Global regulatory circuits in bacteria are most often controlled via transcriptional activators or architectural proteins modulating transcription, such as RpoS, H-NS, or Fis (1). In recent years, a class of diverse regulatory RNAs (often denoted riboregulators) has emerged that regulate expression at the posttranscriptional level, adding additional complexity to the interplay of factors involved in global control. In fact, some posttranscriptionally regulating RNAs affect proteins, which in turn act as posttranscriptional regulators of other genes (2). These regulatory RNAs seem to fine tune cellular responses to stress conditions, integrating environmental signals into global regulation. The biological function and mechanism of action of one of these riboregulators, DsrA, is the subject of a study by Lease and Belfort in this issue of PNAS (3). A striking feature of DsrA and a few other small regulatory RNAs such as OxyS (4) is that they can affect more than one target gene. Moreover, RNAs such as DsrA and OxyS use different mechanisms on different and probably multiple target RNAs either to inhibit or activate. In this commentary, we will discuss general properties of riboregulators, their diverse mechanisms of action, and their functional significance. In our view, a deeper understanding of the biological roles of regulatory RNAs will have to account for common themes as well as instructive differences in the activities displayed.
DsrA.
The small RNA DsrA was first discovered through its regulatory effect on colanic acid capsule synthesis. DsrA acts in trans to activate and repress, respectively, the synthesis of two transcriptional regulators, HN-S, a major histone-like protein responsible for silencing of a number of bacterial genes, and RpoS (σs), the stationary phase σ-factor of RNA polymerase. Regulation of both hns and rpoS by DsrA is mediated by base-pairing interactions between short RNA sequences. In this issue of PNAS, Lease and Belfort (3) show that DsrA RNA affects the stability of both rpoS and hns mRNAs; turnover of hns mRNA is increased, whereas rpoS mRNA is stabilized, either directly or indirectly. Structure probing of DsrA RNA confirms the previously predicted stem-loops 1 and 3, whereas a modified structure is proposed for stem-loop 2. Unexpectedly, structure determination of DsrA in the presence of hns mRNA suggested a second site of interaction between the 3′ end of hns and the loop nucleotides of the second stem–loop structure. Lease and Belfort propose that base pairing between DsrA and the two regions of hns, near the 5′ and 3′ ends of its coding region, results in a contiguous coaxial stack, looping out the middle part of hns mRNA exposed to nucleases.
Regulatory RNAs: Different Modes of Control.
The main function of regulatory RNAs is posttranscriptional regulation of gene expression. By this definition, a small number of untranslated RNAs in bacteria qualify (5, 6). Regulatory RNAs vary in the mechanisms by which they affect their target mRNAs. Some RNAs accomplish regulation by modulating the activity of proteins (6). The vast majority, often referred to as antisense RNAs, act through base pairing with target RNA (7, 8). Below we will focus on the class of riboregulators that regulates by antisense mechanisms.
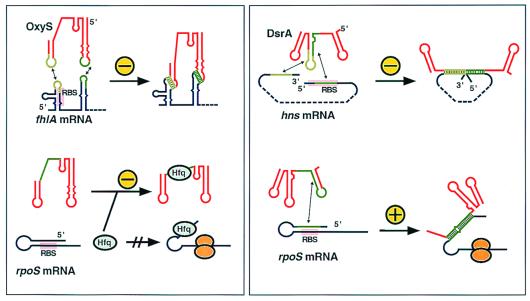
A first interesting observation is that base pairing between two RNAs can have different consequences: negative regulation by antisense RNAs is the rule, but at least two interesting exceptions show that activation is also possible. DrsA, in one of its two regulatory modes, activates translation of the rpoS mRNA by base pairing to a sequence that otherwise forms an inhibitory intramolecularly base-paired structure (refs. 9–11; Fig. 1). A similar mechanism had been proposed for the agr virulence control locus of Staphylococcus aureus. This locus encodes a small RNA, RNAIII, that pairs with an antiribosome-binding site within the hla (α-toxin) mRNA to activate translation (12).
Figure 1.
Regulation of target RNAs by OxyS and DsrA. The left box shows a schematics of inhibition mechanisms of OxyS and the right box inhibition by DsrA. Target RNAs are shown in blue and antisense RNAs in red. Interacting regions are indicated in green. For clarity, different shades of green are used in the bipartite interactions in the upper two drawings. “+” and “−” indicate activation and inhibition, respectively. RBS indicates the approximate position of the ribosome-binding site. Hfq is host factor Q (see text), and orange mushrooms represent translating ribosomes.
