Abstract
The family of serpins is known to fold into a metastable state that is required for the proteinase inhibition mechanism. One of the consequences of this conformational flexibility is the tendency of some mutated serpins to form polymers, which occur through the insertion of the reactive center loop of one serpin molecule into the A-sheet of another. This “A-sheet polymerization” has remained an attractive explanation for the molecular mechanism of serpinopathies. Polymerization of serpins can also take place in vitro under certain conditions (e.g., pH or temperature). Surprisingly, on sodium dodecyl sulfate/polyacrylamide gel electrophoresis, bovSERPINA3-3 extracted from skeletal muscle or expressed in Escherichia coli was mainly observed as a homodimer. Here, in this report, by site-directed mutagenesis of recombinant bovSERPINA3-3, with substitution D371A, we demonstrate the importance of D371 for the intermolecular linkage observed in denaturing and reducing conditions. This residue influences the electrophoretic and conformational properties of bovSERPINA3-3. By structural modeling of mature bovSERPINA3-3, we propose a new “non-A-sheet swap” model of serpin homodimer in which D371 is involved at the molecular interface.
Keywords: serpins, polymerization, mutagenesis, mammalian
Introduction
Serine peptidase inhibitors (serpins) represent a constantly expanding family of structurally related proteins. Most serpins are inhibitory and use a unique suicide mechanism for inactivation of their target proteases. The formation of the non covalent Michaelis complex, the subsequent cleavage of the peptide bond followed by rapid insertion of the serine protease N-terminal part of the loop in the main body of the serpin are well documented.1
Since the first structure of cleaved human α1-proteinase inhibitor,2 X-ray and other structural studies of different serpins and serpins complexes have revealed remarkable similarities of secondary and tertiary structures for all serpins. In the metastable native conformation, serpins contain three antiparallel β-sheets (A, B, and C) and nine α-helices (1–9). β-sheet A is composed of five strands, while β-Sheets B and C are shorter and contained six and four strands, respectively. The reactive center loop (RCL) is exposed outside of the tertiary core of the serpin and contains the residues that, in inhibitory serpins, interact directly with the cognate proteinase.3 More than 85 serpin structures are now resolved. They indicate that inhibitory serpins undergo a dramatic conformational change when they interact with target proteinases. Notably, protease binding leads to the cleavage of the exposed amino-acid sequence RCL. This conformational change allows the serpin to incorporate the RCL into its β-sheet A as strand 4A resulting in a hyperstable six-stranded conformation. There is another type of structure found for uncomplexed serpin, called the latent form. This conformational state was first described for plasminogen activator inhibitor-1 (PAI-1).4 It presents a stronger thermodynamic stability than the normal active form. In this structure, as for the cleaved serpins, β-sheet A has undergone an expansion by insertion of RCL. The major difference is that the uninserted residues of the C-terminal part of the reactive bound, which composed strand 1 of β-sheet C, were pulled away to provide “a return” to the top of the molecule.
An unwanted consequence of this thermodynamically favored conformational change is the tendency of serpins to form polymers. Serpins are able to form large stable multimers by ordered β-sheet linkages leading to intracellular aggregation and diseases known as serpinopathies. These conformational disorders are responsible for thrombosis by mutations in antithrombin,5 still respiratory disease emphysema, liver cirrhosis caused by mutation in α1-antitrypsin (α1AT) or α1-antichymotrypsin (α1ACT)6 or dementia caused by polymerization and tissue deposition of the mutated neuron specific protein, neuroserpin.7 The serpin family has therefore become a model system for understanding the β-sheet expansion disorders collectively known as the conformational diseases, including Alzheimer's, Huntington's, Parkinson's, and prion encephalopathies.
Serpin polymerization was first described for the Z variant of α1AT. In this mutant, a single-nucleotide mutation in the gene is responsible for the substitution E342K. The mutation results in the accumulation of highly stable, inactive polymers of the serpin in the endoplasmic reticulum of liver cells. Patients with this mutation develop emphysema in early adulthood and some develop liver cirrhosis in childhood.8 Besides, two mutants of α1ACT (P229A and L55P) could be responsible for liver retention with hepatocyte inclusions and additional plasma deficiency.9 In a recent study, Yamasaki et al.10 reported the crystallographic structure of stable dimers to explain the molecular basis of serpin polymerization. A domain swapping, involving two strands of the A β-sheet and including the RCL, results in the formation of an antithrombin dimer. To explain the formation of polymers able of incorporating more than two monomers, the authors proposed that the swapping occurred across several molecules.
To complete this study, Whisstock and Bottomley11 proposed a short-lived intermediate between the unfolded protein and the native serpin. This folding intermediate corresponds to a structure in which the region intended to form the 5A β-strand of the A β-sheet, and the externalized RCL are still flexible and unstructured. When folding normally, the last structure to assemble is the 5A β-strand of the A β-sheet. When folding is abnormal, two serpin monomers can swap domains, one monomer donates the 4A β-strand and its neighboring 5A β-strand to replace that of the second monomer. Some mutations induce this process and the folding intermediate begins to polymerize. For example, in the mutant Z, the substitution E342K in the 5A β-strand prevents the interaction of this strand with the 6A β-strand and the incorporation of this segment into the β-sheet A does not take place.8,11 Moreover, in a recent study, Yamasaki et al.12 produced and crystallized a trimeric form of α1AT recognized by an antibody specific for the pathological polymers. This structure reveals another polymeric state mediated by domain swapping the carboxy-terminal 34 residues.
Polymers, observed in cases of some serpinopathies, can also take place in vitro for non mutated serpins. So, wildtype α1ACT forms polymers by interaction between the RCL of one molecule and the β-sheet A of a second molecule at a rate that depends on protein concentration and the temperature of the reaction.13 When analyzed by sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE), some serpins dimerize in the presence of SDS. That is the case for TI, an inhibitor of trypsin isolated from bovine plasma; the N-terminal sequence of this protein TI14 matches with bovSERPINA3. It is also the case for muscle bovSERPINA3-1, a serpin isolated from bovine muscle and which inhibits elastase.15 This serpin is encoded by a gene very similar to the unique human SERPINA3 gene of the α1ACT. Dimer formation was shown to be catalyzed by heating and denaturing agents, such as acidic pH.16–18 In denaturing conditions, the samples are heated in the presence of both SDS and β-mercaptoethanol, a set of conditions necessary and sufficient to induce the formation of the dimers, heating very likely being the primary cause of the process. Unfortunately, the question of why and how do the serpins dimerize in such conditions remains without answer. Does this dimerization imply conformational changes as those proposed by Yamasaki et al.10 and Whisstock and Bottomley11 or mechanisms in which the RCL forms additional β-strands of the β-sheet C and A19–21? Can we suppose other mechanism involved in the process of dimerization?
