Abstract
All acyl carrier protein primary and tertiary structures were gathered into the ThYme database. They are classified into 16 families by amino acid sequence similarity, with members of the different families having sequences with statistically highly significant differences. These classifications are supported by tertiary structure superposition analysis. Tertiary structures from a number of families are very similar, suggesting that these families may come from a single distant ancestor. Normal vibrational mode analysis was conducted on experimentally determined freestanding structures, showing greater fluctuations at chain termini and loops than in most helices. Their modes overlap more so within families than between different families. The tertiary structures of three acyl carrier protein families that lacked any known structures were predicted as well.
Keywords: acyl carrier protein, normal mode analysis, primary structure, protein family, tertiary structure, ThYme
Introduction
Acyl carrier proteins (ACPs) usually have 70 to 100, but occasionally more, amino acid residues, and they are usually linked through an interior serine residue to the terminal phosphate group of a 4′-phosphopantetheine prosthetic group. In turn, the latter binds fatty acids, polyketides, and other moieties by a thioester bond to its terminal thiol group, activating them for reactions that usually produce longer acyl chains, but also many other compounds (Table I).
Table I.
Acyl Carrier Protein Families
| Family | Approximate number of sequencesa | Producing domain | Role | Representative end product (nonexhaustive list) | UniProt accession code | Reference |
|---|---|---|---|---|---|---|
| ACP1 | 2870 | B, E, Ab | Freestanding ACPs: | |||
| in type II fatty acid synthesis | Fatty acids | P0A6A8 | 19 | |||
| in bacterial long-chain fatty acid synthesis | Long-chain fatty acids | P0A2W5 | 20 | |||
| in mitochondria | Fatty acids | P52505 | 21 | |||
| ACP2 | 180 | E | ACP domains of fungal FASs | Fatty acids | P19097 | 22 |
| ACP3 | 190 | E | ACP domains of animal FASs | Fatty acids | P49327 | 23 |
| ACP4 | 50 | B | Freestanding ACPs in polyketide synthesis | Bacillaene | Q7PC63 | 24 |
| ACP5 | 100 | B | Freestanding ACPs in polyketide synthesis | Actinorhodin | Q02054 | 25 |
| Frenolicin | Q54996 | 26 | ||||
| Oxytetracycline | P43677 | 27 | ||||
| ACP6 | 100 | B | ACP domains in PKSs | Mycocerosic acid | Q02251 | 28 |
| Phthioceranic acids | O07798 | 29 | ||||
| ACP7 | 30 | E | ACP domains in PKSs | 6-Methylsalicylic acid | P22367 | 30 |
| ACP8 | 240 | B, E | ACP domains in PKSs | Aflatoxin | Q12053 | 31 |
| Conidial yellow pigment | Q03149 | 32 | ||||
| ACP9 | 80 | E | ACP domain in PKSs/chalcone synthases | Acylpyrones | Q55E72 | 33 |
| ACP10 | 7000 | B, E, A | ACP/PCP domain in PKSs/NRPSs and ACP domain in ferrichrome synthases | Surfactin | P27206 | 34 |
| Erythronolide | Q03131 | 35 | ||||
| Gramicidin | P0C061 | 36 | ||||
| Phthiocerol | Q7TXL8 | 37 | ||||
| Tyrocidine | O30409 | 38 | ||||
| Plipastatin | O31827 | 39 | ||||
| Enniatin | Q00869 | 40 | ||||
| Ferrichrome | Q9P7T1 | 41 | ||||
| Bacillaene | Q05470 | 42 | ||||
| ACP11 | 120 | E | ACP domains in PKSs | Lovastatin | Q9Y8A5 | 43 |
| ACP12 | 280 | B | ACP domains in enterobactin synthases | Enterobactin | P0ADI4 | 44 |
| ACP domains in isochorismatases | Mycobactins | P71717 | 45 | |||
| ACP13 | 220 | B, E | ACP domains in enterobactin synthases | Enterobactin | P11454 | 46 |
| ACP15 | 200 | B | Freestanding ACPs active with malonate decarboxylases | Acetate | O06925 | 47 |
| ACP16 | 300 | B | Freestanding ACPs active with citrate lyases | Acetyl-CoA | P02903 | 48 |
| ACP17 | 250 | B | Freestanding d-alanyl carrier proteins | d-Alanyl lipoteichoic acid | P55153 | 49 |
Present in the ThYme database at the time of writing this article.
A, archaea; B, bacteria; E, eukaryota. Most prevalent producers bolded.
ACP molecules have many more anionic than cationic residues and rather few hydrophobic residues.1 Their tertiary structures feature three generally parallel α-helices (helices I, II, and IV) with a shorter crosswise α-helix (helix III). Helix II has many conserved anionic residues and plays an important role in ACP-enzyme interactions.2 Helix III is not present in all ACP structures, and it displays helix-loop equilibrium conformations.3
ACPs are either independent, freestanding structures, or they are covalently bound as part of multimodular enzymes such as fatty acid synthases (FASs), polyketide synthases (PKSs), and non-ribosomal peptide synthases (NRPSs). Acyl chains attached to freestanding ACPs are held within the hydrophobic pocket formed by the α-helices until they are subjected to reaction,4–7 when they are expelled into the active site of the enzyme catalyzing the reaction.8 The mechanism of delivering an acyl substrate to an enzyme active site from ACP's cavity, which may be accompanied by flexing of the ACP, is not completely understood. Those ACPs that are part of enzymes may not hold acyl chains within their hydrophobic pockets,9 as these chains are less exposed to solvent and to cell membranes.10
Freestanding ACPs, at least, appear to be quite flexible, being found in multiple conformers3, 5, 11 and having very flexible loops and α-helices.12–15
We have gathered all available ACP primary and tertiary structures into the ThYme database.16 There we have classified ACPs into families, following the same techniques that we have used earlier with thioesterases17 and ketoacyl synthases,18 which are described in Computational Methods. In general, members of a protein family have strong sequence similarity. They should also have tertiary structures that can be superimposed with small root mean square deviations (RMSDs) between corresponding amino acid residues. These similarities may imply that members are descended from a common protein ancestor. Members of different families have primary structures with statistically highly significant differences. However, slight similarities in amino acid residue alignments between ACP families may suggest that those without known tertiary structures are related to those with known structures. We describe this work with ACPs for the first time here.
