Abstract
Protein polymers are repetitive polypeptides produced by ribosomal biosynthetic pathways; furthermore, they offer emerging opportunities in drug and biopharmaceutical delivery. As for any polymer, biodegradation is one of the most important determinants affecting how a protein polymer can be utilized in the body. This study was designed to characterize the proteolytic biodegradation for a library of protein polymers derived from the human tropoelastin, the Elastin-like polypeptides (ELPs). ELPs are of particular interest for controlled drug delivery because they reversibly transition from soluble to insoluble above an inverse phase transition temperature (Tt). More recently, ELP block copolymers have been developed that can assemble into micelles; however, it remains unclear if proteases can act on these ELP nanoparticles. For the first time, we demonstrate that ELP nanoparticles can be degraded by two model proteases and that comparable proteolysis occurs after cell uptake into a transformed culture of murine hepatocytes. Both elastase and collagenase endopeptidases can proteolytically degrade soluble ELPs. To our surprise, the ELP phase transition was protective against collagenase but not to elastase activity. These findings enhance our ability to predict how ELPs will biodegrade in different physiological microenvironments and are essential to develop protein polymers into biopharmaceuticals.
Keywords: elastin-like polypeptides, protein polymers, inverse phase transition temperature, biodegradation, hepatocytes, proteolysis, critical micelle temperature, nanoparticles, micelles, elastase
Introduction
Proteolysis has been adapted for various purposes in the fields of drug delivery and biomaterials.1 Many proteases maintain biological homeostasis; however, protease misregulation also causes upregulation of cathepsins and matrix metalloproteinases observed in solid tumors.2 As such, the proteolytic environment can be used to design peptide-based prodrugs that facilitate targeted drug activity.3 The purpose of this study is to identify the effects of some of these proteases on an emerging class of protein-based polymers called Elastin-like polypeptides (ELPs).4, 5
ELPs are unique biopolymers inspired from human tropoelastin that consist of pentameric repeats of (Val-Pro-Gly-Xaa-Gly)n where Xaa is a “guest” residue. ELPs undergo reversible inverse phase transition wherein they are soluble below their transition temperature (Tt) and cluster into aggregates above their Tt.6 Changing the ELP length, n, and the guest residue, Xaa, enables accurate and reproducible control of Tt. ELPs with different Tt can be fused into block copolymers that assemble micelle nanoparticles.4 Block copolymers are emerging as effective drug delivery vehicles because of their ability to self-assemble into nanostructures ranging in scale from tens to thousands of nanometers.7 Similarly, amphiphilic ELP diblock copolymers can form stable spherical micelles.8 ELP micelles may be promising drug carriers, as they have been used successfully to deliver chemotherapeutic cargo to a murine colon carcinoma model.4 ELPs have also been explored as tissue engineering substrates.9 For applications in drug delivery or tissue engineering, the mechanism, and efficiency of proteolytic biodegradation is expected to be a key determinant of their success.
The rate of biodegradation controls accumulation/retention at the target site as well as clearance from the body. The liver plays a central role in clearing endogenous and exogenous peptide-based fragments from the body as well as nanoparticles10; therefore, we decided to determine if ELP nanoparticles can be biodegraded inside a monolayer of transformed murine hepatocytes. We further investigate the possibility that the ELP phase separation provides a general blockade against proteases, as has been suggested previously.11 To explore this question, we compared the biodegradation of di and monoblock ELPs with low and high Tt, under incubation with two model proteases. The ELPs employed had different lengths and hydrophobicities; furthermore, we also modulated their phase separation using salt, sodium sulfate. We observed that ELP assembly provides no protection against elastase; however, ELP assembly does protect against collagenase. These findings support the contention that ELP therapeutics can be substrates for hepatic biodegradation and clearance.
Results
Characterization of ELPs
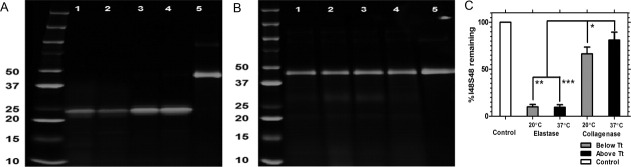
A small library of ELPs was prepared, purified and characterized (Table I) for the purpose of observing the degradation trends for both soluble, insoluble, and nanoparticle ELPs. ELPs remain soluble below their transition temperatures (Tt).12 ELP nanoparticles assemble water-soluble micelles above their critical micelle temperature (CMT). The phase diagram for the S48I48 diblock copolymer is presented [Fig. 1(A)] and the formation of micelles was confirmed using Dynamic Light Scattering (DLS) [Fig. 1(B)]. S48I48 displays two transition temperatures, one at ∼27°C and the other at ∼75°C. The lower Tt represents a CMT indicative of the assembly of the isoleucine block, and the second Tt,bulk is a bulk phase separation caused by association of the serine block. As the temperature was increased from 10 to 37°C, the diblock monomers clearly assemble monodisperse nanoparticles (Rh = 23.7 ± 0.7SD nm) [Fig. 1(C)].8, 13
Table I.
