Abstract
An intracellular second messenger unique to bacteria, c-di-GMP, has gained appreciation as a key player in adaptation and virulence strategies, such as biofilm formation, persistence, and cytotoxicity. Diguanylate cyclases containing GGDEF domains and phosphodiesterases containing either EAL or HD-GYP domains have been identified as the enzymes controlling intracellular c-di-GMP levels, yet little is known regarding signal transmission and the sensory targets for this signaling molecule. Although limited in number, identified c-di-GMP receptors in bacteria are characterized by prominent diversity and multilevel impact. In addition, c-di-GMP has been shown to have immunomodulatory effects in mammals and several eukaryotic c-di-GMP sensors have been proposed. The structural biology of c-di-GMP receptors is a rapidly developing field of research, which holds promise for the development of novel therapeutics against bacterial infections. In this review, we highlight recent advances in identifying bacterial and eukaryotic c-di-GMP signaling mechanisms and emphasize the need for mechanistic structure–function studies on confirmed signaling targets.
Keywords: signaling, c-di-GMP, biofilm formation, protein structure, phosphodiesterase, diguanylate cyclase, c-di-GMP receptors
Introduction
In 1987, Benziman and colleagues published a landmark article describing the identification of a novel, nucleotide-based allosteric activator of Gluconacetobacter xylinus cellulose synthase activity, c-di-GMP. The initial discovery was accompanied by the detection of an enzymatic activity responsible for c-di-GMP production, a c-di-GMP binding protein module regulating cellulose synthesis, and an operon controlling c-di-GMP turnover in the bacterium.1,2
Fifteen years later, c-di-GMP has gained appreciation as a second messenger unique to the bacterial world, which functions as a global regulatory molecule to trigger a plethora of physiological responses. Examples include but are not limited to cell differentiation, changes in motility and surface adhesiveness, secretion of extracellular polysaccharides and proteinaceous fimbriae, host cell cytotoxicity, and virulence gene expression (Fig. 1).3–6 Signaling cascades employing this small molecule show evidence for multilayer impact, which includes control at the transcriptional, translational, and posttranslational levels. Proteins involved in c-di-GMP mediated signal transduction are often characterized by multidomain architecture with such modularity to allow for a variety of regulatory inputs and/or signal ramifications.7–9 This complexity is in stark contrast with canonical two-component transduction systems, where upon signal generation a sensor histidine kinase phosphorylates its cognate response regulator to alter the expression of a typically limited number of genes.10
Figure 1.
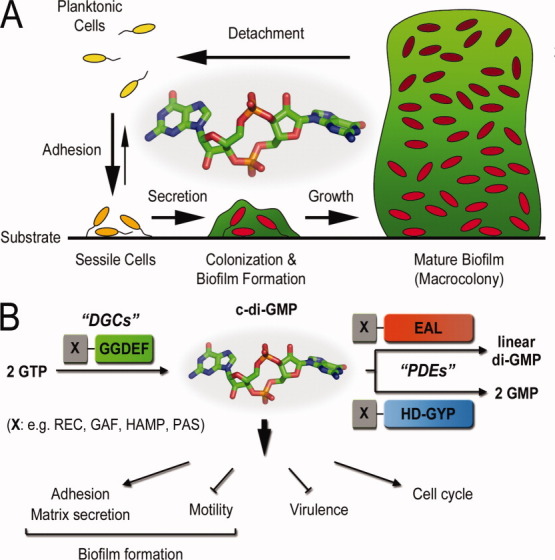
Overview of c-di-GMP signaling. (A) Biofilm formation. High levels of c-di-GMP in bacterial cells are often associated with cell adhesion, matrix secretion, and biofilm formation. (B) C-di-GMP signaling. Opposing activities of diguanylate cyclases (DGCs) with GGDEF domains and phosphodiesterases (PDEs) with EAL or HD-GYP domains control cellular c-di-GMP levels. The domains are often linked to regulatory domains, which control overall enzyme activity and cellular localization. In addition, several cellular programs have been shown to be under c-di-GMP control.
The identification of a diguanylate cyclase in Caulobacter crescentus, responsible for c-di-GMP production during cell cycle-dependent polar differentiation, paired with bioinformatics analyses making use of the increasing number of sequenced genomes have paved the way for many of the discoveries on c-di-GMP signaling in the past decade.8,11–13 The metabolic function of a number of c-di-GMP signaling modules has thus been successfully identified based on conserved sequence motifs, predicted secondary structure or domain organization, inter- and intraoperon genetic environment, and phylogenetic patterns of cross-species evolution.8
It is now well established that c-di-GMP is generated from two GTP molecules by GGDEF domain-containing diguanylate cyclases, whereas phosphodiesterases containing either EAL or HD-GYP protein domains provide selective signal degradation [Fig. 1(B)].14–18 Comparative cross-genome sequence alignments have characterized GGDEF, EAL, and HD-GYP domains as some of the most abundant protein modules encoded by bacterial genomes, with their nomenclature being a direct result of such bioinformatic analyses: GGDEF, EAL, and HD-GYP correspond to characteristic conserved amino acid motifs found in the corresponding protein domains.8,11–13 Interestingly, the number of these signaling modules per species roughly correlates with the organism's adaptational capacities.8,19 As an important example, opportunistic pathogens, which are often required to adapt to different ecological niches, typically show the highest number of c-di-GMP metabolizing enzymes per genome.
In general, increased intracellular c-di-GMP levels resulting from higher diguanylate cyclase activity lead to enhanced biofilm formation and inhibit flagellar and pilus-mediated motility.4,20 Conversely, low levels of the nucleotide associated with active phosphodiesterase catalysis suppress the maintenance of extracellular adhesins and promote biofilm dispersion and bacterial virulence.21,22 Although overexpression studies suggest a redundancy in overall effect, different diguanylate cylases or phosphodiesterases in a given genome often have enzyme specific physiological effects.15,23 Oftentimes proteins with similar domain architecture or enzymatic activity trigger distinct physiological responses.14,24 This is particularly surprising if one assumes that, as a small hydrophilic molecule, c-di-GMP is freely diffusible in the cell. A number of studies argue instead that once generated, c-di-GMP is a sequestered, rather than general, diffusive signal.4,25 Measurements of the intracellular levels of c-di-GMP in several bacterial species indicate concentrations in the micromolar range or lower, without taking into account probable local fluctuations. Based on the fact that most identified c-di-GMP receptors and phosphodiesterases have lower affinity constants for the nucleotide, it has been hypothesized that cellular c-di-GMP exists primarily in a protein-bound form.15,25 It is also a reasonable hypothesis that enzymes that produce or degrade c-di-GMP function in close proximity to the effector systems, ensuring signaling specificity and local regulation.
The complexity of c-di-GMP signaling becomes even more apparent if the nucleotide's known signaling targets are taken into account (Fig. 2). Whether functioning as intermediaries in nucleotide signal relay or as final effectors in signaling cascades, the few c-di-GMP receptors identified to date are characterized by an obvious diversity and multilevel impact [Fig. 3(A–D)].
Figure 2.
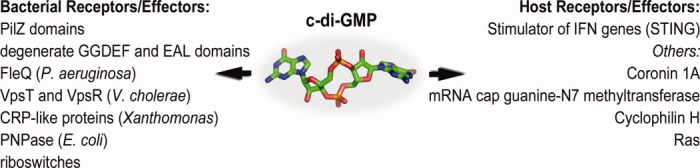
Effectors or receptors for c-di-GMP. Modules and proteins for c-di-GMP binding are diverse. While several bacterial effectors have been studied extensively, only one well-validated target on host cells has been discovered. Others await further characterization.
Figure 3.
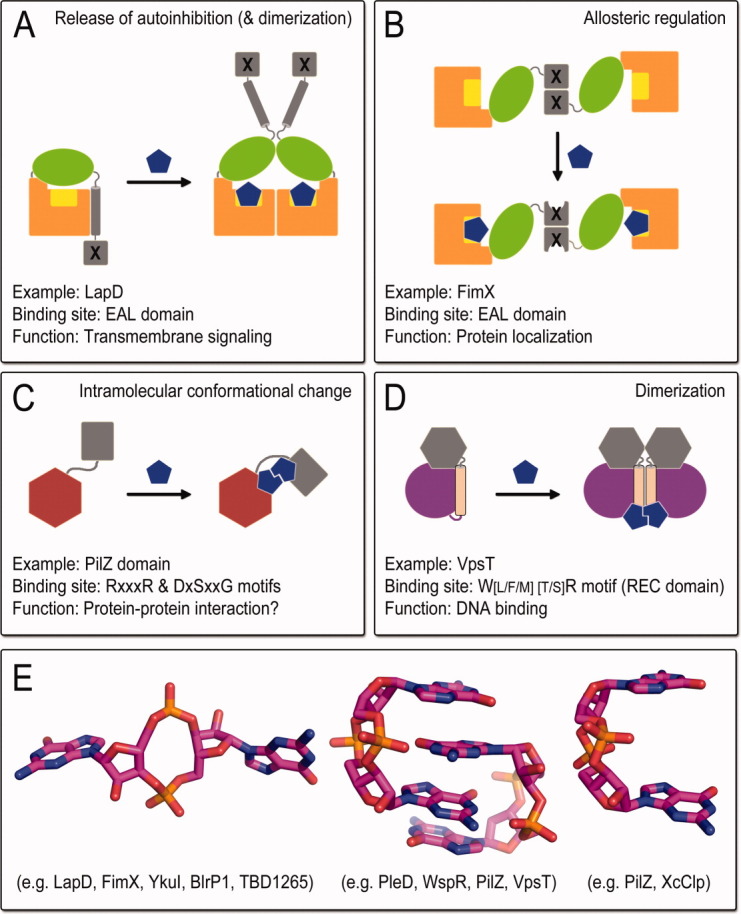
Modes of c-di-GMP effector function. Based on structural studies several modes of c-di-GMP action on effector proteins have been described, including the release of autoinhibitory interactions (A), allosteric regulation (B), major structural rearrangements (C) and/or dimerization (D). In addition, c-di-GMP itself can adopt several distinct conformations when bound to proteins.