This article is an account also of two further efforts: (1) a normal mode analysis of experimentally determined tertiary structures of freestanding apo-ACPs to describe their dynamic structures; and (2) the attempted computational prediction of tertiary structures of freestanding ACPs in three families with no known experimentally determined structures.
Results and Discussion
ACP families
Following the protocol described in the Computational Methods section led to 16 ACP families being defined. A multiple sequence alignment of representative members of these families shows very limited sequence similarity (Fig. 1). Only the serine residue at position 39 (ACP1 numbering), to which the prosthetic group is attached, is almost completely conserved (except for ACP15). In addition, there is substantial conservation at positions 35 (glycine), 38 (aspartate), and 67 (threonine). A number of positions have almost exclusively hydrophobic residues. In ACP15, the only residue that is completely conserved near the position otherwise occupied by a serine residue is Thr40 (ACP1 numbering).
Figure 1.

Multiple sequence alignment of single members of each of the 16 ACP families, roughly arranged in order of similarity to each other. ACP1: Borrelia burgdorferi; ACP2: Schizosaccharomyces pombe; ACP3: Gallus gallus; ACP4: Bacillus subtilis; ACP5: Streptomyces rimosus; ACP6: Mycobacterium tuberculosis; ACP7: Penicillium patulum; ACP8: Aspergillus nidulans; ACP9: Dictyostelium discoideum; ACP10: Saccharopolyspora erythraea; ACP11: Aspergillus terreus; ACP12: Escherichia coli; ACP13: E. coli; ACP15: Klebsiella pneumoniae; ACP16: K. pneumoniae; ACP17: Lactobacillus rhamnosus.
The same representative sequences of ACP families were subjected to pairwise sequence alignments to identify common residues between two sequences in different families. Even though there is low sequence similarity over the 16 families, there is substantial similarity from one family to the next (Supporting Information Table S1 and Fig. S1). The relationship of the families to each other is shown by two versions of a phylogenetic tree (Figs. 2 and Supporting Information Fig. S2). They show ACP15 and ACP16 peripheral to the other ACP families, as they are in Figure 1.
Figure 2.
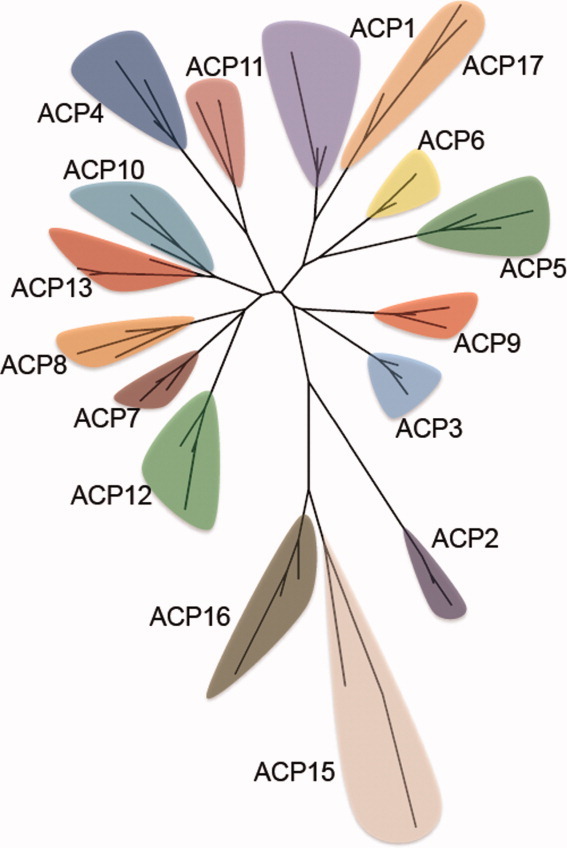
Phylogenetic tree of ACP families.
Table I summarizes the 16 ACP families, showing (1) approximate numbers of sequences in each family; (2) domains of life that produce each family; (3) whether ACP families are composed of freestanding proteins or are parts of multidomain enzymes; (4) the end products of enzymes with which ACP families interact; (5) representative UniProt accession codes of the ACPs; and (6) related literature.
Families ACP1 through ACP3 are involved with fatty acid synthesis. ACP1 members are freestanding ACPs, present in type II fatty acid synthesis, bacterial long-chain fatty acid synthesis, and mitochondrial fatty acid synthesis. ACP2 and ACP3 proteins are parts of multidomain FASs, involved with fungal and animal type I fatty acid synthesis, respectively. A substantial amount of research has been conducted on these ACPs; much of it is covered in the exhaustive review of Chan and Vogel.10
Proteins in ACP4 and ACP5 are freestanding ACPs involved with polyketide synthesis. In ACP4 is AcpK, part of the pksX pathway of Bacillus subtilis in making bacillaene.24 ACP5 includes ACPs involved with the synthesis of the antibiotics actinorhodin,25 frenolicin,26 and oxytetracycline27 in various Streptomyces species.
ACP6 through ACP9 and also ACP11 include the ACP domains of large multidomain PKSs. Among ACP6 sequences are the ACP domains of larger enzymes involved in making complex lipids found in Mycobacterium cell walls, including mycocerosic acid,28 and other sulfolipids.29 ACP7 includes the ACP domain of fungal PKS 6-methylsalicylic acid synthase.30 ACP8 includes mainly fungal sequences; among them are the ACP domains of PKSs involved with aflatoxin production,31 and the ACP domains of naphthopyrone PKSs that make the yellow pigment in conidia.32 ACP9 includes the ACP domain of PKS or chalcone synthase stlA, which produces acylpyrones.33 ACP11 includes the ACP domains of lovastatin synthases.43
ACP10, the family with the most members, includes the ACP domains of many PKSs, the peptide carrier protein domain of NRPSs, hybrid PKS/NRPS enzymes, and ferrichrome synthases.34–42 The enzymes in this family make a variety of natural products from secondary metabolism. A representative sample of them is shown in Table I.
Families ACP12 and ACP13 include the ACP domains of enterobactin synthases,44, 46 isochorismatases,45 and mycobactin synthases.45 ACP14 has been merged into ACP10 and no longer exists.