Multiple Targets.
The majority of antisense RNAs were identified in plasmids, transposons, and bacteriophages. In all of these cases, the RNA-encoding gene overlaps with the target gene (cis encoded). In this class, complete complementarity between the RNAs is therefore ensured, and only one target gene is regulated. By contrast, only a small number of antisense RNA genes have been identified in bacterial chromosomes. Most of these are trans encoded, at genetic loci other than their target genes. Therefore, antisense-target complementarity is incomplete, and regulation must be achieved by formation of partial and imperfect duplexes. In Escherichia coli four small RNAs with these characteristics, DicF, MicF, OxyS, and DsrA, have been suggested to regulate gene expression (5). Because these RNAs are trans encoded and contain only short regions of target complementarity, this could facilitate recognition of multiple targets. The example of two riboregulators, OxyS and DsrA, may serve to illustrate this property. OxyS is a small RNA that is induced in response to oxidative stress. It acts as a global regulator by affecting the expression of multiple genes and protecting against DNA damage (4). As schematically indicated in Fig. 1, both OxyS and DsrA carry separate determinants for regulation of each of their two different target RNAs. OxyS inhibits the translation of fhlA, a transcriptional activator for formate metabolism by an antisense mechanism (13), and rpoS translation is repressed indirectly by titration of Hfq, a host factor required for melting out of an inhibitory mRNA structure (14). As indicated in Fig. 1 (Left, light and dark green lines), different regions of OxyS are involved in these activities. DsrA uses an antisense mechanism for both up-regulation of rpoS and down-regulation of hns. Mutational analyses have defined different segments of DsrA as the modules involved in target interactions (refs. 10 and 11; Fig. 1 Right).
Bipartite Interactions.
To add further complexity, both OxyS and DsrA bind their target mRNAs, fhlA and hns, respectively, by bipartite interactions. Short antisense sequences exposed in two loops of OxyS contact two separate target RNA regions more than 40 nucleotides apart. Recent results have indicated the importance of both interactions for OxyS-target binding as well as for blocking ribosome access to the fhlA translation initiation site (15). By contrast, one extended sequence of DsrA base pairs with two regions within its target RNA, one located at the start of the coding region and one at its end (3). This results in a structure in which the two contiguous antisense regions in DsrA align the two target regions to form a coaxial stack, looping out the hns mRNA body (Fig. 1). This model is tentatively supported by structure probing of the hns-DsrA complex in vitro and by computer-aided complementarity searches; Lease and Belfort show that two other suspected targets, ilvI and argR mRNAs, could form similar coaxially stacked structures. So far, it is unclear whether these latter two are regulated by DsrA. As yet, Lease and Belfort's attractive structure model awaits an in vivo confirmation of the biological importance of the second (DsrA/hns-3′-end) base-pairing interaction.
A challenging question also concerns the primary effect of DsrA-hns mRNA binding. DsrA induction has now been demonstrated to decrease the half life of hns mRNA, as well as to render rpoS mRNA more stable. However, as the authors point out, it cannot be decided whether altered mRNA degradation rates simply reflect the protective presence or absence of translating ribosomes, or whether the proposed looped-out structure of hns by itself exposes the RNA for accelerated decay.
Base Pairing: How Much Is Needed?
OxyS and DsrA carry out their functions by forming a relatively modest number of base pairs as determined by the region of complementarity [8 + 11 (for both helices; see Fig. 1), DsrA-hns; 7 + 9, OxyS-fhlA]. The stereotypical copy number regulators of, e.g., plasmids ColE1, R1, pIP501 and others (7), are completely complementary to their targets over 90–140 nucleotides. At first glance, one might expect the more the better. However, association rates, not the thermodynamic stability (ΔG) of a complex between antisense and target RNA, constitute the most important parameters for inhibitory efficiency (16). OxyS forms complexes with fhlA mRNA characterized by a dissociation constant of 25 nM (15). The association rate constant was determined to be ≈6 × 105 M−1⋅s−1, i.e., similar to that of the plasmid antisense systems. This implies that formation of the presumed inhibitory kissing complex is very rapid, dissociation occurs at a half life of >1 min, and the Kd suggests that as few as 100 molecules per cell will inhibit the majority of the RNA targets. Thus, relatively short sequences can certainly suffice for efficient binding and regulation, provided they can interact rapidly with exposed target sequences. Furthermore, results obtained in recent years have shown clearly that even antisense RNAs that were expected to hybridize fully with targets fail to do so. For both plasmid R1 (17) and ColIb-P9 (18), the inhibitory complex is a peculiar cruciform-like structure stabilized by an additional intermolecular helix. Thus, the topological difficulties of unwinding stable stem–loop structure may have selected solutions in which more or less extensive kissing complexes are used for regulation (19).
