Abstract
Nanoparticle tracking analysis permits the determination of both the size distribution and relative concentration of microvesicles, including exosomes, in the supernatants of cultured cells and biological fluids. We have studied the release of microvesicles from the human lymphoblastoid T-cell lines Jurkat and CEM. Unstimulated, both cell lines release microvesicles in the size range 70–90 nm, which can be depleted from the supernatant by ultracentrifugation at 100 000 g, and by anti-CD45 magnetic beads, and which by immunoblotting also contain the exosome-associated proteins Alix and Tsg101. Incubation with known potentiators of exosome release, the ionophores monensin and A23187, resulted in a significant increase in microvesicle release that was both time and concentration dependent. Mass spectrometric analysis of proteins isolated from ultracentrifuged supernatants of A23187-treated cells revealed the presence of exosome-associated proteins including heat-shock protein 90, tubulin, elongation factor α1, actin and glyceraldehyde 3-phosphate dehydrogenase. Additionally, treatment of peripheral blood monocyte-derived dendritic cells with bacterial lipopolysaccharide displayed an increase in secreted microvesicles. Consequently, nanoparticle tracking analysis can be effectively applied to monitor microvesicle release from cells of the immune system.
Keywords: cell processes/associated and molecules, dendritic cells, T cells
Introduction
Microvesicles, including exosomes, in the size range 50–200 nm are secreted by a wide range of cell types, including those of the immune system, and display a wide range of biological activities.1,2 Exosomes in particular have been the focus of much recent research. They are derived from a specialized compartment of the endosomal–lysosomal pathway, in the form of multvesicular bodies, which functionally and phenotypically allows them to be distinguished from vesicles of plasma membrane origin.3
Exosomes appear to function in a range of intercellular communication processes, principally based on receptor–ligand interactions mediated by polypeptides incorporated into the exosome membrane, but they can also deliver a complex cargo of materials to recipient cells including polypeptides, micro-RNA and mRNA.4–6 One area where research has taken place is in the context of immunomodulation. Dendritic cell-derived exosomes (sometimes called dexasomes) in particular can exhibit both immunostimulatory or immunosuppressive effects, depending on how they are prepared.7,8 Dendritic cell-derived exosomes are therefore of interest as clinically relevant cell-free therapeutics, and several clinical trials using them as anti-cancer vaccines have already taken place.9–11 In addition, neuron-specific targeting of modified exosomes has recently allowed the delivery of short interfering RNA into brain tissue in vivo, further highlighting potential therapeutic applications.12
As a result of their size, microvesicles pose some unique problems for their study. Commonly used methodologies to study them include isolation by ultracentrifugation, immuno-isolation by antibodies, immunoblotting, adsorption to latex beads followed by flow cytometry, and electron microscopy.13 Flow cytometry of non-adsorbed free vesicles has also been reported, but is problematical with substrates below 300 nm in size.14–16 Recently, a novel technique in the form of nanoparticle tracking analysis (NTA) has been adapted for the study of microvesicles.16 NTA uses the light-scattering characteristics of laser light on particles undergoing Brownian motion when in solution. A video recording of the particles is analysed, particles are tracked and a mean squared displacement is calculated for each particle, allowing a hydrodynamic radius to be determined and displayed as a particle size distribution. Here, we show the use of NTA in the detection and quantification of microvesicles from two human lymphoblastoid T-cell lines undergoing enhanced microvesicle release by treatment with ionophores.
Materials and methods
Cell lines, antibodies and reagents
The human lymphoblastoid T-cell line CEM was a gift from Antony Antoniou (UCL, London, UK). The human T-cell line Jurkat was obtained from the UK Health Protection Agency (line 88042803; HPA Culture Collections, Porton Down, UK). Both lines were maintained in RPMI-1640 supplemented with 5% fetal bovine serum (both from Invitrogen, Paisley, UK) at 37° and 5% CO2 in a humidified incubator. Monocyte-derived dendritic cells were obtained from human monocytes cultured in 50 ng/ml interleukin-4 and 50 ng/ml granulocyte–macrophage colony-stimulating factor for 5 days. Antibodies recognizing ALIX, Tsg101 and CD45 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-MHC class I mouse monoclonal antibody HC10 was a gift from Jacques Neefjes (Netherlands Cancer Institute, Amsterdam, the Netherlands), and rabbit anti-ERp57 was a gift from Neil Bulleid (University of Glasgow, UK). Monensin, A23187, ammonium chloride, chloroquine and lipopolysaccharide (LPS) were obtained from Sigma-Aldrich (Poole, UK).
