Abstract
Interleukin-5 (IL-5), a major hematopoietin, stimulates eosinophil proliferation, migration, and activation, which have been implicated in the pathogenesis of allergic inflammatory diseases, such as asthma. The specific IL-5 receptor (IL-5R) consists of the IL-5 receptor α subunit (IL-5RA) and the common receptor β subunit (βc). IL-5 binding to IL-5R on target cells induces rapid tyrosine phosphorylation and activation of various cellular proteins, including JAK1/JAK2 and STAT1/STAT5. Here, we report the crystal structure of dimeric IL-5 in complex with the IL-5RA extracellular domains. The structure revealed that IL-5RA sandwiches the IL-5 homodimer by three tandem domains, arranged in a “wrench-like” architecture. This association mode was confirmed for human cells expressing IL-5 and the full-length IL-5RA by applying expanded genetic code technology: protein photo-cross-linking experiments revealed that the two proteins interact with each other in vivo in the same manner as that in the crystal structure. Furthermore, a comparison with the previously reported, partial GM-CSF•GM-CSFRA•βc structure enabled us to propose complete structural models for the IL-5 and GM-CSF receptor complexes, and to identify the residues conferring the cytokine-specificities of IL-5RA and GM-CSFRA.
Keywords: interleukin (IL), common receptor β subunit (βc), cytokine, eosinophilic, asthma
Introduction
Acquired immune responses are involved in the late phase of infection and the generation of immunological memory, and are mediated by a specialized group of lymphocytes, including T and B cells. These lymphocytes recognize antigens via cell surface antigen receptors. The activated T cells release cytokines and chemokines, which activate phagocytosis by the innate lymphocyte cells, and provide increased protection against pathogens.1
Interleukin-5 (IL-5) is a conserved hematopoietic cytokine. IL-5 is mainly produced by Th2 cells after stimulation with antigens, and by mast cells upon stimulation with an allergen/IgE complex or a calcium ionophore.2–6 The overexpression of IL-5 in vivo significantly increases the numbers of eosinophils and B cells, while mice lacking the functional gene for IL-5 display various developmental and functional impairments in the B cell and eosinophil lineages. In humans, the biological effects of IL-5 have been best characterized for eosinophils. The recent expansion of research on eosinophil development and the activation and pathogenesis of eosinophil-dependent inflammatory diseases has led to enhanced therapeutic options.7 The intravenous administration of a humanized anti-IL-5 monoclonal antibody reduces the baseline of bronchial mucosal eosinophils in mild asthma, thus providing important implications for strategies to inhibit the actions of IL-5, for treating asthma and other allergic diseases.8
IL-5 is a dimeric glycoprotein with a four-helix bundle motif, and it acts on target cells by binding to its specific IL-5 receptor (IL-5R), which consists of an IL-5 receptor α subunit (IL-5RA) and a common receptor β subunit (βc).9–12 IL-5RA specifically binds IL-5, and induces the recruitment of βc to IL-5R.13–17 The βc subunit is a signal-transducing molecule shared with two receptors for monomeric cytokines, interleukin-3 (IL-3), and granulocyte macrophage colony stimulation factor (GM-CSF) (IL-3R and GM-CSFR, respectively).18–21 Both IL-5RA and βc have a membrane proximal proline-rich sequence (PPXP motif) in their cytoplasmic domains, which is also essential for IL-5-induced signal transduction.22–25
IL-5 stimulation induces rapid tyrosine phosphorylation of various cellular proteins, including βc, the SH2/SH3-containing proteins Vav, HS1, Shc, and so forth. Btk, the Btk-associated molecules, and the JAK1/JAK2, STAT1/STAT5, PI3K, and MAP kinases, activates downstream signaling molecules,24, 26–28 and leads to the maintenance of the survival and functions of B cells and eosinophils. The activation of JAK2 and STAT5 is essential for IL-5-dependent signal transduction.29–31 Especially, IL-5 binding to IL-5R activates the JAK kinases through the associations of IL-5RA with JAK2 and of βc with JAK1, by means of their cytoplasmic domains.28 On the other hand, GM-CSF binding to GM-CSFR induces JAK2 activation via the formation of a unique dodecameric receptor complex.32 The activation of JAK2/STAT5 by the mature GM-CSF receptor has been well characterized, in both normal hematopoiesis and disease states, where aberrant signaling has been shown to contribute to dysregulated myelopoiesis. In this way, the JAK/STAT signaling pathways differ between IL-5 and GM-CSF, although they share the βc subunit.
In this study, we determined the crystal structure of the 1:1 complex between dimeric IL-5 and the IL-5RA extracellular region, consisting of three domains (D1-D3). The D2 and D3 domains, as well as the D1 domain, of IL-5RA contribute to IL-5 binding. In the complex, the two subunits of the IL-5 dimer interact with the IL-5RA molecule in different manners. Moreover, the application of an expanded genetic code to the full-length IL-5RA protein revealed that the two proteins exhibit the same interaction mode on the cell surface. A comparison with the previously reported GM-CSF•GM-CSFRA•βc complex structure enabled us to define the residues determining the cytokine-specificities of the two receptors.
Results
Structure of IL-5 bound to IL-5RA
We expressed the mature IL-5 (residues 23–134) and IL-5RA ectodomain (residues 21–335) fragments by cell-free protein synthesis. These fragments precipitated during synthesis, but were prepared as a soluble protein complex by co-refolding. The purified complex of IL-5 and the IL-5RA ectodomain (designated hereafter as the IL-5•IL-5RA complex) was successfully crystallized. The crystals contain one protein complex, with a stoichiometry of 2:1 (one IL-5 dimer plus one receptor).
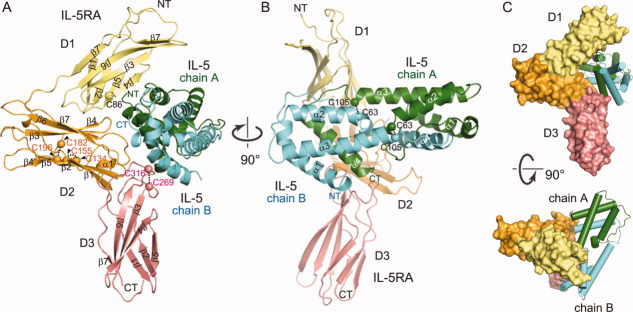
We determined the crystal structure of the IL-5•IL-5RA complex at 2.7 Å resolution by the single-wavelength anomalous dispersion (SAD) method, using the crystal of the selenomethionine-labeled proteins. The crystallographic data are summarized in Table I. In the complex, IL-5 forms a homodimer, and IL-5RA sandwiches the IL-5 dimer [Fig. 1(A,B)]. The IL-5 dimer and IL-5RA in the complex are arranged in a nearly perpendicular manner [Fig. 1(B)]. Similar to the other class I cytokine receptors,33 IL-5RA is composed of three tandem domains (D1, D2, and D3). The N-terminal D1 domain adopts the fibronectin III-like (FnIII) domain fold. The D2 and D3 domains constitute the canonical cytokine receptor homology module (CRM),34 which is also composed of a pair of FnIII domains arranged orthogonally [Fig. 1(A)]. Each domain of IL-5RA includes seven anti-parallel β strands (D1: β1–β7, D2: β1–β7, D3: β1–β7) [Fig. 1(A)]. The D1 domain contains one cysteine (Cys86) on the β5 strand, which is not involved in disulfide bond formation. The D2 domain, consisting of about 100 amino acid residues, contains two conserved disulfide bonds (Cys134–Cys155 and Cys182–Cys196), while the D3 domain, including the conserved WSXWS motif,35 contains one (Cys269–Cys316). On the other hand, the IL-5 protomer is composed of four α-helices (α1–α4), connected by three loops. The IL-5 dimer is stabilized by two intermolecular disulfide bonds, Cys63–Cys105 and Cys105–Cys63 (chain A–chain B) [Fig. 1(B)]. The structures of the IL-5 dimer alone10 and in complex with IL-5RA superpose well (root-mean-square deviation [rmsd] 1.0 Å for Cα atoms), except for the α2–α3 loop of IL-5 chain A and the α3–α4 loop of IL-5 chain B (represented as an arrowhead) [Fig. 2(A)]. In the complex structure, α4 of IL-5 chain A is accommodated in a large concave surface of IL-5RA, and is covered by the three α-helices (α1–α3) of IL-5 chain B (discussed below).
Table I.
X–Ray Data Collection, Phasing, and Refinement Statistics of the IL-5•IL-5RA Complex
| Native | Se-Met | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 58.6, 88.7, 126.7 | 58.5, 89.3, 126.1 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Wavelength (Å) | 1.0000 | 0.9789 |
| Resolution (Å) | 50–2.7 (2.80–2.70) | 50–3.0 (3.11–3.00) |
| Unique reflections | 18667 | 13915 |
| Redundancy | 6.7 | 7.2 |
| Completeness (%) | 99.5 (99.9) | 99.4 (98.5) |
| I / σ (I) | 20.1 (3.9) | 18.5 (7.2) |
| Rsyma (%) | 7.7 (47.9) | 9.6 (52.6) |
| SAD analysis | ||
| Resolution (Å) | 3.5 | |
| No. of sites | 4 | |
| FOMb | 0.66 | |
| Refinement | ||
| Resolution (Å) | 43.0–2.7 | |
| No. of reflections | 18615 | |
| No. of protein atoms | 4205 | |
| No. of water molecules | 28 | |
| Rwork (%) | 23.5 | |
| Rfree (%)c | 28.7 | |
| r.m.s.d. bond length (Å) | 0.003 | |
| r.m.s.d. bond angles (°) | 0.7 | |
| Ramachandran plot | ||
| Most favored regions (%) | 96.9 | |
| Additional allowed regions (%) | 3.0 | |
| Generously allowed regions (%) | 0.1 | |
| Disallowed regions (%) | 0.0 |
All numbers in parentheses represent last outer shell statistics.
