Abstract
The last gene in the genome of the bacteriophage HK97 encodes the protein gp74. We present data in this article that demonstrates, for the first time, that gp74 possesses HNH endonuclease activity. HNH endonucleases are small DNA binding and digestion proteins characterized by two His residues and an Asn residue. We demonstrate that gp74 cleaves lambda phage DNA at multiple sites and that gp74 requires divalent metals for its endonuclease activity. We also present intrinsic tryptophan fluorescence data that show direct binding of Ni2+ to gp74. The activity of gp74 in the presence of Ni2+ is significantly decreased below neutral pH, suggesting the presence of one or more His residues in metal binding and/or DNA digestion. Surprisingly, this pH-dependence of activity is not seen with Zn2+, suggesting a different mode of binding of Zn2+ and Ni2+. This difference in activity may result from binding of a second Zn2+ ion by a putative zinc finger in gp74 in addition to binding of a Zn2+ ion by the HNH motif. These studies define the biochemical function of gp74 as an HNH endonuclease and provide a platform for determining the role of gp74 in life cycle of the bacteriophage HK97.
Keywords: HNH endonucleases, bacteriophage HK97, divalent metals, DNA digestion, Zn2+ finger
Introduction
The genome of the bacteriophage HK97 is 39.7 kb in size and has a total of 61 open reading frames.1 The organization of the HK97 genome is similar to that of other lambda-like bacteriophages2 and functions have been ascribed for many of the HK97 genes. Like many phages, the HK97 genes are arranged in clusters related by structure or function,1 with genes encoding the terminase at the 5′ end, followed by genes encoding structural proteins such as the head, connector, and tail proteins.2 Following the structural genes are genes encoding for enzymes and proteins involved in processes such as DNA transcription1 and repair,3 integration of phage DNA into the host genome during the lysogenic phase,1 and lysis of the host bacterial cell.4 While the functions of the three right most genes, positioned between the holins (or lysis genes5, 6) and the cos site (which is the site of DNA circularization in the host cell7), are unknown, the final gene, gp74, possesses sequence identity with the HNH endonuclease family of proteins (pfam01844).
The HNH motif is approximately 35 amino acids long and is characterized by the presence of two highly conserved His residues and one Asn residue.8 The motif, which has been identified in greater than 1000 proteins from bacteria, archaea, and eukaryotes,9 has been found to bind to nucleic acids and to possess endonuclease activity.8 The largest group of HNH motif-containing proteins of known function is the site-specific homing endonucleases,10 such as I-HmuI and I-HmuII from the Bacillus subtilis phages SPO1 and SP82, respectively.11 HNH motifs are also present in bacterial toxins such as the E7 and E9 colicins12, 13 and the S1 and S2 pyocins,14 as well as in DNA restriction enzymes, such as KpnI,15 PacI,16 and Hpy99I.17 The specificity of HNH motif-containing endonucleases varies, with proteins that mediate non-specific DNA cleavage, such as the colicins, comprised primarily of an HNH motif and lacking additional DNA-binding sites. In contrast, the homing endonucleases, which have high sequence-specificity, contain DNA-recognition domains in addition to the HNH motif.8
The three-dimensional structures of several HNH motif-containing endonucleases have been solved. These include the homing endonuclease I-HmuI,18 the bacterial colicins E7 and E9,19–23 and the restriction enzymes PacI16 and Hpy99I.17 The HNH motif consists of a two-stranded antiparallel β-sheet that is flanked on one side by an α-helix, and is often referred to as the ββα-metal fold. The HNH motif is embedded within a folded domain that varies between the different HNH endonucleases.8 A divalent metal is bound to the center of the structure by interactions with the side chain of the second conserved His residue of the HNH motif, as well as the residue N-terminal to the HNH motif. In I-HmuI, this N-terminal amino acid is an Asp residue,18 while in colicins E7 and E9 it is another His residue.19–23 The first conserved His residue of the HNH motif acts as a general base to activate a water molecule for nucleophilic attack of the DNA backbone, and the Asn determines the orientation of this His residue. The residues that are involved in the HNH structural motif are highly conserved among members of this protein family. HNH motifs generally bind the DNA backbone in the minor groove and additional residues outside the conserved HNH residues mediate interactions with the DNA.
HNH endonucleases have been identified in a number of bacteriophages and have been shown to play a variety of roles in the phage life cycle. For example, I-HmuI and I-HmuII endonucleases are encoded within the intron of the DNA polymerase genes of related Bacillus phages and cleave DNA on a single strand on both intron-containing and intronless targets. Further, each endonuclease prefers DNA of the heterologous phage and the activity of I-HmuII results in exclusion of I-HmuI-containing introns from the progeny of mixed infections.24 The free-standing (i.e. non-intron encoded) HNH endonuclease mobE, which is encoded by bacteriophage T4, introduces strand-specific nicks in the non-coding region of the nrdB gene of T2 phage.25 The activity of mobE promotes mobility of the neighboring I-TevIII, an inactive HNH endonuclease encoded within the intron of the nrdB gene in T4 phage, thus facilitating the inheritance of I-TevIII in progeny phages. Although gp74 displays distant sequence similarity to a number of proteins from bacteria19, 22 and other phages,11, 18 a function has not been ascribed to this particular subclass of HNH proteins in bacteriophages. Thus, characterizing the function of gp74 was the focus of this work.
In this article, we demonstrate that gp74 cleaves lambda phage DNA at multiple sites, and that the endonuclease activity of gp74 requires the presence of divalent metal ions. Gp74 was active with all metals tested (Ni2+, Zn2+, and Mg2+), although the endonuclease efficiency depends on the divalent metal. Binding studies with Ni2+ were conducted using intrinsic tryptophan fluorescence and indicated that gp74 binds metals in the absence of DNA, but with low affinity (∼165 μM). In keeping with the mechanism of HNH endonucleases and the role of His residues involved in metal binding and catalysis, the activity of gp74 in the presence of Ni2+ was significantly reduced below neutral pH. Surprisingly, however, this pH-dependence was not observed when Zn2+ was used as the metal cofactor, which may suggest a different mode of metal binding for Zn2+ compared with Ni2+ that may result from the presence of a zinc finger that binds an additional Zn2+ ion. Taken together, these data indicate the gp74 from the HK97 bacteriophage functions as a previously unidentified HNH endonuclease. These studies provide a platform for determining the role of gp74 in the life cycle of the bacteriophage HK97.