Isolation of microvesicles
Microvesicles, including exosomes, were obtained by sequential centrifugation. Briefly, cells were incubated at between 1 × 106 and 4 × 106 cells/ml for the indicated times and with the indicated ionophores. Cells were then removed by centrifugation at 300 g for 5 min. The supernatant was then depleted of debris by centrifugation at 10 000 g for 20 min. Microvesicles were then isolated by centrifugation of the supernatant at 100 000 g for 2 hr. Because this preparation is likely to contain a mixture of both exosomes and other microvesicles, we have used the generic term microvesicles in this study to include the exosome pool.
Nanoparticle tracking analysis
Samples from the 10 000 g centrifugation step were used for NTA. Briefly, approximately 0·3 ml supernatant was loaded into the sample chamber of an LM10 unit (Nanosight, Amesbury, UK) and three videos of either 30 or 60 seconds were recorded of each sample. Data analysis was performed with both NTA 1·1 and 2·1 software (Nanosight). In NTA the paths of unlabelled particles (i.e. microvesicles) acting as point scatterers, undergoing Brownian motion in a 0·25-ml chamber through which a 635-nm laser beam is passed, is determined from a video recording with the mean squared displacement determined for each possible particle. The diffusion coefficient and sphere-equivalent hydrodynamic radius are then determined using the Stokes–Einstein equation, and results are displayed as a particle size distribution. Samples were analysed using the basic control settings, which resulted in shutter speeds of 30, 6 and 1 milliseconds for the 100-, 200- and 400-nm control beads, respectively (with zero camera gain), and for biological samples the shutter speeds were 30 or 15 milliseconds, with camera gains of between 280 and 560. Software settings for analysis were: Detection Threshold: 5–10; Blur: auto; Minimum expected particle size: 50 nm. Data are presented as the average and standard deviation of the three video recordings. NTA is most accurate between particle concentrations in the range 2 × 108 to 20 × 108/ml. When samples contained higher numbers of particles, they were diluted before analysis and the relative concentration was then calculated according to the dilution factor. Control 100 and 400 nm beads were supplied by Duke Scientific (Palo Alto, CA).
SDS–PAGE, immunoblotting and immunodepletion
Pellets from the 100 000 g centrifugation step were resuspended in lysis buffer (1% nonidet-P40, 150 mm NaCl, 10 mm Tris–HCl pH 7·6, 1 mm PMSF). Then, 10–20 μg protein was analysed by SDS–PAGE and either stained with SimplyBlue reagent (Invitrogen) before excision of bands for mass spectrometric identification, or electrophoretically transferred to nitrocellulose (BA85; Whatman, Maidstone, UK) and probed with relevant antibodies overnight and developed with horseradish peroxidase-coupled anti-mouse or anti-rabbit IgG (Jackson Immunoresearch, Westgrove, PA) and Femto-chemiluminescent reagents (Perbio, Cramlington, UK). Mass spectrometry for protein identification was performed at the University of St Andrews Mass Spectrometry Facility.
Results
Nanoparticle tracking analysis detects microvesicles in conditioned medium
Jurkat and CEM T-cell lines were incubated overnight in serum free RPMI-1640 medium, and the resulting supernatants were processed at 300 g, and then 10 000 g. The samples were analysed by NTA in comparison to serum-free RPMI alone and control 100-nm and 400-nm latex beads. Screen shots (Fig. 1a) indicate the presence of particles in the control 100-nm beads, and in Jurkat-conditioned and CEM-conditioned media, whereas serum-free RPMI alone generates no signal. The NTA gives accurate predictions of the sizes for the control beads, and particles with peak intensities of 76 and 83 nm for Jurkat and CEM, respectively (Fig. 1b). The mean peak of detected particles from Jurkat and CEM cells ranged from c.70 to 160 nm during these experiments, as indicated in the figure legends. Further centrifugation of the Jurkat-conditioned and CEM-conditioned media at 100 000 g, followed by immunoblotting compared with whole cell lysates, revealed the presence of the characteristic exosome-associated proteins Alix and Tsg101 (Fig. 1c). CD45 and MHC class I molecules were also detected in the purified vesicle population (although CEM cells express very low levels of HLA-B molecules). The normally endoplasmic-reticulum-resident oxidoreductase ERp57 was not detected in the exosome samples. Centrifugation of Jurkat-conditioned medium at 100 000 g also removed the NTA signal (Fig. 2b). Hence, NTA can detect the presence of microvesicles, which include exosomes, contained in T-cell-line-conditioned medium.