ELP Protein Polymers Subjected to Proteolysis
| Label | Amino acid sequence | aPurity (%) | CMT (°C) | bTt,bulk (°C) | cExpected mass (g mol−1) | dObserved mass (g mol−1) |
|---|---|---|---|---|---|---|
| I96 | G(VPGIG)96Y | 97.7 | n.a. | 11.5 | 40,895 | 40,638 ± 25 |
| V96 | G(VPGVG)96Y | 97.3 | n.a. | 31.9 | 39,548 | 39,514 ± 37 |
| S96 | G(VPGSG)96Y | 92.4 | n.a. | 62.6 | 38,391 | 38,220 ± 3 |
| I48S48 | G(VPGIG)48(VPGSG)48Y | 99.4 | 26 | 87.4 | 39,643 | 39,575 ± 28 |
| S48I48 | G(VPGSG)48(VPGIG)48Y | 99.2 | 27 | 75.7 | 39,643 | 39,541 ± 20 |
| V48 | G(VPGVG)48Y | 98.7 | n.a. | 42.4 | 20,024 | 20,009 ± 11 |
Polypeptide purity was assessed using SDS-PAGE and subsequent densitometry of copper chloride stained gel.
Inverse transition temperatures, Tt, were analyzed using optical density (350 nm) measurements at a polypeptide concentration of 25 μM.
Expected mass of polypeptide (Mean ± SD in Da).
Data collected using matrix assisted laser desorption ion time of flight mass spectrometry.
Figure 1.
Assembly and biodegradation of elastin-like polypeptide nanoparticles in a hepatocyte monolayer culture. A: Optical density measurements (350 nm) were used to define the temperature-concentration phase diagram for the polypeptide block copolymer S48I48 in phosphate buffered saline. Both the critical micelle temperature (CMT) and bulk transition temperature (Tt,bulk) followed a log-linear correlation with concentration. R2 = 0.903, 0.999 respectively. B: Dynamic light scattering confirmed that these block copolymers assemble monodisperse polypeptide nanoparticles (Rh = 23.7 ± 0.7 nm, mean ± SD) above 27°C. C: Particle distribution histogram depicting monodispersity of S48I48 polymer (10, 25°C) and nanoparticles (37°C). D: These micelles are biodegradable in cell culture. A rhodamine-conjugated ELP, Rh-S48I48, was incubated with a monolayer of transformed murine hepatocytes for intervals from 0 to 48 h. Fluorescently labeled ELPs associated with the cell monolayer were extracted and resolved using SDS-PAGE; furthermore, the disappearance of the ∼40 kDa band over time with respect to a nonincubated control suggests biodegradation of the nanoparticles. The dye front for free rhodamine is also indicated.
Cellular uptake and degradation of the ELP nanoparticles
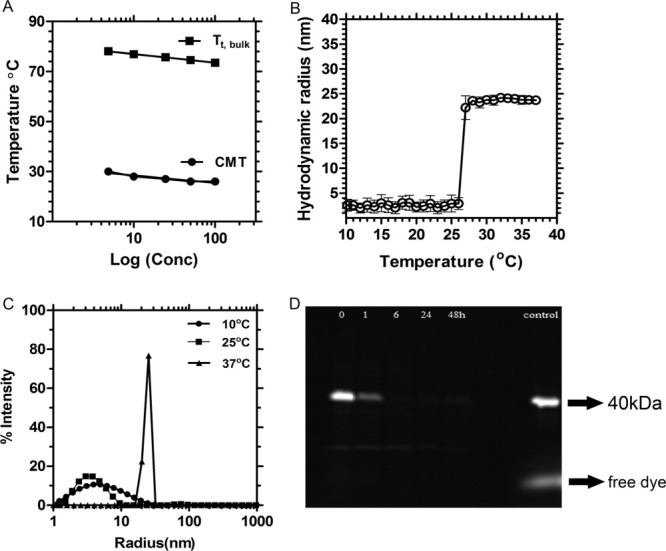
When incubated with cultured mouse hepatocytes, the 40 kDa band of Rh-S48I48 disappears over time, which suggests biodegradation [Fig. 1(D)]. To further explore this possibility, this incubation was observed directly using live cell microscopy (Fig. 2). There was a significant amount of uptake of Rh-S48I48 after 2 h (presence of red punctate spots within the cell); however, some S48I48 remains bound on the surface. Lysotracker green was used to stain the low pH compartments inside the cells. The ELP colocalized with lysotracker green, which suggests that it may be in contact with the proteolytically active lysosomal environment. As the incubation time increased, the amount of material bound at the surface continued to decrease, which suggests continued cellular uptake. At 24 h, the signal from the Rh-S48I48 colocalized with low pH compartments also decreased, which suggests intracellular proteolysis and clearance of the dye.