Results
HK97 gp74 is a member of the HNH-endonuclease family
A PSI-BLAST search performed using the gp74 sequence as a query revealed that it is a member of the HNHc superfamily of proteins, which are found in viruses, archaea, eubacteria, and eukaryotes.8, 10 Gp74 is most similar to HNH endonucleases from bacteria and other phages (Supporting Information Fig. S1), and analysis of the sequence suggests that gp74 is a member of either the 5th or the 8th subclasses of HNHc endonucleases.26 Like other proteins in subclasses 5 and 8, gp74 contains the hallmark His and Asn residues in the HNH motif (His43, Asn73, and His82 in gp74), as well as a CXXC motif (Cys26-Val27-Met28-Cys29) N-terminal to the HNH motif (Supporting Information Fig. S1). Gp74 also contains a CXXH sequence (Cys78-Lys79-Ala80-His81) within the HNH motif, as is also found in HNH endonucleases of subclasses 5 and 8.
To determine if gp74 is a conserved feature in the Caudovirales family of tailed bacteriophages, the Hidden Markov Model (HMM) for pfam01844 for the HNH superfamily was used to search the complete Caudovirales genomes present in the NCBI database. This group includes the long non-contractile (Siphoviridae), long contractile (Myoviridae), and short (Podoviridae) tailed phages. A total of 182 of the 492 complete phage genomes encode a gene that has detectable sequence identity with gp74. The protein homologues are present in a wide variety of phages that infect both Gram-negative and Gram-positive bacteria, including Pseudomonas, Klebsiella, Xanthomonas, Bacteroides, Mycobacteria, Listeria, Bacillus, and Streptococcus species. The majority of the gp74-like proteins are found in Siphophages, with 47% of fully sequenced Siphoviridae genomes encoding at least one homologue. Both Myophages and Podophages had much lower incidence of proteins hit by the HNH superfamily HMM, with 26% and 8% of the genomes, respectively, encoding a gp74 homologue. The majority of the protein homologues of gp74 share a common genomic position adjacent the terminase and other morphogenetic proteins (Supporting Information Fig. S2). For example, in the Geobacillus phage E2, the first open reading frame is annotated as a putative HNH endonuclease, and the following three open reading frames encode the small terminase subunit, the large terminase subunit, and the portal protein, followed by the rest of the morphogenetic region. The Lactococcus phage p2 genome organization begins with genes encoding the small and large terminase subunits, the putative HNH endonuclease, and the portal protein, followed by genes encoding the structural head proteins. In the case of HK97, while gp74 is the final gene in the linear chromosome, following injection into the host cytoplasm the genome circularizes and the ssDNA ends of the cos site7 are ligated. At this point, gp74 is then located adjacent to the small and large terminase subunits and the portal protein. Thus, the location of the gp74 gene near genes for the terminase subunits and portal protein is conserved in HK97.
Gp74 mediates double-stranded digestion of phage DNA in the presence of divalent metals
To determine whether gp74 possesses endonuclease activity, we examined its ability to cleave the λ chromosome, a linear, double-stranded DNA genome. Purified gp74 was tested for endonuclease activity in the presence of a variety of divalent metal ions and the results were visualized by DNA agarose gel electrophoresis. In the presence of 0.5 mM Ni2+, gp74 cleaves the λ chromosome at a large number of sites, as evidenced by the formation of an abundant number of DNA fragments of varying lengths (Fig. 1). By contrast, incubation of phage λ DNA with gp74 or Ni2+ ions alone does not mediate DNA digestion (Fig. 1). The amount of DNA digestion was proportional to the amount of gp74 present in the reaction, indicating that the observed endonuclease activity was catalyzed by gp74 (Fig. 1).
Figure 1.
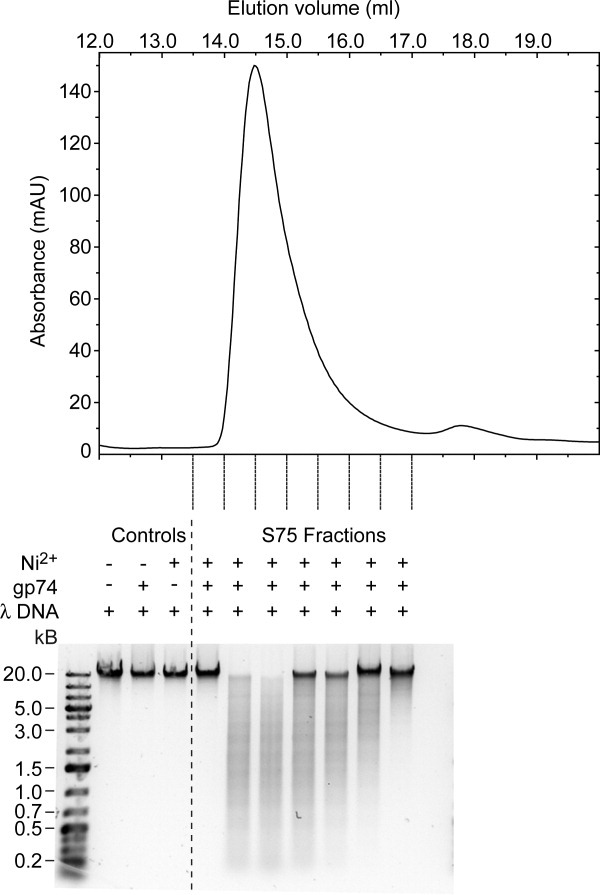
Gp74-mediated digestion of lambda phage DNA in the presence of Ni2+ ions. Fractions containing gp74 from the Superdex 75 gel filtration column were assayed for endonuclease activity in the presence of Ni2+. The top panel shows the elution profile of gp74 from the Superdex 75 column. Fractions between 13.5 and 17.0 mL, indicated by dashed lines at the bottom of the trace, were tested. The bottom panel shows the agarose gel (1%) electrophoresis analysis of the digestion reactions from each fraction. Digestion reactions were allowed to proceed for 6 h. The concentrations of Ni2+ ions and lambda (λ) DNA in the reactions were 0.5 mM and 25 μg/mL, respectively. In each reaction, 26.3 μL of the gp74 fraction was used in a total reaction volume of 200 μL. A volume of 26.3 μL was chosen, so that gp74 concentration would be 24 μg/mL in the reaction corresponding to the elution fraction with the highest gp74 concentration. For the control reaction containing λ DNA and gp74 only, 26.3 μL from the fraction with the greatest amount of gp74 was used.