Figure 1.
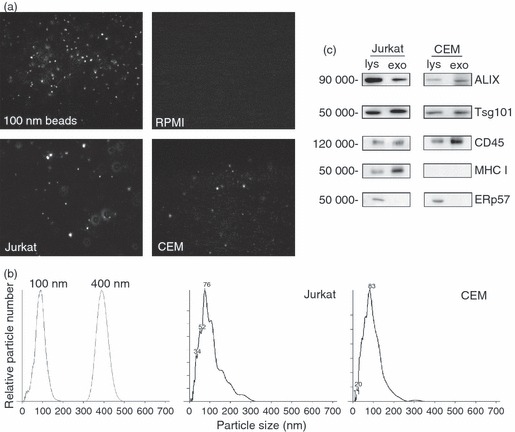
Nanoparticle tracking analysis (NTA) and immunoblotting analysis of microvesicles secreted by Jurkat and CEM cells. (a) Screenshots of NTA of 100-nm beads, serum-fee RPMI-1640, and post-10 000 g conditioned medium from Jurkat and CEM cells. Image size recorded by video is approximately 100 μm × 80 μm. (b) Size and particle distribution plots of 100-nm and 400-nm control beads, and post-10 000 g conditioned medium from Jurkat (peak at 76 nm) and CEM (peak at 83 nm) cells. (c) Immunoblot analysis of detergent cell lysates (lys) and exosome and microvesicles (exo, post-100 000 g pellets) of Jurkat and CEM cells.
Figure 2.
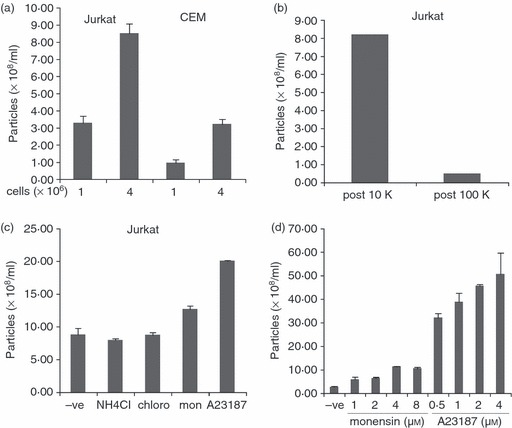
Quantitative determination of microvesicle release by nanoparticle tracking analysis (NTA). (a) 1 × 106 and 4 × 106 Jurkat and CEM cells were incubated in serum-free medium overnight. Post-10 000 g supernatant was analysed by NTA. The mean size of particles detected was 161 nm. (b) Conditioned medium from Jurkat cells incubated overnight in serum-free medium was analysed by NTA after 10 000 g and 100 000 g centrifugation. (c) Jurkat cells were incubated overnight in serum-free medium containing 50 mm NH4Cl, 100 μm chloroquine, 4 μm monensin, or 0·5 μm A23187. Post-10 000 g supernatant was analysed by NTA. The mean size of particles detected was 114 nm. (d) Jurkat cells were incubated overnight with the indicated concentrations of monensin and A23187. Post-10 000 g supernatants were analysed by NTA. The mean size of particles detected was 126 nm.
NTA permits quantification of induced microvesicle release
To determine if NTA allows the detection of increased microvesicle secretion into medium induced by a range of immune cell-modifying agents, we first incubated 1 million and 4 million Jurkat and CEM cells overnight in serum-free medium, then performed NTA on the post-10 000 g supernatant. As shown in Fig. 2(a), more microvesicles were detected from the sample containing more cells, with Jurkat cells releasing more microvesicles in comparison to CEM cells. The amount detected by NTA in the 4 million cell sample was in both cases below a factor of increase of four compared with the 1 million cell sample, which may reflect either incomplete linearity of the NTA detection system, or a balance in the cell cultures between microvesicle release and re-absorption by nearby cells.
We next tested the ability of reagents known to affect the endosomal/lysosomal pathway and also the secretion of exosomes,17 for their ability to be detected by NTA in conditioned medium. Jurkat cells were incubated overnight with the lysosome-influencing agents ammonium chloride, chloroquine and the ionophores monensin and A23187. Only monensin and A23187 produced significant increases upon NTA of the conditioned medium, of approximately 43% and 126%, respectively (Fig. 2c). Secretion of microvesicles was also concentration and time dependent (Fig. 2d and 3a).
