Research Council [U117532048] and by postdoctoral fellowships from the European Commission Marie Curie Programme (J.T. and R.G.-Q.) and the Japan Society for the Promotion of Science (T.W.-A.). Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.080135/-/DC1
References
- Amoyel M., Cheng Y. C., Jiang Y. J., Wilkinson D. G. (2005). Wnt1 regulates neurogenesis and mediates lateral inhibition of boundary cell specification in the zebrafish hindbrain. Development 132, 775–785 [DOI] [PubMed] [Google Scholar]
- Ayala R., Shu T., Tsai L. H. (2007). Trekking across the brain: the journey of neuronal migration. Cell 128, 29–43 [DOI] [PubMed] [Google Scholar]
- Bae Y. K., Shimizu T., Hibi M. (2005). Patterning of proneuronal and inter-proneuronal domains by hairy- and enhancer of split-related genes in zebrafish neuroectoderm. Development 132, 1375–1385 [DOI] [PubMed] [Google Scholar]
- Bally-Cuif L., Hammerschmidt M. (2003). Induction and patterning of neuronal development, and its connection to cell cycle control. Curr. Opin. Neurobiol. 13, 16–25 [DOI] [PubMed] [Google Scholar]
- Berghmans S., Murphey R. D., Wienholds E., Neuberg D., Kutok J. L., Fletcher C. D., Morris J. P., Liu T. X., Schulte-Merker S., Kanki J. P., et al. (2005). tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl. Acad. Sci. USA 102, 407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster R., Lee J., Ruiz i Altaba A. (1998). Gli/Zic factors pattern the neural plate by defining domains of cell differentiation. Nature 393, 579–583 [DOI] [PubMed] [Google Scholar]
- Bylund M., Andersson E., Novitch B. G., Muhr J. (2003). Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat. Neurosci. 6, 1162–1168 [DOI] [PubMed] [Google Scholar]
- Cavodeassi F., Houart C. (2012). Brain regionalization: of signaling centers and boundaries. Dev. Neurobiol. 72, 218–233 [DOI] [PubMed] [Google Scholar]
- Chan J., Mably J. D., Serluca F. C., Chen J. N., Goldstein N. B., Thomas M. C., Cleary J. A., Brennan C., Fishman M. C., Roberts T. M. (2001). Morphogenesis of prechordal plate and notochord requires intact Eph/ephrin B signaling. Dev. Biol. 234, 470–482 [DOI] [PubMed] [Google Scholar]
- Chen B. L., Hall D. H., Chklovskii D. B. (2006). Wiring optimization can relate neuronal structure and function. Proc. Natl. Acad. Sci. USA 103, 4723–4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. C., Amoyel M., Qiu X., Jiang Y. J., Xu Q., Wilkinson D. G. (2004). Notch activation regulates the segregation and differentiation of rhombomere boundary cells in the zebrafish hindbrain. Dev. Cell 6, 539–550 [DOI] [PubMed] [Google Scholar]
- Cooke J. E., Kemp H. A., Moens C. B. (2005). EphA4 is required for cell adhesion and rhombomere-boundary formation in the zebrafish. Curr. Biol. 15, 536–542 [DOI] [PubMed] [Google Scholar]
- Dahmann C., Basler K. (1999). Compartment boundaries: at the edge of development. Trends Genet. 15, 320–326 [DOI] [PubMed] [Google Scholar]
- Diez del Corral R., Olivera-Martinez I., Goriely A., Gale E., Maden M., Storey K. (2003). Opposing FGF and retinoid pathways control ventral neuralpattern, neuronal differentiation, and segmentation during body axis extension. Neuron 40, 65–79 [DOI] [PubMed] [Google Scholar]
- Esain V., Postlethwait J. H., Charnay P., Ghislain J. (2010). FGF-receptor signalling controls neural cell diversity in the zebrafish hindbrain by regulating olig2 and sox9. Development 137, 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale N. W., Holland S. J., Valenzuela D. M., Flenniken A., Pan L., Ryan T. E., Henkemeyer M., Strebhardt K., Hirai H., Wilkinson D. G., et al. (1996). Eph receptors and ligands comprise two major specificity subclasses and are reciprocally compartmentalized during embryogenesis. Neuron 17, 9–19 [DOI] [PubMed] [Google Scholar]
- Geling A., Itoh M., Tallafuss A., Chapouton P., Tannhäuser B., Kuwada J. Y., Chitnis A. B., Bally-Cuif L. (2003). bHLH transcription factor Her5 links patterning to regional inhibition of neurogenesis at the midbrain-hindbrain boundary. Development 130, 1591–1604 [DOI] [PubMed] [Google Scholar]
- Geling A., Plessy C., Rastegar S., Strähle U., Bally-Cuif L. (2004). Her5 acts as a prepattern factor that blocks neurogenin1 and coe2 expression upstream of Notch to inhibit neurogenesis at the midbrain-hindbrain boundary. Development 131, 1993–2006 [DOI] [PubMed] [Google Scholar]
- Gerety S. S., Wilkinson D. G. (2011). Morpholino artifacts provide pitfalls and reveal a novel role for pro-apoptotic genes in hindbrain boundary development. Dev. Biol. 350, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Quevedo R., Lee Y., Poss K. D., Wilkinson D. G. (2010). Neuronal regulation of the spatial patterning of neurogenesis. Dev. Cell 18, 136–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham V., Khudyakov J., Ellis P., Pevny L. (2003). SOX2 functions to maintain neural progenitor identity. Neuron 39, 749–765 [DOI] [PubMed] [Google Scholar]
