Abstract
The osteogenic and oncogenic transcription factor RUNX2 downregulates the RNA polymerase I (RNA Pol I)-mediated transcription of rRNAs and changes histone modifications associated with the rDNA repeat. However, the mechanisms by which RUNX2 suppresses rRNA transcription are not well understood. RUNX2 cofactors such as histone deacetylases (HDACs) play a key role in chromatin remodeling and regulation of gene transcription. Here, we show that RUNX2 recruits HDAC1 to the rDNA repeats in osseous cells. This recruitment alters the histone modifications associated with active rRNA-encoding genes and causes deacetylation of the protein upstream binding factor (UBF, also known as UBTF). Downregulation of RUNX2 expression reduces the localization of HDAC1 to the nucleolar periphery and also decreases the association between HDAC1 and UBF. Functionally, depletion of HDAC1 relieves the RUNX2-mediated repression of rRNA-encoding genes and concomitantly increases cell proliferation and global protein synthesis in osseous cells. Our findings collectively identify a RUNX2–HDAC1-dependent mechanism for the regulation of rRNA-encoding genes and suggest that there is plasticity to RUNX2-mediated epigenetic control, which is mediated through selective mitotic exclusion of co-regulatory factors.
Key words: RUNX2, HDAC1, rRNA, RNA polymerase I, Osseous cell, Nucleolus, Histone acetylation
Introduction
Genetic and epigenetic mechanisms are coordinated by lineage-specific transcription factors to regulate the temporal expression of genes that are required for the fidelity of growth and phenotype (Mayer and Grummt, 2006; Sarge and Park-Sarge, 2009; Stein et al., 2010). The osteogenic transcription factor RUNX2 is a context-dependent activator and suppressor of target genes transcribed by RNA polymerase II (RNA Pol II) that regulate bone cell proliferation and differentiation (Lian et al., 2003; Westendorf, 2006; Young et al., 2007a). RUNX2 functions by interacting with cofactors, such as histone acetyltransferases (HATs), histone deacetylases (HDACs), transducin-like enhancer of split (TLE) proteins and the mammalian homolog of yeast protein Switch Independent (mSIN). as well as effectors of several signaling pathways (Ducy et al., 1997; Durst and Hiebert, 2004; Javed et al., 2000; Jensen et al., 2007; Komori, 2006). Importantly, RUNX2 and a subset of co-regulators also regulate ribosomal RNA (rRNA) genes that are transcribed by RNA Pol I, providing a potential link between cell phenotype and growth control (Ali et al., 2008; Ali et al., 2010; Young et al., 2007a).
Ribosomal RNA (rRNA) gene expression is intimately linked with cell growth and phenotype, and is compromised in cancer cells (Derenzini et al., 2000; Mayer and Grummt, 2006). rRNA expression is stringently controlled by a highly organized and efficient RNA Pol I transcriptional machinery through multiple mechanisms (Grummt, 1999; Grummt, 2007; Russell and Zomerdijk, 2005). Several transcriptional activators [SL1, upstream binding factor (UBF) and Myc] and repressors [DNA methyltransferases (DNMTs), histone deacetylases (HDACs), MyoD, RUNX2, CCAAT-enhancer-binding proteins (C/EBPs) etc.] interact with the RNA Pol I machinery to regulate rRNA transcription and to maintain optimal levels of cellular rRNA (Ali et al., 2008; Arabi et al., 2005; Budde and Grummt, 1999; Grandori et al., 2005; Grummt and Pikaard, 2003; Young et al., 2007a). UBF plays a central role in rRNA activation by interacting with the components of the RNA Pol I complex. The activity of UBF is regulated by interaction with other co-regulators and by post-translational modifications that include phosphorylation and acetylation (Hannan et al., 2003; Pelletier et al., 2000; Stefanovsky et al., 2006; Tuan et al., 1999; Voit et al., 1995).
Our laboratory has demonstrated that the involvement of RUNX2 in rRNA suppression is mediated by interacting with UBF (Young et al., 2007a). However, how the RUNX2 association with UBF regulates rRNA gene expression is not well understood. Here, we show that HDAC1, a co-regulator of RUNX2 (Lee et al., 2006) and known repressor of RNA Pol I transcription (Meraner et al., 2006; Pelletier et al., 2000; Zhou et al., 2002) is recruited to rDNA loci to modulate UBF and histone acetylation in a RUNX2-dependent manner. Our key finding is that RUNX2 and HDAC1 participate in fine-tuning rRNA gene expression to respond to cellular requirements for protein synthesis and growth.
Results
HDAC1 associates with both RUNX2 and UBF at rDNA loci
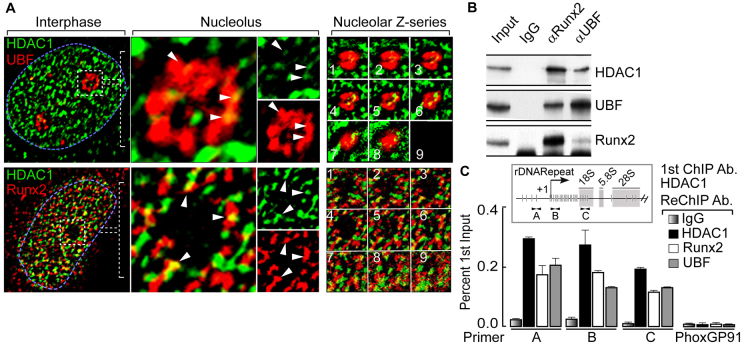
We have shown previously that RUNX2 suppresses rRNA gene transcription in osseous cells, in part by modifying the rDNA-associated histone code (Young et al., 2007a). HDAC1 is a known regulator of rRNA gene expression (Meraner et al., 2006; Young et al., 2007a; Zhou et al., 2002) and interacts with RUNX2 (Lee et al., 2006). Here, we tested the hypothesis that HDAC1 interacts with RUNX2 at the rDNA locus in osseous cells and contributes to RUNX2-mediated downregulation of rRNA gene transcription. We initially performed immunofluorescence microscopy using specific antibodies to assess whether RUNX2 and HDAC1 colocalize at the nucleolus. As expected, we observed punctate staining of both RUNX2 and HDAC1 throughout the nucleus, with limited signal overlap (Fig. 1A, lower panels). Interestingly, we detected relatively consistent and specific overlap of the signals for HDAC1 and RUNX2 at the periphery of the nucleoli (see the z-series images in Fig. 1A, lower-right panel). The nucleolar periphery was identified by staining for UBF, a key nucleolar regulatory factor (Kuhn and Grummt, 1992; Meraner et al., 2006) and this staining also showed signal overlap with HDAC1 (Fig. 1A, upper panels). It is notable that, unlike TLE1 or RUNX2 (Ali et al., 2010; Young et al., 2007a), HDAC1 was not associated with mitotic chromosomes (data not shown). Furthermore, our co-immunoprecipitation (IP) assays using RUNX2 or UBF antibodies revealed that these proteins were present in the same complex with HDAC1 in osseous cells (Fig. 1B; supplementary material Fig. S1A).