To determine if the secreted vesicles being detected by NTA, induced by monensin and A23187, included exosomes, and were not therefore exclusively other types of membrane-derived vesicles, we performed immunblotting and mass spectrometry on vesicles isolated by 100 000 g centrifugation. Immunoblotting revealed an increase in both Alix and Tsg101 signal corresponding to the NTA signal changes (Fig. 3a,b), and mass spectrometric fingerprinting analysis of five bands excised from an SDS–PAGE gel of the 100 000 g microvesicle pellet from A23187-treated Jurkat cells revealed the presence of heat-shock protein 90, tubulin, elongation factor α1, actin and glyceraldehyde 3-phosphate dehydrogenase (labelled 1–5, respectively, in Fig. 3c), all of which have previously been reported to be associated with exosomes, based on searches of the ExoCarta database (http://www.exocarta.org).
Figure 3.
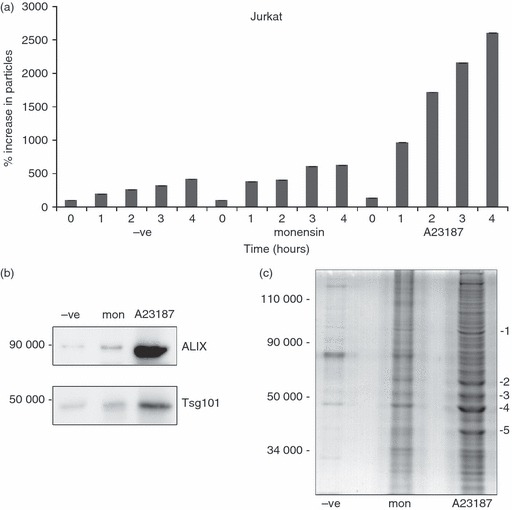
Ionophore treatment of Jurkat cells releases microvesicles containing exosome-associated polypeptides. Jurkat cells were treated for the indicated times with 4 μm monensin or 1 μm A23187. Post-10 000 g supernatants were analysed by nanoparticle tracking analysis (NTA). (b) Supernatants of Jurkat cells treated with the same concentration of ionophores as in (a) for 6 hr were spun at 10 000 g and then 100 000 g, and the latter pellets were analysed by immunoblotting for Alix and Tsg101 expression. The mean size of particles detected was 158 nm (c). Supernatants of Jurkat cells incubated for 6 hr as in (b) were spun at 10 000 g and 100 000 g and the latter pellets were analysed using SDS–PAGE and SimplyBlue protein staining. The indicated bands were excised and mass spectrometric fingerprinting was performed. Proteins identified included 1, heat-shock protein 90; 2, tubulin; 3, elongation factor α1; 4, actin; and 5, glyceraldehyde 3-phosphate dehydrogenase.
Immunodepletion of NTA signal by antibody-coupled magnetic beads
To determine if the microvesicle population could be depleted by the selective use of antibody-coupled microbeads, we incubated post-10 000 g supernatants from Jurkat cells overnight with increasing amounts of anti-human CD45-coupled magnetic beads. Approximately 60% of the NTA signal could be removed by this method (Fig. 4a). Hence, antibody depletion can be used to isolate specific microvesicle populations during NTA of secreted microvesicles.
Figure 4.
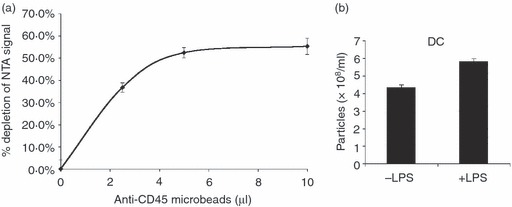
Immunodepletion of microvesicles and nanoparticle tracking analysis (NTA) of lipopolysaccharide (LPS) treated monocyte-derived dendritic cells (DC). (a) Jurkat supernatants were incubated with the indicated amounts of anti-human CD45 magnetic beads overnight, before NTA. Data represent samples measured in triplicate with mean and standard deviation plotted. The data are representative of three separate experiments. (b) Human peripheral blood monocytes cultured in interleukin-4/granulocyte–macrophage colony-stimulating factor for 5 days to generate DC were incubated overnight in serum-free medium containing 50 ng/ml LPS. Post-10 000 g supernatants were analysed by NTA.