Initial in silico prediction identified PilZ domains as protein modules involved in various c-di-GMP signaling pathways and predicted that they might function as direct sensors relaying the second messenger's input.26 Further experimental evidence has corroborated c-di-GMP binding to several PilZ domain-containing proteins, as well as the direct involvement of these sensors in biofilm formation or expression of virulence determinants.27–31 These domains are expressed either alone or as fusions with other modules, including but not restricted to EAL, GGDEF, HD-GYP, PAS, and helix-turn-helix motifs.26 A PilZ domain was also identified as the c-di-GMP binding module in G. xylinus cellulose synthase, where c-di-GMP regulatory function was first reported. Phylogenetic and structural analyses showed that PilZ domains have low sequence conservation with the exception of a few interspersed residues responsible for nucleotide docking.28 In addition, recent structural studies have shown that even highly similar PilZ domain-containing proteins can employ a markedly different molecular readout upon ligand binding.32
Interestingly, many bacteria that utilize c-di-GMP mediated signaling for adaptation lack PilZ domains encoded in their genomes.26 In addition, in some organisms PilZ signaling modules seem to control some, but not all the c-di-GMP dependent processes involved in biofilm formation and pathogenicity. For example, while PilZ domain-containing proteins in Vibrio cholerae are not essential for rugosity and exopolysaccharide production, some of them are required for efficient intestinal colonization during environment-to-host transition.29,33
It is now established that the gamut of c-di-GMP receptors spreads far beyond the family of PilZ protein domains. Identified c-di-GMP sensors include but are not limited to bacterial riboswitches, transcription factors of various domain architectures, divergent c-di-GMP turnover domains, and allosteric sites on active or degenerate diguanylate cyclases (Fig. 2).22,33–41 Interestingly, high-resolution structural data have shown that this structural diversity of c-di-GMP signal recognition is not limited to the nucleotide-sensing modules, but c-di-GMP itself can adopt a variety of stable conformations [Fig. 3(E)].42
Some of the identified c-di-GMP targets belong to protein families generally involved in sensing or metabolizing different nucleotide-based small molecules. For example, FleQ of Pseudomonas aeruginosa and VpsR of V. cholerae belong to the AAA+ superfamily of ATPases, but seem to function as c-di-GMP signal effectors independent of ATP binding or hydrolysis.38 As another example, the Clp proteins of Xanthomonas sp. are classified as catabolite activator-like proteins, whose homolog in Escherichia coli regulates gene expression in a cAMP-dependent manner.40,43 Similarly, c-di-GMP turnover domains are themselves homologous to protein domains involved in the synthesis or hydrolysis of different nucleic acid-based metabolites. For example, while GGDEF domains are evolutionary close to adenylate cyclases,11 HD-GYP domains belong to the larger HD superfamily of phosphohydrolases with nucleotide-based substrates such as dGTP or ppGpp.8 This highlights the functional complexity and rapid evolution of homologous proteins in bacterial signaling networks and raises the question of whether there are a limited number of universal c-di-GMP binding motifs and/or protein folds, or individual c-di-GMP receptors have evolved specific modes of recognition to ensure signal isolation.
Taken together, the huge number of homologous but nonredundant c-di-GMP metabolizing proteins per species, the diversity of receptor protein domain architectures and putative domain-domain interactions, the variety of adopted c-di-GMP conformations, and the widespread effects of c-di-GMP-mediated signaling phenomena complicate prediction-based approaches for the characterization of c-di-GMP signaling pathways and underscore the need for high-resolution structure-function studies on individual signaling targets (Table I).
Table I.
Representative High-Resolution Structures of c-di-GMP Metabolizing Enzymes and Protein Receptors
| Protein | Organism | Function | Conserved domains | PDB code and nucleotide-bound state | References |
|---|---|---|---|---|---|
| C-di-GMP turnover enzymes | |||||
| Diguanylate cyclases | |||||
| PleD | Caulobacter crescentus | C-di-GMP synthesis; polar differentiation | Two CheY-like REC domains; | 1W25 (c-di-GMP/Mg complex); | 41,44 |
| GGDEF domain | 2WB4 (c-di-GMP/ Mg/BeF3 complex); | ||||
| 2V0N (c-di-GMP/Mg/BeF3/ GTP-α-S complex) | |||||
| WspR | Pseudomonas aeruginosa | C-di-GMP synthesis; biofilm formation and persistence | CheY-like REC domain; | 3BRE (c-di-GMP/Mg complex); | 39,45 |
| GGDEF domain | 3I5B (GGDEF domain; apoprotein) | ||||
| Phosphodiesterases | |||||
| YkuI | Bacillus subtilis | C-di-GMP hydrolysis | EAL domain; | 2BAS (apoprotein) | 46 |
| PAS-like domain | 2W27 (c-di-GMP/Ca complex) | ||||
| BlrP1 | Klebsiella pneumoniae | Light-regulated c-di-GMP hydrolysis | BLUF sensor domain; | 3GFX 3GG1 (c-di-GMP/ Ca/FMN complex); | 47 |
| EAL domain | 3GFY (c-di-GMP/FMN complex); | ||||
| 3GFZ, 3GG0 (c-di-GMP/ Mn/FMN complex) | |||||
| Bd1817 | Bdellovibrio bacteriovorus | Unknown | HD-GYP domain | 3TM8, 3TMB, 3TMC, 3TMD (apoprotein) | 48 |
| C-di-GMP protein receptors | |||||
| PilZ domain proteins | |||||
| VCA0042/PlzD | Vibrio cholerae | Potential role in virulence | YcgR-N* domain; | 1YLN (apoprotein); | 28 |
| PilZ domain | 2RDE (c-di-GMP complex) | ||||
| PP4397 | Pseudomonas putida | unknown | YcgR-N* domain; | 3KYF (c-di-GMP complex) | 31 |
| PilZ domain | |||||
| PA4608 | Pseudomonas aeruginosa | Potential role in biofilm formation and/or quorum sensing | PilZ domain | 1YWU (apoprotein); | 30,32 |
| 2L74 (c-di-GMP complex) | |||||
| Transcription factors | |||||
| VpsT | Vibrio cholerae | Global transcription control; biofilm formation | Noncanonical REC domain; | 3KLN (apoprotein); | 37 |
| HTH DNA-binding domain | 3KLO (c-di-GMP complex) | ||||
| Clp | Xanthomonas campestris | Global transcription control; virulence | cNMP-binding domain; | 3IWZ (apoprotein) | 40 |
| HTH DNA-binding domain | |||||
| Degenerate EAL domains | |||||
| FimX | Pseudomonas aeruginosa | Type IV pilus-mediated twitching motility | CheY-like REC domain; | 3HV9 (EAL domain; apoprotein); | 35 |
| PAS domain; GGDEF domain; EAL domain | 3HV8 (EAL domain; c-di-GMP complex); | ||||
| 3HVA (GGDEF domain); | |||||
| 3HVB (GGDEF-EAL module; apoprotein) | |||||
| LapD | Pseudomonas fluorescens | Adhesin maintenance; biofilm formation and dispersal | Periplasmic output domain; | 3PJV (periplasmic domain); | 49 |
| HAMP domain; GGDEF domain; EAL domain | 3PJT, 3PJU (EAL domain; c-di-GMP complex); | ||||
| 3PJW, 3PJX (GGDEF-EAL module; apoprotein) | |||||
| Others | |||||
| PNPase | Escherichia coli | O2-dependent RNA degradation | Two RNase PH domains; | 3CDI (apoprotein); | 50 |
| KH RNA-binding domain; | 3CDJ (PH1-PH2 module; apoprotein) | ||||
| S1 RNA-binding domain | |||||
Conserved domains responsible for c-di-GMP synthesis, degradation, or recognition are underlined.
The Making and Breaking of c-di-GMP
Structure and regulation of diguanylate cyclases
The predicted adenylate cyclase fold of diguanylate cyclases11 was first confirmed experimentally when the structures of a GGDEF domain-containing protein, PleD from C. crescentus, were determined.41,44 PleD consists of two CheY-like phosphoreceiver (REC) domains and a C-terminal GGDEF domain, responsible for GTP to c-di-GMP conversion (Fig. 4). The structures revealed conserved residues within the GGDEF domain that are involved in nucleotide and magnesium binding, as well as residues within the N-terminal REC domain, responsible for phosphate coordination.
Figure 4.
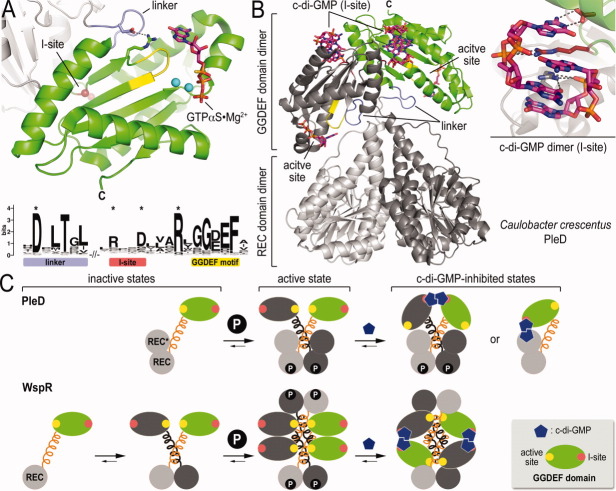
GGDEF domains and diguanylate cyclases. (A) Prototypical GGDEF domain structure. The PleD GGDEF domain bound to GTP-alpha-S is shown (PDB code: 2V0N).44 The GGDEF motif is colored yellow. The position of the I-site is highlighted as small spheres. The sequence logo highlights several conserved motifs extending in to the linker upstream of the GGDEF domain fold. (B) Product-inhibited structure of full-length PleD. The structure of a PleD dimer activated by BeF−3 bound to one REC domain is shown. The enzyme is inhibited by a stacked c-di-GMP dimer that occupies the I-site on the GGDEF domains (inset). (C) Models for the regulatory cycle for PleD and WspR. The models highlight conserved and unique feature that control enzymatic function of the two proteins.
More importantly, structural studies on PleD and a related but not identical diguanylate cyclase from P. aeruginosa, WspR, elucidated many features controlling enzymatic activity.39,44,45,51,52 In particular, dimerization of the GGDEF domains emerged as a key requirement and a regulated step in catalysis. Although WspR contains a single N-terminal REC domain, in both cyclases the response receiver modules were proposed to oligomerize upon phosphorylation by upstream histidine kinases and thus establish the catalytically competent state. In both proteins, an extended helix of the response regulator domain proximal to the GGDEF module contributes to the oligomerization interface, with the catalytic domain resting at the tip of these stalk-like protrusions. In support of the dimerization requirement, a catalytically active diguanylate cyclase was created by replacing all regulatory subunits by a short leucine zipper motif.45
A structurally peculiar feature is the conformation of the loop preceding the GGDEF domain [Fig. 4(A)]. In the majority of GGDEF domain-containing proteins, this loop contains a conserved aspartate residue that forms a hydrogen bond with an equally conserved arginine located just upstream of the GGDEF motif.49 The conformation of this loop is invariable in structures of GGDEF domain-containing proteins determined so far, and the specific arrangement may help to coordinate nucleotide binding given the close proximity of the arginine residue to the guanine moiety of the active site-bound nucleotide.44 Probably equally important, the particular loop conformation and underlying stabilizing interactions may restrict the freedom of the GGDEF domain relative to the adjacent regulatory modules. This likely applies to the majority of diguanylate cyclase proteins, given the prevalent presence of helical secondary structure elements predicted to precede GGDEF domains in bacteria. As a result of such inherent restricted flexibility of the interdomain linkage, conformational changes in the regulatory modules that change the angle or orientation of the stalk-like motifs will have an impact on the GGDEF domain interdistance and hence catalytic activity. Interestingly, this feature is not only limited to active diguanylate cyclases but is also found in degenerate GGDEF domain-containing proteins that function as c-di-GMP receptors.49
The structures of PleD also revealed another very common motif, an inhibitory c-di-GMP binding site (I-site) distinct from the active site and located at the base of the beta-hairpin that displays the conserved GGDEF motif in active diguanylate cyclases [Fig. 4(A,B)].41 The primary I-site is comprised of a conserved RxxD motif, where the arginine forms both hydrogen bonds, as well as π-stacking interactions with two intercalated dinucleotide molecules. The full c-di-GMP binding I-site requires secondary interactions donated by other parts of the proteins. There are at least two distinct ways that the I-site can be complemented by adjacent subunits or structural modules. In the first structure of PleD, a response regulator domain within one molecule contributes the secondary I-site residues—mainly an arginine side chain—to complete the I-site—c-di-GMP interactions. In the structure of BeF−3-activated PleD and of WspR, this second arginine is located at the back of the GGDEF domain, not overlapping with the primary I-site motif.39,41,44,45
Functionally, the I-site establishes a mode of feedback inhibition. Two nonmutually exclusive molecular mechanisms for feedback regulation have been proposed: allosteric inhibition or sequestration of the active site.39,41,44,45,53 The former model is based on molecular dynamics simulations of the nucleotide-bound and unbound state, while the latter is apparent in several structures of c-di-GMP-bound diguanylate cyclases, including that of the leucine-zippered artificial construct.45 In general, experimental data indicate that the main effect of c-di-GMP binding to the I-site is a restriction of conformational changes paired with sequestration of the active site dimer in a split state. Considering the apparent restricted flexibility of the linker segment connecting the regulatory and GGDEF modules, it will be interesting to obtain the structure of a GGDEF domain-containing protein en route to nucleotide cyclization, which has not been achieved to date. Interestingly, a recent study proposed a second mode of substrate inhibition based on the structural analysis of an isolated GGDEF domain bound to c-di-GMP at the active site.42 The putative inhibitory site overlaps significantly with the predicted active site and is conserved in almost all GGDEF domain sequences. It is therefore plausible that this structure could in fact depict the substrate-bound state right after nucleotide cyclization rather than a distinct mode of feedback inhibition.