The bovine serpin A3-3 (bovSERPINA3-3, UniProtKB/Swiss-Prot: Q3ZEJ6) was isolated from bovine muscle as previously described by Brémaud et al.22 In our study, we demonstrate the role of residue D371 in the dimeric state of the bovine serpin A3-3 in denaturing conditions. This residue D371 corresponds to D384 of mature human serpinA3 (SERPINA3, UniProtKB/Swiss-Prot: P01011) and to N378 of mature human serpinA1 (SERPINA1, UniProtKB/Swiss-Prot: P01009). To test the predicted properties of this residue, recombinant NH2-His-tagged bovSERPINA3-3 was expressed to examine its structural behavior in denaturing and native conditions. The role of this residue was assessed by site-directed mutagenesis experiments designed on the basis of a modeling of the dimeric form of bovSERPINA3-3. The results indicated that D371 influences the electrophoretic mobility on denaturing conditions and favors the dimer formation of bovSERPINA3-3.
The polymerization of wild-type α1ACT occurs under physiological conditions and may be important in the deposition of α1ACT in plaques in patients with Alzheimer's disease.12 The bovSERPINA3 could become another model system to determine the structural basis of inter molecular linkages and to develop strategies with human serpins to prevent polymerization-related disorders.
Results
Production and characterization of bovSERPINA3-3s
Wild-type bovSERPINA3-3 and mutant forms of bovSERPINA3-3 (without signal sequences) were expressed in Escherichia coli as NH2-His-tagged fusion proteins. The recombinant proteins were purified by affinity chromatography on Ni-NTA Fast Start column with a yield ∼ 0.1 mg L−1 of induced culture. For each construction (wild-type and mutant forms), the expressed proteins were found in the cytoplasmic soluble fraction.
To assess the functionality of recombinant bovSERPINA3-3s, the inhibitory activity of the purified inhibitors against the serine proteinase trypsin was determined (Table I and Ref.23). According to the experimentally determined association rate constants (kass), wild-type bovSERPINA3-3 and bovSERPINA3-3D371A, expressed in E. coli, show values closed to bovSERPINA3-3 extracted from bovine skeletal muscle.22
Table I.
Association Rate Constants for Inhibition of Serine Proteinase Trypsin
| kass (M−1 s−1) | |
|---|---|
| Muscular skeletal bovSERPINA3-3 | 0.67 × 106 |
| Bacterial recombinant bovSERPINA3-3 | |
| BovSERPINA3-3 | 1.87 × 106 |
| BovSERPINA3-3D371A | 0.78 × 106 |
| BovSERPINA3-3D371E | ND |
| BovSERPINA3-3D371N | ND |
ND, nondetermined.
Electrophoretic mobility of recombinant wild-type bovSERPINA3-3
Migration on reducing SDS/PAGE indicated purified recombinant wild-type bovSERPINA3-3 as a mainly 100 kDa band and as a minor 47 kDa band revealed by Coomassie blue staining [Fig. 1(A), Lane 1] and by Western blot using two antibodies as described in the Materials and methods section [Fig. 1(B), Lanes 1 and 2]. The bovine skeletal muscular bovSERPINA3-3 purified as described by Brémaud et al.,22 was observed as 75 and 150 kDa bands on SDS/PAGE [Fig. 1(A), Lane 2]. Five consensus potential N-glycosylation sites are found in the sequence of bovSERPINA3-3. The glycosylation state of the bovine skeletal muscular bovSERPINA3-3 probably explains the difference in the molecular mass revealed for the bovine extracted protein, when compared with the recombinant serpin expressed in E. coli (75 kDa vs. 47 kDa and 150 kDa vs. 100 kDa).
Figure 1.
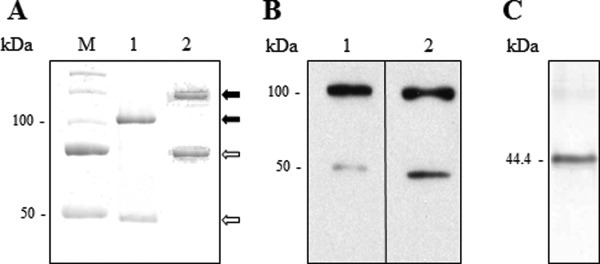
Electrophoretic analyses of bovSERPINA3-3. A: Comparative migration of recombinant bovSERPINA3-3 (Lane 1) and skeletal muscular bovSERPINA3-3 (Lane 2) on discontinuous SDS/PAGE 12% (1 μg of purified protein for each lane). Proteins were revealed by Coomassie blue staining. The two proteins were expressed under monomeric form ( ) and dimeric form (
) and dimeric form ( ). M, molecular weight markers. B: Western blot analyses of recombinant bovSERPINA3-3. The immunodetection of recombinant bovSERPINA3-3 was realized with anti-His6-Peroxidase (Lane 1) or rabbit anti-bovine SERPINA3s serum (Lane 2). C: Native PAGE analysis of recombinant bovSERPINA3-3 on discontinuous PAGE 10% (1 μg of purified protein). Proteins were revealed by Coomassie blue staining. The protein was expressed under monomeric form. The molecular weight (44.4 kDa) was calculated as described in the Materials and methods section.
). M, molecular weight markers. B: Western blot analyses of recombinant bovSERPINA3-3. The immunodetection of recombinant bovSERPINA3-3 was realized with anti-His6-Peroxidase (Lane 1) or rabbit anti-bovine SERPINA3s serum (Lane 2). C: Native PAGE analysis of recombinant bovSERPINA3-3 on discontinuous PAGE 10% (1 μg of purified protein). Proteins were revealed by Coomassie blue staining. The protein was expressed under monomeric form. The molecular weight (44.4 kDa) was calculated as described in the Materials and methods section.
To assign the 47 kDa to bovSERPINA3-3, the 100 kDa band to a homodimer and so, to verify the noncontamination of purified recombinant protein by bacterial proteins of the production system, Mascot query was performed with “all species” taxonomy and then filtered for only bovine and bacterial proteins. Analysis by nano-LC MS/MS of 47 and 100 kDa bands leads to the identification of bovine SERPINA3-3, with a coverage of 39% and 62%, respectively. No bacterial protein was found in the 100 kDa band, confirming that this band is a homodimer of the bovSERPINA3-3.