The Ni2+ ion concentration dependence of the gp74-mediated DNA digestion reaction was also examined as activity has been shown to decrease for other HNH proteins in the presence of high divalent metal concentrations.18, 22, 27, 28 Gp74 was able to efficiently cleave λ DNA within 2 h with Ni2+ ion concentrations ranging from 0.5 to 5 mM (Fig. 2). In the presence of high Ni2+ ion concentrations, while the cleavage reaction occurs as quickly, it appears to be slightly inhibited as evidenced by the presence of full length λ DNA and the absence of small DNA products (i.e. less than 1 kb). The ability of other divalent metals, including Zn2+ (Fig. 3) and Mg2+ (Supporting Information Fig. S3), to catalyze the DNA-cleavage reaction was also assessed. Both Zn2+ and Mg2+ were able to induce numerous fragments in λ DNA, although with varying efficiency. For example, the full length λ DNA is completely digested after 2 h when 0.5 mM Ni2+ is used in the reaction (Fig. 2), whereas it takes approximately 4 h to completely digest the intact λ DNA in the presence of 0.5 mM Zn2+ (Fig. 3). In the case of Mg2+, complete DNA digestion is less efficient when compared with Ni2+ at low concentrations (1 mM), but there is less of a difference at higher concentrations (5 and 10 mM) (Fig. 2 and Supporting Information Fig. S3).
Figure 2.
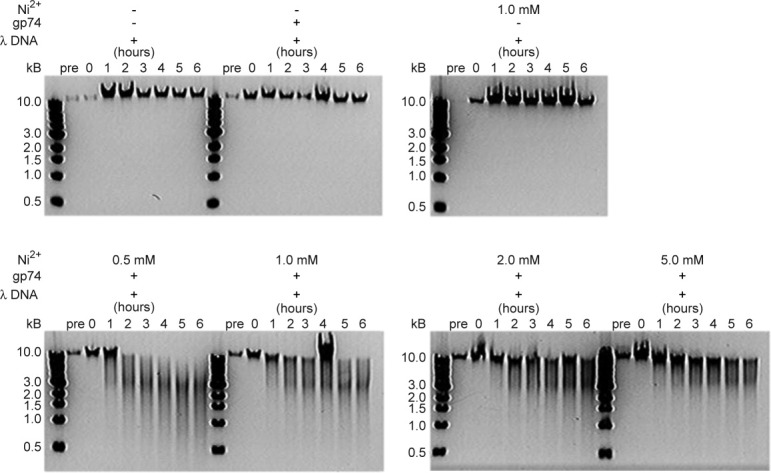
Concentration-dependence of gp74-mediated digestion of lambda phage DNA with Ni2+ ions. Agarose gel (1%) electrophoresis analysis of the time course of the lambda phage DNA digestion reactions is shown. The upper panel shows control reactions in which the DNA (50 μg/mL) was incubated in 20 mM HEPES, pH 7.0 alone, or in the presence of either gp74 (48 μg/mL) or 1 mM Ni2+ ions. There is no digestion of lambda phage under these conditions. The lower panel depicts digestion experiments in which lambda phage DNA was incubated with both gp74 and Ni2+ ions at concentrations of 0.5–5 mM. The smear of DNA in these gels indicates that in the presence of Ni2+, gp74 mediates digests λ DNA at multiple sites.
Figure 3.
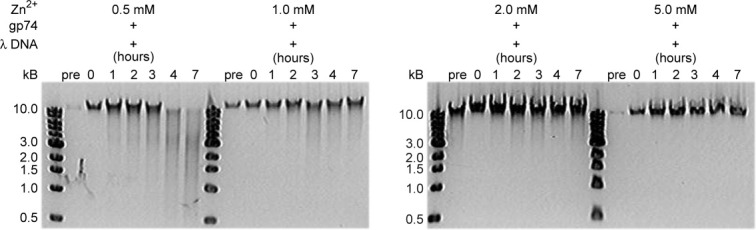
Gp74 also digests lambda phage DNA in the presence of Zn2+. Agarose gel (1%) electrophoresis analysis of the time course of the lambda phage DNA digestion by gp74 in the presence of 0.5–5 mM Zn2+ ions. As with the Ni2+ digestion experiments, the concentration of λ DNA was 50 μg/mL and that of gp74 was 48 μg/mL. Reactions were performed in 20 mM HEPES, pH 7.0. As seen in the presence of Ni2+, gp74 mediates digestion of λ DNA at multiple sites (smear) in the presence of Zn2+, albeit with reduced efficiency.