Comparative analysis of the structures of PleD and WspR indicated that several of the regulatory mechanisms described above are strictly conserved, while others appear to have flexibility in their implementation [Fig. 4(C)]. In particular, the mechanistic details concerning dimerized GGDEF domains in both the active and product-inhibited states appear virtually identical. In contrast to PleD's canonical regulation via REC domain phosphorylation and subsequent dimerization, however, experimental evidence supports the notion that WspR employs a noncanonical dimer-tetramer transition to establish the active state.39,45
An interesting question is whether or not substrate-inhibited diguanylate cyclases can be reactivated by phosphodiesterases as part of their cellular regulation, a phenomenon that has been demonstrated in vitro.39 Such a post-transcriptional mechanism could be controlled by targeted spatial distribution of c-di-GMP synthesis and degradation enzymes, and may provide a fast and localized signaling response. Taken together, the structure–function studies on catalytically active diguanylate cyclases demonstrate sophisticated regulation at multiple levels, suggesting that their activity can be finely tuned in the bacterial cytosol.
Structure and regulation of EAL domain-containing phosphodiesterases
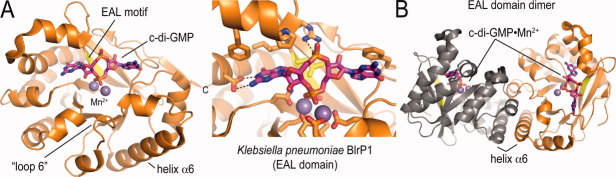
Many of the regulatory principles that establish phosphodiesterase activity have been uncovered using biochemical and biophysical approaches.54,55 Informed by then unpublished EAL domain structures determined by a structural genomics consortium, a modeling approach and its experimental validation revealed several conserved motifs that were important for c-di-GMP cleavage.55 These findings could be used to identify catalytically active EAL domains based on their sequence, which was later confirmed and extended by an independent crystallographic study.56 Another insight came from the identification of a conserved loop in active EAL domains.54 While the initial study established its crucial role for catalysis, crystal structures revealed its location at a EAL domain-dimer interface and assigned its functions as being important for metal coordination [Fig. 5(A)].46,47 This loop was referred to as loop 6 in SadR/RocR,54 β5-α5 loop in the light-regulated phosphodiesterase BlrP1,47 and the switch loop in LapD.49
Figure 5.
Structure of a canonical EAL domain. (A) EAL domain fold. The EAL domain was extracted for the full-length crystal structure of the light-regulated phoshphodiesterase BlrP1 (PDB code: 3GFZ).47 Metal ion and c-di-GMP binding sites are shown. (B) EAL domain dimer. Several structures of EAL domains show a dimeric assembly that is likely relevant for c-di-GMP binding and establishing the catalytically competent state.
The first published crystal structures of an EAL domain-containing protein, YkuI from Bacillus subtilis, documented the triose-phosphate isomerase (TIM)-barrel fold as the building block of EAL-domain containing c-di-GMP-specific phosphodiesterases [Fig. 5(A)].46 Structures were determined both for the apo- and c-di-GMP-bound states, which revealed the nucleotide binding site and residues crucial for catalysis. The conserved EAL motif is presented on a central strand and faces the active site, which in turn is conserved in its location to canonical TIM-barrel-containing proteins.46,57 Calcium, a known inhibitor of EAL domain-containing phosphodiesterases,2,58,59 was co-crystallized with c-di-GMP, resulting in a bound calcium ion at the active site that marked a conserved metal-binding motif.46 Later, two independent structural studies of active phosphodiesterases, those of Klebsiella pneumoniae BlrP1 and Thiobacillus denitrificans TDB1265, established a two-metal ion mechanism for c-di-GMP catabolizing enzymes [Fig. 5(A)].47,56
At the same time, nucleotide binding to the EAL domain fails to trigger any major structural rearrangements except for changes of rotamer conformation in the residues that are involved in c-di-GMP coordination.46 The low extent of conformational changes upon c-di-GMP-binding has been replicated in several other cases, including those of T. denitrificans TDB1265 and the inactive EAL domain-containing protein FimX from P. aeruginosa.35,56 Yet, an interdependence of c-di-GMP binding and EAL domain dimerization was noted in LapD, a receptor for the dinucleotide,49 suggesting that even subtle changes can have a profound effect on the stability of intermolecular interactions.
In several structures of EAL domain-containing proteins, dimerization occurs between equivalent structural motifs within the EAL domain, which include helix α6 and loop 6 mentioned above [Fig. 5(B)].46,47,49,56 In addition, the overall quaternary structure of otherwise very different EAL domain-containing proteins can be conserved, as demonstrated in the comparison between K. pneumoniae BlrP1 and the unrelated protein YkuI from B. subtilis.47 Both proteins were crystallized in a dimeric state, with the former containing a light-sensing BLUF domain in place of the PAS domain of YkuI. While YkuI was inactive in its purified form,46 the structure of BlrP1 highlighted light-induced, allosteric regulation across the dimer interface as a likely mechanism of activation.47,60
In addition to proteins that contain either an EAL or GGDEF domain, dual-domain proteins with a GGDEF-EAL domain module are also widely encoded in bacterial genomes.8 While in the majority of the cases studied to date either one or both domains are inactive,22,35,58 there is an increasing number of dual-activity enzymes being identified.61–63 Yet, their regulation and the functional consequences of coupled opposing activities are poorly understood.
The first structure of a HD-GYP domain
HD-GYP domain-containing proteins were first identified and implicated in c-di-GMP turnover in the plant pathogen Xanthomonas campestris.17,64 Similar to EAL and GGDEF domains, they occur as single-domain proteins or as modules in multidomain proteins.7,8 Furthermore, they may couple directly to GGDEF domains via direct protein-protein interactions.65
The first structural model for a HD-GYP domain was revealed recently based on a crystallographic study on Bdellovibrio bacteriovorus Bd1817, an inactive protein that lacks the active site tyrosine (Fig. 6).48 The HD-GYP domain is composed of seven helices that form a compact fold, roughly resembling other proteins of the HD superfamily [Fig. 6(A)]. In addition to the HD-GYP domain, this protein also contains an N-terminal, less-characterized domain that is connected to the degenerate phosphodiesterase domain via a compact three-helix linker. Importantly, bound metal and phosphate ions mark the active site, which is located within the C-terminal domain of Bd1817 and is lined by the conserved HD-GYP motif [Fig. 6(B)]. Modeling suggested that the fold may accommodate c-di-GMP in an extended conformation similar to that observed bound to EAL domains. More detailed mechanistic insights would be aided by structures of active and/or c-di-GMP-bound HD-GYP domain-containing proteins.
Figure 6.
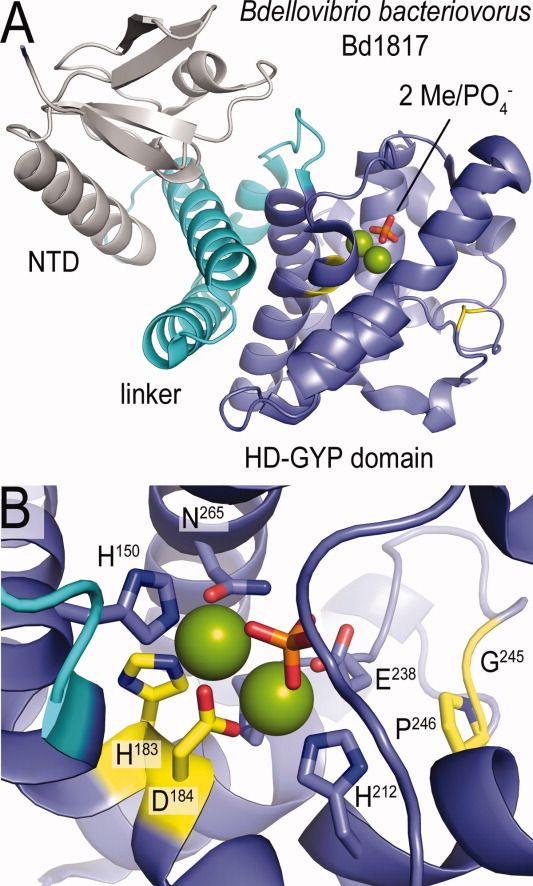
Structure of an HD-GYP domain fold. (A) Overall fold. The modular nature of a full-length HD-GYP domain-containing protein is shown (PDB code: 3TM8).48 (B) Close-up view of the putative nucleotide binding site.
C-di-GMP sensors
PilZ homology domains: The first identified c-di-GMP receptors
Comparative sequence analyses identified PilZ homology domains as widespread protein modules with a phylogenetic distribution pattern similar to those of GGDEF and EAL domain-containing proteins.26 In addition, many of the proteins predicted to contain PilZ domains were also suggested to participate in c-di-GMP mediated signal transduction based on available biochemical, genetic and functional data. For example, a PilZ homology domain was detected in G. xylinus cellulose synthase, where allosteric regulation by c-di-GMP was initially described, as well as in YcgR of E. coli, which controls motility in a way opposite to that of a c-di-GMP specific phosphodiesterase.26,66,67
High-resolution structural data are now available for several PilZ domain-containing proteins in their apo- and/or c-di-GMP-bound form.28,30–32,68 The core of the isolated module consists of a six-stranded antiparallel β-barrel, which is commonly found in hydrophobic ligand-binding proteins. Sequence alignment across protein family members showed low sequence conservation except for a few interspersed residues responsible for nucleotide docking.28 C-di-GMP is primarily coordinated by two consensus motifs—RxxxR and D/NxSxxG—located at the extreme N-terminus and the β2/β3 hairpin, respectively.28 The nucleotide binds as a single molecule in cis conformation (in VCA0042/PlzD of V. cholerae)28 or as an intercalated dimer (in PP4397 and PA4608 of Pseudomonas sp.)30–32,68 through typical H-bonding and arginine-mediated π-π stacking interactions.