Moreover, migration on native PAGE revealed the recombinant wild-type bovSERPINA3-3 as a single form [Fig. 1(C)]. To estimate the molecular mass of this single band, we used the method described by Ferguson24 with a set of referent proteins provided by Sigma (Catalog number MWND500). So, the calculated mass of this form is 44.4 kDa and corresponds to a monomeric bovSERPINA3-3.
Electrophoretic profile of D371 mutant forms of bovSERPINA3-3
In another study,23 we were interested in the crossclass inhibition of bovSERPINA3-3. Possible roles for different residues, especially D371, in the cysteine proteinase inhibitory are examined (data not shown). Thus, three different mutant recombinant bovSERPINA3-3s were expressed.
In bovSERPINA3-3D371N, the asparagine brings an uncharged polar amide group. In bovSERPINA3-3D371E, aspartate was converted to glutamate; the length of the side chain was increased but the negatively electric charge due to the acidic carboxylic group was unchanged. In the third mutant bovSERPINA3-3D371A, aspartate was substituted for alanine, a residue with a small and hydrophobic side chain. Surprisingly, in SDS/PAGE, electrophoretic profiles of these mutant proteins are variable as indicated in Figure 2.
Figure 2.
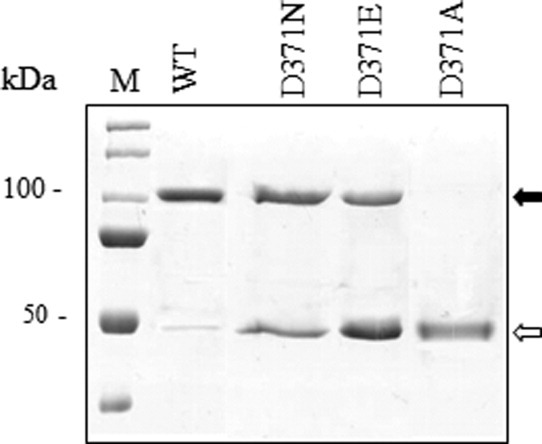
Effect of D371 on the dimerization of bovSERPINA3-3. Purified recombinant wild-type and mutant bovSERPINA3-3s were prepared as described in the Materials and methods section, analyzed by discontinuous SDS/PAGE 12% and revealed by Coomassie blue staining (1 μg of protein for each lane). WT, purified wild-type bovSERPINA3-3; D371N, bovSERPINA3-3D371N; D371E, bovSERPINA3-3D371E; D371A, bovSERPINA3-3D371A. The monomeric ( ) and dimeric (
) and dimeric ( ) forms are indicated by arrows, respectively. M, molecular weight markers.
) forms are indicated by arrows, respectively. M, molecular weight markers.
On reducing SDS/PAGE, it was noted that bovSERPINA3-3D371N was detected as a doublet consisting of a major band of 100 kDa and a lesser band of 47 kDa corresponding to monomeric form. Moreover, nearly equivalent levels of monomeric and dimeric forms were detected in bovSERPINA3-3D371E. Finally, mutagenesis of D to A resulted in the detection of only the monomeric form of bovSERPINA3-3D371A. These data suggest that the length of the side chain and the negatively electric charge of the residue at the position 371 modify the electrophoretic profile and oligomeric state of bovSERPINA3-3. These results highlight the importance of aspartate 371 in the presence of an intermolecular bound between two molecules of bovSERPINA3-3 that could generate a homodimer.
Dimerization of a closely related protein bovSERPINA3-7?
Serpins are able to polymerize such as α1AT25 or PAI-2.26 However, it seems that some of them do not possess this property. In a previous study,27 we have characterized a bovine multigenic SERPINA3 family composed of eight genes and one pseudo gene with an unexpected high degree of conservation. BovSERPINA3-3 is one of the proteins encoded by these genes. Among the seven others members, we also study bovSERPINA3-7 (UniProtKB/Swiss-Prot: A2I7N3).
Wild-type bovSERPINA3-7 (without signal sequence) was expressed in E. coli and purified according to the same method as described above. These two closely related proteins show a different electrophoretic mobility and profile. BovSERPINA3-7 presents a unique monomeric (43.1 kDa) form after migration under native [Fig. 3(C)] but also denaturing [Fig. 3(A), Lane 2] conditions. The absence of dimer was confirmed by Western Blot analysis [Fig. 3(B)]. Moreover, site-directed mutagenesis of bovSERPINA3-7 demonstrates that the substitution E376D does not allow anymore dimer (data not shown).
Figure 3.
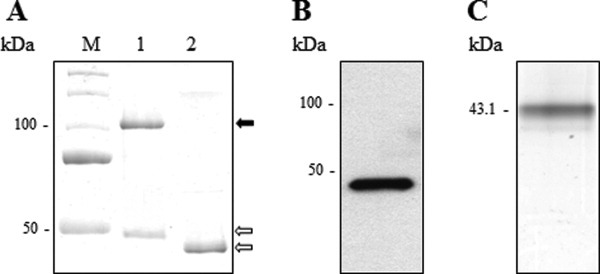
Electrophoretic analyses of recombinant bovSERPINA3-7. A: Comparative migration of recombinant bovSERPINA3-3 (Lane 1) and recombinant bovSERPINA3-7 (Lane 2) on discontinuous SDS/PAGE 12% (1 μg of purified protein for each lane). Proteins were revealed by Coomassie blue staining. The two proteins were expressed under monomeric form ( ). SDS induced dimer formation for bovSERPINA3-3 (
). SDS induced dimer formation for bovSERPINA3-3 ( ). M, molecular weight markers. B: Immunodetection of recombinant bovSERPINA3-7 with anti-His6-Peroxidase. Immunoblot revealed a unique monomeric form. C: Native PAGE analysis of recombinant bovSERPINA3-7 on discontinuous PAGE 10% (1 μg of purified protein). Proteins were revealed by Coomassie blue staining. The protein was expressed under monomeric form. The molecular weight (43.1 kDa) was calculated as described in the Materials and methods section.
). M, molecular weight markers. B: Immunodetection of recombinant bovSERPINA3-7 with anti-His6-Peroxidase. Immunoblot revealed a unique monomeric form. C: Native PAGE analysis of recombinant bovSERPINA3-7 on discontinuous PAGE 10% (1 μg of purified protein). Proteins were revealed by Coomassie blue staining. The protein was expressed under monomeric form. The molecular weight (43.1 kDa) was calculated as described in the Materials and methods section.