Gp74-mediated DNA digestion in the presence of Ni2+ but not Zn2+ is dependent on pH
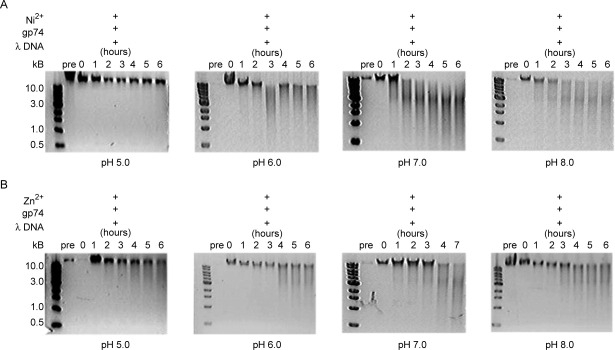
Previously determined structures of HNH endonucleases3, 16–19, 21–23, 29 and accompanying mutagenesis studies17, 18, 22 elucidated that two highly conserved His residues are involved in DNA binding and catalysis. The structures suggest a mechanism in which the first conserved His residue of the HNH motif acts as a general base to generate a nucleophilic hydroxyl ion that attacks a phosphate oxygen in the DNA backbone.18, 22 The metal ion, which is bound by the second conserved His residue, stabilizes the phospho-anion transition state and the leaving group during the reaction. For the His residues of the HNH motif to perform their respective functions, they must be deprotonated. Thus, DNA cleavage efficiency is expected to vary with pH. The λ DNA cleavage studies in the presence of Ni2+ and Zn2+ were repeated at pH values of 5, 6, 7, and 8 in 20 mM HEPES buffer. In the presence of Ni2+ [Fig. 4(A)], gp74-mediated cleavage of λ DNA was equally efficient at pH 7 and 8, but decreased at pH 6 and was completely abrogated at pH 5. Similar results were observed with Mg2+ (data not shown). These results provide evidence for a role of His residues in catalysis (and likely also in metal binding) that is consistent with the identification of gp74 as an HNH endonuclease. Surprisingly, when the pH-dependence of gp74 activity in the presence of Zn2+ was examined, a different profile was observed. DNA digestion was only slightly inhibited at the lower pH values [Fig. 4(B)], suggesting that additional non-His residues in gp74 may be involved in binding and/or catalysis. As low pH may induce a structural change in gp74 and thereby affect its catalytic activity, circular dichroism (CD) spectroscopy was used to assess any pH-dependent structural changes in the protein (Fig. 5). The CD spectrum of gp74 displays minima at 208 nm and 222 nm that are characteristic of a predominantly α-helical protein. CD spectra of gp74 are essentially identical at pH values between 6 and 8, and slightly different at pH 5. The CD spectrum at pH 5 displays the double minima characteristic of a folded α-helical protein, with a slight decrease in mean residue ellipticity, possibly caused by small structural changes from ionization of different groups or precipitation of a small amount of the protein.
Figure 4.
The activity of gp74 in the presence of Ni2+, but not Zn2+, is mediated by pH. Agarose gel (1%) electrophoresis analysis of lambda phage DNA digestion by gp74 in the presence of (A) Ni2+ ions or (B) Zn2+ ions. At pH 5, reactions contained 25 μg/mL of λ DNA, 48 μg/mL of gp74, and 0.5 mM of either Ni2+ or Zn2+ ions. At pH 6–8, reactions contained 50 μg/mL of lambda phage DNA, 48 μg/mL of gp74, and 0.5 mM of either Ni2+ or Zn2+ ions.
Figure 5.
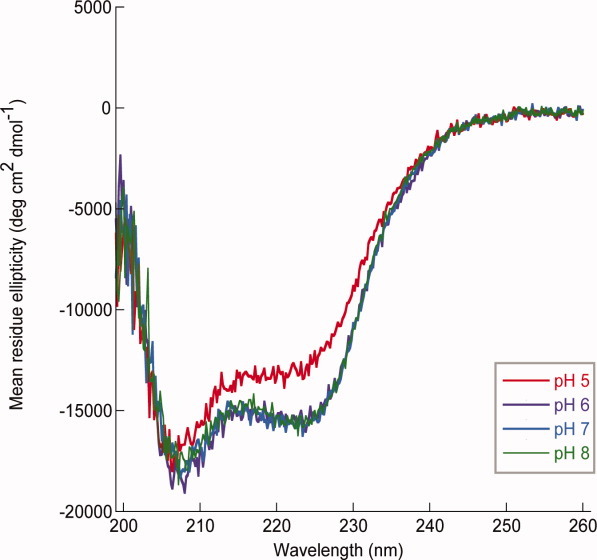
Circular dichroism spectra of gp74. CD spectra were recorded of 2 μM gp74 in 20 mM Na+ phosphate at pH values of 5 (red), 6 (purple), 7 (blue), or 8 (green). CD spectra are an average of 5 scans with absorbance measurements every 0.2 nm. All CD spectra were blank corrected.
Gp74 binds Ni2+ ions with low affinity in the absence of DNA
Previous studies on HNH endonucleases18–23, 30 indicate that residues involved in metal binding are distinct from those involved in catalysis. Therefore, we examined the ability of gp74 to bind metal ions in the absence of DNA using intrinsic tryptophan fluorescence. Gp74 contains four tryptophan residues; the highly conserved Trp12, the conserved hydrophobic positions Trp69 and Trp74, and the non-conserved Trp111. Trp12 and Trp111 are located outside the HNH motif, while Trp69 and Trp74 are located in a loop region and in the second β-strand in the HNH motif, respectively (Supporting Information Fig. S1). As Trp69 and Trp74 are located in close proximity to the conserved Asn73 residue of the HNH motif and the CXXH motif present at residues 78-81, the binding of divalent ions at these sites might be expected to alter the tryptophan emission spectrum. Alternatively, structural changes associated with metal binding could cause global conformational changes, which might alter the emission spectrum of gp74. Figure 6(A) shows the emission spectra for gp74 in the absence and presence of Ni2+, and reveals direct binding of Ni2+ to gp74. Fluorescence titration experiments [Fig. 6(B)] were used to obtain affinities for binding of Ni2+ to gp74. Our Ni2+-binding curves fit well (r2 = 0.995) to an equation assuming that one Ni2+ ion binds one gp74 molecule (see Fluorescence metal-binding studies), but not well to an equation assuming that two (or more) Ni2+ ions can cooperatively bind one gp74 molecule. Our data indicate that one divalent metal Ni2+ ion binds gp74 with relatively low affinity of 165.0 ± 0.4 μM. We expect that the affinity of gp74 for divalent metal ions, such as Ni2+, is higher in the presence of DNA, which is consistent with our observations that gp74 induces digestion of lambda phage DNA in the presence of stoichiometric concentrations of Ni2+ (Supporting Information Fig. S4).
Figure 6.