Rsym=∑ |Iavg–Ii| / ∑Ii, where Ii is the observed intensity and Iavg is the average intensity.
Figure of merit after SAD phasing.
Rfree is calculated for 5% of randomly selected reflections excluded from refinement.
Figure 1.
Structure of the IL-5•IL-5RA complex. IL-5RA is composed of the FnIII domain (D1: yellow, amino acid residues 27–121) and CRM (D2: orange, amino acid residues 122–237 and D3: salmon pink, amino acid residues 238–332). IL-5RA bound to IL-5 (chain A: green and chain B: cyan), presented as a ribbon diagram. The two views (A) and (B) are related by a 90° rotation about the vertical axis. (C) Representation of the IL-5•IL-5RA complex with a wrench-like architecture. The two views are related by a 90° rotation about the horizontal axis.
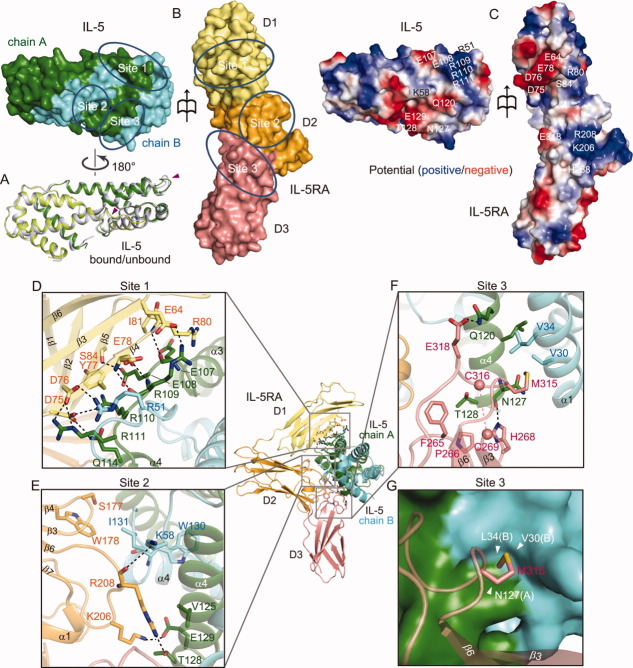
Figure 2.
IL-5 binding to IL-5RA is mediated by three distinct interfaces. A: Superimposition of IL-5 alone and IL-5 in the complex. B: Surface representation of IL-5 and IL-5RA, showing the three binding interfaces. Surface colors are same as in Figure 1. C: Potential mapping of the binding interfaces between IL-5 and IL-5RA. The amino acid residues participating in interactions involving salt bridges and hydrogen bonds are indicated in sites 1, 2, and 3. Positive potential: blue, negative potential: red. D: Details of site 1, composed of the IL-5RA D1 domain and IL-5. E: Details of site 2, composed of the IL-5RA D2 domain and IL-5. F: Details of site 3, composed of the IL-5RA D3 domain and IL-5. G: Surface representation of the IL-5 dimer binding-pocket in site 3, which is recognized by the β6-CT loop of IL-5RA.
Recently, Patino et al. published the crystal structure of human IL-5 in complex with a five-point mutant of IL-5RA (C66A, K72M, L138M, K167M, and L234M) (PDB ID: 3QT2).36 They described the wrapping of the “preformed” architecture of IL-5RA around the four-helix IL-5 as resembling that of a wrench around a nut. Their reported complex and our wild-type complex superimposed almost perfectly (rmsd 0.3 Å for all Cα atoms), and displayed the “wrench-like” architecture [Fig. 1(C)].
IL-5•IL-5RA binding interface
IL-5RA forms an extensive binding interface with IL-5, burying ∼ 1,115 Å2 of the surface area. Approximately 54% of this buried surface area is mediated by the IL-5RA D1 domain. There are three main interaction sites, 1, 2, and 3, at the binding interface [Fig. 2(B)].
Site 1 is composed of the β3–β5 regions of the IL-5RA D1 domain, the α3–α4 loops of IL-5 chain A, and the α1–α2 loops of IL-5 chain B [Fig. 1(A,B), and 2(D)]; these regions are arranged in an anti-parallel manner, and form hydrogen-bonding, hydrophobic, and polar interactions. The 107-Glu-Glu-Arg-Arg-Arg-111 sequence of IL-5 interacts with the IL-5RA D1 domain. Arg109 of IL-5, which is located at the center of the interface, forms a salt bridge with Glu64, as well as two main chain-main chain hydrogen bonds with Glu78 of IL-5RA. Arg110 and Arg111 of IL-5 chain A form a salt bridge with Asp75 and Asp76, respectively, of IL-5RA, while Glu107 and Glu108 of IL-5 chain A hydrogen bond with Arg80 and Ser84, respectively, of IL-5RA. In addition, Gln114 of IL-5 chain A hydrogen bonds with Asp75 of IL-5RA. Interestingly, Asp76 of IL-5RA participates in hydrogen bond formation with both Arg111 of IL-5 chain A and Arg51 of IL-5 chain B. The hydrophobic interactions between Tyr77 and Ile81 (β4) in the IL-5RA D1 domain reinforce the salt bridges and the hydrogen bonds [Fig. 2(D)]. Consequently, these interactions, including many salt bridges, constitute an interface with good charge complementarity between the IL-5RA D1 domain and the α3–α4 loop of IL-5 chain A, as revealed by the potential mapping of the interface of each protein [Fig. 2(C)].
Site 2 consists of the β3–β4 and β5–β6 loops of the IL-5RA D2 domain, which interact with the C termini of IL-5 chains A and B [Fig. 2(E)]. Glu129 of IL-5 chain A forms a salt bridge with Lys206, and is also close to Arg208 from the β5–β6 loop of the IL-5RA D2 domain. The backbone of Arg208 forms a hydrogen bond with Lys58 of IL-5 chain B, and the side chain of Arg208 utilizes its polar and hydrophobic characteristics to interact with Val125 and Thr128 of IL-5 chain A. In the β3–β4 loops of the IL-5RA D2 domain, hydrophobic interactions facilitate the contacts of Ser177 and Trp178 with Ile131 and W130, respectively, of IL-5 chain B.
Site 3 is composed of the β2–β3 and β6–β7 loops of the IL-5RA D3 domain, which contact α4 of IL-5 chains A and B [Fig. 2(F)]. First, the side chain of His268, from the D3 β2–β3 loop of IL-5RA, hydrogen bonds with the main chain oxygen atom of Asn127 (IL-5 chain A). Second, the D3 β6–β7 loop of IL-5RA provides the side chain of Glu318, which hydrogen bonds with Gln120 of IL-5 chain A. These hydrophilic interactions are surrounded by the hydrophobic groups of Phe265 and Pro266 (IL-5RA D3 domain), Met315 (IL-5RA D3 domain), and Thr128 (IL-5 chain A). Furthermore, the other protomer of the IL-5 dimer also interacts with the receptor: Val30 and Leu34 (IL-5 chain B) interact with Met315 (IL-5RA D3 domain) [Fig. 2(G)].
Thus, the three interfaces, sites 1, 2 and 3, exhibit different types of interactions. Site 1 is formed by the sequence of IL-5, which interacts tightly with IL-5RA by three salt bridges, surrounded by hydrophobic and hydrogen-bonding interactions. On the other hand, sites 2 and 3 each contain two contact points between IL-5 and IL-5RA, so that they snugly fit with each other.
Photo-cross-linking between IL-5 and IL-5RA in vivo
We also performed experiments with the full-length IL-5RA and IL-5 molecules in mammalian cells, to gather evidence supporting the determined complex structure of the extracellular domain. A photo-reactive amino acid, p-benzoyl-L-phenylalanine (pBpa), has been site-specifically incorporated into proteins in living cells with an expanded genetic code.37, 38 This non-natural amino acid is activated by exposure to 365-nm light, and covalently links the pBpa-containing protein to an interacting protein, when the pBpa is incorporated at a site adjacent to the binding interface. The substitution of the cross-linker (pBpa) for the residues involved in hydrogen bonding interactions or located in the core of the interface abrogates the protein–protein interactions, and results in marked decreases in the cross-linking efficiency.38, 39 Accordingly, we selected several residues, such as positions 75, 77, 79, 80, and 81 in site 1, as well as in the surrounding region, as candidates for the incorporation sites of the cross-linker, since sites 2 and 3 and their surrounding areas lack suitable sites for the incorporation of the cross-linker. To probe the binding mode between the full-length IL-5RA and IL-5 molecules, we incorporated the cross-linker at these candidate sites (positions 75, 77, 79, 80, and 81) and at another position, 166, in IL-5RA; the former five positions are located within β4–β5 and the surrounding region of the IL-5RA D1 domain, which interact with IL-5 in the determined crystal structure, while the latter position is far from the IL-5 interface, at the β2–β3 loop of the IL-5RA D2 domain [Fig. 3(A)].