The prosthetic group in ACP15 and ACP16 is 2′-(5″-phosphoribosyl)-3′-dephospho-CoA, instead of 4′-phosphopantetheine, linked to an interior serine residue of apo-ACP through its 5″-phospho group and to the acyl molecule with a thioester bond through its terminal thiol group. ACP15 proteins include the ACPs active with malonate decarboxylases in bacteria that convert malonate to acetate and CO2 as an energy source.47 ACP16 enzymes include the ACPs active with citrate lyases that convert citrate to oxaloacetate and acetate in bacteria.48
Members of ACP17 do not carry acyl groups, but instead they are D-alanyl carrier proteins, as the moiety bound by 4′-phosphopantetheine is D-alanine, which is ligated using adenosine triphosphate to poly(ribitol phosphate). These enzymes are involved with the production of D-alanyl lipoteichoic acid.49
The members of all but four families, ACP1, ACP8, ACP10, and ACP13, are produced exclusively by either bacteria or eukaryota. More specifically, virtually all members of ACP2 and ACP8 and all members of ACP7 and AC11 are produced by fungi (the latter three families all by ascomycota), while all ACP3 members come from animals. AC9 members are from slime molds. Members of different bacterial phyla produce different members of ACP4, ACP12, ACP15, ACP16, and ACP17. ACP5 and ACP6 members are all from actinobacteria, with the latter being only from Mycobacterium species.
In summary, ACPs have diverged into different families based on primary structures that have statistically highly significant differences. They are either freestanding or are covalently bound to enzymes, they are specific to different substrates, they are produced by different classes of organisms, and they have sharply defined roles.
Existing ACP tertiary structures
Families ACP1, ACP2, ACP3, ACP5, ACP8, ACP10, ACP12, ACP13, and ACP17 contain members with known tertiary structures (Fig. 3). All but ACP8, ACP12, and ACP13 have more than one known structure. Most known structures and their properties were reviewed by Chan and Vogel.10 All tertiary structures are tabulated, with links to the Protein Data Bank (PDB), in the ACP section of the ThYme database.
Figure 3.
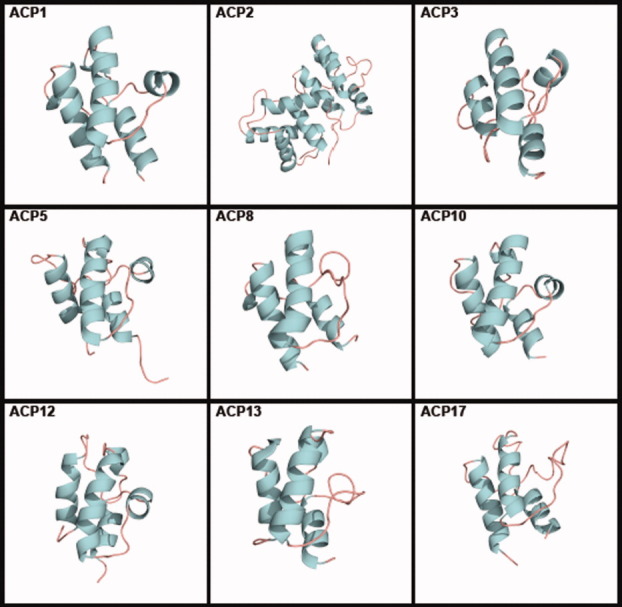
Tertiary structures of single members of ACP families. ACP1: Thermus thermophilus (PDB accession code 1X3O); ACP2: Saccharomyces cerevisiae (2UV8); ACP3: Homo sapiens (2CG5); ACP5: Streptomyces roseofulvus (1OR5); ACP8: Aspergillus parasiticus (2KR5); ACP10: Brevibacillus parabrevis (2JGP); ACP12: E. coli (2FQ1); ACP13: E. coli (2ROQ); and ACP17: Lactobacillus casei (1HQB).
We superimposed one tertiary structure per species within each putative ACP family and calculated their RMSDs using the protocol in the Computational Methods section. ACP2 and ACP17 have multiple known structures of freestanding ACPs or of ACP domains, but they all come from the same species. Therefore, their RMSDs were not calculated, as this would represent structure conservation among the same sequence and not sequences in a family. ACP2 domains in yeast FAS structures contain four extra helices in the C-terminal region,22 not seen in other ACPs, making them about twice as long as the others (Fig. 2). We found ACP3 and ACP10 domains in enzymes containing them by superimposing freestanding ACPs and extracting the former for RMSD calculations. In ACP10, we chose to superimpose the structures in the A/H conformer, very similar to other ACPs, one of three conformers in which ACP10 structures have been found.50 Structures within putative families should have low RMSDave values and high Pave values (average percentage of α-carbon atoms of the amino acid residues between two structures compared for calculations). Shown in Table II are the structures used, RMSDave values (ranging from 1.74 Å to 2.02 Å), and Pave values (ranging from 84.8 to 94.4%). These findings further indicate that the structures represent members in the same families.
Table II.
Acyl Carrier Protein Tertiary Structures
| Family | RMSDave (Å) | Pave (%) | Tertiary structuresa | Total number of PDB accessions in family |
|---|---|---|---|---|
| ACP1 | 2.01 | 86.4 | 1HY8, 1KLP, 1L0H, 1X3O, 2DNW, 2EHS, 2FQ0, 2FVE | 45 |
| 2KOO, 2KW2, 2KWL, 2L0Q, 2L3V, 2L4B, 2QNW | ||||
| ACP2 | — | — | 2PFF | 6b |
| ACP3 | 2.02 | 94.4 | 2CG5c, 2PNG | 4 |
| ACP5 | 1.75 | 84.8 | 1NQ4, 1OR5, 2AF8 | 14 |
| ACP8 | — | — | 2KR5 | 1 |
| ACP10 | 1.74 | 85.9 | 1DNY, 2GDW, 2JGPc, 2JU2, 2VSQc | 9 |
| ACP12 | — | — | 2FQ1 | 1 |
| ACP13 | — | — | 2ROQ | 1 |
| ACP17 | — | — | 1DV5 | 2b |
For ACP1, ACP3, ACP5, and ACP10 all structures listed were superimposed to yield reported RMSDave and Pave values; for others a representative sequence is shown. Only one structure per species within a family was superimposed.
RMSD not calculated, as all resolved ACP domains in existing tertiary structures come from the same species.