Figure 2.
Uptake and degradation of elastin-like polypeptide nanoparticles in transformed hepatocytes. Rhodamine-conjugated S48I48 (red) was incubated with transformed murine hepatocytes for 0, 2, 4, 24 h at 37°C and imaged using live-cell confocal microscopy. Cells were counterstained with lysotracker green to indicate the presence of low pH compartments; where, lysosomal proteases are active. Differential interference contrast (DIC) imaging was used to illustrate cellular morphology. ELP nanoparticles were visible both inside and on the cell surface after 2 h. At later time-points, there was a noticeable decrease in surface staining; furthermore, the intracellular staining also significantly decreased after 24-h incubation. The arrow indicates internalized nanoparticles with the lysosomes. Scale bar: 10 μm.
Proteolytic degradation of ELP nanoparticles
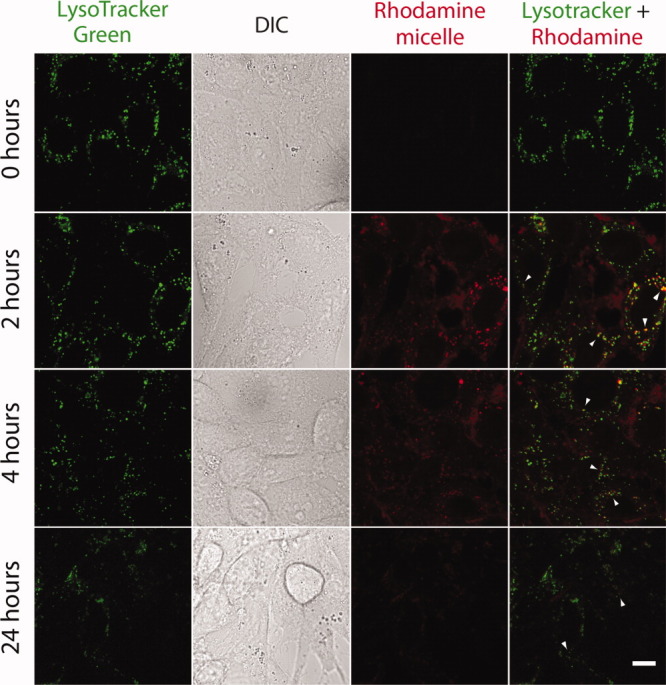
Soluble ELPs are susceptible to degradation by elastase and collagenase;14 therefore, we selected these two model proteases to determine if nanoparticle assembly influences proteolysis. As seen in Figure 3(A), I48S48 was readily degraded by elastase at both 20 and 37°C, which are below and above the CMT (Table I). Micelle assembly had no effect on elastase degradation, with the starting 40 kDa bands disappearing after 2 h. Instead of a single band, the degradation products showed up on the electrophoretic gel as a smear of several different fragments ranging from 10 to 25 kDa. Co-incident with the disappearance of the 40 kDa band, the appearance of a ∼25 kDa raised the possibility that one of the two ELP blocks remained untouched by elastase. Two pieces of data rule out this possibility: i) elastase completely digests both the S96 and I96 monoblock ELP (Fig. 4) at 37°C; and ii) digestion of the V96 band also produces a ∼25 kDa band [Fig. 4(A)].
Figure 3.