Figure 3.
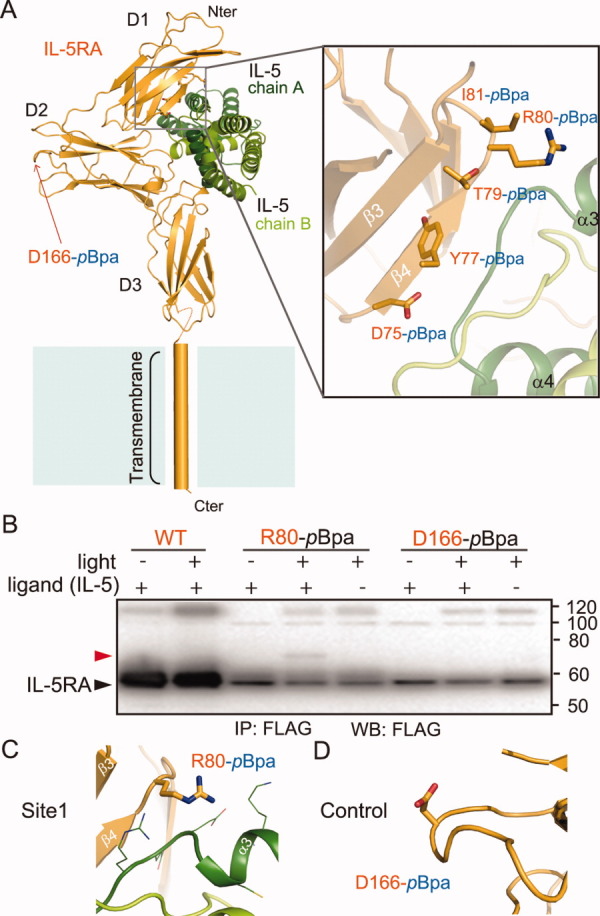
Photo-cross-linking between IL-5 and IL-5RA in mammalian cells. The FLAG-tagged IL-5RA variants, with the photo-reactive amino acid pBpa at the indicated positions in (A), were expressed in HEK293 c-18 cells. A 1 μM concentration of the purified IL-5 was added to the cells expressing IL-5RA on the cell surface, and after the cells were exposed to light, IL-5RA and its photo-cross-linked products were purified by immunoprecipitation with FLAG-M2 agarose. The precipitates were subjected to SDS-PAGE and analyzed by western blotting with anti-FLAG antibodies. B: The cross-linked IL-5 is indicated by the red arrowhead. The IL-5RA variants are indicated by the black arrowhead. WT, wild type IL-5RA; IP, immunoprecipitation; WB: western blotting. C, D: Close-up views of the cross-links between IL-5RA-pBpa and IL-5 and the control residue locations. The residues are represented as orange sticks.
Human embryonic kidney 293 cells, expressing the wild-type and variants of IL-5RA with C-terminal FLAG tags, were incubated with 1 μM of purified IL-5 (lacking the histidine tag) to generate the IL-5–IL-5RA complex on the cell surface, and then were exposed to 365-nm light. The IL-5RA and its crosslinked products were immunoprecipitated with anti-FLAG antibody affinity resin and then detected with an anti-FLAG antibody. For position80-pBpa IL-5RA, a protein band with a molecular mass of ∼ 70 kDa (represented by an arrowhead), which is almost equal to the sum of the molecular masses of the two proteins at a 1:1 stoichiometry, was successfully detected [Fig. 3(B)]. We clearly showed that this band including position80-pBpa is the cross-linked product with IL-5, because it disappeared under ligand (–) and light (+) conditions [Fig. 3(B,C)]. When WT and position 166-pBpa were tested as negative controls, no significant band was detected under the same conditions [Fig. 3(B,D)]. The other tested candidates (positions 75, 77, 79, and 81-pBpa) did not yield any cross-linked product, because the size of pBpa and its orientation in these positions are not appropriate for cross-link formation (data not shown).
Characterization of the IL-5•IL-5RA complex and its family
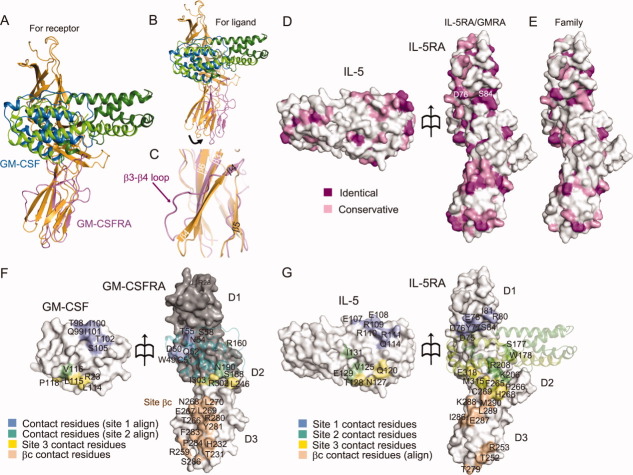
The α receptors for IL-5, IL-3, and GM-CSF (IL-5RA, IL-3RA, and GM-CSFRA, respectively) constitute a subgroup of the class I receptor superfamily. IL-5RA shares less than 30% sequence identity to IL-3RA and GM-CSFRA. As for the previously determined structure of GM-CSFRA in complex with GM-CSF and βc,32 the coordinates of only one of the domains (αD2a), with a homologous sequence to that of the D3 domain of IL-5RA (Supporting Information Fig.S1A), were deposited in the Protein Data Bank (PDB: 3CXE). This structure of αD2a superimposes well with that of the D3 domain of IL-5RA in our IL-5•IL-5RA complex structure [Fig. 4(A)]. In both domain structures, two anti-parallel β-sheets are formed between β1 and β2 and between β3 and β6, involving many identical residues (Supporting Information Fig. S1A). However, the structures between β4 and β5 are significantly different [Fig. 4(C)]. In the IL-5RA D3 domain structure, β4 forms an anti-parallel β sheet with β3. In contrast, in the GM-CSFRA αD2a structure, the region corresponding to β4 of the IL-5RA D3 domain forms a long loop, and then β4 forms an anti-parallel β sheet with β5 [Fig. 4(C)]. This region between β3 and β5 of GM-CSFRA is involved in the interactions with βc (Supporting Information Fig. S1A).32 On the other hand, the GM-CSF monomer and the corresponding part of the IL-5 dimer (α4 of chain A and α1–α3 of chain B) in the complexes also superpose well, except for α3 [Figs. 1(B) and 4(A)]. Because of the dimer formation by IL-5, the orientation of α3 in IL-5 is quite different from that in GM-CSF (Supporting Information Fig. S1C). When we superimposed the complex structures, based on the ligands (IL-5 and GM-CSF), their respective receptors (GM-CSFRA and IL-5RA) have different positions [Fig. 4(B)], indicating that the ligand-binding interfaces slightly differ between the two receptors.
Figure 4.
Structural comparison and surface properties of the binding interface between IL-5•IL-5RA and GM-CSF•GM-CSFRA. Superimposition of GM-CSFRA (light purple) bound to GM-CSF (blue) for the IL-5RA D3 domain (A) and for the IL-5 chain B (B), shown as a ribbon diagram. (C) Close-up view of the superimposition in (A) between the IL-5RA and the GM-CSFRA D3 domains. (D) Residue conservation mapping on the surfaces of IL-5 and IL-5RA. Purple and pink surfaces indicate the locations of identical and similar residues between IL-5•IL-5RA and GM-CSF•GM-CSFRA, respectively, among receptor α of the human βc family (E), according to the sequence alignment in Supporting Information Fig. S1A. (F) Contact residue mapping on the surfaces of GM-CSF and GM-CSFRA. The D1 and D2 domains of GM-CSFRA were modeled based on the structure of IL-5RA by MODELLER, and are colored gray. Blue and green surfaces indicate the locations of putative contact residues with GM-CSF•GM-CSFRA, based on the sequence alignment, the contact residues with sites 1 and 2, and yellow and wheat surfaces indicate the contact residues with site 3 and βc, respectively. (G) Contact residue mapping on the surfaces of IL-5 and IL-5RA. Blue, green, yellow, and wheat surfaces indicate the locations of contact residues with sites 1, 2, and 3, and putative contact residues with βc, respectively, based on the sequence alignment.