NTA detects microvesicle secretion from monocyte-derived dendritic cells
Exosomes from dendritic cells are important immunomodulatory agents.7,8 We isolated blood monocyte-derived dendritic cells using interleukin-4/granulocyte–macrophage colony-stimulating factor treatment, and determined their microvesicle secretion pattern upon stimulation with bacterial LPS. Overnight treatment with LPS induced an increase in the number of microvesicles detected by NTA in post-10 000 g conditioned medium (Fig. 4). Hence, NTA can be used to detect increases in the levels of secretion of exosomes and microvesicles upon stimulation of a variety of cells of the immune system.
Discussion
Microvesicles, including those known as exosomes, are now known to display a wide range of biological activities, for example the capacity to be both immunostimulatory and immunosuppressive,7,8 the ability to show immuno-evasion by causing T-cell apoptosis,14,15 the down-regulation of receptors on natural killer cells,18 and the capacity to promote angiogenesis, potentially increasing tumour metastasis.19 They are therefore of potential importance as biomarkers for disease and as therapeutic agents. The ability to rapidly monitor the presence, size and quantity of microvesicles in both cell cultures and in clinically relevant body fluids would be a great advantage. NTA would appear to be a suitable candidate technology for this purpose.
In this current study we have used two T-cell lines to study the ability of NTA to detect increases in the secretion of microvesicles into tissue culture medium after a variety of stimuli. The ionophores monensin and A23187, because of their ability to raise intracellular calcium levels, and also bacterial LPS have been shown to induce exosome and microvesicle secretion in relevant cells.17 Our data on dendritic cells indicate an LPS-induced increase in microvesicle release, which is in contrast to previous reports,20 and which may indicate significant differences in the measurement techniques used, and requires further study into its implications.
The rapidity of NTA shows distinct advantages over such current methods of microvesicle analysis as absorption to latex beads, followed by flow cytometry, or isolation by ultracentrifugation and analysis by immunoblotting or electron microscopy.13 We have also demonstrated the ability to reduce the NTA signal by immunodepletion of microvesicles, in this case using anti-human CD45 magnetic beads.21 Interestingly, we were only able to deplete approximately 50–60% of the NTA signal in a series of experiments. This may indicate either that the microvesicle population is polymorphic, or it may also demonstrate that, because of the small size of such microvesicles, that not every particle will contain all the canonical markers for that particular subset of microvesicles. Hence, the use of several markers to better define each subset might be required. We attempted to use a similar approach with anti-CD9 and anti-CD63 antibodies, but these were unsuccessful because of the very low levels of these markers expressed in the T-cell lines used in this current study.
The detection of exosomes and microvesicles by NTA is, however, not without its own challenges. Discrimination between the particles being detected, that is, between true exosomes and other membrane microvesicles, would be beneficial. A recent advance in the ability to incorporate fluorescence detection into NTA might provide an answer to this problem. Hence, using either fluorescently coupled antibodies or antibody-conjugated quantum dots might allow the detection of subsets of microvesicles within a sample.16 A further problem that must be taken into consideration is background signal. The use of serum-free medium in this current study was required because serum supplements (such as fetal calf serum) can often contain vesicles, which may therefore require ultracentrifugation before their addition to the culture medium. Aggregates of serum proteins may also provide spurious signals. However, not all cell systems can tolerate significant lengths of time in serum-free conditions without adverse reactions being induced, which could further influence NTA results.
Using NTA may provide other valuable information to those working in the exosome and microvesicle field. For example, in most current systems, the exosomal input into the experiment is usually reported in terms of micrograms protein, which can often range from a few to several hundred micrograms.14,22 The precise number of microvesicles being added in each system is therefore completely unknown. For the first time, NTA in conjunction with protein determinations could be used to report how many microvesicles are present per microgram of a typical sample, and therefore how many microvesicles are being added to the experimental system. Such information is likely to lead to a better standardization of systems. Furthermore, at present it is unknown what physical state the microvesicle preparations are in before their addition to a system. NTA of a small sample of any given preparation would provide information on whether a sample is in a monodispersed or aggregated condition, which may significantly impact on a biological system. Similarly, reporting the average size of the particles detected will also be of use in determining the quality of microvesicle preparations. In this study the particles detected ranged from c.70 to 160 nm, which is the expected size range for exosomes and microvesicles.1,2 Some variation was noted depending on the software settings chosen in the analysis program, and at higher drug treatment concentrations of A23187 we noted morphological appearance changes consistent with the onset of apoptosis. Again, standardization across multiple users will assist in generating standard operating procedures for the use of NTA.
