Abstract
Cynanchum komarovii Al Iljinski is a desert plant that has been used as analgesic, anthelminthic, and antidiarrheal, but also as herbal medicine to treat cholecystitis in people. In this work, an antifungal protein with sequence homology to chitinase was isolated from C. komarovii seeds and named CkChn134. The three-dimensional structure prediction of CkChn134 indicated that the protein has a loop domain formed a thin cleft, which is able to bind molecules and substrates. The protein and CkTLP synergistically inhibited the fungal growth of Verticillium dahliae, Fusarium oxysporum, Rhizoctonia solani, Botrytis cinerea, and Valsa mali in vitro. The full-length cDNA was cloned by RT-PCR and RACE-PCR according to the partial protein sequences obtained by nanoESI-MS/MS. The real-time PCR showed that the transcription level of CkChn134 had a significant increase under the stress of ethylene, NaCl, low temperature, drought, and pathogen infection, which indicates that CkChn134 may play an important role in response to abiotic and biotic stresses. The CkChn134 protein was located in the extracellular space/cell wall by CkChn134::GFP fusion protein in transgenic Arabidopsis. Furthermore, overexpression of CkChn134 significantly enhanced the resistance of transgenic Arabidopsis against V. dahliae. Interestingly, the coexpression of CkChn134 and CkTLP showed substantially greater protection against the fungal pathogen V. dahliae than either transgene alone. The results suggest that the CkChn134 is a good candidate protein or gene, and it had a potential synergistic effect with CkTLP for contributing to the development of disease-resistant crops.
Keywords: antifungal protein, CkChn134, CkTLP, Verticillium dahliae, Cynanchum komarovii, transgenic Arabidopsis
Introduction
Chitin is the main components of fungal cell walls. Chitinases catalyze the degradation of chitin from fungal cell walls to inhibit the growth of fungal mycelium, which mainly hydrolyze β-1,4-linkage of the N-acetylglucosamine polymer of chitin.1 Moreover, chitinases also display lysozymal activity involved in conferring resistance to bacterial pathogens, hypocholesterolemic, and antihypertensive activities.2 Chitinases, being antifungal protein paid close attention on their exploitation, are thought to be more environment-friendly than chemical pesticide. Therefore, many chitinases were isolated from natural sources such as animals, plants, and bacteria. Based on their different hydrolytic mechanisms, chitinases are classified into two categories: family 18 using the mechanism of substrate-assisted catalysis and family 19 demonstrating acid catalysis.3
Chitinases can be used in free or immobilized form to kill microbe in affected soil and also play an important role in genetically modified crops to improve the resistance to disease by constitutive expression of normally inducible defense gene. For example, overexpression of a bean vacuolar chitinase gene in tobacco and canola increased ability to survive in soil infected by Rhizoctonia solani and delayed development of disease symptoms.4 Overexpression of chitinase of barley enhanced resistance to powdery mildew infection in transgenic wheat.5 To effectively inhibit fungal growth and significantly enhance protection against fungal attack in plants, expression of chitinase genes in combination with other PR-proteins has been reported, which indicated synergistic protective interaction of the coexpressed antifungal proteins.6–12 Hybrid tobacco with rice basic chitinase and alfalfa acidic glucanase genes showed at least 75% reduction in the number of lesions produced by Cercospora nicotianae.7 Transgenic tobacco with barley chitinase and β-1,3 glucanase genes showed enhanced resistance to R. solani.8 Combined expression tobacco chitinase and β-1,3 glucanase indicated resistance against Fusarium oxysporum, while transgenic tobacco that transformed the two genes, respectively, did not show resistance to F. oxysporum.9 A modified maize ribosome-inactivating protein and rice chitinase gene in transgenic rice enhanced resistance to sheath blight.11 Coexpression chi11 and glucanase genes in wheat plants could delay Fusarium head blight development.12 The experimental evidences are an encouraging attempt toward genetically engineered disease resistance in chitinase and glucanase in varieties of agriculturally important crops. However, not much is known about the synergistic effect of chitinase with thaumatin-like, which will be further studied by transgenic Arabidopsis with CkChn134, a new protein in chitinase family identified from Cynanchum komarovii.
Cynanchum komarovii Al Iljinski is a desert plant adapted to the dry and barren environment in the desertification process, belonging to the Asclepiadaceae family. The plant has been used as analgesic, anthelminthic, and antidiarrheal and also as herbal medicine to treat cholecystitis in people. In addition, it can provide raw materials for producing pesticides in agriculture. So far, a novel thaumatin-like protein (TLP) was isolated from C. komarovii seeds, which displayed strong antifungal activity and resistance to V. dahliae in transgenic Arabidopsis.13 To screening more antimicrobial proteins, we continue to choose this plant to develop more valuable proteins to control the verticillium wilt of cotton. Cotton losses due to this disease-causing organism are ∼50% in production in China and bring huge economic losses every year.14
In this work, we report the isolation and characterization of an antifungal protein-CkChn134 from C. komarovii seeds. The V. dahliae show delayed and inhibited development when infected Arabidopsis express CkChn134 and coexpress CkChn134 and CkTLP. The results suggest that the CkChn134 and CkTLP can significantly contribute to the development of disease-resistant crops and demonstrate the combinatorial effect of two genes as effective ways for incorporating durable protection against V. dahliae.
Results
Purification of an antifungal protein from C. komarovii seeds
We found that the extractions from C. komarovii seeds had strong activity against several pathogenic fungi such as V. dahliae, F. oxysporum, R. solani, Botrytis cinerea, and Valsa mali. Extracted proteins precipitated with 60–80% ammonium sulfate were separated into two different fractions named F1 and F2 by cation exchange chromatography in SP-sepharose fast flow column [Fig. 1(A)]. The antifungal activity of fractions was assayed on the test fungus V. dahliae. Fraction F2 with antifungal activity was collected and further purified on Resource Q column in FPLC [Fig. 1(B)], and the antifungal activity was associated with P2 fraction. Fraction partially purified concentrate of P2 with Superdex 75 HR10/30 (GE Healthcare) in FPLC [Fig. 1(C)] and isolated antifungal protein H1. The isolated protein showed strong inhibition to the mycelial growth in V. dahliae, F. oxysporum, R. solani, B. cinerea, and V. mali. Antifungal activity of the purified protein against the test pathogenic fungus V. dahliae was displayed in Figure 2. To further examine the combination of CkChn134 and CkTLP from C. komarovii seeds,13 the IC50 values of these fungi were tested. When the two proteins were used in combination, the IC50 values lower than the proteins were used individually (Table I). It indicated that CkChn134 and CkTLP have a synergistic effect on fungal growth.