Degradation of elastin-like polypeptide nanoparticles depends on protease identity. Proteolytic degradation of the ELP block copolymer CFSE-I48S48 was assessed using SDS-PAGE and fluorescent imaging. Digestion was performed above (37°C) and below (20°C) the CMT to determine if self-assembly protects against proteolysis. Images were then quantified for the integrated intensity of the starting band in triplicate for all conditions. A: Proteolysis in presence of elastase after 2 h. Lane 1: 0.35 units at 20°C; 2: 0.7 units at 20°C; 3: 0.35 units at 37°C; 4: 0.7 units at 37°C; 5: control polypeptide. B: Proteolysis in presence of collagenase after 2 h. Lane 1: 3.6 units at 20°C; 2: 6.0 units at 20°C; 3: 3.6 units at 37°C; 4: 6.0 units at 37°C; 5: control polypeptide. C: Image analysis was used to normalize the integrated intensity of the 40 ∼kDa ELP band between the control and proteolytic samples. A one-way ANOVA revealed significant differences in the percent of remaining 40 kDa ELP as a function of temperature and protease identity. Micelle assembly had a statistically significant, albeit small, protective effect against collagenase *P = 4.11 × 10−4. Under these conditions, micelle assembly had no effect on elastase proteolysis; however, elastase was significantly more active than collagenase **P = 9.35 × 10−13, ***P = 9.35 × 10−13.
Figure 4.
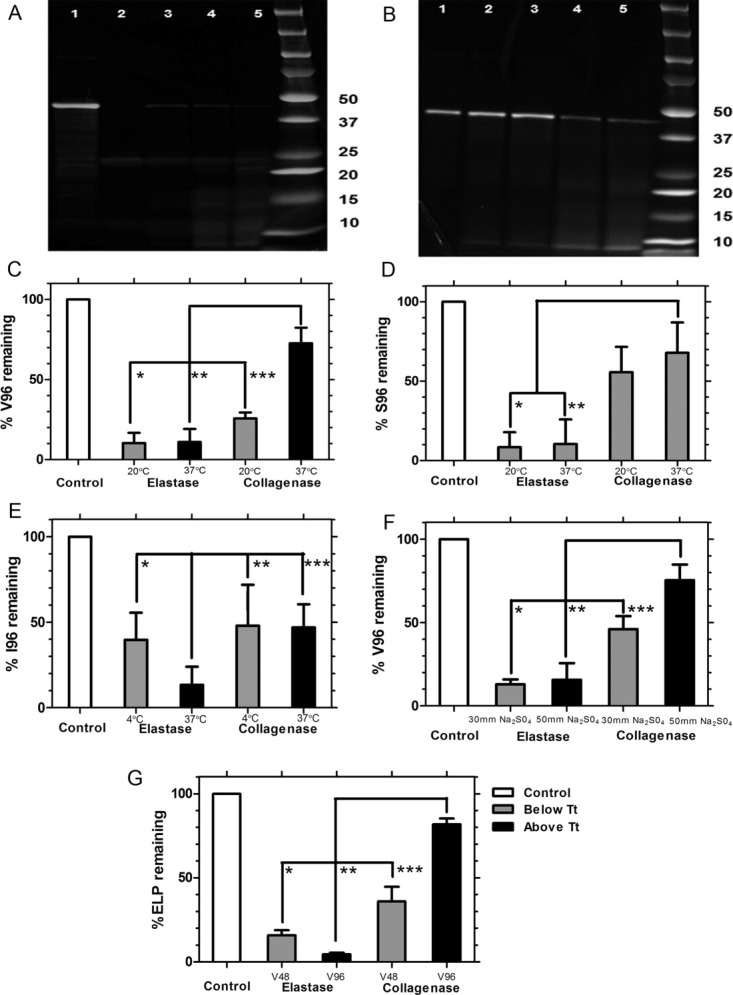
ELP phase separation is protective against degradation by collagenase but not elastase. SDS PAGE was used to resolve the proteolysis of four CFSE conjugated ELP monoblocks (V48, V96, S96, I96) above and below their phase transition temperatures. Digestion and electrophoresis were performed in triplicate and quantified using image analysis. A: Representative gel for V96 incubated with elastase for 2 h: Lane 1: control; 2: 0.35 units at 20°C; 3: 0.7 units at 20°C; 4: 0.35 units at 37°C; 5: 0.7 units at 37°C. B: Representative gel for V96 incubated with collagenase for 2 h: Lane 1: control; 2: 3.6 units at 37°C; 3: 6.0 units at 37°C; 4: 3.6 units at 20°C; 5: 6.0 units at 20°C. C: Quantification of V96 degradation above and below Tt, bulk (31.9°C) shows that elastase is more active than collagenase (*P = 2 × 10−11, **P = 3 × 10−11) and that phase separation protects only against collagenase (***P = 4 × 10−8). D: Quantification of soluble S96 degradation below Tt, bulk (55.7°C) shows that elastase is more active than collagenase (*P = 1.72 × 10−8, **P = 2.11 × 10−9); however, there are no significant differences in enzyme activity between 20 and 37°C. E: Quantification of I96 degradation above and below Tt, bulk (11.5°C). Despite the phase separation, elastase at 37°C degrades more ELP compared to 4°C (*P = 0.04); furthermore, elastase is more active than collagenase both below (**P = 0.004) and above (***P = 0.006) the phase transition temperature. Under these assay conditions, phase separation is not protective against collagenase activity due to the reduced enzyme activity at 4°C. F: Quantification of V96 degradation above and below Tt,bulk (31.9°C) following addition of sodium sulfate. It follows the same trend where elastase is more active than collagenase (*P = 6.9 × 10−6, **P = 1 × 10−5) and that phase separation protects only against collagenase (***P = 0.04). G: Quantification of V48 and V96 degradation at 37°C, which is above the Tt of V96 and below that of V48, shows that elastase degrades both V48 and V96 almost equally (*P = 3.9 × 10−8, **P = 8.3 × 10−9) and that phase separation only protects V96 against collagenase (***P = 1.3 × 10−6).