BovSERPINA3-3 and bovSERPINA3-7 have different behavior in denaturing conditions. Despite the high level of sequence identity (75%), they present major differences within the RCL (only 35% of identity and two additional amino acids in the RCL of bovSERPINA3-7). We can suppose that these differences prevent the exposure of E376 at the interface of the protein and inhibit its dimerization. The conservation of the negative charge (D371 and E376) is necessary but not sufficient for the dimerization of serpin in denaturing conditions.
Structural modeling of dimeric bovSERPINA3-3
If D371 could modify the electrophoretic mobility of wild-type bovSERPINA3-3, in contrast, the equivalent residue E376 has no effect on the mobility of a closely related protein bovSERPINA3-7. The analysis of the homodimeric molecular model computed for the bovSERPINA3-3 indicates that the D371 is involved at the interface of the dimer [Fig. 4(A)]. On the contrary, in the model of bovSERPINA3-7, the equivalent E376 is not involved and points toward the hydrophobic core of the molecule [Fig. 4(B)]. Moreover, in comparison with the recently solved structure of serpins in domain-swapped dimeric state,11 the N378 residue in mature human serpinA1 corresponding to D371 is not involved in the dimer interface and points to the solvent in the middle of each monomer. Thus, our computed model better explains experimental results observed in denaturing conditions.
Figure 4.
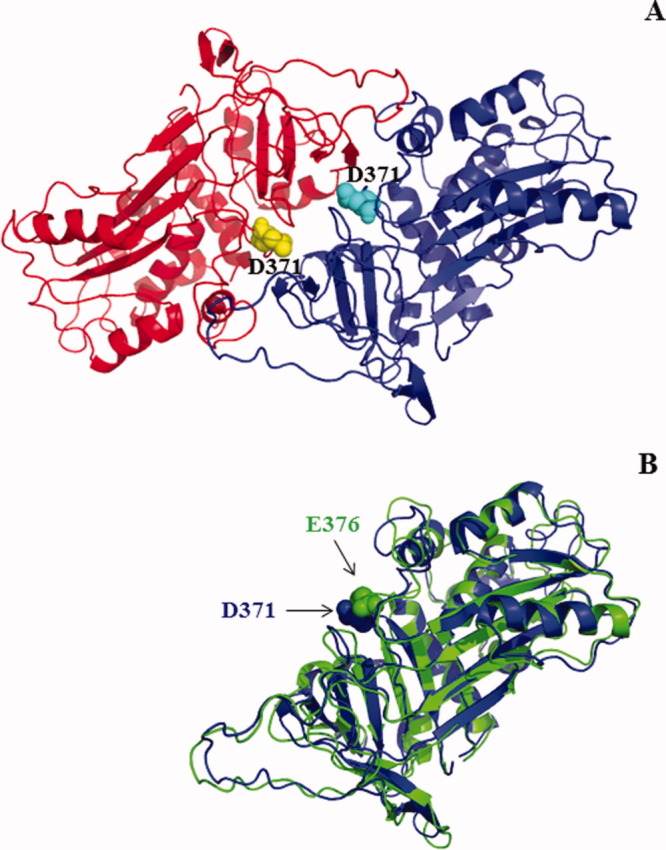
Modeling of bovSERPINA3-3 and bovSERPINA3-7. A: Modeling of dimeric bovSERPINA3-3. The two chains of the dimer are rendered in cartoon mode with in red chain A and blue chain B. The D371 atoms are rendered in sphere and colored in yellow for chain A and in cyan for chain B (Figure drawn with PyMOL 0.99rc6). The aspartate residues of each monomer are involved in the dimeric interface in this model. B: Superposition of the two chains bovSERPINA3-3 (blue) and bovSERPINA3-7 (green). The D371 radical in bovSERPINA3-3 points toward the external face of the molecule. On contrary, the E376 radical in bovSERPINA3-7 points toward hydrophobic core of the molecule.
Discussion
In denaturing conditions, the two SERPINA3-3 preparations, extracted from bovine skeletal muscle22 and produced and purified from E. coli were observed as a homodimer. The difference of molecular mass, 150 versus 100 kDa, respectively, is probably due to post-translational modifications. Consensus N-glycosylation sites (N-X-S/T) predicted that bovSERPINA3-3 may be a glycoprotein with five potential N-linked oligosaccharides. The expression of bovSERPINA3-3 in Saccharomyces cerevisiae as ∼60 kDa protein (Fig. 5) confirmed that this serpin is a glycoprotein. Partial digestion by PNGase F, which removes all N-glycans, was performed on the protein produced and purified from S. cerevisiae. Six states of glycosylation were detected on SDS/PAGE (Fig. 5). They correspond to different molecular masses, 60 kDa for fully glycosylated protein and 57, 54.6, 52.3, 46.6, and 44.1 kDa (unglycosylated state). Also, we can conclude that the five N-glycosylation sites of bovSERPINA3-3 are effectively occupied and can account for the molecular mass of the extracted protein and that predicted from the amino acid sequence.
Figure 5.
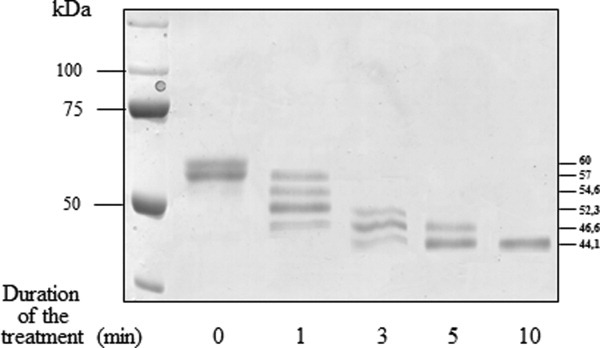
Deglycosylation of S. cerevisiae recombinant bovSERPINA3-3. Kinetic of PNGase F digestion (2.5 Units, 37°C) was performed on bovSERPINA3-3 (6.5 μg) produced and purified from S. cerevisiae. Six states of glycosylation were detected on 12% SDS/PAGE, 44.1 for unglycosylated state until 60 kDa for fully glycosylated protein. Proteins were revealed by Coomassie blue staining.
On native PAGE (10%), recombinant wild-type bovSERPINA3-3 and mutant D371A migrate under a monomeric form exclusively. By heating the same samples in denaturing conditions and, in presence of both SDS and β-mercaptoethanol, we induce the polymerization of the serpins. In our tested conditions, on native PAGE, a typically ladder pattern is visualized for the two proteins (data not shown). This result demonstrated that D371 was not required for the polymerization process.