Figure 1.
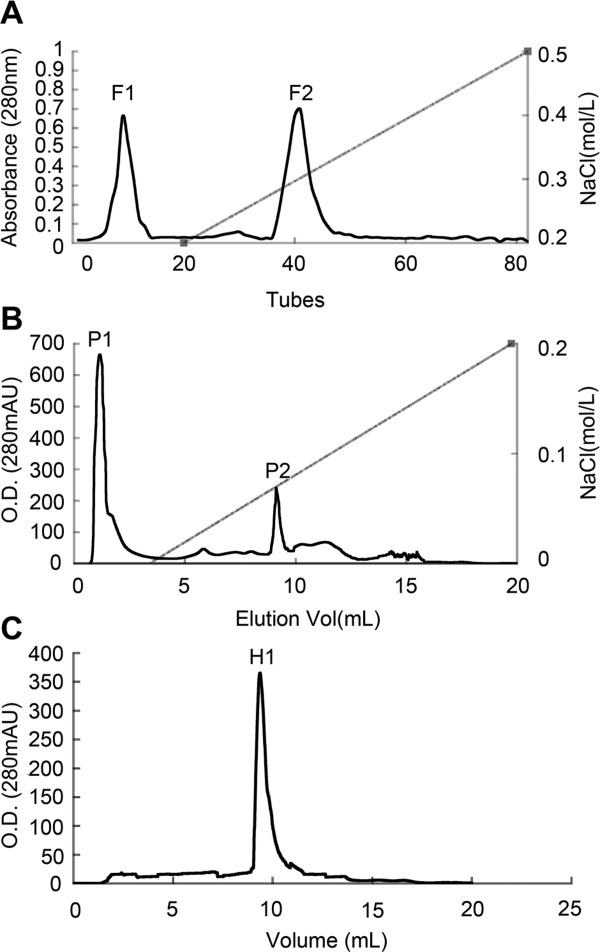
Isolation of antifungal proteins from C. komarovii seeds. A: The sample was loaded on SP-Sepharose Fast Flow (1.6 × 30 cm) with the flow rate of 1 mL/min. B: Antifungal fraction F2 was separated on FPLC-Resource Q column. C: Antifungal fraction P2 was loaded on FPLC-Superdex 75 HR10/30 column.
Figure 2.
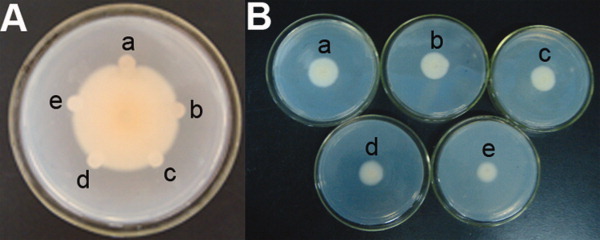
Antifungal activity of CkChn134 against V. dahliae. A: Inhibitory effects of CkChn134 protein on hyphal growth of V. dahliae. (a) 5 μg purified protein; (b) 10-μg extracted protein; (c) phosphate buffer, pH 6.8; (d) boiled CkChn134 protein; (e) 2-μg purified CkChn134 protein. B: Different doses of CkCkChn134 against V. dahliae at 4 days post-inoculation. (a) Phosphate buffer, pH 6.8; (b) 0.3 μM CkChn134; (c) 1.5 μM CkChn134; (d) 7.5 μM CkChn134; (e) 38.5 μM CkChn134. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Table I.
Effect of Purified Proteins on Fungi Growth1
| IC50 (μM) | |||
|---|---|---|---|
| Fungi species | CkChn134 | CkTLP | CkChn134 + CkTLP |
| Verticillium dahliae | 14 | 24 | 11 |
| Fumrium oxysporum | 13 | 20 | 10 |
| Rhizoctonia solani | 11 | 21 | 8 |
| Botrytis cinerea | 6 | 11 | 4 |
| Valsa mali | <3.0 | <3.0 | <3.0 |
Four doses of purified CkChn134, CkTLP, and CkChn134+CkTLP proteins were used for determination the half maximal inhibitory concentration (IC50) of antifungal activity. Buffer was used as a control.
Characterization and identification of antifungal protein from C. komarovii
The analysis of antifungal protein profile showed that the H1 fraction contained only one protein with molecular mass about 26 kDa in SDS–PAGE (Fig. 3). After gel-trypsin digestion, the peptides were identified with nano-ESI–MS/MS analysis and obtained the resulting pattern of fragmentation from MS. The four polypeptide sequence fragments were shown in the inset of Figure 4. The nano-ESI–MS/MS analysis of each polypeptide revealed a long series of the complementary y- and b-type ion-fragments and their correspondence to the amino acid (aa) sequence on spectrum (Supporting Information Fig. S1). The polypeptide sequences had high homology to chitinase genes from Carica papaya (GI: 195927481), Solanum lycopersicum (U30465), Nicotiana tabacum (AB008892), and Solanum tuberosum (X67693).
Figure 3.
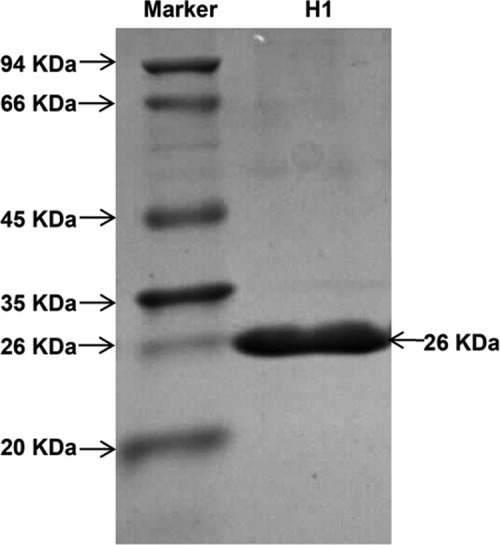
SDS–PAGE analysis of purified protein. SDS–PAGE analysis of H1, antifungal fraction on FPLC-Resource S column.
Figure 4.