- Guthrie S., Lumsden A. (1991). Formation and regeneration of rhombomere boundaries in the developing chick hindbrain. Development 112, 221–229 [DOI] [PubMed] [Google Scholar]
- Hanneman E., Trevarrow B., Metcalfe W. K., Kimmel C. B., Westerfield M. (1988). Segmental pattern of development of the hindbrain and spinal cord of the zebrafish embryo. Development 103, 49–58 [DOI] [PubMed] [Google Scholar]
- Hauptmann G., Gerster T. (1994). Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 10, 266 [DOI] [PubMed] [Google Scholar]
- Heyman I., Faissner A., Lumsden A. (1995). Cell and matrix specialisations of rhombomere boundaries. Dev. Dyn. 204, 301–315 [DOI] [PubMed] [Google Scholar]
- Irvine K. D., Wieschaus E. (1994). fringe, a boundary-specific signaling molecule, mediates interactions between dorsal and ventral cells during Drosophila wing development. Cell 79, 595–606 [DOI] [PubMed] [Google Scholar]
- Irvine K. D., Rauskolb C. (2001). Boundaries in development: formation and function. Annu. Rev. Cell Dev. Biol. 17, 189–214 [DOI] [PubMed] [Google Scholar]
- Irving C., Flenniken A., Alldus G., Wilkinson D. G. (1996). Cell-cell interactions and segmentation in the developing vertebrate hindbrain. Biochem. Soc. Symp. 62, 85–95 [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T., Kobayashi T. (2007). The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134, 1243–1251 [DOI] [PubMed] [Google Scholar]
- Kiecker C., Lumsden A. (2005). Compartments and their boundaries in vertebrate brain development. Nat. Rev. Neurosci. 6, 553–564 [DOI] [PubMed] [Google Scholar]
- Liu K. S., Fetcho J. R. (1999). Laser ablations reveal functional relationships of segmental hindbrain neurons in zebrafish. Neuron 23, 325–335 [DOI] [PubMed] [Google Scholar]
- Louvi A., Artavanis-Tsakonas S. (2006). Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 7, 93–102 [DOI] [PubMed] [Google Scholar]
- Marín O., Valiente M., Ge X., Tsai L. H. (2010). Guiding neuronal cell migrations. Cold Spring Harb. Perspect. Biol. 2, a001834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellet-Many C., Frankel P., Jia H., Zachary I. (2008). Neuropilins: structure, function and role in disease. Biochem. J. 411, 211–226 [DOI] [PubMed] [Google Scholar]
- Qiu X., Xu H., Haddon C., Lewis J., Jiang Y.-J. (2004). Sequence and embryonic expression of three zebrafish fringe genes: lunatic fringe, radical fringe, and manic fringe. Dev. Dyn. 231, 621–630 [DOI] [PubMed] [Google Scholar]
- Qiu X., Lim C. H., Ho S. H., Lee K. H., Jiang Y. J. (2009). Temporal Notch activation through Notch1a and Notch3 is required for maintaining zebrafish rhombomere boundaries. Dev. Genes Evol. 219, 339–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley B. B., Chiang M. Y., Storch E. M., Heck R., Buckles G. R., Lekven A. C. (2004). Rhombomere boundaries are Wnt signaling centers that regulate metameric patterning in the zebrafish hindbrain. Dev. Dyn. 231, 278–291 [DOI] [PubMed] [Google Scholar]
- Tamagnone L., Comoglio P. M. (2004). To move or not to move? EMBO Rep. 5, 356–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Maeda R., Shoji W., Wada H., Masai I., Shiraki T., Kobayashi M., Nakayama R., Okamoto H. (2007). Novel mutations affecting axon guidance in zebrafish and a role for plexin signalling in the guidance of trigeminal and facial nerve axons. Development 134, 3259–3269 [DOI] [PubMed] [Google Scholar]
- Trevarrow B., Marks D. L., Kimmel C. B. (1990). Organization of hindbrain segments in the zebrafish embryo. Neuron 4, 669–679 [DOI] [PubMed] [Google Scholar]
- Westerfield M. (1993). The Zebrafish Book. Eugene, OR: University of Oregon Press; [Google Scholar]
- Xu Q., Wilkinson D. (1998). In situ hybridisation of mRNA with hapten labelled probes. In In Situ Hybridisation: a Practical Approach (ed. Wilkinson D.), pp. 87–106 Oxford: Oxford University Press; [Google Scholar]
- Xu Q., Alldus G., Holder N., Wilkinson D. G. (1995). Expression of truncated Sek-1 receptor tyrosine kinase disrupts the segmental restriction of gene expression in the Xenopus and zebrafish hindbrain. Development 121, 4005–4016 [DOI] [PubMed] [Google Scholar]
- Yu H. H., Moens C. B. (2005). Semaphorin signaling guides cranial neural crest cell migration in zebrafish. Dev. Biol. 280, 373–385 [DOI] [PubMed] [Google Scholar]
- Yu H. H., Houart C., Moens C. B. (2004). Cloning and embryonic expression of zebrafish neuropilin genes. Gene Expr. Patterns 4, 371–378 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.