On denaturing conditions (SDS/PAGE, 12%), recombinant wild-type bovSERPINA3-3 was mainly observed as a dimeric protein. Even when we used a buffer without β-mercapthoethanol and without heating of samples, bovSERPINA3-3 was observed as a dimer. These results would indicate that SDS is responsible for the dimerization of bovSERPINA3-3 and suggest that SDS promotes a conformational change to generate a thermodynamically stable homodimer that is observed as a 100 kDa band on denaturing PAGE.
A mutant form of bovSERPINA3-3 with the substitution D371A was generated by site-directed mutagenesis, expressed as NH2-His-Tagged protein in E. coli and purified by affinity purification on Ni2+ column. The substitution D371A resulted in loss of dimerization on denaturing PAGE. BovSERPINA3-7, one member of bovine multigenic SERPINA3 family, which shares 75% of identity of sequence with bovSERPINA3-3, presents a unique monomeric form after migration under native but also denaturing conditions. The substitution E376D in bovSERPINA3-7, equivalent to the position 371 in bovSERPINA3-3, is not sufficient to induce the dimerization of the protein in denaturing conditions, in presence of SDS. These results indicate that D371 is not the only amino acid of bovSERPINA3-3 involved in the formation of dimer. The analysis of the homodimeric molecular model computed for the bovSERPINA3-3 indicates that D371 is involved with other residues R100, N240, K370, and T372 at the interface of the dimer. The substitution D371A prevents the formation and/or stabilization of this interface. On the contrary, in the model of bovSERPINA3-7 the amino acid at the same position, E376, is not involved and points toward the hydrophobic core of the molecule.
Interestingly, the glycosylated recombinant protein produced in S. cerevisiae remains under a monomeric form after migration in presence of SDS (data not shown). We can hypothesize that N-glycans present on the yeast protein prevent the dimerization of bovSERPINA3-3 unlike the bovSERPINA3-3 purified from skeletal muscles. In fact, the recombinant serpin produced in the yeast (60 kDa) and the extracted bovine serpin (75 kDa) share the same amino acid sequence and differ only by their glycans. This is consistent with glycosylation ability of the yeast, which is recognized as lower than in mammalian cells.
Different studies showed that serpins are a superfamily of proteins susceptible to polymerize. In vivo, this polymerization leads to multiple pathologies including both emphysema and severe liver disease.28,29 The molecular basis of this mechanism remains unknown. However, to understand these misfoldings, crystallographic structures of initial30 and aggregated forms of α1AT31 and antithrombin10 have been resolved. In dimer structure, the two monomers simply exchange parts of their A-sheet including the long β-strands s4A and s5A.10 More recently, a structure of a domain-swapped trimer of α1AT was resolved including strand 1 from β-sheet C and Strands 4 and 5 from β-sheet B.12 In serpins, the amino acid sequence of RCL, within the C-terminus loop, is responsible, for the specificity of protease inhibition. It was also shown that the length of the serpin RCL is critical for its mechanism of protease inhibition.32 Although the RCL was involved in the polymerization process, the P10–P6 region of RCL constitutes only a small part of the extensive domain swap. Indeed, the P8–P6 Asp mutations do not prevent the in vitro and in vivo polymerization.33
Our in vitro study, performed on SERPINA3-3s extracted from the bovine skeletal muscle or produced as a bacterial recombinant protein, allows to propose another modeling of dimerization without swap of β-strands. In this modeling, the D371 residue, localized in the C-terminus loop, is essential for this mechanism of dimerization in denaturing conditions. As previously analyzed, this amino acid is essential but not sufficient. Therefore, the amino acid sequence and the length of RCL (two additional amino acids in bovSERPINA3-7) play central role in this in vitro dimerization process.
Finally, these studies highlight the intermolecular linkages that occurred in vitro. As serpin polymerizations take place in pathological situations, the work on bovSERPINA3s could constitute a model system to develop strategies with human serpins to prevent serpin polymerization and cellular dysfunctions.
Materials and Methods
Site-directed mutagenesis
Substitutions of A, E, and N for D371 of bovSERPINA3-3 and D for E376 of bovSERPINA3-7 were achieved by polymerization chain reaction (PCR) site directed-mutagenesis to generate bovSERPINA3-3D371A, bovSERPINA3-3D371E, bovSERPINA3-3D371N, and bovSERPINA3-7E376D mutants.
The pCR®2.1-TOPO® vector (Invitrogen, Carlsbad, CA) containing the coding sequence without its signal peptide of the bovine SERPINA3-3 and SERPINA3-7 cDNAs (wild-type bovSERPINA3-3 and wild-type bovSERPINA3-7) were directly used for PCR-based mutagenesis. The mutations were performed with the QuickChange™ Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Reactions were performed with 10 ng of vector, 125 ng of sense, and reverse primers listed in Table II and 2.5 units of the PfuTurbo DNA polymerase. Amplification was performed under PCR conditions of 95°C for 30 s followed by 16 cycles of 95°C for 30 s, 55°C for 1 min, and 68°C for 6 min. Parental DNA was removed by treatment with the DpnI restriction endonuclease.
Table II.
Primers Used in this Study
| Primer name | Stranda | Primer sequence |
|---|---|---|
| BovSERPINA3-3 | ||
| SERPINA3-3f | s | 5′-CATATGCTGCCGGAGAATGTGGTGGTGAAGGACCGTCAC-3′ |
| SERPINA3-3r | a | 5′-GGATCCCTAGGCTTCACTGGGGTTGACTTTC-3′ |
| SERPINA3-3D371Af | s | 5′-GTAGTTCTCAAAGCCACCCAGAGCATC-3′ |
| SERPINA3-3D371Ar | a | 5′-GATGCTCTGGGTGGCTTTGAGAACTAC-3′ |
| SERPINA3-3D371Ef | s | 5′-GTAGTTCTCAAAGAAACCCAGAGCATC-3′ |
| SERPINA3-2D371Er | a | 5′-GATGCTCTGGGTTTGTTTGAGAACTAC-3′ |
| SERPINA3-3D371Nf | s | 5′-GTAGTTCTCAAAAACACCCAGAGCATC-3′ |
| SERPINA3-2D371Nr | a | 5′-GATGCTCTGGGTGTTTTTGAGAACTAC-3′ |
| BovSERPINA3-7 | ||
| SERPINA3-7f | s | 5′-CATATGCTGCCGGAGAATGTGACCCCA-3′ |
| SERPINA3-7r | a | 5′-GGATCCCTAGGCTTCCTTGGGGTTGGT-3′ |
| SERPINA3-7E376Df | s | 5′-CTTTCCATATTCTGCAAAGACACTCAGAGCATCATCTTTTTG-3′ |
| SERPINA3-7E376Dr | a | 5′-CAAAAAGATGATGCTCTGAGTGTCTTTGCAGAATATGGAAAG-3′ |
Strand refers to orientation of the primer in the sense (s) or antisense (a) direction.