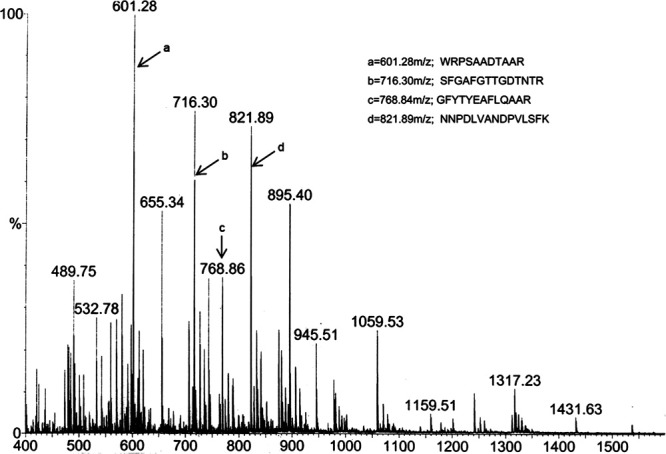
Electrospray ionization mass spectral analysis of antifungal protein. Antifungal protein of 26 kDa was submitted to nanoESI-MS/MS analysis, and polypeptide fragments corresponding to peaks a, b, c, and d are shown in the inset.
Cloning of CkChn134 cDNA
Using RT-PCR and RACE methods, we cloned the full-length cDNA of the CkChn134 gene (GenBank accession number: GU067482). The entire coding region of CkChn134 was analyzed and deduced (Fig. 5). The deduced aa sequence confirmed an identical protein result with four matched polypeptide sequences from nano-ESI–MS/MS. The CkChn134 gene was 961 bp, contained an open-reading frame (ORF) with 756 bp, encoding a protein of 252 aa, pI 8.3, and a molecular weight about 26 kDa. The deduced protein has a typical feature of signal peptide at 1–22 aa. In the BLAST search, CkChn134 shared high identity with the family 19 chitinases from N. tabacum (71%), S. tuberosum (69%), and S. lycopersicum (67%). However, the alignment of CkChn134 with different chitinase sequences of family 19 shows the presence of significant difference that is one deletion in the catalytic domain of CkChn134 (Supporting Information Fig. S2). Phylogenetic analysis of the family CkChn134 was grouped with N. tabacum (AB008892), S. tuberosum (X67693), S. lycopersicum (U30465), and C. papaya (GI: 195927481), which belong to class II chitinases (Fig. 6). The phylogeny suggests that they may have similar features and functions.
Figure 5.

Nucleotide sequence of CkChn134 and deduced amino acid sequence. The signal peptide is underlined. The regions in shadow and bold indicate the four polypeptide fragments obtained from nano-ESIMS/MS of CkChn134.
Figure 6.
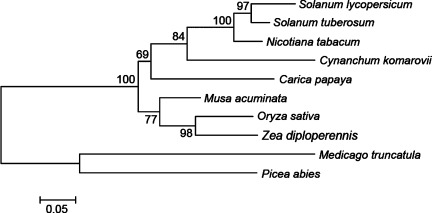
Phylogenetic tree of chitinases. The tree was constructed by the neighbor-joining method, and bootstrap values are indicated at the branches. Amino acid sequences of chitinases come from Cynanchum komarovii (GU067482), Carica papaya (GI:195927481), Solanum lycopersicum (AAB08443), Nicotiana tabacum (BAA33971), Musa acuminata (CAC81812), Solanum tuberosum (X67693), Oryza sativa (EAZ02416), Zea diploperennis (AAT40032), Medicago truncatula (AAR87869), and Picea abies (AAT09426).
Transcript level of CkChn134
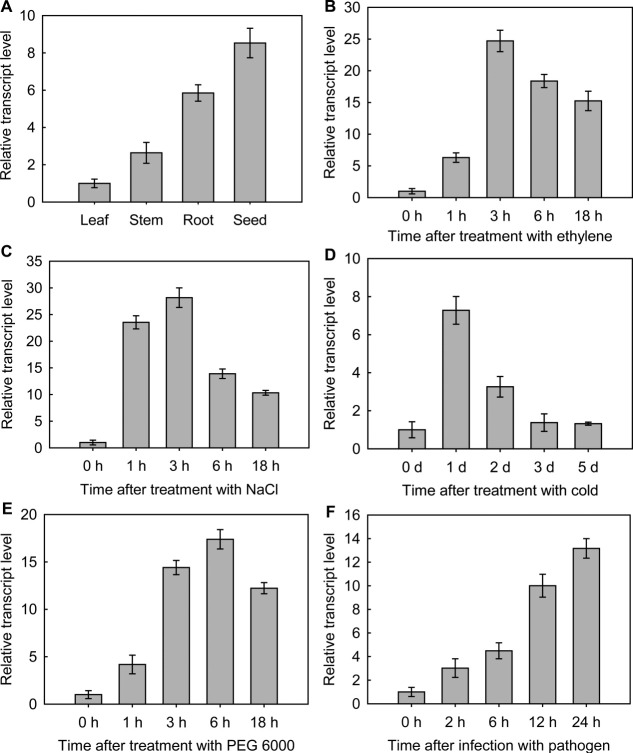
The tissue-specific expression pattern of CkChn134 was detected in root, stem, leaf, and seed [Fig. 7(A)]. CkChn134 mRNA was most abundant in seed (8.53 ± 0.79-fold higher than in leaf), had lower levels in root (5.85 ± 0.44-fold), followed by lower levels in stem (2.64 ± 0.56-fold), and the levels were the lowest in leaf.
Figure 7.
Real-time PCR was used to measure the relative expression of CkChn134. A: Tissue-specific expression pattern of CkChn134 in C. komarovii. B: Ethylene. C: NaCl. D: Low temperature. E: PEG 6000 F: V. dahliae infection. Results are expressed as mean ± SD (n = 3).
To study the transcript levels of CkChn134 in response to abiotic and biotic stresses, C. komarovii seedlings were treated with different inductions. In ethylene treatment, the transcription of CkChn134 was up-regulated at the early stage, until it reached the maximum, 24.71 ± 1.69-fold over basal activity at 3-h post-treatment, and then it was down-regulated at the late stage [Fig. 7(B)]. The transcription of CkChn134 in 300M NaCl treatment was up-regulated and reached 23.54 ± 1.23-fold over basal level at 1-h time point and achieved the maximum at 3 h [Fig. 7(C)]. The transcription of CkChn134 under low-temperature sharp increased to the maximum of 7.28 ± 0.73-fold at 1 day, and the level of transcription rapidly decreased during 2–5 days [Fig. 7(D)]. The accumulation of CkChn134 mRNA in response to polyethylene glycol (PEG 6000) increased to 4.35 ± 0.37-fold at 1 h, then rapidly climbed to the maximum of 17.39 ± 1.02-fold at 6 h, and descended slightly at 18 h [Fig. 7(E)]. When the seedlings were infected with V. dahliae, the transcription level of CkChn134 increased markedly after 12 h and reached the maximum of 13.17 ± 0.83-fold at 24 h [Fig. 7(F)]. These transcript profiles indicate that CkChn134 is responsive to different stresses.