Fig. 1.
HDAC1 colocalizes with RUNX2 and UBF in the interphase nucleolus. (A) Immunofluorescence microscopy in SaOS-2 cells demonstrates HDAC1 colocalization with UBF (upper panels) and RUNX2 (lower panels) at the nucleolar periphery. The blue dotted line represents the nucleus and the white dotted box marks a nucleolus. (B) Co-immunoprecipitation assays show that HDAC1 associates with UBF and RUNX2 in proliferating osteosarcoma cells. (C) ChIP-ReChIP experiments show the binding of HDAC1 together with RUNX2 and UBF at rDNA repeats (primer A, B and C). Primary (1st) ChIP was performed using anti-HDAC1 antibody (Ab) followed by ReChIP (2nd) using anti-HDAC1, anti-RUNX2 and anti-UBF antibodies. A Phox-GP91 gene primer was used as a negative control to demonstrate the specificity of the ChIP-ReChIP assays. In the inset, a single rDNA repeat is shown with the location of the primer sets used in ChIP assays (A, B and C) at different regions of the rDNA. Vertical lines indicate the RUNX2-binding sites at the rDNA repeat. White arrows show the location of primers.
The presence of HDAC1, RUNX2 and UBF, all known regulators of ribosomal gene expression, in a complex at the nucleolar periphery suggests that these proteins co-occupy rDNA repeats. We experimentally addressed this hypothesis by performing chromatin immunoprecipitation (ChIP) assays using an antibody against HDAC1 followed by secondary ChIPs (ReChIP) with anti-RUNX2 and anti-UBF antibodies. UBF, RUNX2 and the co-repressor HDAC1 occupied the same rDNA segments, as analyzed by primers representing three different regions of the rDNA repeat (Fig. 1C, primer set A, B and C). None of the factors was bound to the unrelated Phox-GP91 (CYBB) gene, indicating the specificity of the ChIP–ReChIP assays (Fig. 1C; supplementary material Fig. S1B). Taken together, our results demonstrate that HDAC1, UBF and RUNX2 are in a complex that occupies rDNA repeats in osseous cells and suggest that this complex is involved in regulating rRNA gene expression.
HDAC1 interaction with rDNA chromatin requires the RUNX2 C-terminus
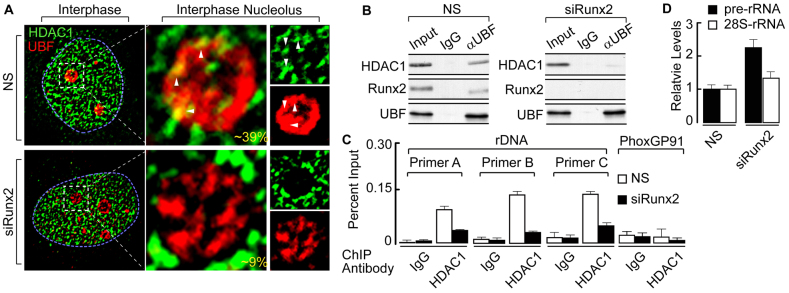
We hypothesized that in osseous cells, HDAC1 is recruited to rDNA and associates with UBF through RUNX2. We depleted endogenous RUNX2 protein levels in SaOS-2 cells using specific small interfering RNA (siRNA) oligonucleotides (siRUNX2) (supplementary material Fig. S2A) and initially performed in situ immunofluorescence microscopy. Our analysis revealed that nearly half (39%) of randomly selected nucleoli exhibited limited, but specific, colocalization of HDAC1 with UBF at the nucleolar periphery in the presence of a control siRNA oligonucleotide (Fig. 2A, upper panels). By contrast, siRUNX2-mediated reduction of RUNX2 levels decreased colocalization of HDAC1 with UBF so that it was only observed in ~9% of nucleoli (Fig. 2A, lower panels). Consistent with these findings, immunoprecipitation assays in parallel samples demonstrated a decreased association of HDAC1 with UBF in the absence of RUNX2 (siRUNX2) compared with that in the control (NS) (Fig. 2B; supplementary material Fig. S2B). To assess whether HDAC1 interaction at rDNA is also altered by depleting RUNX2, we carried out ChIP analysis. Similar to our previous observations with immunofluorescence and immunoprecipitation experiments, our results showed that downregulating RUNX2 levels decreased the level of HDAC1 binding to rDNA, as determined by amplification of three different regions of the locus (Fig. 2C, primer set A, B and C). Importantly, decreased UBF colocalization and association with HDAC1, as well as the reduced recruitment of HDAC1 to the rDNA locus, coincided with an increase in pre-rRNA levels (Fig. 2D). These findings suggest that there is a role for HDAC1 in RUNX2-mediated rRNA gene repression.
Fig. 2.
Decrease in HDAC1 nucleolar recruitment positively regulates rRNA gene expression. (A) Knockdown of RUNX2 by using siRNA oligonucleotides decreases HDAC1 recruitment at the periphery of interphase nucleolus (~9%), when compared with non-specific (NS) control (~39%) in SaOS-2 cells as analyzed by immunofluorescence microscopy. The blue dotted line represents the nucleus and the white dotted box marks an interphase nucleolus. The percentage reflects the proportion of the total nucleoli (randomly selected for analysis) that show colocalization between HDAC1 and UBF at the nucleolus. (B) Immunoprecipitation experiments showing that reduction in RUNX2 levels decreases UBF association with HDAC1 as compared with non-specific control sample. (C) ChIP assays demonstrating that knocking down RUNX2 decreases HDAC1 rDNA binding, as shown by rDNA primer sets A, B and C. HDAC1 shows no binding to the unrelated negative control gene Phox-GP91. (D) RUNX2 knockdown, concomitant with decreased UBF association with HDAC1 and reduced HDAC1 occupancy of rDNA, also decreases the pre-rRNA levels as determined by RT-qPCR. To normalize the values β-actin-encoding mRNA was used as an internal control.
Abrogation of the RUNX2 C-terminus perturbs its nuclear matrix association and interaction with co-regulatory proteins including HDAC1, thus compromising RUNX2 functional activity (Choi et al., 2001; McLarren et al., 2001; Westendorf, 2006). We therefore investigated the role of the RUNX2 C-terminus in formation of the HDAC1–UBF complex. Only WT (wild type) RUNX2, but not the ΔC mutant (Fig. 3A) nor the empty vector control, increased HDAC1 association with UBF in SaOS-2 cells (Fig. 3B; supplementary material Fig. S3B). To examine the involvement of the RUNX2 C-terminus in recruiting HDAC1 at rDNA repeats, ChIP experiments were performed using an antibody specific for HDAC1 in cells expressing WT-RUNX2 or the ΔC mutant. Consistent with our immunoprecipitation results, HDAC1 occupancy at three different regions of the rDNA repeat was increased only in the presence of WT-RUNX2 protein. Neither of the RUNX2 proteins (WT or ΔC) recruited HDAC1 to the unrelated Phox-GP91 gene, showing the specificity of these ChIP experiments (Fig. 3C). Collectively, our results demonstrate an essential role for the RUNX2 C-terminus in interaction with HDAC1 and its recruitment to rDNA repeats and suggest a functional relationship between RUNX2 and HDAC1 in regulation of rRNA gene expression.