Only ACP domain of a larger structure
If different families have members with tertiary structures that can be closely aligned, even though their primary structures are different, then they may have a more distant common protein ancestor and may be gathered into the same clan. To determine this, one tertiary structure each from ACP1 (PDB accession code 1X3O), ACP2 (2UV8), ACP3 (2CG5), ACP5 (1OR5), ACP8 (2KR5), ACP10 (2JGP), ACP12 (2FQ1), ACP13 (2ROQ), and ACP17 (1HQB) (Fig. 3) were superimposed. Of these, ACP2, ACP3, ACP8, ACP10, and ACP12 were domains in larger enzymes. The first four helices of the ACP2 structure were well superimposed on the structures of the other families. The superposition of the structures of these nine families resulted in an RMSDave value of 2.34 Å and a Pave value of 76.2%, indicating that at least those families may descend from a distant common ancestor and may be gathered into the same clan. The moderate percentages of common residues between different pairs of families (Supporting Information Table S1 and Fig. S1) suggest that other ACP families may also have the same common ancestor. However, this conclusion must await determination of tertiary structures of members of these families.
Normal mode analysis of freestanding ACP structures
We subjected seven tertiary structures of freestanding apo-ACPs to normal mode analysis, using the Anisotropic Network Model (ANM) web server.51 We did not attempt to subject ACPs that were part of larger proteins to normal mode analysis, since their structures and fluctuations are most likely affected by their proximity of other parts of the protein. ACP1 members were PDB accession codes 2EHS and 2EHT (two conformers of the same protein) from Aquiflex aeolicus, 1T8K from Escherichia coli, 1X3O from Thermus thermophilus, and 2QNW from Toxoplasma gondii. The one ACP5 member was 2AF8 from Streptomyces coelicolor, while 1HQB in ACP17 was from Lactobacillus rhamnosus. All are produced by bacteria but one; T. gondii is an apicomplexan protozoan.
The residue fluctuations of the five slowest modes of each structure are plotted in Supporting Information Figure S3, and ANM mode visualizations of the slowest mode of each structure are shown in Supporting Information Figure S4.
Chain ends, especially the N-terminal one, and loops were predicted to fluctuate more than the three long and parallel α-helices (helices I, II, and IV), which move very little. The region between helices II and IV, which in ACP1 structures comprises the loop between helices II and III and the crosswise helix III, strongly fluctuated in many structures. In the ACP5 and ACP17 structures, helix III is replaced by a loop. This region was proposed to act as a gatekeeper in the acyl chain delivery process,52 as the acyl chain is exposed through a fissure following a conformational rearrangement. The acyl chain delivery process is unknown; sword-unsheathed or switchblade-like mechanisms have been proposed. Lower fluctuations are seen in the loop between helices I and II. The N-terminal region of helix II, where the 4′-phosphopantetheine prosthetic group is attached, shows relatively higher fluctuations than the rest of helix II. In general, the vibrations of the first five slowest modes in any structure are moderately to strongly correlated in location, although much less so in magnitude.
We compared the normal modes of two conformers (2EHS and 2EHT) of the same structure in ACP1, of two bacterial structures (1T8K and 1X3O) in ACP1, of bacterial (1T8K) and eukaryotic (2QNW) structures in ACP1, of ACP1 (1T8K) and ACP5 (2AF8) structures, of ACP1 (1T8K) and ACP17 (1HQB) structures, and of ACP5 (2AF8) and ACP17 (1HQB) structures. They were analyzed as explained in the Computational Methods section. Results are summarized in Table III, overlap charts are shown in Supporting Information Figure S5, and residue fluctuation comparisons of the three most overlapped modes for each case are shown in Supporting Information Figure S6.
Table III.
Normal Mode Analysis Comparisons
| Structural comparison | PDB designations | Highest overlap between two modes | RMSDf-ave (Å) | Pf-ave (%) |
|---|---|---|---|---|
| Same structure, two conformers | 2EHS, 2EHT | 0.47 | 0.90 | 99.1 |
| Same family, two bacterial structures | 1T8K, 1X3O | 0.42 | 1.51 | 80.9 |
| Same family, bacterial and eukaryotic | 1T8K, 2QNW | 0.47 | 1.05 | 95.7 |
| ACP1 to ACP5 | 1T8K, 2AF8 | 0.65 | 2.70 | 61.2 |
| ACP1 to ACP17 | 1T8K, 1HQB | 0.58 | 2.08 | 85.9 |
| ACP5 to ACP17 | 2AF8, 1HQB | 0.64 | 2.67 | 71.1 |
The two conformers of the same structure showed a very low frame-averaged RMSD (RMSDf-ave, as defined in the Computational Methods section), showing that their structural similarity was conserved throughout the motion of the slowest normal mode. Several of their slowest normal modes showed some overlap (Supporting Information Fig. S5). Residue fluctuations (Supporting Information Fig. S6) show that 2EHT has larger amplitudes than 2EHS in the N-terminal region, while 2EHS has larger amplitudes in the central and C-terminal regions. Their fluctuations appear in very similar residue locations.
Two different structures within ACP1 display lower overlaps than two conformers of the same structure (Supporting Information Fig. S5). Values of RMSDf-ave are higher than between two conformers of the same structure, but still are <2 Å, as would be expected of two structures within a family. In both cases, fluctuations appear conserved in residue location but not so in amplitude (Supporting Information Fig. S6), although they appear more correlated between the bacterial and eukaryotic structures.
Comparing the normal modes between structures in different families showed that the slowest mode of each structure was the most highly overlapped (Supporting Information Fig S5), especially between ACP1 and ACP5 and between ACP5 and ACP17. The slowest modes in each structure display an overall twisting rather than stretching of discrete loops, and overlap highly due to their low flexibility. Their RMSDf-ave values are higher than in previous cases, showing structural differences between two families. However, within the most overlapped normal mode, structural differences are not great. Residue fluctuation for modes other than the most overlapped (Supporting Information Fig. S6) are poorly correlated in both amplitude and location, especially between ACP1 and ACP5 and between ACP5 and ACP17, but less so between ACP1 and ACP17.
Even though overlap charts (Supporting Information Fig. S5) do not differ much for comparisons within and between families, residue fluctuations (Supporting Information Fig. S6) are more conserved between two structures within a family than between two structures in different families. However, fluctuation amplitudes can be quite different in different structures produced by organisms in the same genus and, especially noteworthy, in different conformers of the same structure. Differences in fluctuation amplitudes may be due to slight crystal packing effects that could be present in loops.
Determination of tertiary structures of freestanding ACPs by computation
To determine whether the homology modeling and molecular dynamics (MD) protocol described in the Computational Methods section is a trustworthy method of determining tertiary structures of freestanding ACPs whose structures were previously unknown, we used experimentally determined freestanding structures of two families from ACP1 (2EHS), ACP5 (1OR5), and ACP17 (1HQB) as templates to predict the structure of the third family. We did not use ACP domains as templates or predict unknown ACP domains of large multidomain enzymes, as their folded states may be influenced by nearby sections of the larger protein.