Subcellular localization
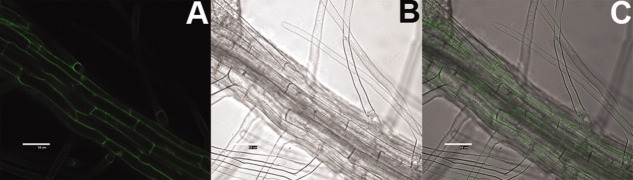
To analyze the subcellular localization of CkChn134, we first used the ProComp version 9.0 program and predicted that CkChn134 would be located in the extracellular space (score = 8.1). Then the subcellular localization of the CkChn134 within plant cell was assessed by the fusion protein CkChn134::GFP. Bright fluorescence was observed in extracellular space of seedling root cells by confocal laser scanning microscopy (Fig. 8). To differentiate between the cytoplasm membrane and cell-wall location, the seedlings were treated with 30% sucrose solution for about 2 h. No difference of fluorescence in the plasmolyzed cell was observed, suggesting that CkChn134 is located in the extracellular space or cell wall.
Figure 8.
Subcellular localization of CkChn134::GFP in Arabidopsis. A: GFP. B: Bright-field. C: Overlap. Scale bars represent 50 μm. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Disease resistance of CkChn134 in transgenic Arabidopsis
Wild-type and transgenic Arabidopsis with CkChn134, CkTLP, and chn-tlp were inoculated with V. dahliae after growing for 3 weeks in the growth chamber. Disease severity (percentage of leaves with wilt symptom) was recorded starting at 6 days postinoculation (dpi) and until 30 dpi (Fig. 9). The wild-type plants showed more prominent symptoms than each transgenic plant, and the disease incidence was 83% at 30 dpi; in the transgenic CkChn134 and CkTLP plants, the rate was 31 and 42%, respectively. Interestingly, the two genes of chn and tlp coexpressing in Arabidopsis showed a more significant degree of resistance to V. dahliae infection than the lines expressing CkChn134 and CkTLP, and the disease severity was 25% at 30 dpi. These results suggest that combination of these two genes obtained a greater protection against the fungal pathogen V. dahliae than either transgene alone. It implicates that the CkChn134 protein is capable of protecting plants against fungal diseases and show synergistic effect with CkTLP.
Figure 9.
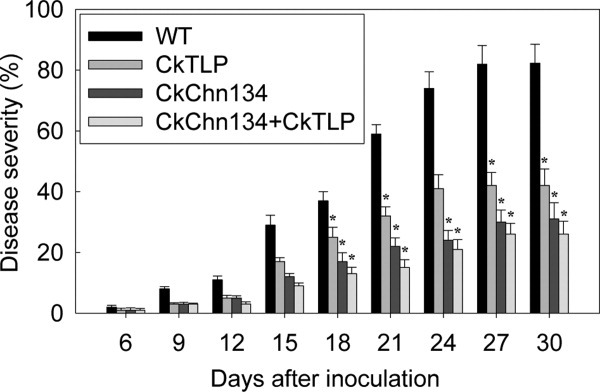
Analysis the resistance of transgenic Arabidopsis against V. dahiae. The image is showing severity of infection in WT, transgenic CkChn134, CkTLP, and coexpressing CkChn134 and CkTLP Arabidopsis seedlings. The vertical bars indicate the standard errors. *Significance at the 0.05 level compared to WT values at the same infection time.
Analysis of the predicted three-dimensional structure of the CkChn134 protein
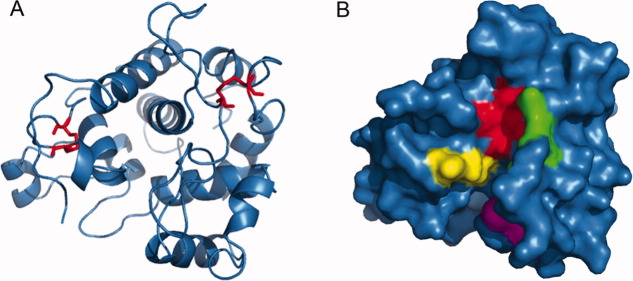
To understand the mechanism of CkChn134 antifungal activity, we predicted the three-dimensional structure by computer modeling (Fig. 10).The model was built using the template protein from C. papaya, which had 66% sequence identity with CkChn134 by SWISS-MODEL server. The CkChn134 model consisted of 12 helices and contained two disulfide bridges by four cysteine residues, 23–85 and 212–245 [Fig. 10(A)], which was important for forming the backbone of the protein. The loop domain formed a thin cleft, which is able to bind molecules and substrates. There are four major substrate-interacting regions in the protein, located around the catalytic E69(H68 and T70), the second catalytic residue E91 in the short segment (E91-R92), the sequence region 107–111 (residues T107, H108, D109, and N111), and the α-helical segment 185–189 (residues I185, N186, G187, and I189). The similar substrate interactions regions have explained the active sites of chitinase and tetra-GlcNAc substrate in C. papaya.15 It suggests that the predicted substrate-binding regions from CkChn134 may be an essential feature for the observed enzyme activity.
Figure 10.
Predicted three-dimensional structure of CkChn134. A: A ribbon diagram of CkChn134. The two disulfide bridges in structure are shown in red. B: Representation of the molecular surface of CkChn134. The active region in cleft is colored. H89-T91 is colored red. E112-R113 is displayed in green. T128-N132 is showed in magenta. I206-I210 is displayed in yellow. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Discussion
In this study, a new antifungal protein (CkChn134) was isolated from C. komarovii seeds with its purification guided by antifungal activity, which belonged to the glycoside hydrolase family 19 chitinase and lysozyme-like superfamily. The protein is unique in which it is different in aa sequences. The Ckchn134 showed typical class II chitinase structure but comparison of the aa sequence with those previously published chitinase from plants indicates that Ckchn134 contains one deletion in the catalytic domain (Supporting Information Fig. S2). Furthermore, the presence of the deletion caused the lack of two cysteine residues, which are important residues for its backbone structure and enzymatic activity.16 These residues establish disulfide bonds, forming loops in the tertiary structure of Ckchn134. The similar deletions in chitinase were shown in Crocus sativus and Dioscorea japonica, which belong to class I.16, 17 Although the loops formed by disulfide bonds of the conserved cysteines are essential for the activity of the chitinases, the first and second-conserved cysteine residues in catalytic domain of chitinase from C. sativus are not essential for its catalytic activity.17 Whether the deletion in CkChn134 affects its catalytic activity remains to be determined.