We have shown that the D3 domains of IL-5RA and GM-CSFRA adopt similar structures. Based on the sequence identities of D1 and D2 between IL-5RA and GM-CSFRA (Supporting Information Fig. S1A), we modeled the D1 and D2 domains of GM-CSFRA, which are missing in the previous GM-CSF•GM-CSFRA•βc ternary complex structure, using the present crystal structure of IL-5RA as a template. From these domain models of GM-CSFRA, we identified putative disulfide bonds between Cys11 and Cys79 in the D1 domain, and between Cys126 and Cys136, and Cys165 and Cys178 in the D2 domain (Supporting Information Fig. S1A). The Cys51 in this model would be unpaired, and possibly plays a similar role to Cys86 in IL-5RA. Based on this model, we compared the residues located in the interface between IL-5 and IL-5RA, with the corresponding residues of GM-CSF and GM-CSFRA. Mapping of the ligand-interacting residues in sites 1, 2, and 3 on each receptor [Fig. 4(F,G)] revealed that they are clustered in similar positions, but the participating amino acid residues are poorly conserved between IL-5RA and GM-CSFRA [Fig. 4(D,E)]. In contrast, the residues corresponding to the βc-interacting residues of GM-CSF are highly conserved in IL-5RA [Fig. 4(D,G)]. On the ligand sides, the receptor-interacting residues reside in slightly different locations, particularly in α4 within sites 2 and 3 [Fig. 4(F,G)]. Moreover, the residues located within the interfaces on the ligands are hardly conserved, as in the cases of their receptors. These divergences in the amino acid residues are likely to determine the ligand specificities of the receptors.
Specificity for ligand binding of the IL-5RA and GM-CSFRA D3 domains
A comparison of site 3 of the IL-5•IL-5RA complex with the corresponding contact site of the GM-CSF•GM-CSFRA complex revealed significant structural differences, formed by the β6-CT loop (the D3 domain) of the receptor α molecules, and in the interacting regions, α1 and α4, of the ligands (Fig. 5). In IL-5RA, Glu318 in the β6-CT loop hydrogen bonds with Gln120 in α4 of IL-5 (chain A). In contrast, the corresponding residue of GM-CSFRA is Ile303, which hydrophobically interacts with Leu115 in α4 of GM-CSF. In the case of GM-CSFRA, the side chain of Arg302 in the β6-CT loop hydrogen bonds with the main chain oxygen atom of Leu115 in α4 of GM-CSF, and the main chain of Arg302 in the β6-CT loop hydrogen bonds with the side chain nitrogen atom of Arg23 in α1 of GM-CSF, while the side chain of the corresponding Arg residue (Arg317) of IL-5RA is rotated away from the interface, and is not involved in IL-5 binding. Instead, in IL-5RA, Met315 in the β6-CT loop (IL-5RA D3 domain) protrudes into the hydrophobic pocket and interacts with Val30 and Leu34 in α1 of IL-5 (chain B) [Fig. 5(A)]. This difference occurred because Cys316 (IL-5RA D3 domain), which is not conserved in GM-CSFRA, forms a disulfide bond with Cys269 (IL-5RA D3 domain) and changes the conformation of the β6-CT loop [Figs. 1(A), 4(F,G), and 5(A)]. Interestingly, the modes of these receptor•ligand interactions in IL-5•IL-5RA and GM-CSF•GM-CSFRA are totally different: IL-5•IL-5RA involves polar and hydrophobic interactions, while GM-CSF•GM-CSFRA involves hydrophobic interactions and hydrogen bonds. This aspect is clearly shown by the electrostatic potential map [Fig. 5(B,C)]. Thus, the two α receptors regulate the ligand specificity by changing the types of interactions and the interactive residues in site 3.
Figure 5.
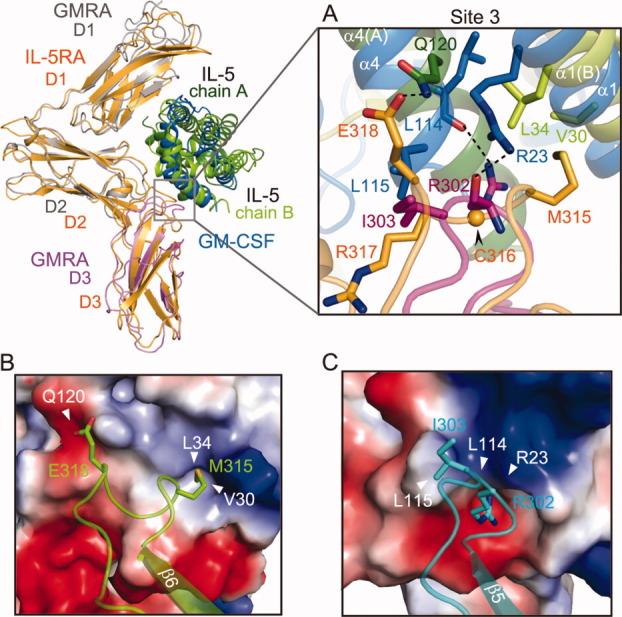
Divergence in ligand and alpha receptor heterodimer formation by the IL-5•IL-5RA complex and the GM-CSF•GM-CSFRA complex in the D3 domains. The superimposed structures of IL-5•IL-5RA and GM-CSF•GM-CSFRA are presented in the upper left. A: Close-up view of the interfaces between the α helices of the ligands and the β6-CT loops of the α receptors. B: Potential mapping of the contact residues between IL-5 and IL-5RA, and GM-CSF and GM-CSFRA (C). The residues are represented as sticks.
Discussion
In the present study, we determined the crystal structure of IL-5RA bound with IL-5. One IL-5RA recognizes three binding sites in the dimeric IL-5. We confirmed, through photo-cross-linking experiments, that the two proteins interact with each other in vivo in the same manner as that in the crystal structure. Previous mutagenesis studies indicated that mutations of the residues in the α3–α4 loop (Glu107, Glu108, Arg109, and Arg110) and α4 (Thr128, Glu129, and Trp130) of IL-5 weaken the ability to bind IL-5RA.40–42 On the other hand, the IL-5RA mutations that diminish the high-affinity binding are located in the D1 domain (Asp75, Asp76, and Glu78), the D2 domain (Lys206 and Arg208), and the D3 domain (Arg317).43 All of these IL-5 and IL-5RA residues, except for Trp130 and Arg317, directly participate in the complex formation in our IL-5•IL-5RA complex structure. The residues in the α3–α4 loop of IL-5 and the D1 domain of IL-5RA constitute site 1, while those in α4 of IL-5 and in the D2 and D3 domains of IL-5RA constitute sites 2 and 3. By contrast, Trp130 of IL-5 does not directly participate in the interaction with IL-5RA in the complex structure, but its side chain maintains the local structure of Glu129 of IL-5 (data not shown). Therefore, the present structure of the IL-5•IL-5RA complex can explain the effects of the previous mutations, with only one exception, Arg317 of IL-5RA, which is not directly involved in the intermolecular interaction. Arg317 does not interact with any of the above-mentioned functional residues, but it supports the side chain of Glu318 of IL-5RA, which hydrogen bonds with Gln120 of IL-5.
Furthermore, we compared the D3 domains in the two complex structures, the IL-5•IL-5RA complex and the GM-CSF•GM-CSFRA complex. A structural comparison of the two D3 domains revealed that in site 3, the interactive residues and the types of interactions are totally different between the IL-5•IL-5RA and GM-CSF•GM-CSFRA complexes (Fig. 5). It is likely that these differences determine the ligand-binding specificities of IL-5RA and GM-CSFRA. On the other hand, the residues that constitute the D1–D3 domains of IL-5RA are well conserved in GM-CSFRA [Supporting Information Fig. 1(A,B)]. Therefore, we utilized our crystal structure of IL-5RA as a template, to perform a modeling of the missing GM-CSFRA D1–D2 domains structure [Fig. 4(F)]. The model structure indicated that the overall architecture of the GM-CSF•GM-CSFRA complex closely resembles that of the IL-5•IL-5RA complex; therefore, GM-CSFRA is also likely to adopt a similar preformed wrench-like architecture as that in IL-5RA. In addition, in site 1, we found differences in the residues located on the ligand•receptor interface between the IL-5•IL-5RA and GM-CSF•GM-CSFRA complexes. For example, within IL-5, residues 107-Glu-Glu-Arg-Arg-Arg-111 interact with the IL-5RA D1 domain, but they are replaced with 98-Thr-Gln-Ile-Ile-Thr-102 in GM-CSF. Similarly, within IL-5RA, residues 75-Asp-Asp-Tyr-Glu-78 and 80-Arg-Ile-81 are replaced with 49-Trp-Asp-Cys-Gln-52 and 54-Asn-Thr-55, respectively, in GM-CSFRA [Fig. 4(F,G), Supporting Information Fig. 1(A,B)]. As in the case of site 2, these differences in the amino acid residues may also determine the ligand specificities of the receptors.
On the other hand, in the context of ligand-binding, IL-5 binds to IL-5RA with a higher binding affinity, due to the 20-fold faster association rate (larger kon value), as compared to that of GM-CSF toward GM-CSFRA.44 Lopez et al. reported that IL-5, IL-3, and GM-CSF cross-compete for binding to the same cell.45 In other words, the fast association can facilitate ligand capture by IL-5RA on the cell surface. Based on our complex structure, the two binding interfaces of the IL-5 homodimer with IL-5RA have more chances to bind to its receptor, as compared with GM-CSF, with only one binding interface with GM-CSFRA. Once in the IL-5RA-bound state, the other binding interface of IL-5 is oriented toward the cell membrane side [Fig. 1(B)], and thus the IL-5 dimer is not capable of binding two IL-5RA molecules.