This similar mode of nucleotide recognition among PilZ domain-containing proteins is in contrast to the markedly diverse modes of conformational switching observed in the various structures. In PA4608, which is a single domain protein of P. aeruginosa, nucleotide recognition does not affect the monomeric oligomerization state but is instead accompanied by significant rearrangements in the N- and C-terminal regions peripheral to the core β-barrel.30,32,68 In apo-PA4608, the N-teminal RxxxR motif, also known as the c-di-GMP “switch,” is unstructured and a C-terminal helix alternatively covers and exposes the hydrophobic surface of the nucleotide-binding pocket. In the presence of c-di-GMP, the “switch” motif wraps around the ligand and in turn ejects the C-terminal lid, creating a highly negative unilateral surface proposed to act in downstream signal transduction.32,68
In contrast to PA4608, two other model PilZ domain-containing proteins, PP4397 of Pseudomonas putida and VCA0042/PlzD of V. cholerae, are found to adopt a dimeric conformation in their nucleotide-free states. C-di-GMP binding is accompanied by dimer-to-monomer transition (for PP4397) or by conformational reorganization within the dimer (for VCA0042/PlzD), underscoring once again the diversity in signaling output.28,31 The two proteins are structurally homologous to the YcgR motility switch of E. coli and contain an additional N-terminal YcgR-N* domain with an overall similar fold to that of the C-terminal PilZ module. In both proteins the c-di-GMP-binding RxxxR motif forms a hinge-like connector between the two domains. While in VCA0042 this “switch” is unstructured in the apo-state, but wraps around the ligand to dramatically change the relative orientation of the two domains in the presence of c-di-GMP, in apo-PP4397 it forms a short helix participating in dimerization contacts, which become restructured upon ligand recognition and in turn contributes to dimer disassembly.
The significant effects on conformation following c-di-GMP binding to YcgR homologs were also employed to design the first c-di-GMP biosensor.69 The fusion of full-length E. coli YcgR between the established FRET pair of cyan and yellow fluorescent proteins70 was successfully applied to monitor cellular c-di-GMP distribution during the cell cycle of C. crescentus and P. aeruginosa. The imaging study also confirmed for the first time spatiotemporal control of c-di-GMP signal transduction, contrary to the more intuitive model of intracellular diffusion due to the second messenger's small size and high solubility.
Taken together, the bioinformatic and structural studies on PilZ domain-containing proteins have revealed a plethora of protein domain architectures and mechanisms for molecular read-out. This complexity of c-di-GMP signaling mechanisms further underscores the need for high-resolution structure–function analyses on individual targets in addition to more limited, homology-based models.
Degenerate c-di-GMP turnover domains
The ensemble of bioinformatic and structural studies on c-di-GMP turnover domains indicates that many of these exist in a catalytically inactive form. However, knockout, overexpression, or mutational studies of proteins containing such “degenerate” domains have strong effects on biofilm formation or virulence. In the current model, these proteins function at the post-translational level to ensure c-di-GMP signal relay through direct protein-protein interactions.
Interplay between the PilZ and FimX proteins
As discussed above, PilZ homology domains were identified as the first bonafide c-di-GMP binding modules. Nevertheless, the PilZ protein itself, originally identified as necessary for biofilm-related Type IV pilus (T4P) biogenesis in P. aeruginosa,71 is incompetent for direct c-di-GMP recognition. Studies on PilZ/PA2960 orthologs from two Xanthomonas species revealed important structural differences from c-di-GMP-binding PilZ homology domains, including lack of the signature RxxxR and D/NxSxxG motifs responsible for ligand coordination.72,73 Even so, the PilZ protein has retained its role in c-di-GMP signal transduction, acting as an adaptor between the c-di-GMP receptor protein FimX and the T4P assembly ATPase PilB.72
FimX is a multidomain protein with an N-terminal CheY-like REC domain followed by a PAS domain of unknown sensitivity, and a GGDEF-EAL tandem domain module. Similarly to PilZ, it was originally identified as involved in the T4P-mediated twitching motility of P. aeruginosa and was found to localize to a single pole of the bacterial cell together with the pilus biosynthetic machinery.74
Although initially described as an active phosphodiesterase,75 FimX was subsequently shown to lack significant c-di-GMP catabolizing activity based on homology modeling, enzymatic assays, and direct structural evidence.35,55 In particular, the crystal structures of the EAL domain showed that while the overall TIM-barrel fold of conventional phosphodiesterases is preserved, the protein lacks key residues for metal ion coordination required for catalytic activity.35,76 In addition, low-resolution SAXS (small angle X-ray scattering) data indicated that the full-length protein likely exists as a dimer through interactions between its N-terminal REC/PAS domains, while the EAL domains remain separated at the distant ends of the extended quaternary structure. Such an assembly excludes EAL domain dimerization through the functionally important “switch” loop discussed above.46,47,49,54
Similarly to the EAL domain, the N-terminal REC domain and the GGDEF module of FimX have also evolved away from their canonical sensory and enzymatic activities, respectively: the REC domain lacks the conserved phosphoreceiver aspartate residue of archetypal response regulators and both the active and inhibitory sites on the GGDEF module show significant sequence deviation from the consensus motifs required for c-di-GMP synthesis or regulatory binding.35
Regardless of the lack of catalytic and phospho-receiver activities, the REC, GGDEF, and EAL domains of FimX have important roles in c-di-GMP signal relay. The second messenger was shown to bind the degenerate phosphodiesterase domain with high affinity, and the crystal structure of the c-di-GMP-bound EAL domain revealed a conserved mode of protein-nucleotide interactions.35 Furthermore, while no major changes in the overall quaternary structure were observed between the apo-protein and its nucleotide-bound form, hydrogen/deuterium exchange experiments revealed that binding of c-di-GMP to the EAL domain triggers a long-range conformational change in the N-terminal REC domain and the adjacent linker.77
Interestingly, residues in this region together with intact signature motifs in the GGDEF and EAL domains of FimX—G346DSIF and E475VL, respectively—have been shown to be crucial for proper unipolar protein localization.74,75,77 It has also been proposed that the C-terminal EAL domain directly binds PilZ, and therefore indirectly the T4P biosynthetic machinery, and that this interaction is stabilized by c-di-GMP.72
Collectively, these data indicate that the various structural modules of FimX have evolved an important and interdependent role in the nucleotide signal relay. Important questions still remain, such as the biological significance of the long-range conformational changes upon nucleotide recognition, and the identities of additional binding partners which may be responsible for regulation of c-di-GMP-dependent T4P assembly and twitching motility.
Inside-out c-di-GMP signaling in Pseudomonas fluorescens: A role for degenerate domain-containing protein LapD
Similar to FimX, many GGDEF and EAL domain-containing proteins possess both types of domains in the same polypeptide chain, with variation among catalytic activities ranging from no, single, or dual catalytic activity preserved among protein family members. In addition, several of these function as bacterial transmembrane receptors and contain an additional HAMP domain as a juxtamembrane signal relay module, located N-terminally to the GGDEF-EAL domain tandem.
One such receptor with HAMP-GGDEF-EAL domain organization is LapD of Pseudomonas fluorescens, which controls cell adhesion during biofilm formation in response to phosphate availability via direct read-out of cellular c-di-GMP (Fig. 7).22,49,78–81 C-di-GMP binding to the degenerate EAL domain in the cytosol is communicated via the HAMP relay module to an N-terminal output domain in the periplasm. In the c-di-GMP bound state, LapG, an otherwise active periplasmic protease, is bound to the output domain of LapD. As a result, cleavage of LapG's substrate, the large adhesin protein LapA, is prevented, leading to biofilm formation.22,49,79
Figure 7.
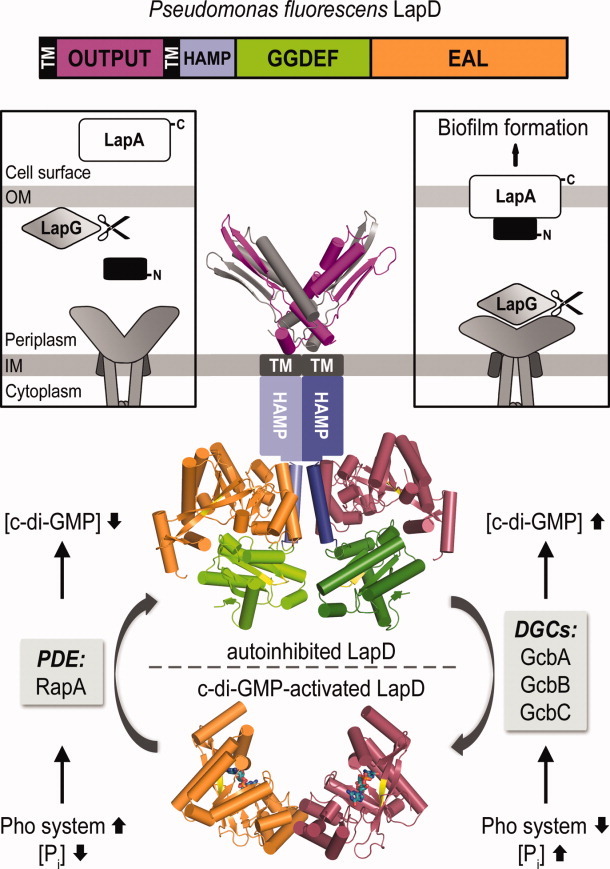
Model for LapD-mediated signaling. The composite structural model highlights the proposed conformational changes of the transmembrane, c-di-GMP effector LapD.49 Switching in the intracellular domains regulates the conformation of its periplasmic output domain, which differentially interacts with a protease, LapG, that in turn controls the stability of a cell surface adhesin, and hence biofilm formation. Intracellular regulators as well as environmental signals are well-established for this system.80,81
Structural analyses of the cytosolic domains of LapD revealed an autoinhibited conformation in the absence of c-di-GMP. In particular, the EAL domain was found to fold back onto a helical extension of the HAMP domain, dubbed as the S- or signaling- helix.49 Interaction of the EAL domain with the S-helix are mostly hydrophobic and extend over the conserved “switch” loop crucial for canonical dimerization and metal ion coordination in active phosphodiesterases. In the closed conformation, the GGDEF domain restricts access of c-di-GMP to the EAL domain though the ligand-binding pocket remains only partially occluded. Increased c-di-GMP levels were therefore proposed to compete out the inhibitory interactions and lead to opening of the cytosolic tandem domain module. Disrupted EAL domain-S-helix interactions are likely compensated by canonical EAL domain dimerization—as observed in crystal structures of the nucleotide-bound isolated module49—as well as molecular motions of the HAMP domain helices securing signal transduction to the periplasmic output domain.22,49
The isolated output domain was shown to be poised for LapG sequestration, as demonstrated in direct protein binding studies. Interestingly, the distal tips of the V-shaped dimer are formed by a conserved GWxQ motif, and mutation of the tryptophan residue alone is sufficient to abolish LapG-output domain binding in vitro, as well as biofilm phenotype rescue in a ΔlapD genetic background in vivo.49 Overall, these data indicate that the periplasmic output domain, and in particular the conserved tryptophan residues at the distal ends of the V-shaped module, act as a molecular caliper for LapG recognition. The observation that the isolated output domain can bind LapG is consistent with a model in which the nucleotide-free intracellular domains hold the receptor in an autoinhibited conformation that relaxes into a LapG-binding state upon c-di-GMP activation.49
Bioinformatic analyses based on the structural model for regulation of the LapD-LapG signaling system identified homologous circuits in a variety of free-living and pathogenic species. Importantly, LapG homologs are likely to have different substrates in systems for which no clear target adhesins can be identified. In particular, proteins that contain regions homologous to the cleavage site in LapG's substrate adhesin include RTX-like bacterial toxins and the majority of such proteins are encoded in close proximity to lapD and lapG homologous genes.49 This indicates that c-di-GMP-dependent inside-out signaling systems might have evolved a common mechanism to control periplasmic proteolysis for toxin secretion or biofilm dispersal depending on bacterial physiology and adaptational needs.