Results are shown in Table IV. The amino acid identities, based on pairwise alignments between individual family members, between template and target structures vary between 20.0 and 23.4%. Two RMSD values between predicted and crystal tertiary structures (RMSDp-c) are reported: first the value of the predicted structure by homology modeling to its crystal structure, and second the predicted structure after MD refinement to its crystal structure. RMSDp-c values of homology modeling predictions vary between 1.17 Å and 2.10 Å, and RMSDp-c values after MD refinement vary between 1.42 Å and 1.96 Å. The predicted structures superimposed to their crystal structures can be seen in Figure 4. As indicated by low RMSDp-c values (<2 Å) and by visual inspection, the protocol described can predict freestanding ACP structures when the template used for homology modeling is at least 20% identical in sequence to the target.
Table IV.
Tertiary Structure Prediction Validation
| Target structure | Template structure | Amino acid identity (%)a | RMSDp-c (homology) (Å) | RMSDp-c (MD) (Å) |
|---|---|---|---|---|
| ACP1 | ACP5 | 23.4 | 1.17 | 1.42 |
| ACP1 | ACP17 | 20.8 | 1.44 | 1.48 |
| ACP5 | ACP1 | 21.7 | 1.97 | 1.87 |
| ACP5 | ACP17 | 20.5 | 2.10 | 1.96 |
| ACP17 | ACP1 | 20.0 | 1.73 | 1.93 |
| ACP17 | ACP5 | 21.3 | 1.78 | 1.88 |
Percentage of same residues in same position between template and target structures divided by the number of residues in the template.
Figure 4.
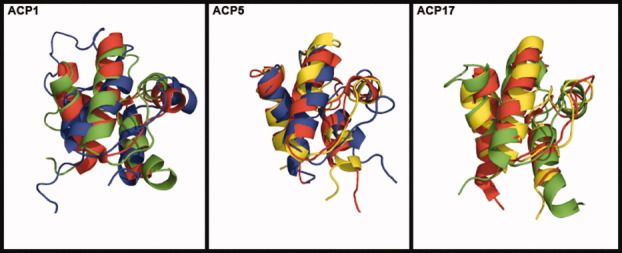
Predicted tertiary structure of an ACP1 member using ACP5 (green) and ACP17 (blue) tertiary structures as templates, compared with a known ACP1 (red) tertiary structure; of an ACP5 member using ACP1 (yellow) and ACP17 (blue) tertiary structures as templates, compared to a known ACP5 (red) tertiary structure; of an ACP17 member using ACP1 (yellow) and ACP5 (green) tertiary structures as templates, compared to a known ACP17 (red) tertiary structure. Known structure templates: ACP1: 2EHS; ACP5: 1OR5; ACP17: 1HQB.
We then attempted to predict tertiary structures for sequences in ACP4, ACP15, and ACP16, which contain freestanding members with no known structures. The structure of Bacillus subtilis AcpK (UniProt ID Q7PC63) in ACP4 was predicted using as a template 2EHS from ACP1, with which it has an amino acid identity of 25.6%. The predicted structure of ACP4 (Fig. 5) is very similar to structures in other ACP families, and it clearly shows a cavity and three long helices (helices I, II, and IV). When the predicted ACP4 structure was superimposed to structures in ACP1 (2EHS), ACP5 (1OR5), and ACP17 (1HQB), the resulting RMSDs were 1.92 Å, 2.12 Å, and 2.21 Å, respectively.
Figure 5.
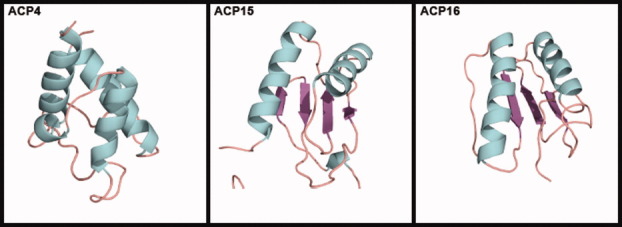
Tertiary structure of ACP4 (Bacillus subtilis AcpK) predicted by homology modeling and MD. Representative models of tertiary structures of ACP15 (Malonomonas rubra malonate decarboxylase ACP) and ACP16 (E. coli citrate lyase ACP) predicted by threading.
Subjecting Malonomonas rubra malonate decarboxylase (UniProt ID: O06925) from ACP15 and E. coli citrate lyase (UniProt ID: P69330) from ACP16 to the homology modeling and MD protocol resulted in unfolded structures that lacked the standard three parallel helices and cavity seen in other ACPs. However, they are most likely not intrinsically unstructured. Ten models were then predicted for each sequence using threading programs. Four of the 10 ACP15 models (1, 5, 6, and 8) displayed two main α-helices and four β-strands (Supporting Information Fig. S7), while seven of the 10 ACP16 models (1, 2, 4–8) showed two main α-helices with three β-strands. Representative models of the four and seven similar structures of ACP15 and ACP16, respectively, are shown in Fig. 5. When superimposed, the four similar models in ACP15 have RMSDave and Pave values of 0.73 Å and 88.7%, and the seven similar models of ACP16 show values of 0.99 Å and 91.3%.
ACP15 and ACP16 carrier proteins are used in reactions that break substrates into smaller units, unlike other ACPs, which are involved in building large molecules from small units. Also, ACP15 and ACP16 do not use the same 4′-phosphopantetheine prosthetic group present in other ACPs. These facts further suggest that ACP15 and ACP16 may be structurally different from other ACP families, and if this is correct, ACP15 and ACP16 members may descend from different ancestors than most or all of the other ACPs. The multiple sequence alignment (Fig. 1), phylogenetic tree (Fig. 2, Supporting Information Fig. S2), and threading predictions (Fig. 6, Supporting Information Fig. S7) support this. However, only experimentally determined structures will show if ACP15 and ACP16 are indeed different from other ACP families.
Concluding Remarks
Considering that ACPs, except those of ACP2, are all of roughly the same length and in nearly all cases they bind and activate acyl chains prior to their being subjected to enzymatic catalysis, it is noteworthy that they can be separated into 16 families by their highly significant differences in primary structure. It is perhaps equally noteworthy that many of these families have very similar tertiary structures, signifying that they may be descended from a common distant ancestor. We predicted the normal vibrational modes of freestanding ACPs, finding them more conserved within families. Also, we extended knowledge of three families possessing freestanding ACPs, one family by predicting its characteristic tertiary structure to be like those of many other ACP families, and two families by apparently showing that they do not have similar structures.