Real-time PCR analysis on the CkChn134 expression patterns of C. komarovii displayed more abundant mRNA in seed and root than stem and leaf [Fig. 7(A)]. Chitinases have different expression constitutively pattern. Class I chitinases expressed mostly in the roots and floral tissues of plants.18 Constitutive expression of class III chitinases from Cucumis sativus and Arabidopsis was found in the vascular bundles, hydathodes, and guard cells.19, 20 Class II chitinase of pepper expression occurs in roots and flowers.18 Moreover, the constitutive expression of chitinase genes increased with the plant's age.19, 21 It was interesting to note that seeds of C. komarovii had the highest transcription levels of CkChn134, which further indicate that the expression pattern is preferentially expressed in seeds. It is also suggested that chitinase may constitute a pre-emptive defense in plant's root and seeds against soil pathogen infection.
The chitinase of plants has a higher transcription level under both biotic and abiotic stresses. For instance, the chitinase gene from Cucurbita sp. Callus and suspension culture was expressed both with and without 2,4-D presence in the nutrient medium.22 In D. japonica callus culture, exogenous ethylene elevated the chitinase activity.23 In addition, jasmonic acid (JA) acts as signal that promotes defense system in plant and regulates the plant's responses to wounding. The expression of the SafchiA from C. sativus was affected by JA and wounding directly, but not by ethylene, abscissic acid or salicylic acid (SA).16 In Brassica juncea, a class I chitinase, BjCHI 1 showed the same behavior.24 In C. komarvii, the transcription level of CkChn134 showed marked increase after treatment with ethylene, but not by SA and methyl jasmonate [Fig. 7(B)]. It indicated that exogenous ethylene can induce the expression of CkChn134 and may establish systemic acquired resistance (SAR) against the pathogens. In addition, the transcription of chitinase genes could be induced by other external stimuli, for example, drought, cold, and high salt. In Cynodon sp., the expression of chitinase class II gene is induced by cold acclimation and dehydration.25 Furthermore, wounding and osmotic stress induce the synthesis of basic extracellular chitinase in Cucuribia sp.22 In high salt, low temperature, and drought treatment, the transcripts of CkChn134 were upregulated [Fig. 7(C–E)]. These results indicated that CkChn134 is involved in C. komarovii response to osmotic stresses. Furthermore, we found that the transcription of CkChn134 increased under pathogen infection [Fig. 7(F)]. It indicated that the expression of CkChn134 was correlated with SAR progress. The result suggests that the CkChn134 may be involved in host defense to biotic stress.
Many chitinases have been isolated from plants and possessed antifungal or antibacterial properties. For instance, the chitinase from barley in E. coli expression demonstrated a broad-spectrum antifungal activity against Botrytis cinerea, Pestalotia theae, Bipolaris oryzae, Alternaria sp., Curuularia lunata, and R. solani.26 The SafchiA from C. sativus inhibited the mycelia growth of Beauveria sp., Phoma sp., and F. oxysporum f. sp tuberose.16 The CkChn134 protein from C. komarovii displayed an obvious inhibitory effect against V. mali, B. cinerea, R. solani, F. oxysporum, and V. dahliae (Table I). Compared to chitinases from barley and C. sativus, the CkChn134 in vitro showed more potent inhibitory activity on R. solani and F. oxysporum.16, 26 Interestingly, we found that the inhibitions of fungal growth by combination of CkChn134 and CkTLP were more effective than applied individually (Table I). It is first time report the synergistic activity of purified CkChn134 and CkTLP in vitro. These results suggested that CkChn134 was a novel antifungal protein and potential application in cotton against V. dahliae. In addition, chitinases were also used as a method to control insect and pest population. We presumed that CkChn134 may have insecticidal activity, which are worthy of further studies.
Verticillium wilt has been the most important disease of cotton with extensive occurrence and maximal harming in the world. Combined expression of both chitinase and β-1,3-glucanase genes was attempted to achieve higher levels of fungal resistance and to broaden the spectrum of resistance against many fungal pathogens. However, in most cases, the genes did not achieve the good inhibition to V. dahliae, leading to the need for using more effective genes combinations to achieve significant levels of cotton resistance. Previous work in our laboratory had shown the enhance tolerance to V. dahliae by overexpression of CkTLP in Arabidopsis.13 In this work, it is the first-time report the development of V. dahliae-resistant transgenic Arabidopsis with the expression of CkChn134 and coexpression of CkChn134 and CkTLP. We found transgenic plants expressed constitutively CkChn134 and CkTLP, which showed more significantly enhanced resistance to V. dahliae than overexpression CkChn134 and CkTLP, respectively (Fig. 9). This result suggested that CkChn134 exhibits antifungal activity and synergistic properties with CkTLP in transgenic Arabidopsis, which was consistent with in vitro experiment by combination of purified CkChn134 and CkTLP. Early research in other plants had indicated synergistic protective interaction of the coexpression PR proteins.6–12 Coexpression chitinase and β-1,3 glucanse by placing both genes in the same T-DNA improved the resistance of tobacco against R. solani.8 Chitinase and β-1,3 glucanse genes coexpressed in carrot reduced the disease severity caused by Alternaria dauci, A. radicina, C. carotae, and Erysiphe heraclei.27 Chitinase and β-1,3 glucanse or thaumatin-like genes expression in wheat delayed the development of the powdery mildew and scab diseases.12, 28 Although, a little known the synergistic mechanism of chitinase and thaumatin-like genes, they degrade the structural components of fungal cell or include alterations in fungal cell membrane integrity to inhibit the fungal cell growth.29–32 The CkTLP protein from C. komarovii was located in the extracellular space/cell wall13 and the CkChn134 had same distribution in plant, it showed the two proteins accumulated in the extracellular space/cell wall in plant to inhibit the invasion and spread of fungal pathogens, which may potentially be a significant reason for synergistic protective interaction of the two proteins in transgenic cotton against V. dahliae.
In summary, these results presented in this work identified a new chitinase from C. komarovii seeds as candidate protein capable of protecting crops against some pathogenic fungi and enhanced resistance to V. dahliae in transgenic Arabidopsis with CkTLP.