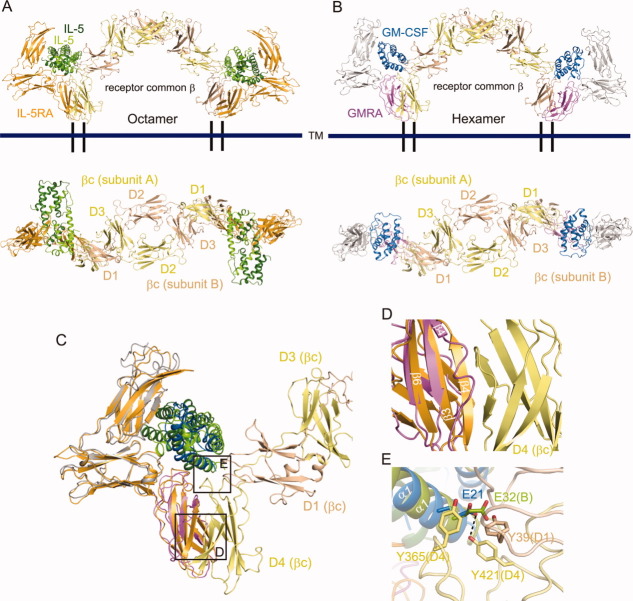
IL-5 and GM-CSF share the βc subunit to induce cytoplasmic signal transduction, as described previously. To clarify the signaling association, we built a structural model for the IL-5•IL-5RA•βc octamer assembly (4:2:2 ratio), by superimposing our IL-5•IL-5RA crystal structure onto the previously determined GM-CSF•GM-CSFRA•βc crystal structure (PDB ID: 3CXE) [Fig. 6(A)]. Similarly, we also reconstituted a structural model for the GM-CSF•GM-CSFRA•βc hexamer assembly (2:2:2 ratio), including our model of the GM-CSFRA D1–D2 domains [Fig. 6(B)]. The models revealed that the IL-5•IL-5RA complex associates with βc in a similar manner to that observed for the GM-CSF•GM-CSFRA complex [Fig. 6(C)]. In our structural model, β4 of IL-5RA D3 domain, which corresponds to the βc-interacting region of GM-CSFRA, sterically hinders the D4 domain of βc [Fig. 6(D)]. This probably occurs because, in GM-CSFRA, the β4 strand has been changed to a loop, to maintain the stable binding interface and to avoid structural hindrance with βc. A similar phenomenon may occur when the IL-5RA D3 domain interacts with βc, considering the high structural conservation of the D3 domain. In the previously determined GM-CSF•GM-CSFRA•βc crystal structure, Glu21 of GM-CSF hydrogen bonds with Tyr421 of βc, and these residues are reportedly essential for GM-CSF binding and function [Fig. 6(E)].46 Our model suggests the existence of structurally equivalent interactions between Tyr421 of βc and Glu32 of IL-5 [Fig. 6(E)], and thus is consistent with the previous mutagenesis studies that identified the essential roles of these residues in IL-5 binding.47 In addition, the βc mutations, Y39S, Y365Q, and Y365S, largely suppressed the IL-5-induced assembly of βc with IL-5RA [Fig. 6(E)].48 Consistently, in our structural model of the IL-5•IL-5RA•βc complex, Tyr39 and Tyr365 of βc are located on the interface with IL-5 in the complex with IL-5RA [Fig. 6(E)]. From these findings, it is likely that the two models of the IL-5•IL-5RA•βc and GM-CSF•GM-CSFRA•βc assemblies share common structural features, with the exception that the IL-5 dimer uses the two protomers for different purposes.
Figure 6.
Assembly and signaling complex models mediated by βc. Comparisons of ternary complex formation (upper side), related by a 90° rotation about the horizontal axis (lower side): IL-5 and IL-5RA (A), GM-CSF and GM-CSFRA (B). Orange: IL-5RA, green: IL-5 chain A, light green: IL-5 chain B, yellow: βc subunit A, beige: βc subunit B, light purple: GM-CSFRA, blue: GM-CSF, gray: modeled GM-CSFRA. C: Superimposition of the IL-5 and GM-CSF ternary complex models. D: Close-up view of the IL-5RA/GM-CSFRA D3 domains and the βc D4 domain. E: Interaction between IL-5/GM-CSF and the βc D1/D4 domain.
IL-5 activates at least three different signaling pathways, including the JAK2/STAT5, Btk, and Ras/ERK pathways. The JAK2 and JAK1 tyrosine kinases are constitutively associated with IL-5RA and βc, respectively, and are activated by IL-5 stimulation.28 According to the structural model of the IL-5•IL-5RA•βc octamer (4:2:2 ratio) [Fig. 6(A), Supporting Information Fig. 2(A)], JAK2 and JAK1 bound to the receptor intracellular domains are located close to each other, thus facilitating their trans phosphorylation and mutual activation. Therefore, it is reasonable to assume that the IL-5•IL-5RA•βc octamer is a functional complex for the JAK2/JAK1 activation by IL-5. On the other hand, JAK2 is reportedly associated with βc, and not GM-CSFRA, and JAK2 activation by GM-CSF requires functional βc dimerization,49 as proposed from a structural model of the GM-CSF•GM-CSFRA•βc dodecamer (4:4:4 ratio), consisting of two hexameric complexes related by a crystallographic two-fold axis [Supporting Information Fig. 2(B)].32 Therefore, despite the conserved architectures of the IL-5•IL-5RA•βc (4:2:2 ratio) and GM-CSF•GM-CSFRA•βc (2:2:2 ratio) complexes, the quaternary structures that are minimally required for their signal transduction may differ, according to the JAKs associated with the receptors.
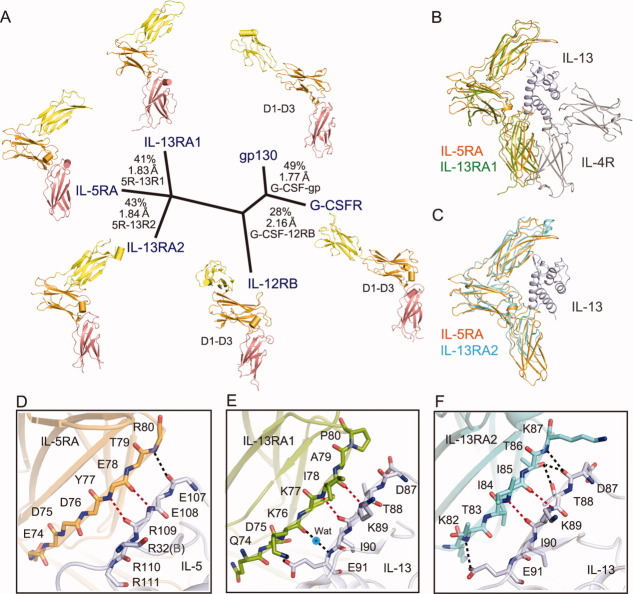
In addition, we compared the ectodomain structure of IL-5RA with those of other members of the cytokine class I receptor family that contain at least one of the following three domains, the Ig-like domain, the FnIII domain, and one CRM. A structure-based alignment indicated that these ectodomains are classified into three groups, according to the dissimilarity of the D1 domain orientation and the angle formed by the D2–D3 hinge [Fig. 7(A)]. It is noteworthy that the ectodomain structures of IL-5RA, IL-13RA1,50 and IL-13RA251 in complex with either the IL-5 dimer or the IL-13 monomer are quite similar, although the amino acid sequence homology is low, with 24% sequence identity to IL-13RA1 and 28% to IL-13RA2, among the family [Fig. 7(B,C), Supporting Information Fig. 3). Furthermore, all three ligand–receptor interactions in the D1 domain (site 1), between the IL-5RA D1 domain and IL-5, IL-13RA1 D1 and IL-13, and IL-13RA2 D1 and IL-13, share conserved bidentate main chain (mc)–main chain (mc) hydrogen bonds [Fig. 7(D–F)]. Interestingly, the IL-5–IL-5RA interaction involving the side chains has more hydrogen bonds, as compared with the IL-13RAs interactions [Fig. 2(D–F)]. In this way, the preformed receptor architecture has thus far only been seen with the rather distantly related cytokine receptors IL-13RA1/2; therefore, it does not necessarily arise from the unique homodimer of IL-5. Since IL-13RA1 retains the wrench-like structure in its complexes with IL-13 and IL-4R [Fig. 7(B)], it is likely that IL-5RA also retains the wrench-like structure in the βc complex.
Figure 7.
Structure-based phylogenetic analysis of the cytokine class I receptor family. Available structures of D1–D3 domains (colored yellow, orange, and salmon pink, respectively) [IL-12RB: 1F45 (PDB ID),63 gp130: 1P9M (PDB ID),64 G-CSFR: 2D9Q (PDB ID),65 IL-13RA1: 3BPO (PDB ID),50 IL-13RA2: 3LB6 (PDB ID)51] were superposed (for all Cα atoms), and a pairwise distance matrix was constructed based on structural similarity, using the programs ClustalW and DALI. Structure-based sequence identity (%) and Cα atom rmsds (Å), calculated with the program Indonesia (http://xray.bmc.uu.se/dennis), are shown for each domain comparison. Superposition of the α receptors between IL-5RA and IL-13RA1/IL-13/IL-4R (B), and IL-5RA and IL-13RA2/IL-13 (C). All three cytokine-receptor [IL-5RA (D), IL-13RA1 (E), and IL-13RA2 (F)] interactions through the D1 domain share conserved main chain (mc)-main chain (mc) hydrogen bonds (indicated by red dotted lines).