Computational Methods
Family identification and phylogeny
Families were identified following the procedures outlined earlier.16–18 In brief, families were based on sequences with evidence at protein level from the UniProt database53 or the literature, and populated by use of BLAST54 (downloadable version 2.2.19) to query the nonredundant (nr) protein sequence database.55 A cutoff E-value of 0.001 (likelihood that similarity between query and compared sequence is due to chance) was used as the exclusion criterion. Families were confirmed with multiple sequence alignments of the retrieved sequences, using MUSCLE 3.6,56 and with tertiary structure superimpositions.
Phylogenetic trees were built using Molecular Evolutionary Genetics Analysis 5 (MEGA5),57 based on a multiple sequence alignment containing three sequences per family made with MUSCLE 3.6. The minimum evolution algorithm58 was used, gaps were treated with pairwise deletion, and an amino acid Jones-Taylor-Thornton matrix59 was chosen as the model. A bootstrap test with 1000 replicates was performed to further verify the results.
Tertiary structure superposition and RMSD calculations
In this study, all tertiary structures were superimposed with MultiProt.60 All RMSD values were calculated between α-carbon atoms using MATLAB, to consider the most possible α-carbon atoms in the calculation; MultiProt reports the RMSD for only aligned residues. Values of RMSD (between two structures), RMSDave (between three or more structures), and Pave (the average percentage of α-carbon atoms of the amino acid residues used to calculate the RMSD between two compared structures) were calculated, as explained in detail in the Supporting Information in Cantu et al.17 When a NMR-resolved structure was superimposed, only the lowest-energy frame was used, chosen following single-point energy calculations with the ff99SB AMBER force field.61
Normal mode analysis
Normal modes of individual ACPs were calculated using the ANM web server,51 by assuming that a molecule's natural vibrations can be predicted by attaching a spring of uniform force constant to each α-carbon atom of the molecule and allowing the system to oscillate. The cutoff distance and distance weights were changed until the best correlation between theoretical and experimental B-factors was achieved. Structures were submitted individually to the ANM server, which computed the vibrational normal modes of each molecule and returned an animated PDB file for the 20 slowest vibrational modes, their associated eigenvectors, and residue fluctuations. No significant crystal packing effects were found when viewing the symmetric molecules of the PDBs submitted.
To compare the normal modes between two structures, overlap charts showing the 20 slowest vibrational modes of two structures were made by taking the dot product of the eigenvectors from both structures, yielding an 20 × 20 overlap chart where each cell ranges in value from 0 (no overlap) to 1 (complete overlap). From this, the highest overlapped mode was chosen, and the animated structure PDB files of such mode were separated into individual PDB files for each frame. Then the frame from one structure was superimposed with the corresponding frame in the other, and RMSD and P values was calculated. This was done for each frame, and the averages among all frames were taken, resulting in values of RMSDf-ave and Pf-ave. This notation is used here to differentiate the RMSD between two structures averaged over different frames in a normal mode (Table III), from RMSDave, which refers to the average RMSD of superimposing three or more structures (Table II).
Tertiary structure determination
A homology modeling and MD protocol was used for structure predictions. Homology modeling was done with the I-TASSER server,62, 63 using other ACP structures as templates without alignment or restraints.
The resulting predicted structures were loaded in AmberTools1.4 using the ff99SB AMBER force field. The structures were placed in a 12-Å TIP3P water box,64 and Na+ ions were added to neutralize system charge.
The system was then simulated with the sander module of AMBER.65 Solvent and ions were minimized first with 1000 steps while restraining the protein with 500 kcal/Å-mol weights. An unrestrained 10,000-step minimization followed. The system was then heated from 0 K to 300 K at constant volume for 25 to 100 ps while weakly restraining the protein with 10 kcal/Å-mol weights. Finally, the system was equilibrated at constant pressure and temperature and run unrestrained for 1.5 to 2 ns.
Sequences were predicted to not be intrinsically unfolded using the IUPred66 server that bases its predictions by calculating inter-residue energy interactions.
Threading predictions were done using the LOMETS67 server, which generates tertiary structures from sequences using eight different known threading programs. Unlike homology modeling, no templates are specified. The 10 best threading models from the output were taken.
RMSDs between predicted structures to experimental crystal structures in the validation runs (Table IV) are labeled as RMSDp-c, to differentiate it from the other RMSDs used here.
Acknowledgments
K.C. from Johns Hopkins University was part of a research experiences for undergraduates program associated with the Engineering Research Center. The authors thank Michael T. Zimmermann (Iowa State University) for helpful discussions about normal mode analysis and Christopher D. Warner (The Scripps Research Institute, Florida) for discussions about crystal packing effects and structure.