Dynamic Structures.
It has been known for years that changing RNA conformations can be used for regulatory processes. The DsrA–rpoS interaction represents such a case in E. coli, as does RNAIII–hla in Staphylococcus aureus. In both cases, the mRNA can adopt two different conformations. One of these, the translation-competent conformation, is induced by binding of the effector RNA. Similar conformational switches were demonstrated in plasmids pT181, pIP501 (and relatives), and the ColE1 family (20–22). In pT181 and pIP501, the antisense RNA induces premature termination of a replication protein mRNA to regulate copy. In ColE1, the antisense RNA induces an altered folding pathway of the preprimer so that its maturation is inhibited. Notably, the shared feature of these mechanisms is an activity window; the antisense RNA has to bind during transcription to achieve inhibition. After the target RNA has been elongated beyond this window antisense binding fails to inhibit. For the two riboregulators DsrA and RNAIII (of agr), it is unknown whether activation of the target RNA also occurs cotranscriptionally, although it appears reasonable to assume that this should be the case.
Even the coaxial stacking model proposed for negative regulation by DsrA-hns mRNA interaction suggests, to us, cotranscriptional initiation of binding. According to the revised structure model of DsrA, the region of complementarity to the hns 3′-region is exposed in the middle loop and might be expected to initiate binding (3). However, at a time point at which the 3′-end has been transcribed, translation of the 5′-portion of the hns message should already have occurred, because bacterial mRNAs are almost always translated while being transcribed. This would render inhibition inefficient and eventually also result in disruption of the DsrA/3′-end interaction through ongoing translation. Therefore, it is parsimonious to assume that the nascent mRNA is bound at its 5′-end site first, probably facilitated by breathing of the DsrA bottom stem near the bulge in the middle stem (ref. 3; Fig. 1). This should block new translational initiation events and subsequently allow for the second interaction as this region of the mRNA emerges from the transcribing polymerase.
Why Use RNAs as Regulators?
An intriguing question with respect to studies of DsrA (3) and other riboregulators is: Why use RNAs instead of proteins? From so far limiting phylogenetic comparisons, it appears that riboregulators often have orthologs in other bacteria, e.g., DsrA (3, 10) and MicF (23). This suggests functional importance but does not address why RNAs are used for control. A commonly used argument is economy: because these RNAs are small and untranslated, the energetic cost of their synthesis is much lower than for a protein whose mRNA has to be synthesized and decoded into a chain of amino acids. Thus, a stable RNA should be more cost effective than a protein. Chromosomally encoded riboregulators are stable RNAs with half lives usually exceeding ≈15 min (e.g., refs. 4 and 10). Even transposase-encoded antisense RNAs such as RNA-OUT (24) are moderately stable. By contrast, almost all copy number regulator RNAs as well as the antisense RNAs that control postsegregational killing are very unstable, with half lives of typically around one minute (25, 26). Hence, it is conceivable that economy is only part of the design principle. The other part could be design for the particular biological role. This implies that riboregulators whose synthesis is induced when required, e.g., during oxidative stress (OxyS; ref. 4), low temperature (DsrA; ref. 9), or various other stress conditions (MicF; ref. 27), should be stable throughout the stress response. Plasmid copy number regulators, on the other hand, have to “count.” To measure plasmid copies and to regulate initiation of replication accordingly, the intracellular concentration of antisense RNAs like, e.g., RNAI of ColE1 and CopA of R1, should at all times be proportional to the concentration of their genes (equivalent to plasmid concentration). Because the number of plasmid copies fluctuates, regulation requires rapid changes in inhibitor (antisense RNA) concentration, i.e., constitutive synthesis and rapid decay (19). Thus, even though it may seem wasteful to synthesize regulatory RNAs that are subject to rapid degradation, the biological function of this class of regulators requires precisely these properties.
Organisms are often exposed to environmental changes and various stress conditions. Being able to respond to those changes and to adapt to changing environments require regulation at many levels. We have seen that small RNAs take their share in the fine tuning of responses, and we suspect that many more regulatory circuits rely on riboregulators. An important task is therefore the development of appropriate screening procedures to discover more members of this category. Concerning the reason why organisms use riboregulation, it seems to us that the diversity of structures, mechanisms, and biological roles displayed by the known regulatory RNAs suggests that the versatility of RNA, rather than a single unifying feature like cost effectiveness, may have led to its ubiquitous use in regulatory processes.