The E. coli TOP10 competent cells (Invitrogen) were directly transformed with the different amplified vectors. Mutations were verified by automated DNA sequencing with fluorescent-labeled dideoxynucleotides and the ABI PRISM® 3130 Genetic Analyzer (Applied Biosystems, CA).
Construction of wild-type and mutant forms of bovSERPINA3-3 and bovSERPINA3-7 for expression in E. coli
For each construction, purified plasmid DNA was digested with NdeI and BamHI restriction endonucleases (New England Biolabs, Beverly, MA) and ligatured in the pET19b (Novagen, Madison, WI) digested with the same enzymes. DNA sequencing of the wild-type and mutant forms of the pET19b/SERPINA3-3 and pET19b/SERPINA3-7 constructs were performed.
Expression and purification of wild-type and mutant forms of recombinant bovSERPINA3-3 and bovSERPINA3-7
Wild-type and mutant forms of bovSERPINA3-3 and bovSERPINA3-7 were transformed in the E. coli strain BL21-CodonPlus® (DE3)-RP competent cells (Stratagene, La Jolla, CA), and expression of recombinant NH2-His tagged proteins were induced by adding 1 mM Isopropyl β-d-1-thiogalactopyranoside for 2 h at 37°C. Purification in native conditions of recombinant proteins was performed by affinity chromatography on Ni-NTA Fast Start column (QIAGEN, Les Ulis, France) according to the manufacturer's recommendations. Briefly, after bacteria lysis, liquid phase was packed on the column before washing twice with the lysis buffer adjusted to 20 mM imidazole, pH 8. The elution was performed in the same buffer adjusted at 250 mM imidazole, and the sample was then dialyzed. The concentration of recombinant protein in eluted fraction was determined by the Bradford method according to the protocol provided by Biorad (Biorad, Marne la Coquette, France). Eluted fractions were analyzed by SDS/PAGE with Coomassie blue staining and Western blot.
Gel electrophoresis
SDS/PAGE was performed as described previously34 under reducing conditions on a 12% acrylamide separating gel. Proteins were mixed with 2× reducing loading buffer (2% SDS, 20% glycerol, 100 mM Tris-HCl, pH 6.8, 0.1% bromophenol blue, 5% β-mercaptoethanol). Molecular masses were estimated by using the Precision Plus Protein Standards calibration kit (Biorad). Proteins were revealed with 0.25% Coomassie brillant R-250 solution.
Electrophoresis in nondenaturing conditions was performed as above on acrylamide separating gels but in nonreducing conditions and in the absence of SDS. The determination of the molecular weights of bovSERPINA3-3 and A3-7 were calculated according to Ferguson's method24 using four protein standards (α lactalbumin from bovine milk, carbonic anhydrase from bovine erythrocytes, albumin from chicken egg white, and albumin monomer from bovine serum) provided by Sigma (Catalog number MWND500). Four gels of different polyacrylamide concentrations (8%, 10%, 12%, and 15%) were used. For each protein, the log of migration distance according to acrylamide concentration was reported, and the molecular weight according to relative retardation coefficient was determined.
Western blot analysis
Anti-bovine SERPINA3 antibodies were performed as previously described,35 raised against purified bovSERPINA3-1, and crossreacted with all isoforms of bovine serpins identified in Pélissier et al.27 and named bovSERPINA3s.
Separated proteins were then transferred 40 min at 200 mA onto a PVDF Western Blotting membrane (Roche Diagnostics, Mannheim, Germany). After overnight saturation, the membrane was first incubated with agitation and rabbit anti-bovine SERPINA3 serum (dilution 1:500) for 1 h at room temperature, and then with a second antibody, a swine anti-rabbit IgG conjugated to horseradish peroxidase (dilution 1:1000) (DAKO, Denmark) for 40 min at room temperature with agitation or with monoclonal anti-His6-Peroxidase (Roche Diagnostics, Mannheim, Germany) raised against the NH2-His tag (dilution 1:500) for 2 h at room temperature with agitation. The immunoblot was processed by chemiluminescence detection (Chemiluminescence Blotting Substrate [POD], Roche Molecular Biochemicals, Mannheim, Germany).
Enzyme activity
To ensure the viability of various purified proteins, second-order association rate constant (kass) of bovSERPINA3s with trypsin were determined as described by Beatty et al.36 using titrated trypsin as serine protease. Titration of bovine pancreatic trypsin was performed using 4-nitrophenyl p-guanidinobenzoate.37 Each purified inhibitor (5 nM) was preincubated with equimolar amounts (5 nM) of enzyme in 50 mM Tris-HCl buffer, pH 8.0, containing 10 mM CaCl2 for given periods of time (0 to 10 min). A total of 50 μL of 100 μM substrate stock solution (N-CBZ-Phe-Arg-NHMec) were then added before measuring residual activity.
In-gel protein digestion
All chemical products for mass spectrometry analysis were purchased from Sigma (St Louis, MO). Sequencing grade modified trypsin used for protein digestion was purchased from Promega (Charbonnières, France).
Bands of interest were excised from the gel and in-gel digestion was performed as described38 with minor modifications. Briefly, bands were destained and dried under vacuum. Proteins were reduced with 10 mM DTT in 100 mM NH4HCO3 for 35 min at 56°C, and alkylated with 55 mM iodoacetamide in 100 mM NH4HCO3 for 30 min at room temperature in the dark. Trypsin digestion was performed overnight at 37°C (20 ng μL−1 in 25 mM NH4HCO3). Resulting peptides were extracted successively by acetonitrile 40%–formic acid 1%, acetonitrile 25%–formic acid 1%, and acetonitrile 60% dried and resuspended in 6 μL of acetonitrile 2%–trifluoroacetic acid 0.05% for mass spectrometry analysis.