Glossary
Abbreviations
- ACP
acyl carrier protein
- AMBER
Assisted Model Building with Energy Refinement
- ANM
Anisotropic Network Model server
- BLAST
Basic Local Alignment Search Tool
- CoA
coenzyme A
- FAS
fatty acid synthase
- I-TASSER
Iterative Threading ASSEmbly Refinement server
- NRPS
nonribosomal peptide synthase
- P
percentage of α-carbon atoms of the amino acid residues between two structures compared for calculations
- PDB
Protein Data Bank
- PKS
polyketide synthase
- RMSD
root mean square deviation
- ThYme
Thioester active enzYme database
- UniProt
Universal Protein resource.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Rock CO, Cronan JE., Jr Re-evaluation of the solution structure of acyl carrier protein. J Biol Chem. 1979;254:9778–9785. [PubMed] [Google Scholar]
- 2.Zhang YM, Marrakachi H, White SW, Rock CO. The application of computational methods to explore the diversity and structure of bacterial fatty acid synthase. J Lipid Res. 2003;44:1–10. doi: 10.1194/jlr.r200016-jlr200. [DOI] [PubMed] [Google Scholar]
- 3.Sharma AK, Sharma SK, Surolia A, Surolia N, Sarma SP. Solution structures of conformationally equilibrium forms of holo-acyl carrier protein (PfACP) from Plasmodium falciparum provides insight into the mechanism of activation of ACPs. Biochemistry. 2006;45:6904–6916. doi: 10.1021/bi060368u. [DOI] [PubMed] [Google Scholar]
- 4.Roujeinikova A, Baldock C, Simon WJ, Gilroy J, Baker PJ, Stuitje AR, Rice DW, Slabas AR, Rafferty JB. X-ray crystallographic studies on butyryl-ACP reveal flexibility of the structure around a putative acyl chain binding site. Structure. 2002;10:825–835. doi: 10.1016/s0969-2126(02)00775-x. [DOI] [PubMed] [Google Scholar]
- 5.Zornetzer GA, Fox BG, Markley JL. Solution structures of spinach acyl carrier protein with decanoate and stearate. Biochemistry. 2006;45:5217–5227. doi: 10.1021/bi052062d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roujeinikova A, Simon WJ, Gilroy J, Rice DW, Rafferty JB, Slabas AR. Structural studies of fatty-(acyl carrier protein) thioesterases reveal a hydrophobic binding cavity that can expand to fit longer substrates. J Mol Biol. 2007;365:135–145. doi: 10.1016/j.jmb.2006.09.049. [DOI] [PubMed] [Google Scholar]
- 7.Chan DI, Stockner T, Tieleman DP, Vogel HJ. Molecular dynamics simulations of the apo-, holo-, and acyl-forms of Escherichia coli acyl carrier protein. J Biol Chem. 2008;283:33620–33629. doi: 10.1074/jbc.M805323200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rafi S, Novichenok P, Kolappan S, Zhang X, Stratton CF, Rawat R, Kisker C, Simmerling C, Tonge PJ. Structure of acyl carrier protein bound to FabI, the FASII enoyl reductase from Escherichia coli. J Biol Chem. 2006;281:39285–39293. doi: 10.1074/jbc.M608758200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ploskon E, Arthur CJ, Evans SE, Williams C, Crosby J, Simpson TJ, Crump MP. A mammalian type I fatty acid synthase acyl carrier protein domain does not sequester acyl chains. J Biol Chem. 2008;283:518–528. doi: 10.1074/jbc.M703454200. [DOI] [PubMed] [Google Scholar]
- 10.Chan DI, Vogel HJ. Current understanding of fatty acid biosynthesis and the acyl carrier protein. Biochem J. 2010;430:1–19. doi: 10.1042/BJ20100462. [DOI] [PubMed] [Google Scholar]
- 11.Kim Y, Prestegard JH. A dynamic model for the structure of acyl carrier protein in solution. Biochemistry. 1989;28:8792–8797. doi: 10.1021/bi00448a017. [DOI] [PubMed] [Google Scholar]
- 12.Andrec M, Hill RB, Prestegard JH. Amide exchange rates in Escherichia coli acyl carrier protein: correlation with protein structure and dynamics. Protein Sci. 1995;4:983–993. doi: 10.1002/pro.5560040518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park SJ, Kim J-S, Son W-S, Lee BJ. pH-Induced conformational transition of H. pylori acyl carrier protein: insight into the unfolding of local structure. J Biochem (Tokyo) 2004;135:337–346. doi: 10.1093/jb/mvh041. [DOI] [PubMed] [Google Scholar]
- 14.Kim Y, Kovrigin EL, Eletr Z. NMR studies of Escherichia coli acyl carrier protein: dynamic and structural differences of the apo- and holo-forms. Biochem Biophys Res Commun. 2006;341:776–783. doi: 10.1016/j.bbrc.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 15.Zornetzer GA, Tanem J, Fox BG, Markley JL. The length of the bound fatty acid influences the dynamics of the acyl carrier protein and the stability of the thioester bond. Biochemistry. 2010;49:470–477. doi: 10.1021/bi9014659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cantu DC, Chen Y, Lemons ML, Reilly PJ. ThYme: A database for thioester-active enzymes. Nucleic Acids Res 39:D342–D346. 2011. Available at: http://enzyme.cbirc.iastate.edu. [DOI] [PMC free article] [PubMed]
- 17.Cantu DC, Chen Y, Reilly PJ. Thioesterases: a new perspective based on their primary and tertiary structures. Protein Sci. 2010;19:1281–1295. doi: 10.1002/pro.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Kelly EE, Masluk RP, Nelson CL, Cantu DC, Reilly PJ. Structural classification and properties of ketoacyl synthases. Protein Sci. 2011;20:1659–1667. doi: 10.1002/pro.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanaman TC, Wakil SJ, Hill RL. The complete amino acid sequence of the acyl carrier protein of Escherichia coli. J Biol Chem. 1968;243:6420–6431. [PubMed] [Google Scholar]
- 20.Brozek KA, Carlson RW, Raetz CRH. A special acyl carrier protein for transferring long hydroxylated fatty acids to lipid A in Rhizobium. J Biol Chem. 1996;271:32126–32136. doi: 10.1074/jbc.271.50.32126. [DOI] [PubMed] [Google Scholar]
- 21.Cronan JE, Fearnleay IM, Walker JE. Mammalian mitochondria contain a soluble acyl carrier protein. FEBS Lett. 2005;579:4892–4896. doi: 10.1016/j.febslet.2005.07.077. [DOI] [PubMed] [Google Scholar]
- 22.Leibundgut M, Jenni S, Frick C, Ban N. Structural basis for substrate delivery by acyl carrier protein in the yeast fatty acid synthase. Science. 2007;316:288–290. doi: 10.1126/science.1138249. [DOI] [PubMed] [Google Scholar]