Fig. 3.
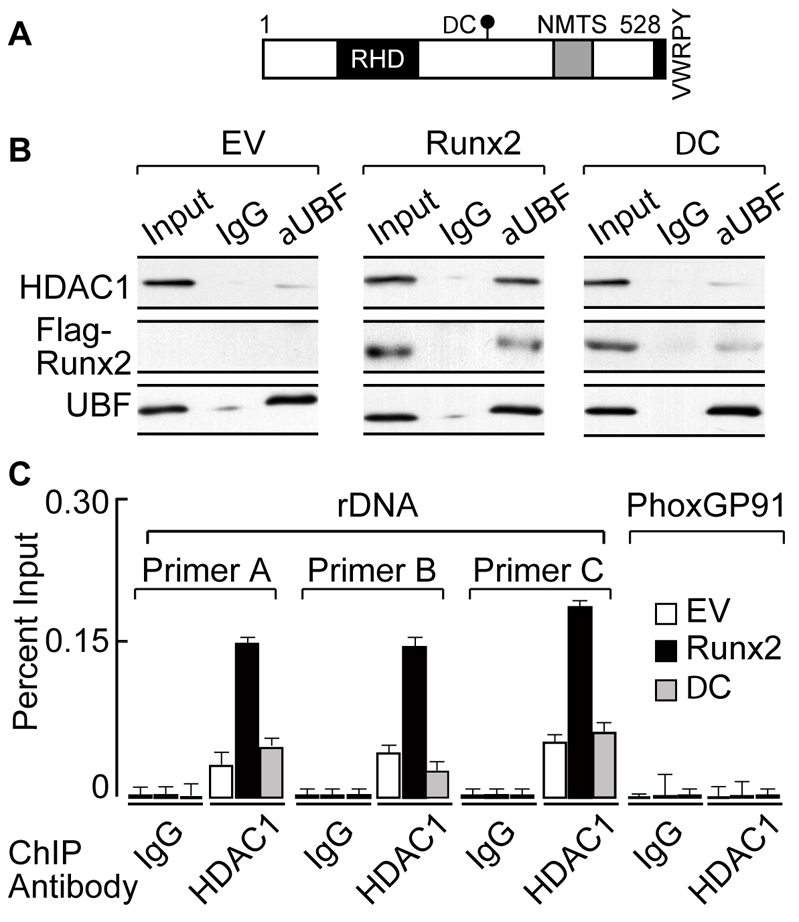
The RUNX2 C-terminus mediates HDAC1 association with UBF and rDNA. (A) A schematic representation of full-length RUNX2 protein indicating the Runt homology domain (RHD) for DNA binding, the nuclear-matrix-targeting signal (NMTS) and the C-terminus, showing the mark for ΔC truncation protein (DC). (B) Immunoprecipitation analysis demonstrates that overexpression of WT-RUNX2, increases UBF association with HDAC1 but not the ΔC mutant (DC), when compared with experiments using the empty vector (EV) control in osteosarcoma cells. (C) To analyze RUNX2-mediated rDNA occupancy of HDAC1, either full-length RUNX2 or the ΔC mutant was expressed in SaOS-2 cells. A ChIP assay shows that overexpression of WT-RUNX2, but not the ΔC mutant, increases HDAC1 occupancy at rDNA, as determined by primer set A, B and C. RUNX2 (WT and ΔC) does not bind to the Phox-GP91 gene, demonstrating the specificity of ChIP experiments.
An HDAC1–RUNX2 complex regulates rRNA gene transcription and affects global protein synthesis
We experimentally addressed the functional relevance of the HDAC1–RUNX2 interaction in regulating rRNA gene expression in osseous cells. Endogenous HDAC1 protein was depleted using three different siRNAs (Fig. 4A, WB) and pre-rRNA levels were analyzed. Downregulation of HDAC1 increased pre-rRNA expression but 28S rRNA levels were not affected (Fig. 4A, bar graphs). These results are consistent with the known role of HDAC1 in suppressing rRNA genes in fibroblasts and kidney cells (Meraner et al., 2006; Meraner et al., 2008). Interestingly, introduction of exogenous RUNX2 into osseous cells in which endogenous HDAC1 had been depleted failed to suppress pre-rRNA expression (Fig. 4B). These findings were further strengthened by the observation that the RUNX2ΔC mutant, which does not interact with HDAC1 (Fig. 3B), did not downregulate pre-rRNA expression (supplementary material Fig. S3C). Taken together, our results demonstrate that HDAC1 is involved in the repressive activity of RUNX2 on the rDNA repeats.
Fig. 4.
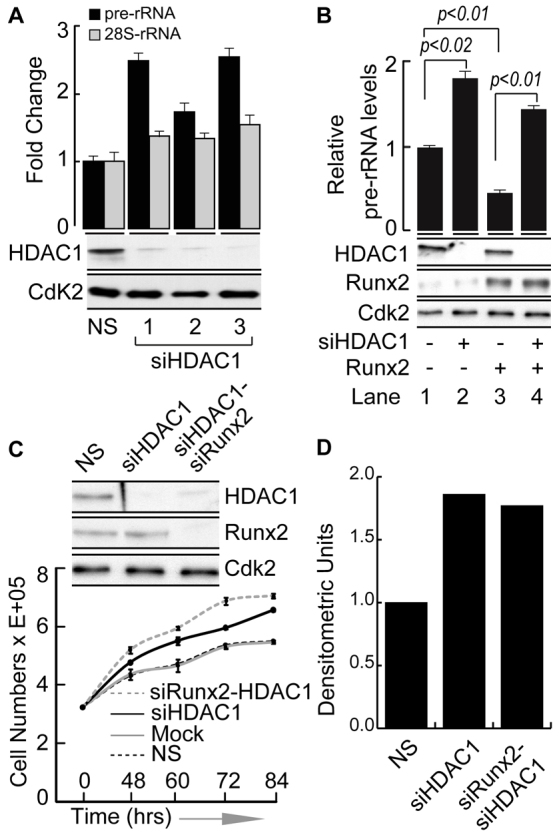
Combined knockdown of HDAC1 and RUNX2 increases cell proliferation and the overall protein synthesis rate. (A) Western blot showing the knockdown of HDAC1 protein by three independent siRNA oligonucleotides after 48 hours. The bar graph demonstrates the increased expression of rRNA genes in SaOS-2 cells as assessed by RT-qPCR. (B) Western blot showing siHDAC1 (lane 2), WT-RUNX2 overexpression (lane 3) and siHDAC1 with RUNX2 overexpression (lane 4) as compared with control (lane 1). In parallel samples, the effect on pre-rRNA expression was determined by RT-qPCR, using β-actin-encoding mRNA as an internal control (bar graphs). (C) Western blot showing the effect of siHDAC1 alone or siHDAC1 in combination with siRUNX2 after 48 hours of treatment with specific oligonucleotides. The line graph represents the increased cell count when either HDAC1 alone or together with RUNX2 was downregulated in SaOS-2 cells. (D) The bar graph shows combined densitometric quantification (Image J) of radioactive proteins from cells metabolically labeled with [35S]methionine. Each bar represents results from two independent experiments in which either HDAC1 alone was depleted or both HDAC1 and RUNX2 were targeted with siRNAs. NS, non-silencing siRNA.