Nanospray liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis and database searching
Five microliters of the tryptic digest were analyzed by nanoLC-MS/MS using a LC Packings system (Dionex, Amsterdam, The Netherlands) coupled with a QTRAP mass spectrometer (Applied Biosystems/MDS Sciex, CA). Peptides were separated on a 75 μm ID × 150 mm C18 Pepmap™ column (LC Packings). Mass data collected during analysis were processed by the Analyst software 1.4.1 (Applied Biosystems/MDS Sciex, CA) and the MS/MS data were used to query Swiss Prot database using the Mascot software (version 2.2, Matrix Science, England) with the following criteria: 0.5 Da for peptide and fragment mass tolerances, one missed trypsin cleavage site allowed, carbamidomethylation of cysteine residues (from iodoacetamide exposure), and methionine oxidation as variable modifications. The protein identification, filtered for bovine and bacterial species, was established if one peptide matches of ion score above 50 or at least two peptides match of ion score above 25 at 95% confidence level.
Homology molecular modeling of bovSERPINA3-3 and bovSERPINA3-7
The bovSERPINA3-3 and bovSERPINA3-7 monomeric molecular models were computed with the Geno3D web server for automatic modeling of protein structure,39 using the template structure PDB ID: 1SEK chain A.40 The dimeric models and their interface analysis were computed with the Swiss-Pdb Viewer software 4.041 from the proposed biological unit structure of 1SEK in PDB42 and PQS43 databases. A maximum interchain interatomic distance threshold of 4 Å was applied to determine which residues are located at the interface of the homodimer.
Acknowledgments
The authors thank Lionel Forestier for his technical assistance.
Glossary
Abbreviations
- kass
association rate constant
- PAGE
polyacrylamide gel electrophoresis
- PAI
plasminogen activator inhibitor
- PNGase F
peptide-N-glycosidase F
- SDS
sodium dodecyl sulfate
References
- 1.Gettins PGW. Serpin structure, mechanism, and function. Chem Rev. 2002;102:4751–4803. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 2.Loebermann H, Tokuoka R, Deisenhofer J, Huber R. Human alpha 1-proteinase inhibitor. Crystal structure analysis of two crystal modifications, molecular model and preliminary analysis of the implications for function. J Mol Biol. 1984;177:531–557. [PubMed] [Google Scholar]
- 3.Whisstock J, Skinner R, Lesk AM. An atlas of serpin conformations. Trends Biochem Sci. 1998;23:63–67. doi: 10.1016/s0968-0004(97)01172-9. [DOI] [PubMed] [Google Scholar]
- 4.Mottonen J, Strand A, Symersky J, Sweet RM, Danley DE, Geoghegan KF, Gerard RD, Goldsmith EJ. Structural basis of latency in plasminogen activator inhibitor-1. Nature. 1992;355:270–273. doi: 10.1038/355270a0. [DOI] [PubMed] [Google Scholar]
- 5.Lomas DA, Belorgey D, Mallya M, Miranda E, Kinghorn KJ, Sharp LK, Phillips RL, Page R, Robertson AS, Crowther DC. Molecular mousetraps and the serpinopathies. Biochem Soc Trans. 2005;33:321–330. doi: 10.1042/BST0330321. [DOI] [PubMed] [Google Scholar]
- 6.Belorgey D, Hägglöf P, Karlsson-Li S, Lomas DA. Protein misfolding and the serpinopathies. Prion. 2007;1:15–20. doi: 10.4161/pri.1.1.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis RL, Shrimpton AE, Carrell RW, Lomas DA, Gerhard L, Baumann B, Lawrence DA, Yepes M, Kim TS, Ghetti B, Piccardo P, Takao M, Lacbawan F, Muenke M, Sifers RN, Bradshaw CB, Kent PF, Collins GH, Larocca D, Holohan PD. Association between conformational mutations in neuroserpin and onset and severity of dementia. Lancet. 2002;359:2242–2247. doi: 10.1016/S0140-6736(02)09293-0. [DOI] [PubMed] [Google Scholar]
- 8.Lomas DA, Evans DL, Finch JT, Carrell RW. The mechanism of Z alpha 1-antitrypsin accumulation in the liver. Nature. 1992;357:605–607. doi: 10.1038/357605a0. [DOI] [PubMed] [Google Scholar]
- 9.Poller W, Faber JP, Weidinger S, Tief K, Scholz S, Fischer M, Olek K, Kirchgesser M, Heidtmann HH. A leucine-to-proline substitution causes a defective α1-antichymotrypsin allele associated with familial obstructive lung disease. Genomics. 1993;17:740–743. doi: 10.1006/geno.1993.1396. [DOI] [PubMed] [Google Scholar]
- 10.Yamasaki M, Li W, Johnson DJ, Huntington JA. Crystal structure of a stable dimer reveals the molecular basis of serpin polymerization. Nature. 2008;455:1255–1258. doi: 10.1038/nature07394. [DOI] [PubMed] [Google Scholar]
- 11.Whisstock JC, Bottomley SP. Structural biology: serpins' mystery solved. Nature. 2008;455:1189–1190. doi: 10.1038/4551189a. [DOI] [PubMed] [Google Scholar]
- 12.Yamasaki M, Sendall TJ, Pearce MC, Whisstock JC, Huntington JA. Molecular basis of α1-antitrypsin deficiency revealed by the structure of a domain-swapped trimer. EMBO Rep. 2011;12:1011–1017. doi: 10.1038/embor.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crowther DC, Serpell LC, Dafforn TR, Gooptu B, Lomas DA. Nucleation of α1-antichymotrypsin polymerization. Biochemistry. 2003;42:2355–2363. doi: 10.1021/bi0259305. [DOI] [PubMed] [Google Scholar]
- 14.Christensen S, Sottrup-Jensen L. Characterization of two serpins from bovine plasma and milk. Biochem J. 1994;303:383–390. doi: 10.1042/bj3030383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tassy C, Herrera-Mendez CH, Sentandreu MA, Aubry L, Brémaud L, Pélissier P, Delourme D, Brillard M, Gauthier F, Levéziel H, Ouali A. Muscle endopin 1, a muscle intracellular serpin which strongly inhibits elastase: purification, characterization, cellular localization and tissue distribution. Biochem J. 2005;388:273–280. doi: 10.1042/BJ20041921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patston PA, Hauert J, Michaud M, Shapira M. Formation and properties of C1-inhibitor polymers. FEBS Lett. 1995;368:401–404. doi: 10.1016/0014-5793(95)00694-5. [DOI] [PubMed] [Google Scholar]