Materials and Methods
Plant and fungal growth conditions and treatments
C. komarovii seeds were planted in a mixture of soil and vermiculite (2:1, w/w), and seedlings were cultivated under a photoperiod of 16-h light/8-h dark at 25 and 22°C, respectively. After 3 weeks, the seedlings of C. komarovii were carefully removed from soil and blotted up water and then placed into the solutions with ethylene (1 mM), NaCl (300 mM), and 10% (w/v) PEG 6000 using three repeated trials, respectively. Control samples were treated with culture solution. Samples were taken at 0, 1, 3, 6, and 18 h after the stress treatments and saved at −80°C. Under low temperature, the seedlings were placed in growth chamber at 4°C in normal light intensity for 1–5 days, sampled at different times, and stored at −80°C. For pathogen-infection treatment, the seedlings were sprayed with fungal spore suspension (5 × 106 conidia/mL) of Verticillium dahliae Kleb. Control plants were sprayed with water. The samples of treatment were harvested at 2, 6, 12, and 24-h postinoculation.
The seeds of Arabidopsis (ecotype Columbia) were treated with 4% NaClO solution and 75% ethanol, sterilized water in turn for surface sterilization, and then sowed on MS culture medium (1 × MS salts, 1 × MS vitamins, 3% sucrose, 1% agar, pH 5.7). After iarovization for 2 days at 4°C, the seeds were cultured in a growth chamber under 16 h light at 23°C and 8 h dark at 18°C, with 80% relative humidity. After 10 days, seedlings of Arabidopsis were transplanted in pots with mixture of soil and vermiculite (1:1, w/w), under the same growth conditions.
The fungal species Verticillium dahliae Kleb, Fusarium oxysporum f. sp. vasinfectum, Rhizoctonia solani Kühn, Botrytis cinerea f. sp. cucumerinum, and Valsa mali Miyabe and Yamada were grown in potato dextrose agar (PDA) plates at 25°C. Four different doses (0.3, 1.5, 7.5, and 38.5 μM) of CkChn134, CkTLP, and CkChn134 + CkTLP proteins and buffer (PBS, pH 6.8) were added in PDA plates at 25°C, and then a small amount of mycelia (0.5 cm in diameter) was placed on the surface of the agar. The antifungal activity assay in vitro and determination of IC50 were performed as previously described.13 The conidial suspensions of V. dahliae were prepared from a culture grown for 5 days at 25°C in Czapek liquid medium (NaNO3 2 g, K2HPO4 1 g, MgSO4.7H2O 0.5 g, KCl 0.5 g, FeSO4 0.01 g, sucrose 30 g, distilled water 1 L, and pH 7.2). All fungi were used for the detection of antifungal proteins in vivo. The analysis of resistance in transgenic Arabidopsis was performed using the conidial suspensions (5 × 106 conidia/mL) of V. dahliae.
Purification of antifungal proteins
One hundred grams of seeds were milled and fully homogenized with two to three volumes extraction buffer (50 mM Tris–HCl, 10 mM KCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 5 mM iodoacetic acid, pH 7.2) at 4°C. After stirring for overnight, the extract was filtered through four layers of gauze to remove undissolved materials, and the filtrate was centrifuged at 25,000 rpm for 30 min at 4°C. The supernatant was precipitated in 60–80% saturated ammonium sulfate solution. The crude proteins were separated by centrifugation (25,000 rpm, 30 min at 4°C), resuspended in 40 mL buffer (50 mM Tris–HCl, pH 7.2), and dialyzed extensively against the buffer using 1-kDa cutoff dialysis membrane. The dialyzed solution was condensed by freeze drying and then passed through an ion-exchange SP-Sepharose Fast Flow (GE Healthcare, NJ) column (1.6 × 30 cm). The column was equilibrated and initially eluted with 10 mM ammonium acetate buffer (pH 4.3). Adsorbed protein was eluted using a linear eluant strength gradient (0.2 –0.5M NaCl) in equilibration buffer. The fraction showed that high inhibitory activity was collected and further purified in AKTA FPLC on Resource Q column (GE Healthcare, NJ). The chromatography was equilibrated with 20 mM Tris–HCl (pH 7.2), and adsorbed protein was eluted in linear gradient using NaCl from 0 to 0.2M in the equilibration buffer. The resulting sample with antifungal activity was further purified on AKTA FPLC Superdex 75 HR10/30 (GE Healthcare, NJ) in 20 mM Tris–HCl (pH 7.2). The identification and purity of the samples were confirmed by SDS–PAGE (12% gel).
Partial amino acid sequencing
The antifungal protein was identified by nano-ESI–MS/MS (Micromass, Manchester, UK). Protein sequence homology searches were applied using BLAST compared the protein sequence with known proteins or translated ORFs of expressed sequence tags in the database at NCBI.
Cloning of CkChn134
Total RNA of C. komarovii seedlings were obtained by an extraction kit (Promega, WI). To obtain cDNA-encoding CkChn134, we designed two degenerate primers: F-5′GGCTTHTATACWTATGNAGCT3′ and R-5′TCCGAGCYGCTGTANCTGCTGC3′, which were based on the determined protein sequences GFYTYE and AADTAAR by nanoESI–MS/MS. The first-strand cDNA was obtained from total RNA using the reversed transcription kit (Takara, Shiga, Japan). The RT-PCR condition was 94°C for 30 s, 50°C for 30 s, 72°C for 1 min, for 33 cycles, and final extension went on at 72°C for 10 min. 3′ RACE was performed with two-nested gene-specific sense primers, GPS1 (5′ AGCCGGGAGAGCAATTGGAGTTG 3′) and GPS2 (5′ CCCAGATTTAGTTGCGAATGA 3′), following the specification of 3′RACE kit (Takara, Shiga, Japan). 5′RACE cDNA was obtained with 5′RACE kit according to the manufacturer's instructions (Invitrogen, CA), and first-strand cDNA synthesis was performed with the antisense primer GPS3 (5′ GAAACTACAGGATCATTCGCAAC 3′). The tailed cDNA was used as template for amplification with the adapter primer and two-nested primers GPS4 (5′ CATGAGTAAGTTGGATAGGACC 3′) and GPS5 (5′ CGATATCTGTCGGAGCTGCCTC 3′). The reaction of RT-PCR started at 94°C for 5 min, and following cycle was repeated 33 times: 94°C for 30 s, 52–55°C for 30 s, 72°C for 1 min, and final extension went on at 72°C for 10 min. The reaction products were purified and cloned into pGEM-T vector (Promega) for sequencing.