Materials and Methods
Protein production
The DNA fragments encoding the full-length ectodomain of human IL-5 (residues 23–134), and human IL-5RA (residues 21–335) were cloned into the TA vector pCR2.1TOPO (Invitrogen), as fusion proteins containing an N-terminal histidine tag and a tobacco etch virus protease cleavage site. These proteins were synthesized using the large scale dialysis mode of the Escherichia coli cell-free reaction.52, 53 Both IL-5 and IL-5RA precipitated during synthesis. The precipitated proteins were denatured in 50 mM Tris-HCl buffer (pH 8.3), containing 8M guanidine hydrochloride and 20 mM dithiothreitol, and were suitable for the subsequent preparation of a soluble protein complex by co-refolding into 100 mM Tris buffer (pH 8.3), containing 1M arginine hydrochloride, 0.2 mM reduced glutathione, and 1 mM oxidized glutathione. After stirring for 36 h at 4°C, the protein solutions were filtered and dialyzed against 20 mM Tris-HCl buffer (pH 8.0), containing 100 mM NaCl, for 18 h at 4°C. The refolded proteins were purified to homogeneity by chromatography on an affinity (His-Trap) column. The histidine tags were removed by overnight digestion with tobacco etch virus protease at 4°C. Because of the location of the tobacco etch virus cleavage site, seven residues from the histidine tag (GSSGSSG) remained at the N termini of the IL-5 and IL-5RA proteins after histidine tag cleavage. For further purification of the IL-5•IL-5RA complex, the proteins were fractionated on a MonoQ anion-exchange column, which was eluted with a linear gradient of NaCl from 0 to 1.0M. The IL-5•IL-5RA complex was concentrated and loaded onto a Superdex 200 HR 10/30 column, equilibrated with 20 mM Tris-HCl buffer (pH 8.0) containing 100 mM NaCl, and the appropriate fractions were collected. All columns were purchased prepacked from GE Healthcare. The eluted protein complex was finally concentrated to about 2.4 mg mL–1 in the size exclusion chromatography (SEC) equilibration buffer, using an Amicon concentrator. The purity of the IL-5•IL-5RA complex was judged by an SDS–PAGE analysis.
Crystallization and X-ray data collection
The initial crystallization conditions were determined by means of the sitting-drop vapor-diffusion method at 293 K, using sparse-matrix screening kits from Hampton Research. A 0.5 μL portion of the protein complex solution was mixed with an equal volume of reservoir solution, and the drop was equilibrated against 100 μL reservoir solution, using 96-well plates. Plate-like micro crystals were obtained using polyethylene glycol as a precipitant in condition No. 36 from Crystal Screen I, condition No. 41 from Crystal Screen I, and condition No. 26 from Crystal Screen II. The initial crystallization conditions were refined by changing the pH and the concentration of the precipitants, and by screening additives. After optimization, the crystals were improved by mixing 1.5 μL protein solution with 0.75 μL reservoir solution. Furthermore, we performed the micro-repeat seeding method, which generated single plate-shaped crystals. The crystals grew to maximum dimensions of 0.7 × 0.15 × 0.05 mm in 7–10 days. Crystals of the SeMet-labeled IL-5•IL-5RA complex were optimized mainly under the conditions that we examined, and they were obtained using 0.1M Na HEPES, pH 7.3, 22% (v/v) PEG 4000, 2% i-propanol, and 6% (w/v) 1,6-hexanediol, by means of the hanging-drop vapor-diffusion method at 293 K. The protein concentration was 3.5 mg/mL. Furthermore, we performed the micro-repeat seeding method, which generated a single, large plate-shaped crystal measuring 0.7 mm in the longest dimension within 7–10 days. X-ray diffraction data were collected on beamline BL41XU at SPring-8 (Harima, Japan). Before the X-ray experiments, crystals of the SeMet-labeled IL-5•IL-5RA complex were soaked in crystallization buffer, containing 27.5% glycerol as a cryoprotectant, at 100 K. The SeMet-labeled crystal belonged to the space group P212121, with unit cell dimensions of a = 58.6 Å, b = 88.7 Å, and c = 126.7 Å. The diffraction data for the molecular replacement (MR) and single-wavelength anomalous dispersion (SAD) methods were processed with HKL2000.54 The crystallographic data and data collection statistics of the IL-5•IL-5RA complex and the SeMet-labeled IL-5•IL-5RA complex are provided in Table I.
Structure determination and refinement
We solved the structure of a single IL-5 homodimer by molecular replacement in PHASER,55 using the reported human 2.4 Å IL-5 structure (Protein Data Bank (PDB) accession code: 1HUL) as a search model. This solution was subjected to one round of simulated annealing refinement in PHENIX,56 and was subsequently completed by manual rebuilding in COOT57 and refinement in PHENIX. The initial maps revealed additional density on one side of the IL-5 homodimer, which indicated the binding site for IL-5RA. To improve the phases, the SAD data set of the SeMet-labeled IL-5•IL-5RA complex crystal was collected. The selenium sites in the asymmetric unit were determined with the crystal structure–determination program SHELXD,58 and were confirmed by their correspondence with the selenomethionine positions in IL-5 and IL-5RA. Four methionine positions, in addition to the predicted disulfide bonds, were used to register the polypeptide in the density and complete the construction of the initial IL-5RA model. The program PHENIX was used for iterative rounds of coordinate and B-factor refinement involving TLS59 refinement, which was incorporated into the later stages of the refinement process with each chain as a separate TLS group, intersected with manual model building in Coot. The electron density map was greatly improved by the combination of SAD phases and the calculated phases from the molecular replacement solution with PHASER. The final model was refined with the native data set to 2.7 Å, with R-factor and R-free values of 23.5% and 28.7%, respectively. The asymmetric unit contains one complex of two IL-5s and one IL-5RA. The program PROCHECK60 was used to assess the geometry of the final model. The CCP4 suite programs, Contact and Areamol, were used to determine the interface contacts and the buried surface area, respectively. All structural figures were generated with the program PyMOL (DeLano Scientific, Palo Alto, CA). The atomic coordinates and the structure factors have been deposited in the Protein Data Bank, with the accession code 3VA2.
Protein photo-cross-linking in mammalian cells
The IL-5RA gene, C-terminally tagged with a FLAG-tagged epitope, was cloned in the pOriP vector,61 as previously described.62 The IL-5RA gene variants with amber codons at positions 75, 77, 79, 80, 81, and 166 were created using a QuikChange site-directed mutagenesis kit (Stratagene). The gene encoding a pBpa-specific variant of E. coli TyrRS or Eco-pBpaRS was cloned in the pSuPT vector61 to create pSuPT-pBpaRS, which allows coexpression of the enzyme and the suppressor tRNA. The HEK 293 c-18 cells (ATCC) were transfected with the IL-5RA expression plasmids together with pSuPT-pBpaRS, using the Lipofectamine 2000 reagent (Invitrogen) in Opti-MEMI growth medium (Invitrogen). Four hours after transfection, the growth medium was replaced by serum-free D-MEM medium (Invitrogen) containing 0.5 mMpBpa, and the cells were further incubated at 37°C for 16 h. The cells were detached from the cell culture dish by gentle pipetting, suspended in Hanks' balanced salt solution (pH 7.4), and then incubated with 1 μM of the purified IL-5 at 4°C for 30 min. The cell suspension was transferred to a 24-well cell culture plate and exposed to 365-nm light for 15 min on ice, for photo-cross-linking. Cell extracts were prepared with lysis buffer [30 mM Tris-HCl (pH 7.4), 10% glycerol, 150 mM NaCl, 5 mM EDTA, 1.5% n-dodecyl-maltoside (DDM), and protease inhibitor cocktail at a 1:100 dilution (Nacalai Tesque, Kyoto, Japan)], and the cross-linked products were purified by a one-hour incubation at 4°C with anti-FLAG M2 affinity gel (Sigma). The gel was washed once with lysis buffer, and twice with wash buffer [30 mM Tris-HCl (pH 7.4), 150 mM NaCl, 5 mM EDTA, and 0.05% DDM], and the products were then eluted by FLAG peptides (Sigma). The eluates were fractionated on 4–12% Nu-PAGE gradient gels (Invitrogen), transferred to PVDF membranes (Millipore), and detected by western blotting with an anti-FLAG antibody (Sigma).