- 17.Devlin GL, Chow MK, Howlett GJ, Bottomley SP. Acid denaturation of α1-antitrypsin: characterization of a novel mechanism of serpin polymerisation. J Mol Biol. 2002;324:859–870. doi: 10.1016/s0022-2836(02)01088-4. [DOI] [PubMed] [Google Scholar]
- 18.Chow MK, Lomas DA, Bottomley SP. Promiscuous β-strand interactions and the conformational diseases. Curr Med Chem. 2004;11:491–499. doi: 10.2174/0929867043455936. [DOI] [PubMed] [Google Scholar]
- 19.Carrell RW, Stein PE, Fermi G, Wardell MR. Biological implications of a 3 Å structure of dimeric antithrombin. Structure. 1994;2:257–270. doi: 10.1016/s0969-2126(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 20.Lomas DA, Elliott PR, Sidhar SK, Foreman RC, Finch JT, Cox DW, Whisstock JC, Carrell RW. Alpha 1-Antitrypsin Mmalton (Phe52-deleted) forms loop-sheet polymers in vivo. Evidence for the C sheet mechanism of polymerization. J Biol Chem. 1995;270:16864–16870. doi: 10.1074/jbc.270.28.16864. [DOI] [PubMed] [Google Scholar]
- 21.Zhou A, Faint R, Charlton P, Dafforn TR, Carrell RW, Lomas DA. Polymerization of plasminogen activator inhibitor-1. J Biol Chem. 2001;276:9115–9122. doi: 10.1074/jbc.M010631200. [DOI] [PubMed] [Google Scholar]
- 22.Brémaud L, Herrera-Mendez CH, Coulis G, Pélissier P, Sentandreu MA, Aubry L, Delourme D, Chambon C, Maftah A, Levéziel H, Ouali A. Purification of the skeletal muscle protein Endopin 1B and characterization of the genes encoding Endopin 1A and 1B isoforms. FEBS Lett. 2006;580:3477–3484. doi: 10.1016/j.febslet.2006.04.099. [DOI] [PubMed] [Google Scholar]
- 23.Blanchet X, Herrera-Mendez CH, Becila S, Pélissier P, Delourme D, Coulis G, Sentandreu MA, Boudjellal A, Brémaud L, Ouali A. Inhibition of human initiator caspase 8 and effector caspase 3 by cross-class inhibitory bovSERPINA3–1 and A3–3. FEBS Lett. 2009;583:2743–2748. doi: 10.1016/j.febslet.2009.07.055. [DOI] [PubMed] [Google Scholar]
- 24.Ferguson KA. Starch-gel electrophoresis. Application to the classification of pituitary proteins and polypeptides. Metabolism. 1964;13:985–1002. doi: 10.1016/s0026-0495(64)80018-4. [DOI] [PubMed] [Google Scholar]
- 25.Lomas DA, Finch JT, Seyama K, Nukiwa T, Carrell RW. Alpha 1-antitrypsin Siiyama (Ser53–>Phe). Further evidence for intracellular loop-sheet polymerization. J Biol Chem. 1993;268:15333–15335. [PubMed] [Google Scholar]
- 26.Mikus P, Ny T. Intracellular polymerization of the serpin plasminogen activator inhibitor type 2. J Biol Chem. 1996;271:10048–10053. doi: 10.1074/jbc.271.17.10048. [DOI] [PubMed] [Google Scholar]
- 27.Pélissier P, Delourme D, Germot A, Blanchet X, Becila S, Maftah A, Leveziel H, Ouali A, Brémaud L. An original SERPINA3 gene cluster: elucidation of genomic organization and gene expression in the Bos taurus 21q24 region. BMC Genom. 2008;9:151–165. doi: 10.1186/1471-2164-9-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knaupp AS, Bottomley SP. Serpin polymerization and its role in disease—the molecular basis of α1-antitrypsin deficiency. IUBMB Life. 2009;61:1–5. doi: 10.1002/iub.127. [DOI] [PubMed] [Google Scholar]
- 29.Knaupp AS, Bottomley SP. Structural change in β-sheet A of Z α1-antitrypsin is responsible for accelerated polymerization and disease. J Mol Biol. 2011;413:888–898. doi: 10.1016/j.jmb.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 30.Elliott PR, Abrahams JP, Lomas DA. Wild-type alpha 1-antitrypsin is in the canonical inhibitory conformation. J Mol Biol. 1998;275:419–425. doi: 10.1006/jmbi.1997.1458. [DOI] [PubMed] [Google Scholar]
- 31.Huntington JA, Pannu NS, Hazes B, Read RJ, Lomas DA, Carrell RW. A 2.6 A structure of a serpin polymer and implications for conformational disease. J Mol Biol. 1999;293:449–455. doi: 10.1006/jmbi.1999.3184. [DOI] [PubMed] [Google Scholar]
- 32.Zhou A, Carrell RW, Huntington JA. The serpin inhibitory mechanism is critically dependent on the length of the reactive center loop. J Biol Chem. 2001;276:27541–27547. doi: 10.1074/jbc.M102594200. [DOI] [PubMed] [Google Scholar]
- 33.Yamasaki M, Sendall TJ, Harris LE, Lewis GMW, Huntington JA. Loop-sheet mechanism of serpin polymerization tested by reactive center loop mutations. J Biol Chem. 2010;285:30752–30758. doi: 10.1074/jbc.M110.156042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Dutaud D, Aubry L, Henry L, Levieux D, Hendil KB, Kueh LN, Bureau JP, Ouali A. Development and evaluation of a sandwich ELISA for quantification of the 20 S proteasome in human plasma. J Immunol Methods. 2002;260:183–193. doi: 10.1016/s0022-1759(01)00555-5. [DOI] [PubMed] [Google Scholar]
- 36.Beatty K, Bieth J, Travis J. Kinetics of association of serine proteinases with native and oxidized α-proteinase and α -1-antichymotrypsin. J Biol Chem. 1980;255:3931–3934. [PubMed] [Google Scholar]
- 37.Chase T, Shaw E. p-nitrophenyl-p′-guanidinobenzoate HCl: a new active site titrant for trypsin. Biochem Biophys Res Commun. 1967;29:508–514. doi: 10.1016/0006-291x(67)90513-x. [DOI] [PubMed] [Google Scholar]
- 38.Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 39.Combet C, Jambon M, Deléage G, Geourjon C. Geno3D: automatic comparative molecular modelling of protein. Bioinformatics. 2002;18:213–214. doi: 10.1093/bioinformatics/18.1.213. [DOI] [PubMed] [Google Scholar]
- 40.Li J, Wang Z, Canagarajah B, Jiang H, Kanost M, Goldsmith EJ. The structure of active serpin 1K from Manduca sexta. Structure. 1999;7:103–109. doi: 10.1016/s0969-2126(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 41.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 42.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 43.Henrick K, Thornton JM. PQS: a protein quaternary structure file server. Trends Biochem Sci. 1998;23:358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]