Sequence and phylogenetic analysis
The detected aa sequence was carried out using DNAMAN tool. Sequence features, such as signal peptide, pI, and molecular mass, were evaluated using protein analysis tools (http://expasy.org/proteomics). Chitinase sequences were selected from NCBI. The mature protein sequences were aligned with Cluster X version 2.0, and gaps were removed from the alignment. The phylogenetic tree of those alignments was calculated by the neighbor-joining method using MEGA 4 program, and bootstrap values from 1000 replicates indicated at the branches.
Real-time PCR
Total RNA was isolated from stored C. komarovii seedlings after different treatments using a RNA extraction kit (Promega). Two microgram total RNA was reverse transcribed to first-strand cDNA using the high-capacity RNA-to-cDNA kit (Applied Biosystems, Foster City, CA) according to manufacturer's specifications. Diluted cDNA was used as template in each well for the real-time PCR analysis. The unique primers of CkChn134 were designed with Chn134-F (5′ GGTCAGACTTCTCACGAAAC 3′) and Chn134-R (5′ TCCAATTGCTCTCCCAGCT 3′). The endogenous control was EF-1-α (GenBank accession number: HQ849463) from C. komarovii and identified with the sense primer EF1α-F (5′ TGCATCCAACTCGAAGGATG 3′) and antisense primer EF1α-R: (5′ CCTTACCAGATCGTCTGTCT 3′). The real-time PCR was performed by 20-μL reaction mixture in 96-well plate with ABI7500 thermocycler (Applied Biosystems), using Power SYBR Green PCR Master Mix (Applied Biosystems). Statistical analysis of real-time PCR date and standard deviation values was performed as previously described by Livak and Schmittgen.33
Subcellular localization
Intracellular localization of CkChn134 was determined by CkChn134::GFP fusion protein in transgenic Arabidopsis. The roots of 10-day-old transgenic seedlings were scanned by confocal laser scanning microscopy (Nikon C1, Tokyo, Japan). Excitation light at 488 and 543 nm was attenuated to 50% transmittance. Detectors were set at 610 nm for chlorophyll and 530 nm for green fluorescent protein (GFP) fluorescence.
Vectors construction and plant transformation
The ORF of CkChn134 was amplified with sense primer (5′-ACCAAGATCTTATGAGGTTTCGGCTTGCA-3′) and antisense primer (5′-AACAACTAGTGGCAAACGGTCTTTGGTTG-3′; restriction endonuclease BglII and SpeI were underlined). The PCR amplified product was digested by BglII and SpeI and inserted into a binary vector-pCAMBIA 1304 containing a hygromycin phosphotransferase (hph) gene, a fusion of β-glucuronidase and GFP gene under the control of CaMV35S promoter (Supporting Information Fig. S3). The CkTLP gene was subcloned into the pCAMBIA1304 as described as previously (Supporting Information Fig. S4).13 The vector of pCAMBIA1300-Chn-tlp harbors two antifungal genes, which are the CkChn134 and a CkTLP gene (GenBank AC. GU067481), each driven by a CaMV35S promoter (Supporting Information Fig. S5). The three constructed vectors were introduced into Arabidopsis (ecotype Colombia) via Agrobacterium tumefaciens (GV3101) transformation, respectively.34 Transgenic CkChn134 and CkTLP genes Arabidopsis seeds were obtained by screening on MS medium with hygromycin (25 mg/L), respectively, transgenic plants with the pCAMBIA1300-Chn-tlp vector were selected by kanamycin (50 mg/L), and the survived seedlings were confirmed by PCR.
Analysis on transgenic Arabidopsis
To test transgenic Arabidopsis resistance against infection of V. dahliae, the bioassay was performed as described previously Wang et al.13 The stable heritable T1 progeny (T2) transgenic plants with CkChn134, CkTLP and the two antifungal genes were chosen, respectively. T2 transgenic plants seeds were sowed in soil and cultured as described earlier. After 3 weeks, the seedlings were gently uprooted and rinsed in sterile water, and then the seedling roots were cut off 1 cm to drench in 100-mL conidial suspensions (5 × 106 conidia/mL) for 5 min. Plants were transplanted immediately into plot after treatment and kept in a high-humidity environment for 12 h at 25°C. Disease severity was detected by the percentage of diseased leaves number over the total leaves per plant and periodically recorded for 30 days after inoculation. The experiment was repeated three times with 15 plants per treatment.