Because the expression of rRNA genes is linked to cell growth and proliferation as well as protein synthesis rates (Arabi et al., 2005; Moss and Stefanovsky, 1995; Reeder, 1999), we examined the effect of HDAC1 depletion on these parameters in osseous cells. HDAC1, alone or in combination with RUNX2, was downregulated in SaOS-2 cells using specific siRNA oligonucleotides (Fig. 4C, WB) and cell proliferation was assayed (Fig. 4C, line graph). Consistent with upregulation of pre-rRNA expression (Fig. 4B), depletion of HDAC1 alone or in combination with RUNX2 increased the rate of cell proliferation (Fig. 4C, line graph). Concomitantly, global protein synthesis, as assessed by [35S]methionine labeling, was also enhanced (Fig. 4D; supplementary material Fig. S4A,B). These findings identify a functional role of an HDAC1–RUNX2 complex in regulating cell proliferation and growth properties in osseous cells.
HDAC1–RUNX2 interaction decreases UBF and histone protein acetylation at rDNA loci in osseous cells
UBF acetylation plays a key role in activating rRNA gene expression and HDAC1-mediated deacetylation counteracts this effect (Meraner et al., 2006; Meraner et al., 2008). We propose that RUNX2–HDAC1 interaction regulates UBF acetylation levels in osseous cells. Immunoprecipitation experiments were performed using antibody specific for acetylated lysine to test the level of UBF acetylation upon the expression of WT-RUNX2 and the ΔC mutant in osteosarcoma cells. Expression of WT-RUNX2 reduced UBF acetylation; however, expression of the ΔC mutant did not change UBF acetylation compared with that of the empty vector control (Fig. 5A, lower panel). As has been shown previously (Jeon et al., 2006; Meraner et al., 2008), samples treated with trichostatin A (TSA), an inhibitor of global HDAC1 activity, exhibited increased acetylation of both UBF and RUNX2 without affecting the cellular levels of either protein (Fig. 5A, upper panel). Consistent with our previous results that demonstrate the role of the RUNX2 C-terminus in suppressing rRNA gene through HDAC1 recruitment, these findings suggest that the RUNX2 C-terminus is also involved in regulating UBF deacetylation.
Fig. 5.
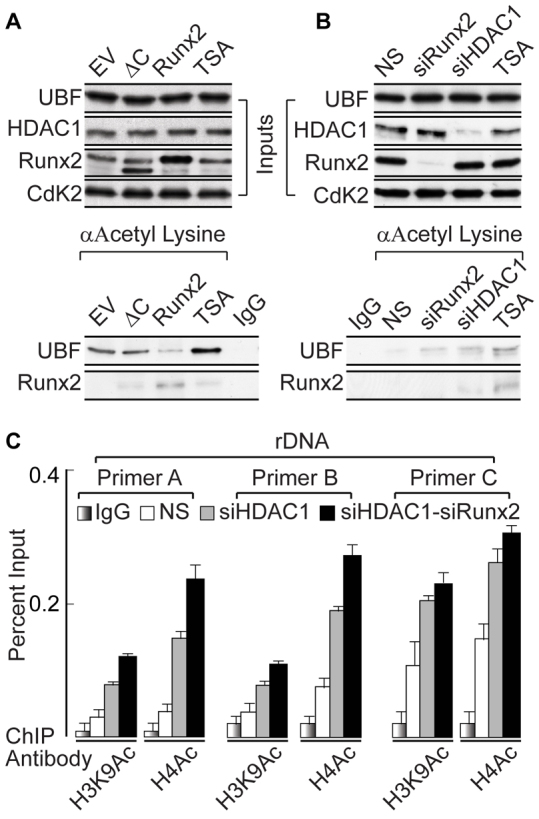
Deacetylation of UBF requires RUNX2-mediated HDAC1 recruitment. (A) Western blots showing the effect of expression of wild-type RUNX2 and the ΔC mutant after 48 hours of lentiviral infection in SaOS-2 cells. Samples were also treated with the histone deacetylase inhibitor TSA as control (upper panel). An immunoprecipitation assay was performed using an antibody against acetylated lysine, demonstrating decreased UBF acetylation in the presence of wild-type RUNX2 but not the ΔC mutant when compared with empty vector (EV) controls (lower panel). (B) Western blots demonstrating the depletion of RUNX2 and HDAC1 by siRNA oligonucleotides and treatment with TSA (upper panel). An antibody against acetylated lysine was used for immunoprecipitation. There is an increase in UBF acetylation in siRUNX2, siHDAC1 and TSA-treated samples, when compared with the non-silencing (NS) control (lower panel). (C) ChIP analysis showing that there is an increase in active histone marks (H3K9Ac and H4Ac) associated with rDNA, in the absence of HDAC1 alone or upon combined depletion of HDAC1 and RUNX2 after 48 hours of siRNA oligonucleotide treatment, as assessed by use of rDNA primer A, B and C.
Furthermore, to address our hypothesis that RUNX2 plays a role in UBF acetylation, we depleted RUNX2 or HDAC1 protein using siRNA oligonucleotides and performed immunoprecipitation assays using an antibody specific for acetylated lysine. Reduction of RUNX2 or HDAC1 levels led to increased UBF acetylation when compared with that in a non-silencing control sample (Fig. 5B, lower panel). In addition, TSA treatment, a positive control for acetylation assays, increased UBF acetylation without changing the protein levels of RUNX2, UBF or HDAC1 (Fig. 5B, upper panel). Importantly, the interaction of RUNX2 and UBF and their recruitment to rDNA loci were retained in the absence of HDAC1 (supplementary material Fig. S4B,C). These findings provide a functional link between pre-rRNA synthesis and RUNX2-mediated deacetylation of UBF by HDAC1.
An important mechanism by which HDACs control gene transcription is the modification of histone acetylation in gene regulatory regions. To assess how the RUNX2–HDAC1 interaction regulates epigenetic histone marks (H3K9Ac and H4Ac) associated with actively transcribed rDNA repeats (Zentner et al., 2011), we carried out ChIP assays using cells deficient for HDAC1 alone or in combination with RUNX2. Our results showed that depletion of HDAC1 increased acetylation of histones H3 at lysine 9 and H4 at the rDNA promoter region (analyzed by primer set A), when compared with results from a non-silencing control oligonucleotide (Fig. 5C). Interestingly, combined downregulation of HDAC1 and RUNX2 further increased the levels of these epigenetic marks at rDNA repeats (Fig. 5C). Collectively, our results have identified two independent, but related, post-translational mechanisms by which HDAC1 contributes to RUNX2-mediated suppression of rRNA gene expression.