Acknowledgments
We acknowledge financial support from the Human Frontier Science Program.
Footnotes
See companion article on page 9919.
References
- 1.Hengge-Aronis R. Curr Opin Microbiol. 1999;2:148–152. doi: 10.1016/S1369-5274(99)80026-5. [DOI] [PubMed] [Google Scholar]
- 2.Nogueira T, Springer M. Curr Opin Microbiol. 2000;3:154–158. doi: 10.1016/s1369-5274(00)00068-0. [DOI] [PubMed] [Google Scholar]
- 3.Lease R A, Belfort M. Proc Natl Acad Sci USA. 2000;97:9919–9924. doi: 10.1073/pnas.170281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 5.Wassarman K M, Zhang A, Storz G. Trends Microbiol. 1999;7:37–45. doi: 10.1016/s0966-842x(98)01379-1. [DOI] [PubMed] [Google Scholar]
- 6.Wagner, E. G. H., Altuvia, S. & Nellen, W. (2000) Encyclopedia of Life Sciences, in press.
- 7.Wagner E G H, Simons R W. Annu Rev Microbiol. 1994;48:713–742. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]
- 8.Zeiler B N, Simons R W. In: RNA Structure and Function. Simons R W, Grunberg-Manago M, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1998. pp. 437–464. [Google Scholar]
- 9.Sledjeski D D, Gupta A, Gottesman S. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 10.Majdalani N, Cunning C, Sledjeski D D, Elliott T, Gottesman S. Proc Natl Acad Sci USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lease R A, Cusick M E, Belfort M. Proc Natl Acad Sci USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morfeldt E, Taylor D, von Gabain A, Arvidson S. EMBO J. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altuvia S, Zhang A, Argaman L, Tiwari A, Storz G. EMBO J. 1998;17:6069–6075. doi: 10.1093/emboj/17.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Argaman L, Altuvia S. J Mol Biol. 2000;300:1103–1114. doi: 10.1006/jmbi.2000.3942. [DOI] [PubMed] [Google Scholar]
- 16.Nordström K, Wagner E G H. Trends Biochem Sci. 1994;19:294–300. doi: 10.1016/0968-0004(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 17.Kolb F A, Malmgren C, Westhof E, Ehresmann C, Ehresmann B, Wagner E G H, Romby P. RNA. 2000;6:311–324. doi: 10.1017/s135583820099215x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asano K, Mizobuchi K. J Biol Chem. 2000;275:1269–1274. doi: 10.1074/jbc.275.2.1269. [DOI] [PubMed] [Google Scholar]
- 19.Wagner E G H, Brantl S. Trends Biochem Sci. 1998;23:451–454. doi: 10.1016/s0968-0004(98)01322-x. [DOI] [PubMed] [Google Scholar]
- 20.Novick R P, Iordanescu S, Projan S J, Kornblum J, Edelman I. Cell. 1989;59:395–404. doi: 10.1016/0092-8674(89)90300-0. [DOI] [PubMed] [Google Scholar]
- 21.Brantl S, Birch-Hirschfeld E, Behnke D. J Bacteriol. 1993;175:4052–4061. doi: 10.1128/jb.175.13.4052-4061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eguchi Y, Itoh T, Tomizawa J. Annu Rev Biochem. 1991;60:631–652. doi: 10.1146/annurev.bi.60.070191.003215. [DOI] [PubMed] [Google Scholar]
- 23.Esterling L, Delihas N. Mol Microbiol. 1994;12:639–646. doi: 10.1111/j.1365-2958.1994.tb01051.x. [DOI] [PubMed] [Google Scholar]
- 24.Pepe C M, Maslea-Gali S, Simons R W. Mol Microbiol. 1994;13:1133–1142. doi: 10.1111/j.1365-2958.1994.tb00504.x. [DOI] [PubMed] [Google Scholar]
- 25.Söderbom F, Wagner E G H. Microbiology. 1998;144:1907–1917. doi: 10.1099/00221287-144-7-1907. [DOI] [PubMed] [Google Scholar]
- 26.Dam Mikkelsen N, Gerdes K. Mol Microbiol. 1997;26:311–320. doi: 10.1046/j.1365-2958.1997.5751936.x. [DOI] [PubMed] [Google Scholar]
- 27.Coyer J, Andersen J, Forst S A, Inouye M, Delihas N. J Bacteriol. 1990;172:4143–4150. doi: 10.1128/jb.172.8.4143-4150.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]