Structural modeling of CkChn134
The three-dimensional structure was carried out by the SWISS-MODEL server (http://swissmodel.expasy.org/), with chitinase (PDB code: 3CQL_A) of C. papaya as template.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Cohen-Kupiec R, Chet I. The molecular biology of chitin digestion. Curr Opin Biotechnol. 1998;9:270–277. doi: 10.1016/s0958-1669(98)80058-x. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharya D, Nagpure A, Gupta RK. Bacterial chitinases: properties and potential. Crit Rev Biotechnol. 2007;27:21–28. doi: 10.1080/07388550601168223. [DOI] [PubMed] [Google Scholar]
- 3.Sharma N, Sharma KP, Gaur RK, Gupta VK. Role of chitinase in plant defense. Asian J Biochem. 2011;6:29–37. [Google Scholar]
- 4.Brogue K, Chet I, Holliday M, Cressman R, Biddle P, Susan K, Mauvais CJ, Broglie R. Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science. 1991;254:1194–1197. doi: 10.1126/science.254.5035.1194. [DOI] [PubMed] [Google Scholar]
- 5.Oldach KH, Becker D, Lörz H. Heterologous expression of genes mediating enhanced fungal resistance in transgenic wheat. Mol Plant Microbe Interact. 2001;14:832–838. doi: 10.1094/MPMI.2001.14.7.832. [DOI] [PubMed] [Google Scholar]
- 6.Mauch F, Mauch-Mani B, Boller T. Antifungal hydrolases in pea tissue: inhibition of fungal growth by combinations of chitinase and β-1,3-glucanase. Plant Physiol. 1988;88:936–942. doi: 10.1104/pp.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Q, Maher EA, Masoud S, Dixon RA, Lamb CJ. Enhanced protection against fungal attach by constitutive co-expression of chitinase and glucanase genes in transgenic tobacco. Nat Biotechnol. 1994;12:807–812. [Google Scholar]
- 8.Jach G, Görnhardt B, Mundy J, Logemann J, Pinsdorf E, Leah R, Schell J, Mass C. Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J. 1995;8:97–109. doi: 10.1046/j.1365-313x.1995.08010097.x. [DOI] [PubMed] [Google Scholar]
- 9.Jongedijk E, Tigelaar H, Van Roeckel JSC, Bres-Vloemans SA, Dekker I, van den Elzen PJM, Cornelissen BJC, Melchers LS. Synergistic activity of chitinases and β-1,3-glucanase enhances fungal resistance in transgenic tomato plants. Euphytica. 1995;85:173–180. [Google Scholar]
- 10.Velazhahan R, Samiyappan R, Vidhyasekaran P. Purification of an elictor-inducible antifungal chitinase from suspension-cultured rice cells. Phytoparasitica. 2000;28:131–139. [Google Scholar]
- 11.Kim JK, Jang IC, Wu R, Zuo WN, Boston RS, Lee YH, Ahn IP, Nahm BH. Co-expression of a modified maize ribosome inactivating protein and a rice chitinase gene in transgenic rice plants confers enhanced resistance to sheath blight. Transgenic Res. 2003;12:475–484. doi: 10.1023/a:1024276127001. [DOI] [PubMed] [Google Scholar]
- 12.Anand A, Zhou T, Trick HN, Gill BS, Bockus WW, Muthukrishnan S. Greenhouse and field testing of transgenic wheat plants stably expressing genes for thaumatin-like protein, chitinase and glucanase against Fusarium graminearum. J Exp Bot. 2003;54:1101–1111. doi: 10.1093/jxb/erg110. [DOI] [PubMed] [Google Scholar]
- 13.Wang Q, Li F, Zhang X, Zhang Y, Hou Y, Zhang S, Wu Z. Purification and characterization of a CkTLP protin from Cynanchum komarovii seeds that confers antifungal activity. PLoS ONE. 2011;6:e16930. doi: 10.1371/journal.pone.0016930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao S, Xiao SH, Liu JG, Wu QJ, Lin L, Di JC, Xu NY, Chen XS. Breeding of a new germplasm cotton resistance to verticillium wilt. China Cotton. 2007;1:12–14. [Google Scholar]
- 15.Huet J, Azarkan M, Looze Y, Villeret V, Wintjens R. Crystallization and preliminary X-ray analysis of a family 19 glycosyl hydrolase from Carica papaya latex. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;F64:371–374. doi: 10.1107/S1744309108007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.López RC, Gómez-Gómez L. Isolation of a new fungi and wound-induced chitinase class in corms of Crocus sativus. Plant Physiol Bioch. 2009;47:426–434. doi: 10.1016/j.plaphy.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Araki T, Funatsu J, Kuramoto M, Konno H, Torikata T. The complete amino acid sequence of yam (Dioscorea japonica) chitinase. A newly identified acidic class I chitinase. J Biol Chem. 1992;267:19944–19947. [PubMed] [Google Scholar]
- 18.Kasprzewska A. Plant chitinases-regulation and function. Cell Mol Biol Lett. 2003;8:809–824. [PubMed] [Google Scholar]
- 19.Lawton KA, Beck J, Potter S, Ward E, Ryals J. Regulation of cucumber class III chitinase gene expression. Mol Plant-Microbe Interact. 1994;7:48–57. doi: 10.1094/mpmi-7-0048. [DOI] [PubMed] [Google Scholar]
- 20.Samac DA, Shah DM. Developmental and pathogen-induced activation of the Arabidopsis acidic chitinase promoter. Plant Cell. 1991;3:1063–1072. doi: 10.1105/tpc.3.10.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samac DA, Hironaka CM, Yallaly PE, Shah DM. Isolation and characterisation of the genes encoding basic and acidic chitinase in Arabidopsis thaliana. Plant Physiol. 1990;93:907–914. doi: 10.1104/pp.93.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arie M, Hikichi K, Takahashi K, Esaka M. Characterisation of a basic chitinase which is secreted by cultured pumpkin cells. Physiol Plantarum. 2000;110:232–239. [Google Scholar]
- 23.Büttner M, Singh KB. Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA binding protein interacts with an ocs element binding protein. PNAS. 1997;94:5961–5966. doi: 10.1073/pnas.94.11.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao JK, Chye ML. Methyl jasmonate induces expression of a novel Brassica juncea chitinase with two chitin-binding domains. Plant Mol Biol. 1999;40:1009–1018. doi: 10.1023/a:1006266407368. [DOI] [PubMed] [Google Scholar]
- 25.de los Reyes BG, Taliaferro CM, Anderson MP, Anderson JA, Melcher U, McMaugh S. Induce expression of the class II chitinase gene during cold acclimation and dehydration of bermudagrass (Cynodon sp.) Theor Appl Genet. 2001;103:297–306. [Google Scholar]
- 26.Kirubakaran SI, Sakthivel N. Cloning and overexpression of antifungal barley chitinase gene in Escherichia coli. Protein Exp Purif. 2007;52:159–166. doi: 10.1016/j.pep.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Melchers LS, Stuiver MH. Novel genes for disease-resistance breeding. Curr Opin Plant Biol. 2000;3:147–152. doi: 10.1016/s1369-5266(99)00055-2. [DOI] [PubMed] [Google Scholar]
- 28.Bliffeld M, Mundy J, Potrykus I, Fütterer J. Genetic engineering of wheat for increased resistance to powdery mildew disease. Theor Appl Genet. 1999;98:1079–1086. [Google Scholar]
- 29.Schlumbaum A, Mauch F, Vogeli V, Boller T. Plant chitinases are potent inhibitors of fungal growth. Nature. 1986;324:365–367. [Google Scholar]
- 30.Velazhahan R, Samiyappan R, Vidhyasekaran P. Purification of an elicitor-inducible antifungal chitinase from suspensioncultured rice cells. Phytoparasitica. 2000;28:131–139. [Google Scholar]
- 31.Roberts WK, Selitrennikoff CP. Zeamatin, an antifungal protein from maize with membrane-permeabilizing activity. J Gen Microbiol. 1990;136:1771–1778. [Google Scholar]
- 32.Abad LR, D'Urzo MP, Liu D, Narasimhan ML, Reuveni M, Zhu JK, Niu X, Singh NK, Hasegawa PM, Bressan RA. Antifungal activity of tobacco osmotin has specificity and involves plasma membrane permeabilization. Plant Sci. 1996;118:11–23. [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabdiopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.