Discussion
In this study we show that RUNX2-mediated HDAC1 recruitment to rDNA loci modifies histone marks and UBF acetylation in osseous cells. Importantly, the RUNX2–HDAC1 interaction decreases rRNA gene expression, with a concomitant effect on protein synthesis and cell proliferation. On the basis of these findings, we propose a mechanism for suppression of rRNA genes that involves RUNX2–HDAC1-dependent deacetylation of UBF and histone proteins.
Binding of UBF to rDNA is crucial for the transcriptional initiation of rRNA gene expression. UBF interacts with and recruits components of the RNA Pol I transcriptional machinery to rDNA, thus facilitating assembly of the pre-initiation complex required for active rRNA transcription (Bell et al., 1988; Schnapp et al., 1994). Our study demonstrates that HDAC1, RUNX2 and UBF associate in a complex at rDNA to regulate rRNA gene expression in osseous cells. In addition to the known roles of the RUNX2 C-terminus in protein–protein interactions and subnuclear targeting of the protein (Choi et al., 2001; Javed et al., 2000; McLarren et al., 2001; Westendorf, 2006), we show that recruitment of HDAC1 to rDNA loci and its association with UBF requires the C-terminus of RUNX2. Interestingly, the truncation of the C-terminus does not change UBF acetylation and fails to suppress pre-rRNA levels, consistent with the requirement of the RUNX2 C-terminus for biological activity (Choi et al., 2001; Lian et al., 2004). These results complement the finding that RUNX2 does not suppress pre-rRNA in the absence of endogenous HDAC1 in osseous cells. Taken together, we have identified an important role for the RUNX2 C-terminus in mediating UBF deacetylation and rRNA gene suppression by recruitment of HDAC1.
A key mechanism that is involved in the repression of rRNA genes is the establishment of the nucleolar repressor complex (NoRC) (Santoro et al., 2002), which modifies the epigenetic code associated with rDNA loci, resulting in a repressive chromatin conformation (Santoro and Grummt, 2005; Strohner et al., 2004). NoRC also recruits HDAC1 to rRNA genes (Zhou et al., 2002). However, it remains unclear whether NoRC functions to silence active copies of rRNA genes or maintains silencing of inactive copies or both (Grummt, 2007; Guetg et al., 2010; McStay and Grummt, 2008). Our results have revealed a unique regulatory mechanism independent of NoRC by which a RUNX2–HDAC1 complex dynamically downregulates actively transcribed rRNA genes. HDAC1 is a ubiquitous protein, with many cellular targets. Modulation of its activity by either its overexpression or by using RNA interference might lead to pleiotropic effects. It is therefore intriguing that in osseous cells HDAC1 alters rRNA gene expression through an association with RUNX2. Because RUNX2 is an osteoblast-related transcription factor and is not ubiquitously expressed, the specificity observed here might stem from the combined regulatory activity of the RUNX2–HDAC1 complex on rRNA genes. Interestingly, HDAC1 also interacts with other cell-fate-determining factors during myogenesis and adipogenesis (Mal et al., 2001; Yoo et al., 2006). Furthermore, we have shown previously that MyoD and myogenin also associate with UBF in the muscle lineage and suppress rRNA genes (Ali et al., 2008). We propose that the control of HDAC1 on rRNA gene expression through lineage-related transcription factors is a common mechanism for cell-type-specific regulation of protein synthesis.
We have previously identified RUNX2 as a component of a epigenetic mechanism by which transcription factors associate with target genes during mitosis to poise them for regulation following cell division (Young et al., 2007a; Young et al., 2007b; Zaidi et al., 2010). We have also shown that TLE1, a RUNX2 co-repressor, interacts with RUNX2 at target gene loci during mitosis (Ali et al., 2010). In this study, we find that the HDAC1 co-repressor does not associate with RUNX2 at mitotic chromosomes. These results are consistent with the exclusion of HDAC1 from mitotic chromosomes in neuroblastoma cells (Kim et al., 2003; Kruhlak et al., 2001); in fact, many DNA-binding factors and co-regulators are displaced from condensed chromatin during cell division (Martínez-Balbás et al., 1995; Muchardt et al., 1996). Our finding that HDAC1 is absent from mitotic chromosomes distinguishes rRNA regulation by the RUNX2–HDAC1 interaction from epigenetic bookmarking of rRNA genes by the RUNX2–TLE1 complex. Plasticity to RUNX2-mediated epigenetic control of rRNA gene expression is suggested by selective mitotic exclusion of co-regulatory factors for histone modification that might be re-established post-proliferation.
Acetylation of nucleosomal histones and transcription factors plays a key role in the control of gene expression (Furumatsu and Asahara, 2010; Kawai et al., 2011; Muth et al., 2001; Strahl and Allis, 2000). Findings reported here demonstrate that both of these activities in osseous cells involve the RUNX2–HDAC1 complex; a combined depletion of RUNX2 and HDAC1 increases acetylation levels of UBF, as well as of histones associated with rDNA repeats. UBF acetylation is involved in activating rRNA gene expression and HDAC1-mediated deacetylation counters UBF activity (Meraner et al., 2006; Pelletier et al., 2000). Similarly HATs and HDACs regulate acetylation-based epigenetic codes and transcription of rRNA genes (Lawrence and Pikaard, 2004; Santoro and Grummt, 2005). We conclude that RUNX2 functions through HDAC1 to regulate rRNA gene transcription by modifying UBF and histone acetylation.
Materials and Methods
Cell culture and in situ immunofluorescence microscopy
Human osteosarcoma (SaOS-2) cells from American Type Culture Collection (ATCC; Manassas, VA) were cultured in Hyclone McCoy's 5A medium containing 15% fetal bovine serum (Atlanta Biological, Lawrenceville, CA), 2 mM L-glutamine and a penicillin-streptomycin cocktail (Invitrogen, Carlsbad, CA). SaOS-2 cells were grown on gelatin-coated coverslips and processed for immunofluorescence microscopy as described previously (Young et al., 2007a). Antibodies used in IF (diluted in 0.5% BSA in PBS) were against: RUNX2 (1:100; 8G5, MBL International Corporation, Woburn, MA), UBF (1:700; F-9, Santa Cruz Biotechnology) and HDAC1 (1:600; H-51, Santa Cruz Biotechnology). Secondary antibodies used were goat anti-mouse-IgG conjugated to Alexa Fluor 594 (1:800) and goat anti-rabbit-IgG conjugated to Alexa Fluor 488 (1:800) from Invitrogen. Nuclear DNA was visualized by 4′, 6-diamidino-2-phenylindole (DAPI) staining. Immunostaining analysis was recorded using an Epifluorescence Zeiss Axioplan 2 microscope (Zeiss MicroImaging Inc., New York, NY) equipped with a charged coupled device. Images were captured and processed by MetaMorph Imaging Software (Molecular Devices, Downington, PA).