Degenerate GGDEF domains as c-di-GMP receptors
Degenerate GGDEF domains capable of c-di-GMP sensing and signal relay through a preserved I-site motif (RxxD) form another class of functionally important c-di-GMP receptors. Examples of proteins with degenerate GGDEF domains that function as c-di-GMP receptors are PelD of P. aeruginosa,36 CdgG of V. cholerae,33 the multidomain histidine kinase SgmT of Myxococcus xanthus,82 MxdA of Shewanella oneidensis,83 and PopA of C. crescentus.84 These proteins play roles in diverse physiological processes including biofilm formation, motility, and cell cycle progression. Insight in the mechanisms of c-di-GMP signal transduction is mostly limited to genetic studies, inferred protein-protein interactions, sequence alignments, or homology modeling on the isolated GGDEF domain. High-resolution structure-function analyses in the context of full-length proteins and macromolecular assemblies would thus complement the genetic and molecular studies already done, and shed light on the target-specific mechanisms of nucleotide signal transduction.
C-di-GMP sensing transcription factors
The global picture of c-di-GMP mediated signaling, where bacteria carefully monitor their environment to control motility, growth, secretion, and biofilm formation in order to rapidly adapt to changing conditions, makes regulators of gene expression likely candidates for c-di-GMP-responsive signal effectors. To date, several protein transcription factors (e.g. FleQ of P. aeruginosa, Clp of Xanthomonas sp., VpsT and VpsR of V. cholerae, MrkH of K. pneumoniae), as well as two distinct riboswitch classes in messenger RNAs, have been identified to directly sense c-di-GMP and regulate gene expression at the transcription initiation and post-transcriptional levels, respectively.34,38,40,43,85–88
VpsT of V. cholerae: A noncanonical response regulator
V. cholerae, causative agent of the eponymous diarrheal disease, is an opportunistic pathogen with an impressive arsenal of c-di-GMP turnover domains encoded by its genome (41 GGDEF, 22 EAL, and 9 HD-GYP protein modules). In contrast, only a few and strikingly diverse protein- and RNA-based sensors for the nucleotide have been so far identified.
Initial biofinformatic searches identified five PilZ domain-containing proteins, as well as two distinct riboswitch sequences as putative targets for the second messenger. Nevertheless, the regulatory role for the c-di-GMP-sensitive RNA aptamers is limited to the expression of their immediately downstream open reading frames and quintuple knock-out of all PilZ domain-containing proteins fails to disrupt colonial rugosity and biofilm formation,33,34 pointing toward additional targets.
In V. cholerae, these c-di-GMP-mediated adaptational strategies are strictly dependent on the expression of Vibrio polysaccharide (vps) genes, which in turn are under the control of two positive transcription regulators, VpsT and VpsR.85,89 This led to the hypothesis that the two could serve as direct c-di-GMP sensor-effectors, which was later corroborated by direct experimental evidence and, in the case of VpsT, by high-resolution structural data.37,90
VpsT folds into an N-terminal receiver (REC) domain for nucleotide recognition and a C-terminal helix-turn-helix (HTH) domain for DNA binding.37 It belongs to the FixJ/LuxR/CsgD family of response regulators, which are typically effectors in two-component signal transduction systems and depend on phosphoryl transfer from upstream histidine kinases for signaling input. Interestingly, both VpsT and its homolog in Escherichia and Salmonella spp., CsgD, show divergent receiver domains where only half of the residues required for phosphotransfer are conserved and no cognate kinases have been identified to date.
In addition, the crystal structure of VpsT showed that the canonical (α/β)5-fold of the receiver domain is extended by an additional helix, α6, which was identified as a common motif among available structures of LuxR-like response regulators, regardless of their input mode for regulation (Malashkevich et al., unpublished).37 Receiver domain oligomerization also differs significantly from that of canonical response regulators (Fig. 8). Rather than utilizing the α4-β5-α5 structural motif for dimerization,95 VpsT utilizes two different and nonoverlapping dimerization interfaces.
Figure 8.
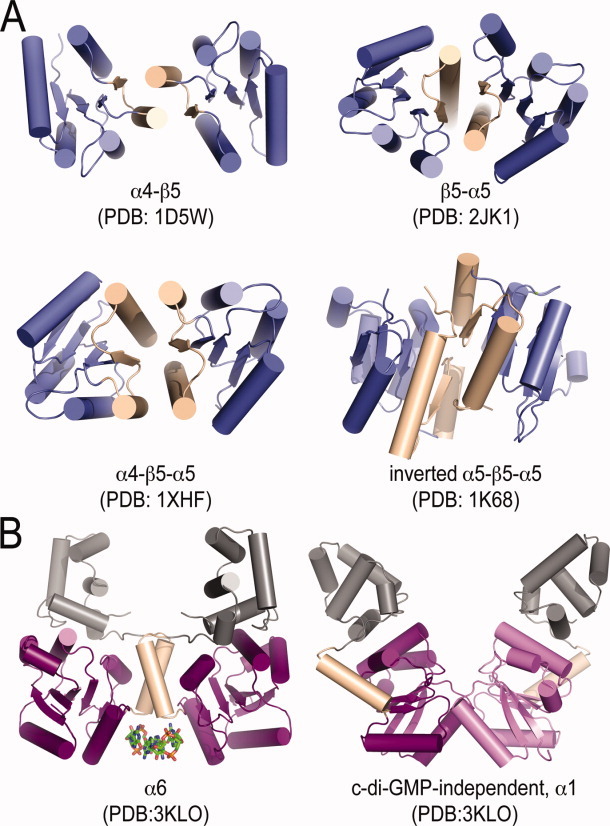
Overview of REC domain dimerization. (A) Canonical REC domains.91–94 (B) VpsT dimerization.37
The first dimerization interface is independent of nucleotide recognition and involves a hydrophobic groove and polar interactions formed between residues of α1, α5, and a binding pocket that extends into the canonical phosphorylation site. Point mutations disruptive to this interface were shown to have an overall stimulating effect on VpsT activity as a positive regulator of biofilm formation and a negative regulator of flagellar motility, emphasizing its biological significance.37
The second dimerization interface is specific to the additional helix α6 and is necessary for and stabilized by c-di-GMP recognition. The nucleotide-binding pocket is formed primarily by four consecutive residues located at the base of α6 in each protomer and conserved as a W[F/L/M][T/S/P]R motif in closely related homologs. C-di-GMP binds as an intercalated dimer and its purine rings form π-stacking interactions with the side chains of the conserved tryptophans and arginines, while the threonine residues in VpsT form hydrogen bonds with the phosphate moieties of the nucleotide. The biological relevance of this second dimerization interface was confirmed in experiments showing that binding of wild-type VpsT to promoter sequences of a vps gene occurred only in the presence of c-di-GMP, while mutants defective for nucleotide binding or α6 dimerization were uncapable of DNA recognition and had markedly different phenotypes.37
The relative orientation of the DNA-binding and receiver domains was preserved between the crystal structures of apo-VpsT and the c-di-GMP bound protein. Interestingly, if the orientation of the HTH motifs is preserved in solution as well, it would require significant changes in DNA architecture for promoter binding to occur. This, together with the fact that VpsT bound to relatively remote fragments of the studied vps gene promoter, led to the hypothesis that promoter binding is accompanied by DNA loop formation, similar to the regulatory mechanisms at play at the lac, gal, and ara operons.37,96,97 To determine whether this is actually the case, what particular oligomerization state VpsT adopts at each target promoter, and what the consensus DNA sequence for VpsT recognition is, would nevertheless require further structure-function studies on the required c-di-GMP—VpsT—DNA interactions.
As mentioned above, VpsT is homologous to CsgD from E. coli and Salmonella typhimurium, a protein whose expression and function are tightly intertwined with various c-di-GMP signaling pathways and are required for the secretion of key biofilm determinants.98,99 Similarly to VpsT, CsgD has been postulated to be incompetent for phosphotransfer and to instead depend on small molecule binding for efficient gene expression regulation.99,100 Nevertheless, CsgD is unlikely to act as a c-di-GMP sensing transcription factor, as it has a divergent and highly conserved YF[T/S]Q motif at the putative ligand-binding site and direct binding experiments have ruled out c-di-GMP recognition.37,101
A recent study has proposed that CsgD could nevertheless be regulated by phosphorylation, as the protein autophosphorylates in the presence of high concentrations (10mM) of acetyl phosphate in vitro.101 While this phosphorylation appears to inhibit protein binding to its cognate DNA, it remains unclear whether this is due to a distinct functional state or is an artifact of nonspecific binding and reduced protein stability. In support of the latter, acetyl phosphate does not affect CsgD function in vivo and a loss-of-function asparagine substitution of the conserved phosphoreceiver aspartate (D59N) preserves phosphate binding to the protein in vitro. In addition, a phosphomimetic aspartate to glutamate substitution (D59E) appears to inactivate the protein, but the mutant is indeed characterized by markedly reduced protein stability.101 Based on the crystal structures of homolog VpsT, the phosphoreceiver aspartate (D59) is likely proximal to a dimerization interface, disruption of which might explain the mutant's altered stability and function and account for the observed effects in vitro and in vivo.
Although there are little data available on the regulation of CsgD in vivo, it is clear that the FixJ/LuxR/CsgD family of response regulators has evolved an array of diverse regulatory sensitivities. Although far from complete, the structural studies and comparative sequence analyses on VpsT identified several conserved structural features (α6-dimerization, ligand-coordinating residues, conserved amino acids connecting the canonical phosphoreceiver site with the nucleotide-binding pocket), which could shed light on the evolution from archetypal two-component phosphotransfer to small molecule recognition and facilitate the prediction of additional second messenger targets.
VpsR of V. cholerae and FleQ of P. aeruginosa: AAA+ domains as putative c-di-GMP sensors
VpsR of V. cholerae is another c-di-GMP-responsive positive regulator of biofilm formation and rugosity, which functions in concert and shares an overall similar regulon with VpsT.89,90,102 Interestingly, VpsR is homologous to one of the first identified protein sensors for c-di-GMP, FleQ of P. aeruginosa.38 The two proteins contain a relatively divergent N-terminal response receiver domain, followed by a more conserved AAA+ σ54-interaction domain and a C-terminal helix-turn-helix motif.
In FleQ, the N-terminal receiver domain has been proposed to participate in regulatory protein-protein interactions, such as binding of the active ATPase FleN, which is also a known antagonist of FleQ function.38,103 Little is known of the structural requirements for c-di-GMP recognition, other than that it occurs independently of the N-terminal receiver domain or ATP hydrolysis. This may suggest that rather than acting as a P-loop NTPase for energy-dependent transcription regulation, the AAA+ domain of FleQ (and possibly VpsR) has evolved an altered nucleotide specificity for c-di-GMP sensing.