In summary, we have successfully demonstrated the ability of NTA to follow increases in the release of microvesicles in the culture medium of immune cells undergoing activation. This technique is likely to be of significant advantage in the monitoring of exosomes and microvesicles in a variety of immune-based experimental systems.
Acknowledgments
This work was supported in part by the Chief Scientist Office (CSO) of the Scottish Government (E.C.C), by the Scottish Universities Life Science Alliance (SULSA), and by Wolfson Foundation support to C.Y.S.
Disclosures
No competing financial interests exists.
References
- 1.Gyorgy B, Szabo TG, Pasztoi M, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68:2667–88. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–93. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 3.Murk JL, Stoorvogel W, Kleijmeer MJ, Geuze HJ. The plasticity of multivesicular bodies and the regulation of antigen presentation. Semin Cell Dev Biol. 2002;13:303–11. doi: 10.1016/s1084952102000605. [DOI] [PubMed] [Google Scholar]
- 4.Clayton A, Mitchell JP, Court J, Mason MD, Tabi Z. Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res. 2007;67:7458–66. doi: 10.1158/0008-5472.CAN-06-3456. [DOI] [PubMed] [Google Scholar]
- 5.Lehmann BD, Paine MS, Brooks AM, McCubrey JA, Renegar RH, Wang R, Terrian DM. Senescence-associated exosome release from human prostate cancer cells. Cancer Res. 2008;68:7864–71. doi: 10.1158/0008-5472.CAN-07-6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 7.Kim SH, Lechman ER, Bianco N, et al. Exosomes derived from IL-10-treated dendritic cells can suppress inflammation and collagen-induced arthritis. J Immunol. 2005;174:6440–8. doi: 10.4049/jimmunol.174.10.6440. [DOI] [PubMed] [Google Scholar]
- 8.Viaud S, Terme M, Flament C, et al. Dendritic cell-derived exosomes promote natural killer cell activation and proliferation: a role for NKG2D ligands and IL-15Rα. PLoS ONE. 2009;4:e4942. doi: 10.1371/journal.pone.0004942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai S, Wei D, Wu Z, Zhou X, Wei X, Huang H, Li G. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol Ther. 2008;16:782–90. doi: 10.1038/mt.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Escudier B, Dorval T, Chaput N, et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of the first phase I clinical trial. J Transl Med [electronic resource] 2005;3:10. doi: 10.1186/1479-5876-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morse MA, Garst J, Osada T, et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J Transl Med [electronic resource] 2005;3:9. doi: 10.1186/1479-5876-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–5. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 13.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006 doi: 10.1002/0471143030.cb0322s30. ; Chapter 3:Unit 3 22. [DOI] [PubMed] [Google Scholar]
- 14.Abusamra AJ, Zhong Z, Zheng X, Li M, Ichim TE, Chin JL, Min WP. Tumor exosomes expressing Fas ligand mediate CD8+ T-cell apoptosis. Blood Cells Mol Dis. 2005;35:169–73. doi: 10.1016/j.bcmd.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Andreola G, Rivoltini L, Castelli C, et al. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. J Exp Med. 2002;195:1303–16. doi: 10.1084/jem.20011624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dragovic RA, Gardiner C, Brooks AS, et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine. 2011;7:780–8. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savina A, Furlan M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem. 2003;278:20083–90. doi: 10.1074/jbc.M301642200. [DOI] [PubMed] [Google Scholar]
- 18.Clayton A, Mitchell JP, Court J, Linnane S, Mason MD, Tabi Z. Human tumor-derived exosomes down-modulate NKG2D expression. J Immunol. 2008;180:7249–58. doi: 10.4049/jimmunol.180.11.7249. [DOI] [PubMed] [Google Scholar]
- 19.Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thery C, Regnault A, Garin J, Wolfers J, Zitvogel L, Ricciardi-Castagnoli P, Raposo G, Amigorena S. Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J Cell Biol. 1999;147:599–610. doi: 10.1083/jcb.147.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coren LV, Shatzer T, Ott DE. CD45 immunoaffinity depletion of vesicles from Jurkat T cells demonstrates that exosomes contain CD45: no evidence for a distinct exosome/HIV-1 budding pathway. Retrovirology. 2008;5:64. doi: 10.1186/1742-4690-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70:9621–30. doi: 10.1158/0008-5472.CAN-10-1722. [DOI] [PubMed] [Google Scholar]