Co-immunoprecipitation and western blot analyses
Actively proliferating osteosarcoma cells were washed with phosphate-buffered saline (PBS) and harvested on ice. A cell pellet was suspended in sonication buffer (50 mM NaCl, 50 mM Tris-HCl pH 8.0, 1% NP-40, 25 mM MG132 and 1×protease inhibitor cocktail; Roche Diagnostics, Indianapolis, IN). Samples were sonicated with nine pulses at 10% power for a total time of 90 seconds, to disrupt membranes, using a Fisher Scientific Sonic Dismembrator 550 fitted with a 1.6-mm tip (Fisher Scientific. Pittsburgh, PA). Lysates were centrifuged to remove debris and incubated overnight at 4°C with 4 μg of anti-RUNX2 (M 70) or anti-UBF (H-300) rabbit polyclonal antibody (Santa Cruz Biotechnology). Immuno-complexes were then isolated by incubation with protein A or G beads (Santa Cruz Biotechnology) for 2 hours, followed by four washes with cold wash buffer (150 mM NaCl, 20 mM Tris-HCl pH 8.3, 0.5% Na-10 deoxycholate, 0.5% Nonidet P-40, 2 mM EDTA with freshly added 25 μM MG132 and 1× protease inhibitor cocktail). Immunoprecipitated samples were resolved by SDS/PAGE and analyzed by western blotting (WB). Antibody dilutions used were: RUNX2 (8G5), 1:2000; UBF (F-9), 1:1000; HDAC1, 1:1000 (Upstate Biotechnology Inc., NY); CDK2 (M2, Santa Cruz Biotechnology), 1:3000; Nucleophosmin (B23, Zymed Laboratories San Francisco, CA), 1:13,000. Secondary horseradish-peroxidase-conjugated goat anti-mouse-IgG used for WB (1:2000) was from Jackson ImmunoResearch Laboratories, West Grove; PA. Protein bands were visualized by chemiluminescence detection reagent (PerkinElmer Life Sciences, Waltham, MA).
Assays for UBF acetylation
To determine UBF acetylation levels, SaOS-2 cells were either treated with small interfering RNA oligonucleotides (siRNA) against RUNX2 and HDAC1 using Oligofectamine (Invitrogen) or the wild-type lentiviral RUNX2 or its ΔC mutant were expressed for 48 hours using 4 μg/ml of polybrene (Sigma, WI, USA). Cells were also treated with trichostatin-A (TSA; 100ng/ml) from Sigma for 36 hours to inhibit global HDAC activity. Lysates were processed for immunoprecipitation assays following the protocol described above. To assess the level of acetylated lysine associated with UBF in different conditions (i.e. siHDAC1, siRUNX2, WT-RUNX2, ΔC mutant and TSA treatment) lysates were immunoprecipitated overnight at 4°C, using an antibody specific for acetylated lysine (Abcam, Cambridge, MA). Protein samples were analyzed by performing western blot analysis as described above.
Chromatin immunoprecipitation and ChIP-ReChIP
ChIP assays were performed as described previously (Young et al., 2007a). ChIP enrichment was determined as percentage of input. Primer sets A, B and C were used to assess the binding of RUNX2, HDAC1 and UBF at human rDNA repeats (Ali et al., 2010). ChIP experiments were carried out essentially as described previously (Young et al., 2007b). Briefly, the immunoprecipitates from the first ChIP were eluted in 10 mM DTT buffer for 30 minutes at 37°C, diluted with ChIP dilution buffer (10% SDS, 10% Triton X100, 0.5 M EDTA, Tris-HCl pH 8.0, 300 mM NaCl) and subjected to a secondary immunoprecipitation (i.e. ReChIP) using 6 μg of antibodies against HDAC1 (Abcam), RUNX2 (M-70), UBF (H-300) or normal IgG purchased from Santa Cruz Biotechnology.
RNA isolation and pre-rRNA expression analysis
Total cellular RNA was isolated from osteosarcoma cells using TRIzol reagent (Invitrogen) and purified with the DNase-free RNA kit (Zymo Research Corp, Orange, CA). cDNA was generated from RNA using SuperScript First Strand kit from Invitrogen and subjected to real-time quantitative PCR (RT-qPCR) using SYBR-green chemistry (Applied Biosystems, Inc., Foster, CA). Transcript levels were normalized using β-actin-encoding mRNA as the internal control. The primer sequences used in this study are provided in supplementary material Table S1.
RNA interference and cell proliferation assays
RUNX2 and HDAC1 protein levels were depleted using small interfering RNA (siRNA) transfection with Oligofectamine reagent from Invitrogen. SaOS-2 cells were grown in 60-mm plates and transfected at 70% confluence with either RUNX2 smart pool siRNA or three independent predesigned HDAC1 siRNAi oligonucelotides (Dharmacon Inc., Lafayette, CO) at 70% confluent. Cells were transfected with 40 nmol of specific or non-specific control oligonucleotides for 48 hours to downregulate protein expression of RUNX2 and HDAC1 alone or in combination. To study proliferation rates of osteosarcoma cells were harvested after every 12 hours by trypsinizing cells and suspending in McCoy's medium. Equal volumes (20 μl) of cell suspension were loaded on Nexcelom Cellometer glass slides and counted using a Cellometer™ Auto T4 Cellcounter (Nexcelom Bioscience LLC, Lawrence, MA).
RUNX2 lentiviral constructs
Wild-type and ΔC mutant RUNX2 lentiviral particles were generated by utilizing the Lentiviral Gateway System (Invitrogen, Carlsbad, CA). Mouse wild-type RUNX2 and ΔC mutant cDNA were cloned into pENTR4-FLAG vector and then recombined with lentiviral destination pLenti-CMV-Blast-DEST vector by using Gateway LR Clonase™ enzyme mix according to the manufacturer's instructions (Invitrogen). Lentiviral particles were generated in HEK-293T cells by transient transfection of RUNX2 constructs along with packaging plasmids. Infected particles were collected after 48 hours of transfection and used for infection. For expression study, 250 μl of viral supernatant (WT and ΔC) per 100-mm plate was used for 48 hours.
Metabolic labeling
Osteosarcoma cells were transfected with siRNAs to target RUNX2 and HDAC1 alone or in combination using Oligofectamine (see above) to determine the effect of these factors on global protein synthesis. After 48 hours of siRNA treatment, cultures were incubated at 37°C in methionine- and cysteine-free DMEM (GIBCO, Grand Island, NY) with 10% dialyzed serum for 1 hour followed by addition of EasyTagTM Express [35S]-protein labeling mix (200 μCi/ml) for 45 minutes (PerkinElmer, Waltham, MA). Then cells were harvested in an equal volume of direct lysis buffer [2 M urea, 2% SDS, 10 mM DTT, 10% glycerol, 10 mM Tris HCl pH 6.8, 0.2 mg/mL Bromophenol Blue, 1× Complete Protease inhibitor (Roche Diagnostic, Indanapolis, IN) and 25 μM MG132]. Proteins were analyzed by using SDS/PAGE. Gels were dried and exposed at −70°C to scientific imaging film BioMax MR (Kodak Company, Rochester, NY).