When bound to DNA, FleQ acts as a master activator of flagellar gene expression, as well as a repressor for the pel exopolysaccharide synthesis operon.38,104 C-di-GMP recognition serves as an “off” switch for DNA binding and thus leads to motility inhibition and biofilm formation. Interestingly, c-di-GMP binding to the protein has been shown to have a much greater effect on promoters that FleQ represses (e.g. of the pel operon), compared to those it activates (e.g. of flagellar biosynthesis genes).38 The main difference between the two types of promoters is the requirement for the alternative RNA polymerase sigma factor σ54 during FleQ-mediated transcription activation, which likely interacts directly with the nucleotide-sensing module.38
CRP-like proteins (Clp): Ditching cAMP sensitivity for c-di-GMP
Clp proteins, homologs of the cAMP receptor protein (CRP, also known as catabolite activator protein) of E. coli, have now been identified as c-di-GMP responsive transcription regulators in several species.40,43,87,105
In Xanthomonas sp., Clp regulates approximately 300 genes involved in plant pathogenesis. Interestingly, clp deletion mutants show markedly decreased virulence potential, similarly to mutants lacking one or more c-di-GMP-specific phosphodiesterases.106,107 This, together with the fact that Clp can bind promoter DNA with high affinity in the absence of any effector, lead to the hypothesis that Clp-dependent virulence gene expression can be inhibited by direct c-di-GMP recognition.43 Such a regulatory mechanism was subsequently validated in at least two different Xanthomonas species, and high-resolution structural data for the nucleotide-free Clp of X. campestris (XcClp) is now available.40,43,87
XcClp consists of an N-terminal cNMP-binding domain and a C-terminal, HTH-based DNA-binding domain, linked by a dimerization module consisting of a long α-helix. The protein adopts a symmetrical dimeric structure, roughly similar to the structure of E. coli CRP in its EcCRP-cAMP-DNA complex.40,108 Specifically, the structure of ligand-free XcClp is characterized by a decreased gap between the N- and C-terminal domains (“closed” form), defined as intrinsically active for DNA recognition. Detailed structural analysis indicated that regardless of the high sequence identity between XcClp and EcCRP (44%), differences of key residues in at least three regions are responsible for a constitutive DNA-binding conformation and altered nucleotide specificity of XcClp.40
First, residue S84 of EcCRP, which alone positions cAMP through five direct or indirect H-bonds in the EcCR-cAMP complex, is substituted by a larger and negatively charged glutamate (E99), whose side chain is flipped away from the ligand-binding site. This single serine-to-glutamate substitution appears sufficient to render the protein insensitive to cAMP and has been even proposed to serve as a sole marker for cAMP- versus c-di-GMP-ligand specificity.40,109,110
Two additional regions in XcClp were shown to contain key amino acid substitutions, likely responsible for the constitutive DNA-binding activation. These include the gap region between the N- and C-terminal domains, where a single glutamine (Q175 in EcClp) to arginine (R195 in XcClp) substitution serves to reduce the gap dimensions, as well as several amino acid substitutions, which are proximal to the DNA-binding motif and known to result in cAMP-independent transcription when introduced in EcClp.40,111
Although no direct structural data are available for a XcClp-c-di-GMP complex, molecular modeling and mutational analysis identified a putative ligand-binding site on the protein.40 In the docked model, c-di-GMP is bound as a single molecule with its purine bases oriented parallel to each other. Similarly to other c-di-GMP binding motifs, the nucleotide is stabilized mainly by side-chain interactions with aspartate (D70, D170) and arginine (R150, R154) residues, including hydrogen bonds, salt bridges and π-π stacking interactions.40 The binding pocket in XcClp is distinct from the one for cAMP recognition in EcCRP but is nevertheless found in a region known to strongly affect the DNA-binding strength of the latter.111
The structural and functional analyses on XcClp indicate that the protein has evolved an altered nucleotide- and DNA-binding propensity, where c-di-GMP recognition serves to block constitutive DNA binding and rapidly switch off virulence gene expression.
C-di-GMP dependent RNA processing: A role for E. coli polynucleotide phosphorylase (PNPase)
The role of c-di-GMP in regulating RNA transactions was initially highlighted by the discovery of c-di-GMP-sensitive riboswitches in messenger RNAs of various species (for review see Ref.112). In addition, a recent study showed that c-di-GMP can also regulate RNA turnover by direct binding to PNPase, an important 3′ to 5′ exonuclease of E. coli.113 The enzyme was found to associate in a megadalton-sized RNA “degradosome” complex with two direct O2 sensors, the diguanylate cyclase DosC and the c-di-GMP-specific phosphodiesterase DosP. The two enzymes are activated in anaerobic and aerobic conditions, respectively, offering a spatially restricted mechanism for regulation of available c-di-GMP levels and PNPase-dependent RNA processing in response to environmental oxygen stimuli. While high-resolution structural data for the c-di-GMP-free PNPase are already available in the literature and the PDB database,50 further structure-function analyses are necessary to explain its c-di-GMP-dependent mode of regulation.
Eukaryotic c-di-GMP receptors
Although c-di-GMP appears to have ubiquitous effects on the survival and virulence strategies adopted by most bacterial species, enzymes for its synthesis or degradation are not found in any other domain of life. In the context of infectious diseases, the presence of c-di-GMP would thus serve as a unique beacon for microbial pathogenesis and recent findings suggest that the mammalian immune system has developed dedicated machinery for c-di-GMP recognition and subsequent pathogen elimination.
It is important to note that c-di-GMP's immunostimulatory effects in mammals also hold promise for the development of novel adjuvants, vaccines, or other immunotherapeutics. Indeed, one early report demonstrated that exogenous c-di-GMP can inhibit the growth of human colon cancer cells without cytotoxic effects against non-cancerous cells in culture.114 More recently, studies from two different groups indicated that pretreatment of mice with c-di-GMP prior to challenge with K. pneumoniae or Staphylococcus aureus resulted in suppressed pathogen burdens,115,116 and that the immune response in mice was enhanced when an intranasal influenza vaccine was supplemented with c-di-GMP.117 Deciphering the mechanisms of c-di-GMP recognition and signaling effects in mammals can thus prove instrumental for the prevention and treatment of a variety of human diseases, which further underscores the need for diligent structure–function analyses.
STING: A mammalian receptor for c-di-GMP
The presence of c-di-GMP is known to elicit host Type I interferon response, which is typically induced by viral and bacterial pathogens and contributes to diverse outcomes toward successful pathogen elimination in vivo.118,119 Interestingly, while the transcription response to c-di-GMP in mammalian cells is virtually identical to the one triggered by cytosolic presence of pathogen DNA, c-di-GMP recognition was found to be independent from known DNA or RNA cytosolic sensors.118
These puzzling effects were recently resolved by the discovery that STING (Stimulator of InterferoN Genes) serving as an essential signaling adaptor between cytosolic DNA detection and interferon gene induction, can moonlight as a direct target for c-di-GMP stimulation.120 Bioinformatic analyses indicate that STING is a transmembrane protein with several membrane-spanning segments at the N-terminus and a globular carboxy-terminal domain. Mutational analysis confirmed that residues important for c-di-GMP binding by the C-terminal domain are separate from those important for bacterial DNA recognition. Interestingly, competitive binding studies indicated that STING is also a likely sensor for another RNA dinucleotide established as a prokaryote-specific second messenger, c-di-AMP.
In terms of structural classification, STING does not share homology with other immunosensory proteins in mammals and thus appears as a stand-alone representative of a novel class of c-di-GMP receptors. Surprisingly, part of the protein is homologous to the diadenylate cyclase Dac1 of Listeria monocytogenes, an intracellular pathogen known to elicit STING-dependent Type I interferon response through efflux of cyclic dinucleotides.120–122 This raises the interesting hypothesis that the mammalian immune system has hijacked bacterial c-di-NMP signaling elements to evolve an effective and highly specific system for pathogen recognition.
Apart from STING, initial binding studies have pointed toward several additional mammalian proteins as putative c-di-GMP receptors. Examples include p21/Ras small GTPases, cytoskeleton remodeling protein coronin 1A, RNA-capping and processing proteins RNMT and cyclophilin H, and others.123,124 Although potentially quite interesting, these candidates will require further structural and functional studies to confirm c-di-GMP binding specificity and physiologically relevant effects and to distinguish specific binding from potentially promiscuous affinity for RNA-based ligands.
Outlook
According to recent World Health Organization reports bacterial infections causing gastroenteric diseases, obstructive pulmonary conditions, and AIDS-derived opportunistic infections comprise some of the leading mortality causes worldwide. Given the long battle that medicine has fought against virulent microbes, such a grim outlook inevitably raises concerns about the effectiveness of conventional prevention and treatment, and underscores the pressing need for alternative therapeutic approaches.
Traditional antibiotics are designed to kill susceptible bacteria and thus select for naturally occurring drug-resistant forms. This selective enhancement of resistant bacteria is especially rampant in multiple or prolonged exposures, often associated with hospital settings or chronic diseases. Mechanistic studies of nonessential virulence determinants, such as enzymes and protein sensors involved in c-di-GMP signal transduction and biofilm formation, could thus provide targets for the design of novel anti-infectives that are aimed at jamming bacterial communication and dispersal of the pathogenic crowds without associated risks of evolutionary pressure and resistance development. We highlighted here recent advances in the structure–function studies on such important c-di-GMP signal transduction proteins highlighting the functional complexity and idiosyncracies of individual targets. It is important to note, that the general lack of redundancy between c-di-GMP signaling mechanisms across or even within species holds the additional promise for selective targeting of parasitic and virulent microbes while preserving beneficial symbiotic bacteria. It remains to be seen whether the rapidly developing field of structural biology dedicated to c-di-GMP signaling research will be able to provide the necessary molecular blueprints and mechanistic insight for the design of such therapeutics.