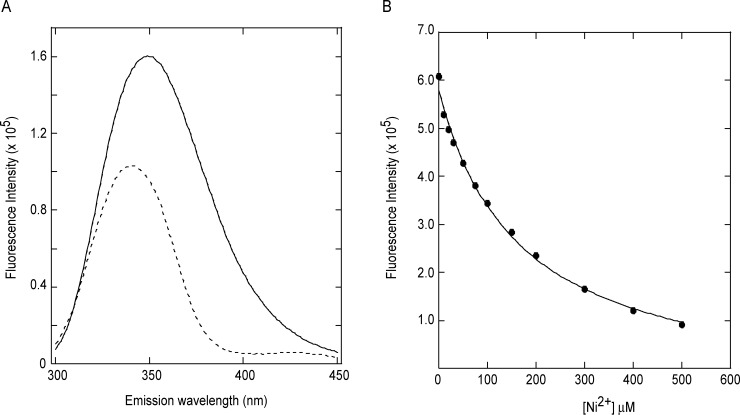
Intrinsic tryptophan fluorescence data indicates direct binding of Ni2+ to gp74. (A) Emission spectra of 1 μM gp74 in the absence (solid line) and presence of 1M Ni2+ ions (dotted line). (B) Binding of Ni2+ ions to gp74 as monitored by intrinsic tryptophan fluorescence. The data (solid circles) were fit assuming a 1:1 complex (solid line),42 as described in Fluorescence metal-binding studies.
Discussion
The data presented in this article, for the first time, demonstrate that gp74 from the bacteriophage HK97 functions as an HNH endonuclease and contains all of the hallmarks of this group of proteins. A sequence alignment of HK97 gp74 and related proteins reveals that gp74 contains the canonical HNH motif,8 which contains an invariant His residue (His43 in gp74) that is part of an Asp-His or a His-His dyad, as seen in the bacteriophage homing endonuclease I-HmuI18 or the bacterial colicins E719 and E9,13, 22 respectively. Like I-HmuI, gp74 contains an Asp-His dyad, with His43 acting as the catalytic base and Asp42 forming part of the metal-binding site. While Asp42 likely mediates contact with the metal ion in the active site, this residue is not defined as part of the HNH motif because it is not highly conserved in other HNH endonucleases.16–20, 23, 29 The second hallmark of the HNH motif is an invariant Asn residue (Asn73 in gp74) that is usually followed by a Leu residue.8 While gp74, like most of the proteins identified in our BLASTp search, has a Trp in this position instead of a Leu, the large hydrophobic nature of this position is maintained. The final residue defining the HNH motif is a His residue (His82 in gp74), which is involved in metal binding. Often the metal-binding His residue is followed by another His, which is three or four residues C-terminal to the metal-binding His residue and is located in a loop outside the ββα-metal fold. Gp74 contains an additional His residue, but this His is at position 110, 29 residues C-terminal of His82 and hence, may or may not be involved in metal binding.
As with other HNH proteins, the activity of gp74 requires a divalent metal ion cofactor and gp74 is active with many metals, although to varying degrees. The difference in gp74 activity may be due to the different chelation properties of the metals in the enzyme and the different orientation of the HNH residues that result. This has previously been observed in colicin E9, for which X-ray structures reveal that a Zn2+ ion bound to the E9 DNase•dsDNA complex displays a tetrahedral coordination, while a bound Mg2+ ion adopts an octahedral geometry.23 These differences in the coordination geometry result in a different orientation for the protein ligands with respect to the scissile bond of the DNA backbone and likely results in a different mechanism of DNA digestion.22 In colicin E9, this different arrangement of residues may be the reason for different activity of the protein toward different substrates. In the presence of Ni2+, colicin E9 displays a greater activity for single-stranded DNA, whereas it primarily mediates digestion of double-stranded DNA in the presence of Mg2+. It is expected that different metals would be coordinated in a similar manner in gp74, with Ni2+ (and Zn2+) adopting a tetrahedral coordination geometry and Mg2+ displaying an octahedral coordination, as seen for most metalloproteins.31, 32 Differences in metal binding in gp74 may also result in differences in the concentration-dependence of the activity.
Other differences in the activity of gp74 with different metals may result from additional binding sites outside the HNH motif for particular metal ions, which would alter its activity. Gp74 also possesses a CXXC motif N-terminal to the ββα-metal fold as well a CXXH motif as part of the HNH motif. CXXC and CXXH motifs have been previously implicated in binding metal ions and form the basis of the Zn2+-binding site in zinc finger domain proteins.33 There are a number of HNH motif-containing proteins that also possess CXXC and CXXH motifs which bind Zn2+. For example, in the structure of the type II restriction endonuclease, Hpy99I, a Zn2+ ion is coordinated by two CXXC motifs.17 When the sequence of Hyp99I is compared with that of gp74, the Zn2+ binding CXXC motifs in Hpy99I align with the CXXC and CXXH motifs found between residues 26–29 and 78–81, respectively, of gp74. The restriction enzymes KpnI15 and PacI16 each also contain a zinc finger in addition to the HNH motif. Although the location of the first CXXC motif in the primary sequence varies between these two proteins and the Hpy99I HNH endonuclease, the location of the zinc finger is close to the HNH motif in the structure and is necessary for protein stability, DNA binding, and DNA digestion.34 Therefore, gp74 may also coordinate a Zn2+ ion via its CXXC and CXXH motifs, which would result in a different orientation for the HNH structural motif. Because the CXXH motif in gp74 is located just N-terminal to His82, it is expected that Cys78 and His81 are located in the α-helix of the ββα-metal fold, as seen in other HNH endonuclease domain family members. Thus, binding a Zn2+ ion at this site may result in a different orientation for this α-helix and could explain the greatly decreased endonuclease activity observed at Zn2+ concentrations above 0.5 mM. The CXXC and CXXH motifs are unlikely to coordinate Ni2+ ions as analysis of high resolution crystal structures of metalloproteins reveals that Ni2+ ions are most often coordinated by multiple His residues.31, 32 This is consistent with our finding that indicates that only one Ni2+ ion binds gp74 and stoichiometric amounts of Ni2+ are sufficient for endonuclease activity.