Supplementary Material
Acknowledgements
We thank our research group, especially Jonathan Gordon, for sharing reagents, Karthiga Gokul, Matthew Mandeville and Marissa Johnson for technical support and Judy Rask for expert assistance with manuscript preparation.
Footnotes
Funding
This work was supported by the National Institutes of Health [grant numbers P01AR48818 to G.S.S., P01CA82834, including an ARRA Supplement to G.S.S., AR049069 to A.J.W., AR39588 to G.S.S., NIDCR R37DE012528 to J.B.L.]. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.100909/-/DC1
References
- Ali S. A., Zaidi S. K., Dacwag C. S., Salma N., Young D. W., Shakoori A. R., Montecino M. A., Lian J. B., van Wijnen A. J., Imbalzano A. N., et al. (2008). Phenotypic transcription factors epigenetically mediate cell growth control. Proc. Natl. Acad. Sci. USA 105, 6632-6637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S. A., Zaidi S. K., Dobson J. R., Shakoori A. R., Lian J. B., Stein J. L., van Wijnen A. J., Stein G. S. (2010). Transcriptional corepressor TLE1 functions with Runx2 in epigenetic repression of ribosomal RNA genes. Proc. Natl. Acad. Sci. USA 107, 4165-4169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi A., Wu S., Ridderstråle K., Bierhoff H., Shiue C., Fatyol K., Fahlén S., Hydbring P., Söderberg O., Grummt I., et al. (2005). c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 7, 303-310 [DOI] [PubMed] [Google Scholar]
- Bell S. P., Learned R. M., Jantzen H. M., Tjian R. (1988). Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science 241, 1192-1197 [DOI] [PubMed] [Google Scholar]
- Budde A., Grummt I. (1999). p53 represses ribosomal gene transcription. Oncogene 18, 1119-1124 [DOI] [PubMed] [Google Scholar]
- Choi J.-Y., Pratap J., Javed A., Zaidi S. K., Xing L., Balint E., Dalamangas S., Boyce B., van Wijnen A. J., Lian J. B., et al. (2001). Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc. Natl. Acad. Sci. USA 98, 8650-8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenzini M., Trerè D., Pession A., Govoni M., Sirri V., Chieco P. (2000). Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. J. Pathol. 191, 181-186 [DOI] [PubMed] [Google Scholar]
- Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. (1997). Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89, 747-754 [DOI] [PubMed] [Google Scholar]
- Durst K. L., Hiebert S. W. (2004). Role of RUNX family members in transcriptional repression and gene silencing. Oncogene 23, 4220-4224 [DOI] [PubMed] [Google Scholar]
- Furumatsu T., Asahara H. (2010). Histone acetylation influences the activity of Sox9-related transcriptional complex. Acta Med. Okayama 64, 351-357 [DOI] [PubMed] [Google Scholar]
- Grandori C., Gomez-Roman N., Felton-Edkins Z. A., Ngouenet C., Galloway D. A., Eisenman R. N., White R. J. (2005). c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 7, 311-318 [DOI] [PubMed] [Google Scholar]
- Grummt I. (1998). Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol. 62, 109-154 [DOI] [PubMed] [Google Scholar]
- Grummt I. (2007). Different epigenetic layers engage in complex crosstalk to define the epigenetic state of mammalian rRNA genes. Hum. Mol. Genet. 16, R21-R27 [DOI] [PubMed] [Google Scholar]
- Grummt I., Pikaard C. S. (2003). Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell Biol. 4, 641-649 [DOI] [PubMed] [Google Scholar]
- Guetg C., Lienemann P., Sirri V., Grummt I., Hernandez-Verdun D., Hottiger M. O., Fussenegger M., Santoro R. (2010). The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 29, 2135-2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan K. M., Brandenburger Y., Jenkins A., Sharkey K., Cavanaugh A., Rothblum L., Moss T., Poortinga G., McArthur G. A., Pearson R. B., et al. (2003). mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol. Cell. Biol. 23, 8862-8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed A., Guo B., Hiebert S., Choi J.-Y., Green J., Zhao S.-C., Osborne M. A., Stifani S., Stein J. L., Lian J. B., et al. (2000). Groucho/TLE/R-esp proteins associate with the nuclear matrix and repress RUNX (CBF(α)/AML/PEBP2(α)) dependent activation of tissue-specific gene transcription. J. Cell Sci. 113, 2221-2231 [DOI] [PubMed] [Google Scholar]
- Jensen E. D., Nair A. K., Westendorf J. J. (2007). Histone deacetylase co-repressor complex control of Runx2 and bone formation. Crit. Rev. Eukaryot. Gene Expr. 17, 187-196 [DOI] [PubMed] [Google Scholar]
- Jeon E. J., Lee K. Y., Choi N. S., Lee M. H., Kim H. N., Jin Y. H., Ryoo H. M., Choi J. Y., Yoshida M., Nishino N., et al. (2006). Bone morphogenetic protein-2 stimulates Runx2 acetylation. J. Biol. Chem. 281, 16502-16511 [DOI] [PubMed] [Google Scholar]
- Kawai Y., Garduño L., Theodore M., Yang J., Arinze I. J. (2011). Acetylation-deacetylation of the transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2) regulates its transcriptional activity and nucleocytoplasmic localization. J. Biol. Chem. 286, 7629-7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. M., Liu H., Tazaki M., Nagata M., Aoki F. (2003). Changes in histone acetylation during mouse oocyte meiosis. J. Cell Biol. 162, 37-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T. (2006). Regulation of osteoblast differentiation by transcription factors. J. Cell. Biochem. 99, 1233-1239 [DOI] [PubMed] [Google Scholar]
- Kruhlak M. J., Hendzel M. J., Fischle W., Bertos N. R., Hameed S., Yang X. J., Verdin E., Bazett-Jones D. P. (2001). Regulation of global acetylation in mitosis through loss of histone acetyltransferases and deacetylases from chromatin. J. Biol. Chem. 276, 38307-38319 [DOI] [PubMed] [Google Scholar]
- Kuhn A., Grummt I. (1992). Dual role of the nucleolar transcription factor UBF: trans-activator and antirepressor. Proc. Natl. Acad. Sci. USA 89, 7340-7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence R. J., Pikaard C. S. (2004). Chromatin turn ons and turn offs of ribosomal RNA genes. Cell Cycle 3, 878-881 [PubMed] [Google Scholar]
- Lee H. W., Suh J. H., Kim A. Y., Lee Y. S., Park S. Y., Kim J. B. (2006). Histone deacetylase 1-mediated histone modification regulates osteoblast differentiation. Mol. Endocrinol. 20, 2432-2443 [DOI] [PubMed] [Google Scholar]
- Lian J. B., Stein J. L., Stein G. S., van Wijnen A. J., Montecino M., Javed A., Gutierrez S., Shen J., Zaidi S. K., Drissi H. (2003). Runx2/Cbfa1 functions: diverse regulation of gene transcription by chromatin remodeling and co-regulatory protein interactions. Connect. Tissue Res. 44 Suppl 1, 141-148 [PubMed] [Google Scholar]