References
- 1.Ross P, Mayer R, Benziman M. Cellulose biosynthesis and function in bacteria. Microbiol Rev. 1991;55:35–58. doi: 10.1128/mr.55.1.35-58.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 3.O'Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 4.Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 5.Furukawa S, Kuchma SL, O'Toole GA. Keeping their options open: acute versus persistent infections. J Bacteriol. 2006;188:1211–1217. doi: 10.1128/JB.188.4.1211-1217.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 7.Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 9.Sondermann H, Shikuma NJ, Yildiz FH. You've come a long way: c-di-GMP signaling. Curr Opin Microbiol. 2012;15:140–146. doi: 10.1016/j.mib.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albright LM, Huala E, Ausubel FM. Prokaryotic signal transduction mediated by sensor and regulator protein pairs. Annu Rev Genet. 1989;23:311–336. doi: 10.1146/annurev.ge.23.120189.001523. [DOI] [PubMed] [Google Scholar]
- 11.Pei J, Grishin NV. GGDEF domain is homologous to adenylyl cyclase. Proteins. 2001;42:210–216. doi: 10.1002/1097-0134(20010201)42:2<210::aid-prot80>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 12.Hecht GB, Newton A. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J Bacteriol. 1995;177:6223–6229. doi: 10.1128/jb.177.21.6223-6229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galperin MY, Natale DA, Aravind L, Koonin EV. A specialized version of the HD hydrolase domain implicated in signal transduction. J Mol Microbiol Biotechnol. 1999;1:303–305. [PMC free article] [PubMed] [Google Scholar]
- 14.Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, Cohen A, Sapir S, Ohana P, Benziman M. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol. 1998;180:4416–4425. doi: 10.1128/jb.180.17.4416-4425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simm R, Morr M, Kader A, Nimtz M, Romling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 16.Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol. 2009;7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 17.Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He YW, Zhang LH, Heeb S, Camara M, Williams P, Dow JM. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci USA. 2006;103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.D'Argenio DA, Miller SI. Cyclic di-GMP as a bacterial second messenger. Microbiology. 2004;150:2497–2502. doi: 10.1099/mic.0.27099-0. [DOI] [PubMed] [Google Scholar]
- 19.Romling U, Gomelsky M, Galperin MY. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 20.Wolfe AJ, Visick KL. Get the message out: cyclic-Di-GMP regulates multiple levels of flagellum-based motility. J Bacteriol. 2008;190:463–475. doi: 10.1128/JB.01418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tischler AD, Camilli A. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect Immun. 2005;73:5873–5882. doi: 10.1128/IAI.73.9.5873-5882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newell PD, Monds RD, O'Toole GA. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0–1. Proc Natl Acad Sci USA. 2009;106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YK, McCarter LL. ScrG, a GGDEF-EAL protein, participates in regulating swarming and sticking in Vibrio parahaemolyticus. J Bacteriol. 2007;189:4094–4107. doi: 10.1128/JB.01510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′–5′)-cyclic-GMP in virulence. Proc Natl Acad Sci USA. 2006;103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinhouse H, Sapir S, Amikam D, Shilo Y, Volman G, Ohana P, Benziman M. c-di-GMP- binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett. 1997;416:207–211. doi: 10.1016/s0014-5793(97)01202-7. [DOI] [PubMed] [Google Scholar]
- 26.Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 27.Ryjenkov DA, Simm R, Romling U, Gomelsky M. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem. 2006;281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 28.Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007;26:5153–5166. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pratt JT, Tamayo R, Tischler AD, Camilli A. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J Biol Chem. 2007;282:12860–12870. doi: 10.1074/jbc.M611593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramelot TA, Yee A, Cort JR, Semesi A, Arrowsmith CH, Kennedy MA. NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein. Proteins. 2007;66:266–271. doi: 10.1002/prot.21199. [DOI] [PubMed] [Google Scholar]
- 31.Ko J, Ryu KS, Kim H, Shin JS, Lee JO, Cheong C, Choi BS. Structure of PP4397 reveals the molecular basis for different c-di-GMP binding modes by Pilz domain proteins. J Mol Biol. 2010;398:97–110. doi: 10.1016/j.jmb.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Habazettl J, Allan MG, Jenal U, Grzesiek S. Solution structure of the PilZ domain protein PA4608 complex with cyclic di-GMP identifies charge clustering as molecular readout. J Biol Chem. 2011;286:14304–14314. doi: 10.1074/jbc.M110.209007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beyhan S, Odell LS, Yildiz FH. Identification and characterization of cyclic diguanylate signaling systems controlling rugosity in Vibrio cholerae. J Bacteriol. 2008;190:7392–7405. doi: 10.1128/JB.00564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navarro MV, De N, Bae N, Wang Q, Sondermann H. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure. 2009;17:1104–1116. doi: 10.1016/j.str.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol. 2007;65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De N, Pirruccello M, Krasteva PV, Bae N, Raghavan RV, Sondermann H. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol. 2008;6:e67. doi: 10.1371/journal.pbio.0060067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chin KH, Lee YC, Tu ZL, Chen CH, Tseng YH, Yang JM, Ryan RP, McCarthy Y, Dow JM, Wang AH, Chou SH. The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J Mol Biol. 2010;396:646–662. doi: 10.1016/j.jmb.2009.11.076. [DOI] [PubMed] [Google Scholar]
- 41.Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci USA. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z, Gaffney BL, Jones RA. c-di-GMP displays a monovalent metal ion-dependent polymorphism. J Am Chem Soc. 2004;126:16700–16701. doi: 10.1021/ja0449832. [DOI] [PubMed] [Google Scholar]
- 43.Leduc JL, Roberts GP. Cyclic di-GMP allosterically inhibits the CRP-like protein (Clp) of Xanthomonas axonopodis pv. citri. J Bacteriol. 2009;191:7121–7122. doi: 10.1128/JB.00845-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wassmann P, Chan C, Paul R, Beck A, Heerklotz H, Jenal U, Schirmer T. Structure of BeF3- -modified response regulator PleD: implications for diguanylate cyclase activation, catalysis, and feedback inhibition. Structure. 2007;15:915–927. doi: 10.1016/j.str.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 45.De N, Navarro MV, Raghavan RV, Sondermann H. Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR. J Mol Biol. 2009;393:619–633. doi: 10.1016/j.jmb.2009.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minasov G, Padavattan S, Shuvalova L, Brunzelle JS, Miller DJ, Basle A, Massa C, Collart FR, Schirmer T, Anderson WF. Crystal structures of YkuI and its complex with second messenger cyclic Di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains. J Biol Chem. 2009;284:13174–13184. doi: 10.1074/jbc.M808221200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature. 2009;459:1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- 48.Lovering AL, Capeness MJ, Lambert C, Hobley L, Sockett RE. The structure of an unconventional HD-GYP protein from Bdellovibrio reveals the roles of conserved residues in this class of cyclic-di-GMP phosphodiesterases. MBio. 2011;2:e00163–11. doi: 10.1128/mBio.00163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Navarro MV, Newell PD, Krasteva PV, Chatterjee D, Madden DR, O'Toole GA, Sondermann H. Structural basis for c-di-GMP-mediated inside-out signaling controlling periplasmic proteolysis. PLoS Biol. 2011;9:e1000588. doi: 10.1371/journal.pbio.1000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi Z, Yang WZ, Lin-Chao S, Chak KF, Yuan HS. Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation. RNA. 2008;14:2361–2371. doi: 10.1261/rna.1244308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paul R, Abel S, Wassmann P, Beck A, Heerklotz H, Jenal U. Activation of the diguanylate cyclase PleD by phosphorylation-mediated dimerization. J Biol Chem. 2007;282:29170–29177. doi: 10.1074/jbc.M704702200. [DOI] [PubMed] [Google Scholar]
- 53.Christen B, Christen M, Paul R, Schmid F, Folcher M, Jenoe P, Meuwly M, Jenal U. Allosteric control of cyclic di-GMP signaling. J Biol Chem. 2006;281:32015–32024. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- 54.Rao F, Qi Y, Chong HS, Kotaka M, Li B, Li J, Lescar J, Tang K, Liang ZX. The functional role of a conserved loop in EAL domain-based cyclic di-GMP-specific phosphodiesterase. J Bacteriol. 2009;191:4722–4731. doi: 10.1128/JB.00327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rao F, Yang Y, Qi Y, Liang ZX. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J Bacteriol. 2008;190:3622–3631. doi: 10.1128/JB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tchigvintsev A, Xu X, Singer A, Chang C, Brown G, Proudfoot M, Cui H, Flick R, Anderson WF, Joachimiak A, Galperin MY, Savchenko A, Yakunin AF. Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases. J Mol Biol. 2010;402:524–538. doi: 10.1016/j.jmb.2010.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagano N, Orengo CA, Thornton JM. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J Mol Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 58.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem. 2005;280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 59.Schmidt AJ, Ryjenkov DA, Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol. 2005;187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khrenova M, Domratcheva T, Grigorenko B, Nemukhin A. Coupling between the BLUF and EAL domains in the blue light-regulated phosphodiesterase BlrP1. J Mol Model. 2011;17:1579–1586. doi: 10.1007/s00894-010-0842-1. [DOI] [PubMed] [Google Scholar]
- 61.Bharati BK, Sharma IM, Kasetty S, Kumar M, Mukherjee R, Chatterji D. A full length bifunctional protein involved in c-di-GMP turnover is required for long term survival under nutrient starvation in Mycobacterium smegmatis. Microbiology. 2012 doi: 10.1099/mic.0.053892-0. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 62.Levet-Paulo M, Lazzaroni JC, Gilbert C, Atlan D, Doublet P, Vianney A. The atypical two-component sensor kinase Lpl0330 from Legionella pneumophila controls the bifunctional diguanylate cyclase-phosphodiesterase Lpl0329 to modulate bis-(3′–5′)-cyclic dimeric GMP synthesis. J Biol Chem. 2011;286:31136–31144. doi: 10.1074/jbc.M111.231340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu N, Xu Y, Hossain S, Huang N, Coursolle D, Gralnick JA, Boon EM. Nitric oxide regulation of cyclic di-GMP synthesis and hydrolysis in Shewanella woodyi. Biochemistry. 2012;51:2087–2099. doi: 10.1021/bi201753f. [DOI] [PubMed] [Google Scholar]
- 64.Slater H, Alvarez-Morales A, Barber CE, Daniels MJ, Dow JM. A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol Microbiol. 2000;38:986–1003. doi: 10.1046/j.1365-2958.2000.02196.x. [DOI] [PubMed] [Google Scholar]
- 65.Ryan RP, Dow JM. Intermolecular interactions between HD-GYP and GGDEF domain proteins mediate virulence-related signal transduction in Xanthomonas campestris. Virulence. 2010;1:404–408. doi: 10.4161/viru.1.5.12704. [DOI] [PubMed] [Google Scholar]
- 66.Ko M, Park C. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J Mol Biol. 2000;303:371–382. doi: 10.1006/jmbi.2000.4147. [DOI] [PubMed] [Google Scholar]
- 67.Ross P, Aloni Y, Weinhouse C, Michaeli D, Weinberger-Ohana P, Meyer R, Benziman M. An unusual guanyl oligonucleotide regulates cellulose synthesis in Acetobacter xylinum. FEBS Lett. 1985;186:191–196. doi: 10.1016/0014-5793(85)80706-7. [DOI] [PubMed] [Google Scholar]