Several HNH endonucleases, such as the bacteriophage endonucleases I-HmuI18 and mobE,25 and the restriction enzyme PacI16 recognize and cleave-specific sequences of DNA. Structures of the different HNH proteins bound to DNA indicate that those that cleave specific DNA sequences possess DNA recognition domains in addition to the catalytic HNH motif. The extent of digestion of the λ DNA into very small fragments suggests that gp74 functions to cleave DNA in a promiscuous manner similar to the bacterial colicins E7 and E9.20, 22 However, there may be additional phage proteins that mediate recognition of specific DNA sequences, or host factors that act as co-nucleases as previously observed for the bacteriophage P1 HNH endonuclease and E. coli RecA.35 Sequence analysis of gp73, which is encoded by a gene located just 5′ of the gene for gp74, indicates the presence of a KilA-N domain, a conserved DNA-binding domain found in many bacterial and eukaryotic DNA viruses.8, 10 As phage genes are clustered according to their structure or function,1 it is possible the gp73 modulates the activity and/or specificity of gp74. Studies of gp74 activity in the presence of gp73 are necessary to establish any functional relationship between the two proteins.
HNH endonucleases are present in many phages and play a variety of roles in the phage life cycle.11, 24, 25 Although the biological role of gp74 is currently not known, the conservation of the location of gp74 in the HK97 genome suggests a possible role for gp74. The location of gp74 in the HK97 genome next to the terminase gene is conserved among other phage HNH endonucleases that also contain CXXC/CXXH motifs, such as gp111 from c2 phage and gp13 from the Lactococcal bacteriophage biL170,36 suggesting that the HNH endonuclease genes are in the same functional unit as the terminase gene and thus may be involved in DNA packaging.36 In HK97, the functional unit may also include gp73 to mediate sequence-specific DNA digestion. The current study, which defines the biochemical role for gp74, provides a platform to test this hypothesis for the role of gp74 in DNA packaging and ultimately phage maturation.
Materials and Methods
Sequence alignment
A search of sequences related to gp74 using BLAST37, 38 indicated that gp74 from the HK97 bacteriophage is a possible member of the HNHc-endonuclease family of enzymes found among bacteria and viruses.8, 10 A diverse set of sequences from bacteria and phage were selected with sequence identities of greater than 60% with gp74. Sequences were aligned in Clustal W39, 40 and putative residues for the HNH motif were identified.
Expression and purification of gp74 from the bacteriophage HK97
The full length gp74 protein from the HK97 bacteriophage was expressed a fusion protein with an N-terminal 6xHis tag using a pET15b-derived expression vector. Proteins were expressed in E. coli BL21 (DE3) STAR cells grown in M9 minimal media. Cells were grown at 37°C until an OD600 of 0.7 was reached, at which point the incubating temperature was reduced to 16°C and gene expression was induced with isopropyl β-d-thiogalactoside (IPTG) at a concentration of 1 mM. After 16–20 h, cells were harvested by centrifugation (∼4500 × g, 15 min) and pellets were stored at –20°C.
Protein purification was conducted at 4°C. Cell pellets were resuspended in 15 mL of lysis buffer (20 mM Tris HCl, pH 7.9, 150 mM NaCl, 2 mM β-mercaptoethanol, 5 mM imidazole, 150 μM PMSF, 5 mM benzamidine, 1 mg/lysozyme, 2 mg/mL deoxycholic acid) per unit OD600 per liter of culture. The cells were lysed by brief sonication on ice and centrifuged at 10,000 × g for 30 min. The soluble 6xHis-gp74 was loaded onto a 5 mL Fast Flow Ni2+ column (GE Healthcare) that was pre-equilibrated with buffer A (20 mM tris HCl, pH 7.9, 500 mM NaCl, 20 mM imidazole, and 2 mM β-mercaptoethanol). Non-specifically bound proteins were washed with 6 column volumes of buffer A and the 6xHis-gp74 fusion protein was eluted with buffer A containing 400 mM imidazole and 5 mM DTT instead of β-mercaptoethanol.
The elution fractions containing 6xHis-gp74 were pooled and dialyzed against 50 mM Na+ phosphate, pH 7.0, 50 mM NaCl, 5 mM β-mercaptoethanol, 5 mM EDTA. TEV protease (1 mg TEV/ 40 mg protein) was added to the sample to cleave the 6xHis-tag from gp74 during dialysis. The gp74 was then purified to homogeneity by size exclusion chromatography using a Superdex 75 column (GE Healthcare) in either 50 mM Na+ phosphate or 20 mM HEPES, pH 7.0, with 150 mM NaCl, 1 mM 6-aminocaproic acid, 5 mM benzamidine, 1 mM PMSF, 5 mM β-mercaptoethanol. Elution fractions containing gp74 were pooled and dialyzed against 20 mM HEPES, pH 7.0, 5 mM β-mercaptoethanol, and stored at 4°C until used. Protein concentration was determined by amino acid analysis and by A280 in 8M urea with an extinction coefficient of 25,840 M−1cm−1.41
DNA digestion assay
Digestion assays were performed with 48 μg/mL (3.4 μM) of gp74 and 50 μg/mL lambda DNA (New England Biolabs), unless otherwise stated. Digestion experiments were conducted in 20 mM HEPES, pH 7.0. A stock solution of gp74 in 20 mM HEPES, pH 7.0, 5 mM β-mercaptoethanol was diluted at least 100-fold in these reactions, so that the final concentration of β-mercaptoethanol was less than 0.05 mM.
A variety of divalent metal ions (Ni2+, Mg2+, and Zn2+) at concentrations of 0.5 to 10 mM were tested as cofactors, and the pH of the solution was varied from 5 to 8. Every 60 min, 20 μL of sample was removed and the reaction was stopped by addition of EDTA to a final concentration of 0.025 mM and DNA loading buffer (New England Biolabs). Samples were analyzed by 1% agarose gel electrophoresis.