While elastase efficiently acts upon these nanoparticles, collagenase is less active under the studied conditions. Densitometric analysis indicated a clear difference in the percentage of starting material remaining compared to an untreated control for all samples [Fig. 3(C)]. On treatment with collagenase, SDS-PAGE showed partial degradation for the ELP nanoparticles [Fig. 3(B)]; furthermore, there was a statistically significant difference above and below the CMT, which suggests that micelle assembly has a marginally protective effect against collagenase (p = 4 × 10−4). These findings demonstrate that collagenase activity tends to be slightly moderated by micelle formation; however, elastase activity remains equivalent regardless of incubation temperature. As observed in the hepatocyte cultures, these findings confirm that ELP nanoparticles are proteolytically biodegradable, and also suggest that ELP-mediated assembly may provide protection against select proteases.
Proteolytic digestion of ELP monoblocks
After confirming that ELP nanoparticles can be degraded in low-pH intracellular compartments within a hepatic cell line (Figs. 1 and 2) and also by model proteases (Fig. 3), we investigated the component ELP monoblocks to determine how ELP sequence and phase separation control proteolysis. Unlike the diblock copolymer I48S48, which forms stable nanoparticles, the ELP monoblocks V48, I96, V96, and S96 phase separate into micron-sized aggregates that settle in water. As observed for the diblock copolymer, elastase degrades V96 almost completely at 20 and 37°C (p = 2 × 10−11, p = 3 × 10−11); however, for V96 there is a significant difference (p = 4 × 10−8) between collagenase digestion above and below the Tt (31.9°C) [Fig. 4(C)]. Similar results were obtained when the V96 phase separation was induced by the addition of salt [Fig. 4(F)]. Likewise, the same trend was observed for degradation of V48 and V96 at 37°C, which was between their respective Tt of 42.4 and 31.9°C [Fig. 4(G)]. Due to the surprisingly efficient elastase digestion of phase V96 under phase separation, we further quantified the kinetics of proteolysis using Michaelis-Menten analysis, which yielded a Vmax of 0.24 × 10−3 s−1 and KM of 1.14 μM (Supplemental Figure 3). In contrast, both incubation temperatures for S96 were below Tt (55.7°C). Elastase almost completely degrades S96 at 20 and 37°C (p = 9 × 10−13 and 9 × 10−13) when compared to the ∼50% degradation caused by collagenase at 37°C. Unlike V96, there was no enhancement of collagenase proteolysis for S96 at 20 vs. 37°C [Fig. 4(D)]. In contrast, I96 has a very low Tt, (11.5°C); therefore, degradation was explored below and above its Tt at 4 and 37°C respectively. Elastase digests I96 more extensively at 37°C than at 4°C (p = 0.04), which is opposite to the observation in V96 and S96 [Fig. 4(E)]. This may result from reduced proteolytic activity at 4°C; however, even at a suboptimal temperature, I96 was still readily degraded by elastase. In total, these data suggest that all of these ELP monoblocks are readily degraded by elastase at 37°C, whereas for ELPs with a Tt near physiological temperature (V96) the phase separation partially protects against collagenase.
Discussion
Previous studies have shown that ELPs that contain lysine are substrates for trypsin, collagenase, and elastase;14, 15 furthermore, non-ionic soluble ELPs can be degraded in fresh mouse serum over a period of several days.16 Despite these reports, there have been no comprehensive studies to determine if ELP phase separation is a complete, partial, or ineffective inhibitor of proteolysis. Biodegradation of ELP nanoparticles is complicated because they contain both soluble and insoluble ELP blocks, both of which must biodegrade in therapeutic applications. To answer this question, this report describes the biodegradation of a small library of ELPs with approximately the same molecular weight with different physical properties (Table I).