Acknowledgments
The authors thank M. Inoue, K. Ishii, and K. Honda for experimental assistance. They are grateful to the beam-line staff at the BL41XU of SPring-8 for assistance in data collection.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Pasare C, Medzhitov R. Control of B-cell responses by Toll-like receptors. Nature. 2005;438:364–368. doi: 10.1038/nature04267. [DOI] [PubMed] [Google Scholar]
- 2.Kinashi T, Harada N, Severinson E, Tanabe T, Sideras P, Konishi M, Azuma C, Tominaga A, Bergstedt-Lindqvist S, Takahashi M, et al. Cloning of complementary DNA encoding T-cell replacing factor and identity with B-cell growth factor II. Nature. 1986;324:70–73. doi: 10.1038/324070a0. [DOI] [PubMed] [Google Scholar]
- 3.Plaut M, Pierce JH, Watson CJ, Hanley-Hyde J, Nordan RP, Paul WE. Mast cell lines produce lymphokines in response to cross-linkage of Fc epsilon RI or to calcium ionophores. Nature. 1989;339:64–67. doi: 10.1038/339064a0. [DOI] [PubMed] [Google Scholar]
- 4.Takatsu K, Tanaka K, Tominaga A, Kumahara Y, Hamaoka T. Antigen-induced T cell-replacing factor (TRF). III. Establishment of T cell hybrid clone continuously producing TRF and functional analysis of released TRF. J Immunol. 1980;125:2646–2653. [PubMed] [Google Scholar]
- 5.Takatsu K, Tominaga A, Hamaoka T. Antigen-induced T cell-replacing factor (TRF). I. Functional characterization of a TRF-producing helper T cell subset and genetic studies on TRF production. J Immunol. 1980;124:2414–2422. [PubMed] [Google Scholar]
- 6.Takatsu K, Tominaga A, Harada N, Mita S, Matsumoto M, Takahashi T, Kikuchi Y, Yamaguchi N. T cell-replacing factor (TRF)/interleukin 5 (IL-5): molecular and functional properties. Immunol Rev. 1988;102:107–135. doi: 10.1111/j.1600-065x.1988.tb00743.x. [DOI] [PubMed] [Google Scholar]
- 7.Haldar P, Brightling CE, Hargadon B, Gupta S, Monteiro W, Sousa A, Marshall RP, Bradding P, Green RH, Wardlaw AJ, Pavord ID. Mepolizumab and exacerbations of refractory eosinophilic asthma. N Engl J Med. 2009;360:973–984. doi: 10.1056/NEJMoa0808991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koike M, Nakamura K, Furuya A, Iida A, Anazawa H, Takatsu K, Hanai N. Establishment of humanized anti-interleukin-5 receptor alpha chain monoclonal antibodies having a potent neutralizing activity. Hum Antibodies. 2009;18:17–27. doi: 10.3233/HAB-2009-0198. [DOI] [PubMed] [Google Scholar]
- 9.Harada N, Takahashi T, Matsumoto M, Kinashi T, Ohara J, Kikuchi Y, Koyama N, Severinson E, Yaoita Y, Honjo T, et al. Production of a monoclonal antibody useful in the molecular characterization of murine T-cell-replacing factor/B-cell growth factor II. Proc Natl Acad Sci USA. 1987;84:4581–4585. doi: 10.1073/pnas.84.13.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milburn MV, Hassell AM, Lambert MH, Jordan SR, Proudfoot AE, Graber P, Wells TN. A novel dimer configuration revealed by the crystal structure at 2.4 A resolution of human interleukin-5. Nature. 1993;363:172–176. doi: 10.1038/363172a0. [DOI] [PubMed] [Google Scholar]
- 11.Mita S, Tominaga A, Hitoshi Y, Sakamoto K, Honjo T, Akagi M, Kikuchi Y, Yamaguchi N, Takatsu K. Characterization of high-affinity receptors for interleukin 5 on interleukin 5-dependent cell lines. Proc Natl Acad Sci USA. 1989;86:2311–2315. doi: 10.1073/pnas.86.7.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takatsu K. Interleukin-5. Curr Opin Immunol. 1992;4:299–306. doi: 10.1016/0952-7915(92)90080-x. [DOI] [PubMed] [Google Scholar]
- 13.Kitamura T, Hayashida K, Sakamaki K, Yokota T, Arai K, Miyajima A. Reconstitution of functional receptors for human granulocyte/macrophage colony-stimulating factor (GM-CSF): evidence that the protein encoded by the AIC2B cDNA is a subunit of the murine GM-CSF receptor. Proc Natl Acad Sci USA. 1991;88:5082–5086. doi: 10.1073/pnas.88.12.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitamura T, Sato N, Arai K, Miyajima A. Expression cloning of the human IL-3 receptor cDNA reveals a shared beta subunit for the human IL-3 and GM-CSF receptors. Cell. 1991;66:1165–1174. doi: 10.1016/0092-8674(91)90039-2. [DOI] [PubMed] [Google Scholar]
- 15.Murata Y, Takaki S, Migita M, Kikuchi Y, Tominaga A, Takatsu K. Molecular cloning and expression of the human interleukin 5 receptor. J Exp Med. 1992;175:341–351. doi: 10.1084/jem.175.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takaki S, Tominaga A, Hitoshi Y, Mita S, Sonoda E, Yamaguchi N, Takatsu K. Molecular cloning and expression of the murine interleukin-5 receptor. EMBO J. 1990;9:4367–4374. doi: 10.1002/j.1460-2075.1990.tb07886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tavernier J, Devos R, Cornelis S, Tuypens T, Van der Heyden J, Fiers W, Plaetinck G. A human high affinity interleukin-5 receptor (IL5R) is composed of an IL5-specific alpha chain and a beta chain shared with the receptor for GM-CSF. Cell. 1991;66:1175–1184. doi: 10.1016/0092-8674(91)90040-6. [DOI] [PubMed] [Google Scholar]
- 18.Miyajima A, Kitamura T, Harada N, Yokota T, Arai K. Cytokine receptors and signal transduction. Annu Rev Immunol. 1992;10:295–331. doi: 10.1146/annurev.iy.10.040192.001455. [DOI] [PubMed] [Google Scholar]
- 19.Takaki S, Mita S, Kitamura T, Yonehara S, Yamaguchi N, Tominaga A, Miyajima A, Takatsu K. Identification of the second subunit of the murine interleukin-5 receptor: interleukin-3 receptor-like protein, AIC2B is a component of the high affinity interleukin-5 receptor. EMBO J. 1991;10:2833–2838. doi: 10.1002/j.1460-2075.1991.tb07832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takaki S, Murata Y, Kitamura T, Miyajima A, Tominaga A, Takatsu K. Reconstitution of the functional receptors for murine and human interleukin 5. J Exp Med. 1993;177:1523–1529. doi: 10.1084/jem.177.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carr PD, Gustin SE, Church AP, Murphy JM, Ford SC, Mann DA, Woltring DM, Walker I, Ollis DL, Young IG. Structure of the complete extracellular domain of the common beta subunit of the human GM-CSF, IL-3, and IL-5 receptors reveals a novel dimer configuration. Cell. 2001;104:291–300. doi: 10.1016/s0092-8674(01)00213-6. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe S, Mui AL, Muto A, Chen JX, Hayashida K, Yokota T, Miyajima A, Arai K. Reconstituted human granulocyte-macrophage colony-stimulating factor receptor transduces growth-promoting signals in mouse NIH 3T3 cells: comparison with signalling in BA/F3 pro-B cells. Mol Cell Biol. 1993;13:1440–1448. doi: 10.1128/mcb.13.3.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takaki S, Kanazawa H, Shiiba M, Takatsu K. A critical cytoplasmic domain of the interleukin-5 (IL-5) receptor alpha chain and its function in IL-5-mediated growth signal transduction. Mol Cell Biol. 1994;14:7404–7413. doi: 10.1128/mcb.14.11.7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouro T, Kikuchi Y, Kanazawa H, Hirokawa K, Harada N, Shiiba M, Wakao H, Takaki S, Takatsu K. Critical proline residues of the cytoplasmic domain of the IL-5 receptor alpha chain and its function in IL-5-mediated activation of JAK kinase and STAT5. Int Immunol. 1996;8:237–245. doi: 10.1093/intimm/8.2.237. [DOI] [PubMed] [Google Scholar]
- 25.Moon BG, Yoshida T, Shiiba M, Nakao K, Katsuki M, Takaki S, Takatsu K. Functional dissection of the cytoplasmic subregions of the interleukin-5 receptor alpha chain in growth and immunoglobulin G1 switch recombination of B cells. Immunology. 2001;102:289–300. doi: 10.1046/j.1365-2567.2001.01196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takaki S, Takatsu K. Reconstitution of the functional interleukin-5 receptor: the cytoplasmic domain of alpha-subunit plays an important role in growth signal transduction. Int Arch Allergy Immunol. 1994;104:36–38. doi: 10.1159/000236746. [DOI] [PubMed] [Google Scholar]
- 27.Sato S, Katagiri T, Takaki S, Kikuchi Y, Hitoshi Y, Yonehara S, Tsukada S, Kitamura D, Watanabe T, Witte O, Takatsu K. IL-5 receptor-mediated tyrosine phosphorylation of SH2/SH3-containing proteins and activation of Bruton's tyrosine and Janus 2 kinases. J Exp Med. 1994;180:2101–2111. doi: 10.1084/jem.180.6.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogata N, Kouro T, Yamada A, Koike M, Hanai N, Ishikawa T, Takatsu K. JAK2 and JAK1 constitutively associate with an interleukin-5 (IL-5) receptor alpha and betac subunit, respectively, and are activated upon IL-5 stimulation. Blood. 1998;91:2264–2271. [PubMed] [Google Scholar]