- Lian J. B., Javed A., Zaidi S. K., Lengner C., Montecino M., van Wijnen A. J., Stein J. L., Stein G. S. (2004). Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit. Rev. Eukaryot. Gene Expr. 14, 1-42 [PubMed] [Google Scholar]
- Mal A., Sturniolo M., Schiltz R. L., Ghosh M. K., Harter M. L. (2001). A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 20, 1739-1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Balbá M. A., Dey A., Rabindran S. K., Ozato K., Wu C. (1995). Displacement of sequence-specific transcription factors from mitotic chromatin. Cell 83, 29-38 [DOI] [PubMed] [Google Scholar]
- Mayer C., Grummt I. (2006). Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 25, 6384-6391 [DOI] [PubMed] [Google Scholar]
- McLarren K. W., Theriault F. M., Stifani S. (2001). Association with the nuclear matrix and interaction with Groucho and RUNX proteins regulate the transcription repression activity of the basic helix loop helix factor Hes1. J. Biol. Chem. 276, 1578-1584 [DOI] [PubMed] [Google Scholar]
- McStay B., Grummt I. (2008). The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 24, 131-157 [DOI] [PubMed] [Google Scholar]
- Meraner J., Lechner M., Loidl A., Goralik-Schramel M., Voit R., Grummt I., Loidl P. (2006). Acetylation of UBF changes during the cell cycle and regulates the interaction of UBF with RNA polymerase I. Nucleic Acids Res. 34, 1798-1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraner J., Lechner M., Schwarze F., Gander R., Jesacher F., Loidl P. (2008). Cell cycle dependent role of HDAC1 for proliferation control through modulating ribosomal DNA transcription. Cell Biol. Int. 32, 1073-1080 [DOI] [PubMed] [Google Scholar]
- Moss T., Stefanovsky V. Y. (1995). Promotion and regulation of ribosomal transcription in eukaryotes by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol. 50, 25-66 [DOI] [PubMed] [Google Scholar]
- Muchardt C., Reyes J.-C., Bourachot B., Leguoy E., Yaniv M. (1996). The hbrm and BRG-1 proteins, components of the human SNF/SWI complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO J. 15, 3394-3402 [PMC free article] [PubMed] [Google Scholar]
- Muth V., Nadaud S., Grummt I., Voit R. (2001). Acetylation of TAF(I)68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J. 20, 1353-1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier G., Stefanovsky V. Y., Faubladier M., Hirschler-Laszkiewicz I., Savard J., Rothblum L. I., Côté J., Moss T. (2000). Competitive recruitment of CBP and Rb-HDAC regulates UBF acetylation and ribosomal transcription. Mol. Cell 6, 1059-1066 [DOI] [PubMed] [Google Scholar]
- Reeder R. H. (1998). Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog. Nucleic Acid Res. Mol. Biol. 62, 293-327 [DOI] [PubMed] [Google Scholar]
- Russell J., Zomerdijk J. C. (2005). RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem. Sci. 30, 87-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R., Grummt I. (2005). Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol. Cell. Biol. 25, 2539-2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R., Li J., Grummt I. (2002). The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 32, 393-396 [DOI] [PubMed] [Google Scholar]
- Sarge K. D., Park-Sarge O. K. (2009). Mitotic bookmarking of formerly active genes: keeping epigenetic memories from fading. Cell Cycle 8, 818-823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp G., Santori F., Carles C., Riva M., Grummt I. (1994). The HMG box-containing nucleolar transcription factor UBF interacts with a specific subunit of RNA polymerase I. EMBO J. 13, 190-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovsky V., Langlois F., Gagnon-Kugler T., Rothblum L. I., Moss T. (2006). Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol. Cell 21, 629-639 [DOI] [PubMed] [Google Scholar]
- Stein G. S., van Wijnen A. J., Imbalzano A. N., Montecino M., Zaidi S. K., Lian J. B., Nickerson J. A., Stein J. L. (2010). Architectural genetic and epigenetic control of regulatory networks: compartmentalizing machinery for transcription and chromatin remodeling in nuclear microenvironments. Crit. Rev. Eukaryot. Gene Expr. 20, 149-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B. D., Allis C. D. (2000). The language of covalent histone modifications. Nature 403, 41-45 [DOI] [PubMed] [Google Scholar]
- Strohner R., Németh A., Nightingale K. P., Grummt I., Becker P. B., Längst G. (2004). Recruitment of the nucleolar remodeling complex NoRC establishes ribosomal DNA silencing in chromatin. Mol. Cell. Biol. 24, 1791-1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan J. C., Zhai W., Comai L. (1999). Recruitment of TATA-binding protein-TAFI complex SL1 to the human ribosomal DNA promoter is mediated by the carboxy-terminal activation domain of upstream binding factor (UBF) and is regulated by UBF phosphorylation. Mol. Cell. Biol. 19, 2872-2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voit R., Kuhn A., Sander E. E., Grummt I. (1995). Activation of mammalian ribosomal gene transcription requires phosphorylation of the nucleolar transcription factor UBF. Nucleic Acids Res. 23, 2593-2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorf J. J. (2006). Transcriptional co-repressors of Runx2. J. Cell. Biochem. 98, 54-64 [DOI] [PubMed] [Google Scholar]
- Yoo E. J., Chung J. J., Choe S. S., Kim K. H., Kim J. B. (2006). Down-regulation of histone deacetylases stimulates adipocyte differentiation. J. Biol. Chem. 281, 6608-6615 [DOI] [PubMed] [Google Scholar]
- Young D. W., Hassan M. Q., Pratap J., Galindo M., Zaidi S. K., Lee S. H., Yang X., Xie R., Javed A., Underwood J. M., et al. (2007a). Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature 445, 442-446 [DOI] [PubMed] [Google Scholar]
- Young D. W., Hassan M. Q., Yang X.-Q., Galindo M., Javed A., Zaidi S. K., Furcinitti P., Lapointe D., Montecino M., Lian J. B., et al. (2007b). Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc. Natl. Acad. Sci. USA 104, 3189-3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S. K., Young D. W., Montecino M. A., Lian J. B., van Wijnen A. J., Stein J. L., Stein G. S. (2010). Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat. Rev. Genet. 11, 583-589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner G. E., Saiakhova A., Manaenkov P., Adams M. D., Scacheri P. C. (2011). Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res. 39, 4949-4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Santoro R., Grummt I. (2002). The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 21, 4632-4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.