Our main finding is that above and below their assembly temperatures, ELPs are excellent substrates for elastase; however, another enzyme, collagenase, appeared to be partially inhibited by the ELP phase separation. Under in vitro conditions, elastase degraded greater than 90% of the starting protein polymer, a pattern confirmed across all the ELPs treated at physiological temperatures. A similar pattern of biodegradation was observed in cellulo, where there are reports of lysosomal homologues of elastases.17 Under similar conditions, collagenase was less active towards both I48S48 (CMT = 26°C) and V96 (Tt, bulk = 31.9°C). Both of these ELPs are significantly (p < 0.05) protected at 37°C compared to incubation below their assembly temperature. This trend was also observed when comparison was made between different lengths of ELPs (V48 and V96) and when Tt was manipulated with different strengths of sodium sulfate. A possible explanation is that the ELP phase separation is coupled with a change in secondary structure18 and reduced proteolytic accessibility.11 Differences in proteolysis may also correlate to differences in amino acid sequence; however, the repetitive motif Val-Pro-Gly-Xaa-Gly provides ample cleavage sites for both elastase and collagenase. Elastase cleavage occurs on the carboxy end of small and neutral hydrophobic amino acids, including Gly and Val.19 Similarly, collagenase cleavage occurs between Yaa-Gly bond where Yaa can be any amino acid.20 Our current study is unable to distinguish between these possibilities; however, it is clear that the ELP phase separation does not provide protection against all proteases.
Another novel finding of this report, ELP nanoparticles are clearly biodegradable in a monolayer of murine hepatocyte-derived cells following cellular uptake. These nanoparticles are internalized, co-localized with low pH compartments (Fig. 2), and are eliminated over a period of 2 to 24 hours [Figs. 1(D) and 2]. The fact that ELP incubation with a purified elastase produces a digested product and there are elastases present in lysosomes (Ela2, Human polymorphonuclear leukocyte elastase), suggests that lysosomal elastases may play a role in the hepatic clearance of therapeutic ELPs. Additional in vivo studies will be required to confirm this hypothesis; however, these results support the assertion that ELPs with a wide range of physico-chemico properties are biodegradable under physiological conditions.
Materials and methods
Synthesis of ELPs
Elastin like-polypeptides are composed from pentapeptide repeats of (VPGXG)n where X can be any amino acid and n is the number of repeats. For this study, we selected different ELP libraries where X was valine with 48 and 96 repeats whereas serine, and isoleucine consisted of 96 repeats each and a diblock copolymer consisting of 48 repeats of isoleucine and serine respectively (Table I). These polypeptides were synthesized from E.coli and purified using a procedure called inverse transition cycling.6 ELP genes were synthesized using a procedure called plasmid reconstruction directional ligation in pET25b+ vector in TOP 10 cells (Invitrogen, Carlsbad, CA) followed by protein expression in BLR cells (Novagen, Madison, WI).21 The purified batches were then characterized (Table I) by confirming their molecular weight using MALDI-TOF (Kratos AXIMA CFR, Shimadzu Corporation, Kyoto, Japan) and purity determination by CuCl2 SDS-PAGE analysis. ELP phase diagrams were characterized in phosphate buffered saline by raising the temperature at 1°C/min on a DU800 UV-vis spectrophotometer (Beckman Coulter, Brea, CA). The transition temperature was defined at the maximum first derivative of the optical density at 350 nm. Finally, the particle size of the S48I48 diblock copolymer was measured using dynamic light scattering (DLS) [DynaPro, Wyatt Technology, Santa Barbara, California, USA].
Enzyme degradation assay
ELPs were labeled with fluorophores (Carboxyfluorescein succinimidyl ester (CFSE) & Rhodamine (Rh)) using N-hydroxysuccinimide chemistry (Thermo Scientific, Rockford, IL). Briefly, 3 μl of labeled ELPs (50 μM) was mixed with different enzyme concentrations, 0.35 and 0.70 units of elastase from porcine pancreas and 3.6 and 6.0 units of type III collagenase from Clostridium histolyticum (EMD Chemicals, Cincinnati, OH) at 20 and 37°C for all ELPs except I96, which was incubated at 4 and 37°C. The lower incubation temperature was selected because I96 has a lower transition temperature than V96 (Table I). To study the effect of salt-induced phase separation at constant temperature, sodium sulfate was added to V96 at 28°C in small increments until the Tt was reached, after which it was incubated with elastase (0.7 U) or collagenase (6 U). To explore ELP proteolysis at constant temperature and ionic strength, we further compared two molecular weight ELPs, one that is soluble (V48) and one that phase separates (V96) at 37°C. All digestions and electrophoretic gels were prepared in triplicate. There were no differences as a function of enzyme concentrations; therefore, data were averaged for statistical comparison. To quantify the Michaelis-Menten kinetics of ELP under phase separation, rhodamine labeled V96 at various concentrations was incubated with 0.50U of elastase for periods from 5 to 40 minutes at 40°C. The samples were resolved on a 4-20% SDS-PAGE (Lonza, Rockland, ME) and the fluorescent bands were detected using a Typhoon 8600 variable mode imager (Amersham Biosciences Corp., Sunnyvale, CA USA. The integrated intensity of the ∼40 kDa bands was analyzed using ImageJ (NIH, Bethesda, MD). In addition to the sample intensities, Isample, the undegraded positive control, Icontrol and a background negative intensity, Ibackground was obtained on each gel. For each digestion, the integrated intensity was then normalized as follows:
| 1 |
Where F%remaining, represents the amount of undegraded ELP.