- 68.Shin JS, Ryu KS, Ko J, Lee A, Choi BS. Structural characterization reveals that a PilZ domain protein undergoes substantial conformational change upon binding to cyclic dimeric guanosine monophosphate. Protein Sci. 2011;20:270–277. doi: 10.1002/pro.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Christen M, Kulasekara HD, Christen B, Kulasekara BR, Hoffman LR, Miller SI. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science. 2010;328:1295–1297. doi: 10.1126/science.1188658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ohashi T, Galiacy SD, Briscoe G, Erickson HP. An experimental study of GFP-based FRET, with application to intrinsically unstructured proteins. Protein Sci. 2007;16:1429–1438. doi: 10.1110/ps.072845607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alm RA, Bodero AJ, Free PD, Mattick JS. Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J Bacteriol. 1996;178:46–53. doi: 10.1128/jb.178.1.46-53.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guzzo CR, Salinas RK, Andrade MO, Farah CS. PILZ protein structure and interactions with PILB and the FIMX EAL domain: implications for control of type IV pilus biogenesis. J Mol Biol. 2009;393:848–866. doi: 10.1016/j.jmb.2009.07.065. [DOI] [PubMed] [Google Scholar]
- 73.Li TN, Chin KH, Liu JH, Wang AH, Chou SH. XC1028 from Xanthomonas campestris adopts a PilZ domain-like structure without a c-di-GMP switch. Proteins. 2009;75:282–288. doi: 10.1002/prot.22330. [DOI] [PubMed] [Google Scholar]
- 74.Huang B, Whitchurch CB, Mattick JS. FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J Bacteriol. 2003;185:7068–7076. doi: 10.1128/JB.185.24.7068-7076.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kazmierczak BI, Lebron MB, Murray TS. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol Microbiol. 2006;60:1026–1043. doi: 10.1111/j.1365-2958.2006.05156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao R, Stock AM. Catalytically incompetent by design. Structure. 2009;17:1038–1040. doi: 10.1016/j.str.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qi Y, Chuah ML, Dong X, Xie K, Luo Z, Tang K, Liang ZX. Binding of cyclic diguanylate in the non-catalytic EAL domain of FimX induces a long-range conformational change. J Biol Chem. 2011;286:2910–2917. doi: 10.1074/jbc.M110.196220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hinsa SM, O'Toole GA. Biofilm formation by Pseudomonas fluorescens WCS365: a role for LapD. Microbiology. 2006;152:1375–1383. doi: 10.1099/mic.0.28696-0. [DOI] [PubMed] [Google Scholar]
- 79.Newell PD, Boyd CD, Sondermann H, O'Toole GA. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol. 2011;9:e1000587. doi: 10.1371/journal.pbio.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Newell PD, Yoshioka S, Hvorecny KL, Monds RD, O'Toole GA. Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf0–1. J Bacteriol. 2011;193:4685–4698. doi: 10.1128/JB.05483-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Monds RD, Newell PD, Gross RH, O'Toole GA. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0–1 biofilm formation by controlling secretion of the adhesin LapA. Mol Microbiol. 2007;63:656–679. doi: 10.1111/j.1365-2958.2006.05539.x. [DOI] [PubMed] [Google Scholar]
- 82.Petters T, Zhang X, Nesper J, Treuner-Lange A, Gomez-Santos N, Hoppert M, Jenal U, Sogaard-Andersen L. The orphan histidine protein kinase SgmT is a c-di-GMP receptor and regulates composition of the extracellular matrix together with the orphan DNA binding response regulator DigR in Myxococcus xanthus. Mol Microbiol. 2012;84:147–165. doi: 10.1111/j.1365-2958.2012.08015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rakshe S, Leff M, Spormann AM. Indirect modulation of the intracellular c-Di-GMP level in Shewanella oneidensis MR-1 by MxdA. Appl Environ Microbiol. 2011;77:2196–2198. doi: 10.1128/AEM.01985-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Casper-Lindley C, Yildiz FH. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J Bacteriol. 2004;186:1574–1578. doi: 10.1128/JB.186.5.1574-1578.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tao F, He YW, Wu DH, Swarup S, Zhang LH. The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di-GMP effectors. J Bacteriol. 2010;192:1020–1029. doi: 10.1128/JB.01253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wilksch JJ, Yang J, Clements A, Gabbe JL, Short KR, Cao H, Cavaliere R, James CE, Whitchurch CB, Schembri MA, Chuah ML, Liang ZX, Wijburg OL, Jenney AW, Lithgow T, Strugnell RA. MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 2011;7:e1002204. doi: 10.1371/journal.ppat.1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Beyhan S, Yildiz FH. Smooth to rugose phase variation in Vibrio cholerae can be mediated by a single nucleotide change that targets c-di-GMP signalling pathway. Mol Microbiol. 2007;63:995–1007. doi: 10.1111/j.1365-2958.2006.05568.x. [DOI] [PubMed] [Google Scholar]
- 90.Srivastava D, Harris RC, Waters CM. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J Bacteriol. 2011;193:6331–6341. doi: 10.1128/JB.05167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Birck C, Mourey L, Gouet P, Fabry B, Schumacher J, Rousseau P, Kahn D, Samama JP. Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure. 1999;7:1505–1515. doi: 10.1016/s0969-2126(00)88341-0. [DOI] [PubMed] [Google Scholar]
- 92.Davies KM, Lowe ED, Venien-Bryan C, Johnson LN. The HupR receiver domain crystal structure in its nonphospho and inhibitory phospho states. J Mol Biol. 2009;385:51–64. doi: 10.1016/j.jmb.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 93.Toro-Roman A, Mack TR, Stock AM. Structural analysis and solution studies of the activated regulatory domain of the response regulator ArcA: a symmetric dimer mediated by the alpha4-beta5-alpha5 face. J Mol Biol. 2005;349:11–26. doi: 10.1016/j.jmb.2005.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Benda C, Scheufler C, Tandeau de Marsac N, Gartner W. Crystal structures of two cyanobacterial response regulators in apo- and phosphorylated form reveal a novel dimerization motif of phytochrome-associated response regulators. Biophys J. 2004;87:476–487. doi: 10.1529/biophysj.103.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gao R, Stock AM. Molecular strategies for phosphorylation-mediated regulation of response regulator activity. Curr Opin Microbiol. 2010;13:160–167. doi: 10.1016/j.mib.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Matthews KS. DNA looping. Microbiol Rev. 1992;56:123–136. doi: 10.1128/mr.56.1.123-136.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schleif R. DNA looping. Annu Rev Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- 98.Kader A, Simm R, Gerstel U, Morr M, Romling U. Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2006;60:602–616. doi: 10.1111/j.1365-2958.2006.05123.x. [DOI] [PubMed] [Google Scholar]
- 99.Romling U. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell Mol Life Sci. 2005;62:1234–1246. doi: 10.1007/s00018-005-4557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chirwa NT, Herrington MB. CsgD, a regulator of curli and cellulose synthesis, also regulates serine hydroxymethyltransferase synthesis in Escherichia coli K-12. Microbiology. 2003;149:525–535. doi: 10.1099/mic.0.25841-0. [DOI] [PubMed] [Google Scholar]
- 101.Zakikhany K, Harrington CR, Nimtz M, Hinton JC, Romling U. Unphosphorylated CsgD controls biofilm formation in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2010;77:771–786. doi: 10.1111/j.1365-2958.2010.07247.x. [DOI] [PubMed] [Google Scholar]
- 102.Yildiz FH, Dolganov NA, Schoolnik GK. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J Bacteriol. 2001;183:1716–1726. doi: 10.1128/JB.183.5.1716-1726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dasgupta N, Ramphal R. Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol. 2001;183:6636–6644. doi: 10.1128/JB.183.22.6636-6644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Arora SK, Ritchings BW, Almira EC, Lory S, Ramphal R. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J Bacteriol. 1997;179:5574–5581. doi: 10.1128/jb.179.17.5574-5581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fazli M, O'Connell A, Nilsson M, Niehaus K, Dow JM, Givskov M, Ryan RP, Tolker-Nielsen T. The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol Microbiol. 2011;82:327–341. doi: 10.1111/j.1365-2958.2011.07814.x. [DOI] [PubMed] [Google Scholar]
- 106.He YW, Ng AY, Xu M, Lin K, Wang LH, Dong YH, Zhang LH. Xanthomonas campestris cell-cell communication involves a putative nucleotide receptor protein Clp and a hierarchical signalling network. Mol Microbiol. 2007;64:281–292. doi: 10.1111/j.1365-2958.2007.05670.x. [DOI] [PubMed] [Google Scholar]
- 107.He YW, Boon C, Zhou L, Zhang LH. Co-regulation of Xanthomonas campestris virulence by quorum sensing and a novel two-component regulatory system RavS/RavR. Mol Microbiol. 2009;71:1464–1476. doi: 10.1111/j.1365-2958.2009.06617.x. [DOI] [PubMed] [Google Scholar]
- 108.Passner JM, Steitz TA. The structure of a CAP-DNA complex having two cAMP molecules bound to each monomer. Proc Natl Acad Sci USA. 1997;94:2843–2847. doi: 10.1073/pnas.94.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.de Crecy-Lagard V, Glaser P, Lejeune P, Sismeiro O, Barber CE, Daniels MJ, Danchin A. A Xanthomonas campestris pv. campestris protein similar to catabolite activation factor is involved in regulation of phytopathogenicity. J Bacteriol. 1990;172:5877–5883. doi: 10.1128/jb.172.10.5877-5883.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang LH. A novel C-di-GMP effector linking intracellular virulence regulon to quorum sensing and hypoxia sensing. Virulence. 2010;1:391–394. doi: 10.4161/viru.1.5.12487. [DOI] [PubMed] [Google Scholar]
- 111.Kim J, Adhya S, Garges S. Allosteric changes in the cAMP receptor protein of Escherichia coli: hinge reorientation. Proc Natl Acad Sci USA. 1992;89:9700–9704. doi: 10.1073/pnas.89.20.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smith KD, Strobel SA. Interactions of the c-di-GMP riboswitch with its second messenger ligand. Biochem Soc Trans. 2011;39:647–651. doi: 10.1042/BST0390647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA. Cyclic di-GMP activation of polynucleotide phosphorylase signal-dependent RNA processing. J Mol Biol. 2011;407:633–639. doi: 10.1016/j.jmb.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 114.Karaolis DK, Cheng K, Lipsky M, Elnabawi A, Catalano J, Hyodo M, Hayakawa Y, Raufman JP. 3′,5′-Cyclic diguanylic acid (c-di-GMP) inhibits basal and growth factor-stimulated human colon cancer cell proliferation. Biochem Biophys Res Commun. 2005;329:40–45. doi: 10.1016/j.bbrc.2005.01.093. [DOI] [PubMed] [Google Scholar]
- 115.Karaolis DK, Newstead MW, Zeng X, Hyodo M, Hayakawa Y, Bhan U, Liang H, Standiford TJ. Cyclic di-GMP stimulates protective innate immunity in bacterial pneumonia. Infect Immun. 2007;75:4942–4950. doi: 10.1128/IAI.01762-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Karaolis DK, Means TK, Yang D, Takahashi M, Yoshimura T, Muraille E, Philpott D, Schroeder JT, Hyodo M, Hayakawa Y, Talbot BG, Brouillette E, Malouin F. Bacterial c-di-GMP is an immunostimulatory molecule. J Immunol. 2007;178:2171–2181. doi: 10.4049/jimmunol.178.4.2171. [DOI] [PubMed] [Google Scholar]
- 117.Madhun AS, Haaheim LR, Nostbakken JK, Ebensen T, Chichester J, Yusibov V, Guzman CA, Cox RJ. Intranasal c-di-GMP-adjuvanted plant-derived H5 influenza vaccine induces multifunctional Th1 CD4+ cells and strong mucosal and systemic antibody responses in mice. Vaccine. 2011;29:4973–4982. doi: 10.1016/j.vaccine.2011.04.094. [DOI] [PubMed] [Google Scholar]
- 118.McWhirter SM, Barbalat R, Monroe KM, Fontana MF, Hyodo M, Joncker NT, Ishii KJ, Akira S, Colonna M, Chen ZJ, Fitzgerald KA, Hayakawa Y, Vance RE. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J Exp Med. 2009;206:1899–1911. doi: 10.1084/jem.20082874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Crimmins GT, Herskovits AA, Rehder K, Sivick KE, Lauer P, Dubensky TW, Jr, Portnoy DA. Listeria monocytogenes multidrug resistance transporters activate a cytosolic surveillance pathway of innate immunity. Proc Natl Acad Sci USA. 2008;105:10191–10196. doi: 10.1073/pnas.0804170105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Amikam D, Steinberger O, Shkolnik T, Ben-Ishai Z. The novel cyclic dinucleotide 3′–5′ cyclic diguanylic acid binds to p21ras and enhances DNA synthesis but not cell replication in the Molt 4 cell line. Biochem J. 1995;311:921–927. doi: 10.1042/bj3110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Abdul-Sater AA, Grajkowski A, Erdjument-Bromage H, Plumlee C, Levi A, Schreiber MT, Lee C, Shuman H, Beaucage SL, Schindler C. The overlapping host responses to bacterial cyclic dinucleotides. Microbes Infect. 2012;14:188–197. doi: 10.1016/j.micinf.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]