- 29.Alam R, Pazdrak K, Stafford S, Forsythe P. The interleukin-5/receptor interaction activates Lyn and Jak2 tyrosine kinases and propagates signals via the Ras-Raf-1-MAP kinase and the Jak-STAT pathways in eosinophils. Int Arch Allergy Immunol. 1995;107:226–227. doi: 10.1159/000236985. [DOI] [PubMed] [Google Scholar]
- 30.Kagami S, Nakajima H, Kumano K, Suzuki K, Suto A, Imada K, Davey HW, Saito Y, Takatsu K, Leonard WJ, Iwamoto I. Both stat5a and stat5b are required for antigen-induced eosinophil and T-cell recruitment into the tissue. Blood. 2000;95:1370–1377. [PubMed] [Google Scholar]
- 31.Horikawa K, Kaku H, Nakajima H, Davey HW, Hennighausen L, Iwamoto I, Yasue T, Kariyone A, Takatsu K. Essential role of Stat5 for IL-5-dependent IgH switch recombination in mouse B cells. J Immunol. 2001;167:5018–5026. doi: 10.4049/jimmunol.167.9.5018. [DOI] [PubMed] [Google Scholar]
- 32.Hansen G, Hercus TR, McClure BJ, Stomski FC, Dottore M, Powell J, Ramshaw H, Woodcock JM, Xu Y, Guthridge M, McKinstry WJ, Lopez AF, Parker MW. The structure of the GM-CSF receptor complex reveals a distinct mode of cytokine receptor activation. Cell. 2008;134:496–507. doi: 10.1016/j.cell.2008.05.053. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi T. Cytokine signaling through nonreceptor protein tyrosine kinases. Science. 1995;268:251–255. doi: 10.1126/science.7716517. [DOI] [PubMed] [Google Scholar]
- 34.Bazan JF. Structural design and molecular evolution of a cytokine receptor superfamily. Proc Natl Acad Sci USA. 1990;87:6934–6938. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baumgartner JW, Wells CA, Chen CM, Waters MJ. The role of the WSXWS equivalent motif in growth hormone receptor function. J Biol Chem. 1994;269:29094–29101. [PubMed] [Google Scholar]
- 36.Patino E, Kotzsch A, Saremba S, Nickel J, Schmitz W, Sebald W, Mueller TD. Structure analysis of the IL-5 ligand-receptor complex reveals a wrench-like architecture for IL-5Ralpha. Structure. 2011;19:1864–1175. doi: 10.1016/j.str.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 37.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 38.Hino N, Okazaki Y, Kobayashi T, Hayashi A, Sakamoto K, Yokoyama S. Protein photo-cross-linking in mammalian cells by site-specific incorporation of a photoreactive amino acid. Nature Meth. 2005;2:201–206. doi: 10.1038/nmeth739. [DOI] [PubMed] [Google Scholar]
- 39.Kusano S, Kukimoto-Niino M, Hino N, Ohsawa N, Okuda K, Sakamoto K, Shirouzu M, Shindo T, Yokoyama S. Structural basis for extracellular interactions between calcitonin receptor-like receptor and receptor activity-modifying protein 2 for adrenomedullin-specific binding. Protein Sci. 2012;21:199–210. doi: 10.1002/pro.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graber P, Proudfoot AE, Talabot F, Bernard A, McKinnon M, Banks M, Fattah D, Solari R, Peitsch MC, Wells TN. Identification of key charged residues of human interleukin-5 in receptor binding and cellular activation. J Biol Chem. 1995;270:15762–15769. doi: 10.1074/jbc.270.26.15762. [DOI] [PubMed] [Google Scholar]
- 41.Morton T, Li J, Cook R, Chaiken I. Mutagenesis in the C-terminal region of human interleukin 5 reveals a central patch for receptor alpha chain recognition. Proc Natl Acad Sci USA. 1995;92:10879–10883. doi: 10.1073/pnas.92.24.10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu SJ, Li J, Tsui P, Cook R, Zhang W, Hu Y, Canziani G, Chaiken I. Randomization of the receptor alpha chain recruitment epitope reveals a functional interleukin-5 with charge depletion in the CD loop. J Biol Chem. 1999;274:20479–20488. doi: 10.1074/jbc.274.29.20479. [DOI] [PubMed] [Google Scholar]
- 43.Ishino T, Pasut G, Scibek J, Chaiken I. Kinetic interaction analysis of human interleukin 5 receptor alpha mutants reveals a unique binding topology and charge distribution for cytokine recognition. J Biol Chem. 2004;279:9547–9556. doi: 10.1074/jbc.M309327200. [DOI] [PubMed] [Google Scholar]
- 44.Ishino T, Harrington AE, Zaks-Zilberman M, Scibek JJ, Chaiken I. Slow-dissociation effect of common signaling subunit beta c on IL5 and GM-CSF receptor assembly. Cytokine. 2008;42:179–190. doi: 10.1016/j.cyto.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez AF, Elliott MJ, Woodcock J, Vadas MA. GM-CSF, IL-3 and IL-5: cross-competition on human haemopoietic cells. Immunol Today. 1992;13:495–500. doi: 10.1016/0167-5699(92)90025-3. [DOI] [PubMed] [Google Scholar]
- 46.Woodcock JM, Zacharakis B, Plaetinck G, Bagley CJ, Qiyu S, Hercus TR, Tavernier J, Lopez AF. Three residues in the common beta chain of the human GM-CSF, IL-3 and IL-5 receptors are essential for GM-CSF and IL-5 but not IL-3 high affinity binding and interact with Glu21 of GM-CSF. EMBO J. 1994;13:5176–5185. doi: 10.1002/j.1460-2075.1994.tb06848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tavernier J, Tuypens T, Verhee A, Plaetinck G, Devos R, Van der Heyden J, Guisez Y, Oefner C. Identification of receptor-binding domains on human interleukin 5 and design of an interleukin 5-derived receptor antagonist. Proc Natl Acad Sci USA. 1995;92:5194–5198. doi: 10.1073/pnas.92.11.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zaks-Zilberman M, Harrington AE, Ishino T, Chaiken IM. Interleukin-5 receptor subunit oligomerization and rearrangement revealed by fluorescence resonance energy transfer imaging. J Biol Chem. 2008;283:13398–13406. doi: 10.1074/jbc.M710230200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perugini M, Brown AL, Salerno DG, Booker GW, Stojkoski C, Hercus TR, Lopez AF, Hibbs ML, Gonda TJ, D'Andrea RJ. Alternative modes of GM-CSF receptor activation revealed using activated mutants of the common beta-subunit. Blood. 2010;115:3346–3353. doi: 10.1182/blood-2009-08-235846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, Heller NM, Keegan AD, Garcia KC. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell. 2008;132:259–272. doi: 10.1016/j.cell.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lupardus PJ, Birnbaum ME, Garcia KC. Molecular basis for shared cytokine recognition revealed in the structure of an unusually high affinity complex between IL-13 and IL-13Ralpha2. Structure. 2010;18:332–342. doi: 10.1016/j.str.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kigawa T, Yabuki T, Matsuda N, Matsuda T, Nakajima R, Tanaka A, Yokoyama S. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J Struct Funct Genomics. 2004;5:63–68. doi: 10.1023/B:JSFG.0000029204.57846.7d. [DOI] [PubMed] [Google Scholar]
- 53.Kigawa T, Matsuda T, Yabuki T, Yokoyama S. Bacterial cell-free system for highly efficient protein synthesis. In: Spirin AS, Swartz JR, editors. Cell-free protein synthesis. Weinheim, Germany: Wiley-VCH; 2007. pp. 83–97. [Google Scholar]
- 54.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 55.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 58.Sheldrick GM. A short history of SHELX. Acta Cryst. 2008;A64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 59.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Cryst. 2006;D62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 60.Laskowski R, MacArthur M, Moss D, Thornton J. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 61.Hino N, Oyama M, Sato A, Mukai T, Iraha F, Hayashi A, Kozuka-Hata H, Yamamoto T, Yokoyama S, Sakamoto K. Genetic incorporation of a photo-crosslinkable amino acid reveals novel protein complexes with GRB2 in mammalian cells. J Mol Biol. 2011;406:343–353. doi: 10.1016/j.jmb.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 62.Hino N, Hayashi A, Sakamoto K, Yokoyama S. Site-specific incorporation of non-natural amino acids into proteins in mammalian cells with an expanded genetic code. Nature Protoc. 2006;1:2957–2962. doi: 10.1038/nprot.2006.424. [DOI] [PubMed] [Google Scholar]
- 63.Yoon C, Johnston SC, Tang J, Stahl M, Tobin JF, Somers WS. Charged residues dominate a unique interlocking topography in the heterodimeric cytokine interleukin-12. EMBO J. 2000;19:3530–3541. doi: 10.1093/emboj/19.14.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boulanger MJ, Chow DC, Brevnova EE, Garcia KC. Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science. 2003;300:2101–2104. doi: 10.1126/science.1083901. [DOI] [PubMed] [Google Scholar]
- 65.Tamada T, Honjo E, Maeda Y, Okamoto T, Ishibashi M, Tokunaga M, Kuroki R. Homodimeric cross-over structure of the human granulocyte colony-stimulating factor (GCSF) receptor signaling complex. Proc Natl Acad Sci USA. 2006;103:3135–3140. doi: 10.1073/pnas.0511264103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.