The constants of kinetic equations were determined by curve fitting using GraphPad Prism (GraphPad Software, San Diego, CA).
Cellular uptake and degradation in a live-cell hepatocyte monolayer
Transformed mouse hepatocytes (PTENloxp/loxp; Alb-Cre-)22 were seeded onto 35 mm glass coverslip-bottomed dishes at 1x105 cells per dish in Dulbecco's Modified Eagle Medium (DMEM) (with 4.5 g/L glucose) (Thermo Scientific, Rockford, IL) containing 10% fetal bovine serum, 5 μg/ml insulin, and 0.02 μg/ml epidermal growth factor (EGF). When the cells reached 75% confluence, they were washed with warm fresh DMEM followed by incubation with 10 μM Rh-S48I48 at 37oC for 30 min. After this incubation pulse, the cells were rinsed with warm DMEM twice, incubated with 75 nM LysoTracker® green [Invitrogen, Carlsbad, CA, USA], and imaged using confocal microscopy with a Zeiss LSM 510 Meta NLO system equipped with Argon, green HeNe and red HeNe lasers. For the degradation experiment, transformed mouse hepatocytes were incubated with 30 μM Rh-S48I48 at 37oC. After washing, the cell lysates were resolved by SDS PAGE and imaged using a Bio-Rad model 3000 VersaDoc Gel Imaging System (Bio-Rad Laboratories, Hercules, CA).
Statistical analysis
The percent of starting material remaining, F% remaining, on each gel was averaged and expressed as the Mean ± SD. Comparisons across the different treatment groups were made using one-way ANOVA followed by Tukey's post hoc test.
Conclusions
Protein polymers inspired by human tropoelastin (ELPs) are emerging as a new class of environmentally-responsive drug carriers; however, their mechanism of biodegradation has not been studied comprehensively. In response, this communication describes the proteolytic biodegradation for a small library of ELPs with distinct self-assembly behavior. Surprisingly, ELP-mediated assembly is mildly protective against some (collagenase), but not all enzymes (elastase). In addition, ELP nanoparticles are internalized and biodegraded within low pH-compartments in a monolayer culture of transformed cells originating from the liver. As hepatic lysosomes are a likely destination for nanoparticle therapeutics of this radius, this study supports the contention that ELP nanoparticles are a viable platform for developing biodegradable therapeutics.