CD spectroscopy
Circular dichroism (CD) spectra of gp74 were recorded from 195 nm to 260 nm in 0.2 nm steps at 25°C on an Aviv 250 CD spectrometer with a bandwidth of 1.0 nm using a 1 cm path length quartz cell. Each CD spectrum was an average of five scans. Samples contained 2 μM gp74 in 20 mM Na+ phosphate, 50 mM NaCl at pH 5.0, 6.0, 7.0, or 8.0.
Fluorescence metal-binding studies
Divalent metal binding was probed using intrinsic tryptophan fluorescence of gp74. Gp74 has four tryptophan residues, at least one of which is near the metal-binding site. Fluorescence emission spectra were recorded on a Horiba-Jovin Fluoromax 4 fluorimeter at 25°C using an excitation wavelength of 280 nm and a slit width of 5 nm. Emission spectra were recorded from 300 nm to 450 nm using an emission slit width of 5 nm. For Ni2+ titration experiments, fluorescence emission was monitored at 346 nm, the wavelength at which the difference in fluorescence between the free and Ni2+-bound gp74 is at a maximum. Binding experiments were conducted with a Hamilton Microlab 541C automated titrator. Affinities were measured in 20 mM HEPES, pH 7.0 with gp74 concentrations of 1 μM. Ni2+ ions were added at concentrations of 0 to 0.5 mM. Titration data were fit to the following equation:
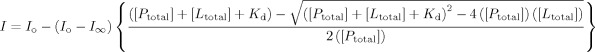 |
where I is the fluorescence intensity at a given total concentration of Ni2+ ions, [Ltotal], I∞ is the fluorescence intensity at saturation, Io is the fluorescence intensity in absence of ligand, Kd is the dissociation constant, and [Ptotal] is the total concentration of gp74 in the reaction. This equation assumes a 1:1 complex.42
Conclusion
The protein gp74 from the bacteriophage HK97 possesses an HNH motif. HNH motifs are formed from two His residues and an Asn residue, and bind divalent metals. We demonstrate that gp74 cleaves DNA at multiple sites in the presence of divalent metals. We also show a difference in the activity of gp74 at acidic pH in the presence of Ni2+ and Zn2+, which suggests that gp74 has an additional metal-binding site. These studies show that gp74 is an HNH endonuclease and provide a platform for determining the function of gp74 in the bacteriophage.
Acknowledgments
The authors wish to acknowledge John L. Rubinstein, Jorge P. Lopez-Alonso, and Paul D. Sadowski for critically reading the manuscript and for useful discussion.
Glossary
Abbreviations
- CD
circular dichroism
- CXXC motif
metal-binding site formed by two Cys residues
- DTT
dithiothreitol
- E. coli
Escherichia coli
- EDTA
ethylenediaminetetraacetic acid
- HMM
Hidden Markov Model
- HNH endonuclease
an endonuclease with an active site formed by two His residues and an Asn residue
- IPTG
isopropyl β-d-thiogalactoside
- KilA-N domain
DNA-binding domain
- λ DNA
lambda phage DNA
- PMSF
phenylmethylsulfonyl fluoride
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Juhala RJ, Ford ME, Duda RL, Youlton A, Hatfull GF, Hendrix RW. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol. 2000;299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- 2.Cardarelli L, Lam R, Tuite A, Baker LA, Sadowski PD, Radford DR, Rubinstein JL, Battaile KP, Chirgadze N, Maxwell KL, Davidson AR. The crystal structure of bacteriophage HK97 gp6: defining a large family of head-tail connector proteins. J Mol Biol. 2010;395:754–768. doi: 10.1016/j.jmb.2009.10.067. [DOI] [PubMed] [Google Scholar]
- 3.Mahdi AA, Sharples GJ, Mandal TN, Lloyd RG. Holliday junction resolvases encoded by homologous rusA genes in Escherichia coli K-12 and phage 82. J Mol Biol. 1996;257:561–573. doi: 10.1006/jmbi.1996.0185. [DOI] [PubMed] [Google Scholar]
- 4.Chang CY, Nam K, Young R. S gene expression and the timing of lysis by bacteriophage lambda. J Bacteriol. 1995;177:3283–3294. doi: 10.1128/jb.177.11.3283-3294.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young R, Blasi U. Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev. 1995;17:191–205. doi: 10.1111/j.1574-6976.1995.tb00202.x. [DOI] [PubMed] [Google Scholar]
- 7.Szybalski EH, Szybalski W. A comprehensive molecular map of bacteriophage lambda. Gene. 1979;7:217–270. doi: 10.1016/0378-1119(79)90047-7. [DOI] [PubMed] [Google Scholar]
- 8.Keeble AH, Mate MJ, Kleanthous C. HNH Endonucleases. Nucleic Acids Mol Biol. 2005;16:49–65. [Google Scholar]
- 9.Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Jones S, Howe KL, Marshall M, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoddard BL. Homing endonuclease structure and function. Q Rev Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 11.Landthaler M, Lau NC, Shub DA. Group I intron homing in Bacillus phages SPO1 and SP82: a gene conversion event initiated by a nicking homing endonuclease. J Bacteriol. 2004;186:4307–4314. doi: 10.1128/JB.186.13.4307-4314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chak KF, Kuo WS, Lu FM, James R. Cloning and characterization of the ColE7 plasmid. J Gen Microbiol. 1991;137:91–100. doi: 10.1099/00221287-137-1-91. [DOI] [PubMed] [Google Scholar]