Acknowledgments
The authors would also like to thank Dr. Sarah F. Hamm-Alvarez for her wonderful guidance and support.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Law B, Tung C-H. Proteolysis: A biological process adapted in drug delivery, therapy, and imaging. Bioconjugate Chem. 2009;20:1683–1695. doi: 10.1021/bc800500a. [DOI] [PubMed] [Google Scholar]
- 2.Berdowska I. Cysteine proteases as disease markers. Clinica Chim Acta. 2004;342:41–69. doi: 10.1016/j.cccn.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 3.Rao MB, Tanksale AM, Ghatge MS, Deshpande VV. Molecular and biotechnological aspects of microbial proteases. Microbiol Mol Biol Rev. 1998;62:597–635. doi: 10.1128/mmbr.62.3.597-635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacKay AJ, Chen M, McDaniel JR, Liu W, Simnick AJ, Chilkoti A. Self-assembling chimeric polypeptide-doxorubicin conjugate nanoparticles that abolish tumours after a single injection. Nat Mater. 2009;8:993–999. doi: 10.1038/nmat2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aluri S, Janib SM, Mackay JA. Environmentally responsive peptides as anticancer drug carriers. Adv Drug Deliv Rev. 2009;61:940–952. doi: 10.1016/j.addr.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer DE, Chilkoti A. Purification of recombinant proteins by fusion with thermally-responsive polypeptides. Nat Biotech. 1999;17:1112–1115. doi: 10.1038/15100. [DOI] [PubMed] [Google Scholar]
- 7.Tyrrell ZL, Shen Y, Radosz M. Fabrication of micellar nanoparticles for drug delivery through the self-assembly of block copolymers. Prog Poly Sci. 2010;35:1128–1143. [Google Scholar]
- 8.Dreher MR, Simnick AJ, Fischer K, Smith RJ, Patel A, Schmidt M, Chilkoti A. Temperature triggered self-assembly of polypeptides into multivalent spherical micelles. J Am Chem Soc. 2007;130:687–694. doi: 10.1021/ja0764862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chilkoti A, Christensen T, MacKay JA. Stimulus responsive elastin biopolymers: Applications in medicine and biotechnology. Curr Opin Chem Biol. 2006;10:652–657. doi: 10.1016/j.cbpa.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MaHam A, Tang Z, Wu H, Wang J, Lin Y. Protein-based nanomedicine platforms for drug delivery. Small. 2009;5:1706–1721. doi: 10.1002/smll.200801602. [DOI] [PubMed] [Google Scholar]
- 11.Liu W, Dreher MR, Furgeson DY, Peixoto KV, Yuan H, Zalutsky MR, Chilkoti A. Tumor accumulation, degradation and pharmacokinetics of elastin-like polypeptides in nude mice. J Control Release. 2006;116:170–178. doi: 10.1016/j.jconrel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 12.Urry DW. Physical chemistry of biological free energy transduction as demonstrated by elastic protein-based polymers. J Phys Chem B. 1997;101:11007–11028. [Google Scholar]
- 13.Sun G, Hsueh P, Janib SM, Hamm-Alvarez SF, MacKay JA. Design and cellular internalization of genetically engineered polypeptide nanoparticles displaying adenovirus knob domain. J Control Release. 2011;155:218–226. doi: 10.1016/j.jconrel.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nettles DL, Chilkoti A, Setton LA. Applications of elastin-like polypeptides in tissue engineering. Adv Drug Deliv Rev. 2010;62:1479–1485. doi: 10.1016/j.addr.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ong SR, Trabbic-Carlson KA, Nettles DL, Lim DW, Chilkoti A, Setton LA. Epitope tagging for tracking elastin-like polypeptides. Biomaterials. 2006;27:1930–1935. doi: 10.1016/j.biomaterials.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 16.Liu W, Dreher MR, Furgeson DY, Peixoto KV, Yuan H, Zalutsky MR, Chilkoti A. Tumor accumulation, degradation and pharmacokinetics of elastin-like polypeptides in nude mice. J Control Release. 2006;116:170–178. doi: 10.1016/j.jconrel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 17.Gabay JE, Almeida RP. Antibiotic peptides and serine protease homologs in human polymorphonuclear leukocytes: defensins and azurocidin. Curr Opin Immunol. 1993;5:97–102. doi: 10.1016/0952-7915(93)90087-9. [DOI] [PubMed] [Google Scholar]
- 18.Yamaoka T, Tamura T, Seto Y, Tada T, Kunugi S, Tirrell DA. Mechanism for the phase transition of a genetically engineered elastin model peptide (VPGIG)(40) in aqueous solution. Biomacromolecules. 2003;4:1680–1685. doi: 10.1021/bm034120l. [DOI] [PubMed] [Google Scholar]
- 19.Narayanan AS, Anwar RA. The specificity of purified porcine pancreatic elastase. Biochem J. 1969;114:11–17. doi: 10.1042/bj1140011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.French MF, Mookhtiar KA, Van Wart HE. Limited proteolysis of type I collagen at hyperreactive sites by class I and II Clostridium histolyticum collagenases: Complementary digestion patterns. Biochemistry. 1987;26:681–687. doi: 10.1021/bi00377a004. [DOI] [PubMed] [Google Scholar]
- 21.McDaniel JR, Mackay JA, Quiroz FG, Chilkoti A. Recursive directional ligation by plasmid reconstruction allows rapid and seamless cloning of oligomeric genes. Biomacromolecules. 2010;11:944–952. doi: 10.1021/bm901387t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stiles BL, Kuralwalla-Martinez C, Guo W, Gregorian C, Wang Y, Tian J, Magnuson MA, Wu H. Selective deletion of Pten in pancreatic Î2 cells leads to increased islet mass and resistance to STZ-induced diabetes. Mol Cell Biol. 2006;2:2772–2781. doi: 10.1128/MCB.26.7.2772-2781.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.