- 13.Pommer AJ, Wallis R, Moore GR, James R, Kleanthous C. Enzymological characterization of the nuclease domain from the bacterial toxin colicin E9 from Escherichia coli. Biochem J. 1998;334:387–392. doi: 10.1042/bj3340387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sano Y, Matsui H, Kobayashi M, Kageyama M. Molecular structures and functions of pyocins S1 and S2 in Pseudomonas aeruginosa. J Bacteriol. 1993;175:2907–2916. doi: 10.1128/jb.175.10.2907-2916.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saravanan M, Bujnicki JM, Cymerman IA, Rao DN, Nagaraja V. Type II restriction endonuclease R.KpnI is a member of the HNH nuclease superfamily. Nucleic Acids Res. 2004;32:6129–6135. doi: 10.1093/nar/gkh951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen BW, Heiter DF, Chan SH, Wang H, Xu SY, Morgan RD, Wilson GG, Stoddard BL. Unusual target site disruption by the rare-cutting HNH restriction endonuclease PacI. Structure. 2010;18:734–743. doi: 10.1016/j.str.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sokolowska M, Czapinska H, Bochtler M. Crystal structure of the beta beta alpha-Me type II restriction endonuclease Hpy99I with target DNA. Nucleic Acids Res. 2009;37:3799–3810. doi: 10.1093/nar/gkp228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen BW, Landthaler M, Shub DA, Stoddard BL. DNA binding and cleavage by the HNH homing endonuclease I-HmuI. J Mol Biol. 2004;342:43–56. doi: 10.1016/j.jmb.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 19.Cheng YS, Hsia KC, Doudeva LG, Chak KF, Yuan HS. The crystal structure of the nuclease domain of colicin E7 suggests a mechanism for binding to double-stranded DNA by the H-N-H endonucleases. J Mol Biol. 2002;324:227–236. doi: 10.1016/s0022-2836(02)01092-6. [DOI] [PubMed] [Google Scholar]
- 20.Wang YT, Yang WJ, Li CL, Doudeva LG, Yuan HS. Structural basis for sequence-dependent DNA cleavage by nonspecific endonucleases. Nucleic Acids Res. 2007;35:584–594. doi: 10.1093/nar/gkl621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang H, Yuan HS. The conserved asparagine in the HNH motif serves an important structural role in metal finger endonucleases. J Mol Biol. 2007;368:812–821. doi: 10.1016/j.jmb.2007.02.044. [DOI] [PubMed] [Google Scholar]
- 22.Pommer AJ, Cal S, Keeble AH, Walker D, Evans SJ, Kuhlmann UC, Cooper A, Connolly BA, Hemmings AM, Moore GR, James R, Kleanthous C. Mechanism and cleavage specificity of the H-N-H endonuclease colicin E9. J Mol Biol. 2001;314:735–749. doi: 10.1006/jmbi.2001.5189. [DOI] [PubMed] [Google Scholar]
- 23.Mate MJ, Kleanthous C. Structure-based analysis of the metal-dependent mechanism of H-N-H endonucleases. J Biol Chem. 2004;279:34763–34769. doi: 10.1074/jbc.M403719200. [DOI] [PubMed] [Google Scholar]
- 24.Goodrich-Blair H, Shub DA. Beyond homing: competition between intron endonucleases confers a selective advantage on flanking genetic markers. Cell. 1996;84:211–221. doi: 10.1016/s0092-8674(00)80976-9. [DOI] [PubMed] [Google Scholar]
- 25.Wilson GW, Edgell DR. Phage T4 mobE promotes trans homing of the defunct homing endonuclease I-TevIII. Nucleic Acids Res. 2009;37:7110–7123. doi: 10.1093/nar/gkp769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehta P, Katta K, Krishnaswamy S. HNH family subclassification leads to identification of commonality in the His-Me endonuclease superfamily. Protein Sci. 2004;13:295–300. doi: 10.1110/ps.03115604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ku WY, Liu YW, Hsu YC, Liao CC, Liang PH, Yuan HS, Chak KF. The zinc ion in the HNH motif of the endonuclease domain of colicin E7 is not required for DNA binding but is essential for DNA hydrolysis. Nucleic Acids Res. 2002;30:1670–1678. doi: 10.1093/nar/30.7.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drouin M, Lucas P, Otis C, Lemieux C, Turmel M. Biochemical characterization of I-CmoeI reveals that this H-N-H homing endonuclease shares functional similarities with H-N-H colicins. Nucleic Acids Res. 2000;28:4566–4572. doi: 10.1093/nar/28.22.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raaijmakers H, Vix O, Toro I, Golz S, Kemper B, Suck D. X-ray structure of T4 endonuclease VII: a DNA junction resolvase with a novel fold and unusual domain-swapped dimer architecture. EMBO J. 1999;18:1447–1458. doi: 10.1093/emboj/18.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galburt EA, Chevalier B, Tang W, Jurica MS, Flick KE, Monnat RJ, Jr, Stoddard BL. A novel endonuclease mechanism directly visualized for I-PpoI. Nat Struct Biol. 1999;6:1096–1099. doi: 10.1038/70027. [DOI] [PubMed] [Google Scholar]
- 31.Dokmanic I, Sikic M, Tomic S. Metals in proteins: correlation between the metal-ion type, coordination number and the amino-acid residues involved in the coordination. Acta Cryst. 2008;D64:257–263. doi: 10.1107/S090744490706595X. [DOI] [PubMed] [Google Scholar]
- 32.Rulisek L, Vondrasek J. Coordination geometries of selected transition metal ions (Co2+, Ni2+, Cu2+, Zn2+, Cd2+, and Hg2+) in metalloproteins. J Inorg Biochem. 1998;71:115–127. doi: 10.1016/s0162-0134(98)10042-9. [DOI] [PubMed] [Google Scholar]
- 33.Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saravanan M, Vasu K, Kanakaraj R, Rao DN, Nagaraja V. R.KpnI, an HNH superfamily REase, exhibits differential discrimination at non-canonical sequences in the presence of Ca2+ and Mg2+ Nucleic Acids Res. 2007;35:2777–2786. doi: 10.1093/nar/gkm114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gruenig MC, Lu D, Won SJ, Dulberger CL, Manlick AJ, Keck JL, Cox MM. Creating directed double-strand breaks with the Ref protein: a novel RecA-dependent nuclease from bacteriophage P1. J Biol Chem. 2011;286:8240–8251. doi: 10.1074/jbc.M110.205088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crutz-Le Coq AM, Cesselin B, Commissaire J, Anba J. Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiology. 2002;148:985–1001. doi: 10.1099/00221287-148-4-985. [DOI] [PubMed] [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 40.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Viguera AR, Arrondo JL, Musacchio A, Saraste M, Serrano L. Characterization of the interaction of natural proline-rich peptides with five different SH3 domains. Biochemistry. 1994;33:10925–10933. doi: 10.1021/bi00202a011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.