- 23.Jayakumar A, Tai MH, Huang WY, Al-Feel W, Hsu M, Abu-Elheiga L, Chirala SS, Wakil SJ. Human fatty acid synthase: properties and molecular cloning. Proc Natl Acad Sci USA. 1995;92:8695–8699. doi: 10.1073/pnas.92.19.8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calderone CT, Kowtoniuk WE, Kelleher NL, Walsh CT, Dorrestein PC. Convergence of isoprene and polyketide biosynthetic machinery: isoprenyl-S-carrier proteins in the pksX pathway of Bacillus subtilis. Proc Natl Acad Sci USA. 2006;103:8977–8982. doi: 10.1073/pnas.0603148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez-Moreno MA, Martinez E, Boto L, Hopwood DA, Malpartida F. Nucleotide sequences and deduced functions of a set of cotranscribed genes of Streptomyces coelicolor A3(2) including the polyketide synthase for the antibiotic actinorhodin. J Biol Chem. 1992;25:19278–19290. [PubMed] [Google Scholar]
- 26.Bibb MJ, Sherman DJ, Omura S, Hopwood DA. Cloning, sequencing and deduced functions of a cluster of Streptomyces genes probably encoding biosynthesis of the polyketide antibiotic frenolicin. Gene. 1994;142:31–39. doi: 10.1016/0378-1119(94)90351-4. [DOI] [PubMed] [Google Scholar]
- 27.Kim E-S, Bibb MJ, Butler MJ, Hopwood DA, Sherman DH. Sequences of the tetracycline polyketide synthase-encoding otc genes from Streptomyces rimosus. Gene. 1994;141:141–142. doi: 10.1016/0378-1119(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 28.Mathur M, Kolattukudy PE. Molecular cloning and sequencing of the gene for mycocerosic acid synthase, a novel fatty acid elongating multifunctional enzyme, from Mycobacterium tuberculosis var. bovis Bacillus Calmette-Guerin. J Biol Chem. 1992;267:19388–19395. [PubMed] [Google Scholar]
- 29.Sirakova TD, Thirumala AK, Dubey VS, Sprecher H, Kolattukugy PE. The Mycobacterium tuberculosis pks2 gene encodes the synthase for the hepta- and octamethyl-branched fatty acids required for sulfolipid synthesis. J Biol Chem. 2001;276:16833–16839. doi: 10.1074/jbc.M011468200. [DOI] [PubMed] [Google Scholar]
- 30.Beck J, Ripka S, Siegner A, Schiltz E, Schweizer E. The multifunctional 6-methylsalicylic acid synthase gene of Penicillim patulum. Eur J Biochem. 1990;192:487–498. doi: 10.1111/j.1432-1033.1990.tb19252.x. [DOI] [PubMed] [Google Scholar]
- 31.Feng GH, Leonard TJ. Characterization of the polyketide synthase gene (pksL1) required for aflatoxin biosynthesis in Aspergillus parasiticus. J Bacteriol. 1995;177:6246–6254. doi: 10.1128/jb.177.21.6246-6254.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe A, Fujii I, Sankawa U, Mayorga ME, Timberlake WE, Ebizuka Y. Re-identification of Aspergillus nidulans wA gene to code for a polyketide synthase of naphthopyrone. Tetrahedron Lett. 1999;40:91–94. [Google Scholar]
- 33.Austin MB, Saito T, Bowman ME, Haydock S, Kato A, Moore BS, Kay RR, Noel JP. Biosynthesis of Dictyostelium discoideum differentiation-inducing factor by a hybrid type I fatty acid-type III polyketide synthase. Nat Chem Biol. 2006;2:494–502. doi: 10.1038/nchembio811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakano MM, Marahiel MA, Zuber P. Identification of a genetic locus required for the biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J Bacteriol. 1988;170:5662–5668. doi: 10.1128/jb.170.12.5662-5668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donadio S, Staver MJ, McAlpine JB, Swanson SJ, Katz L. Modular organization of genes required for complex polyketide biosynthesis. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 36.Stein T, Kluge B, Vater J, Franke P, Otto A, Wittmann-Liebold B. Gramicidin S synthase 1 (phenylalanine racemase), a prototype of amino acid racemases containing the cofactor 5′-phosphopantetheine. Biochemistry. 1995;34:4633–4642. doi: 10.1021/bi00014a017. [DOI] [PubMed] [Google Scholar]
- 37.Azad AK, Sirakova TD, Fernandes ND, Kolattukudy PE. Gene knockout reveals a novel gene cluster for the synthesis of a class of cell wall lipids unique to pathogenic mycobacteria. J Biol Chem. 1997;272:16741–16745. doi: 10.1074/jbc.272.27.16741. [DOI] [PubMed] [Google Scholar]
- 38.Mootz HD, Marahiel MA. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J Bacteriol. 1997;179:6843–6850. doi: 10.1128/jb.179.21.6843-6850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsuge K, Ano T, Hirai M, Nakamura Y, Shoda M. The genes degQ, pps, and lpa-8 (sfp) are responsible for the conversion of Bacillus subtilis. Antimicrob Agents Chemother. 1999;43:2183–2192. doi: 10.1128/aac.43.9.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hacker C, Glinski M, Hornbogen T, Doller A, Zocher R. Mutational analysis of the N-methyltransferase domain of the multifunctional enzyme enniatin synthase. J Biol Chem. 2000;275:30826–30832. doi: 10.1074/jbc.M002614200. [DOI] [PubMed] [Google Scholar]
- 41.Schrettl M, Winkelmann G, Hass H. Ferrichrome in Schizosaccharomyces pombe—an iron transport and iron strorage compound. BioMetals. 2004;17:647–654. doi: 10.1007/s10534-004-1230-z. [DOI] [PubMed] [Google Scholar]
- 42.Butcher RA, Schroeder FC, Fischbach MA, Straight PD, Kolter R, Walsh CT, Clardy J. The identification of bacillaene, the product of the PksX megacomplex in Bacillus subtilis. Proc Natl Acad Sci USA. 2007;104:305–310. doi: 10.1073/pnas.0610503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hendrickson L, Davis CR, Roach C, Nguyen DK, Aldrich T, McAda PC, Reeves CD. Lovastatin biosynthesis in Aspergillus terreus: characterization of blocked mutants, enzyme activities and a multifunctional polyketide synthase gene. Chem Biol. 1999;6:429–439. doi: 10.1016/s1074-5521(99)80061-1. [DOI] [PubMed] [Google Scholar]
- 44.Gehring AM, Bradley KA, Walsh CT. Enterobactin biosynthesis in Escherichia coli: isochorismate lyase (EntB) is a bifunctional enzyme that is phosphopanteteinylated by EntD and then acylated by EntE using ATP and 2,3-dihydroxybenzoate. Biochemistry. 1997;36:8495–8503. doi: 10.1021/bi970453p. [DOI] [PubMed] [Google Scholar]
- 45.Quadri LEN, Sello J, Keating TA, Weinreb PH, Walsh CT. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for the assembly of the virulence-conferring siderophore mycobactin. Chem Biol. 1998;5:631–645. doi: 10.1016/s1074-5521(98)90291-5. [DOI] [PubMed] [Google Scholar]
- 46.Rusnak F, Sakaitani M, Drueckhammer D, Reichert J, Walsh CT. Biosynthesis of the Escherichia coli siderophore enterobactin: sequence of the entF gene, expression and purification of EntF, and analysis of covalent phosphopantetheine. Biochemistry. 1991;30:2916–2927. doi: 10.1021/bi00225a027. [DOI] [PubMed] [Google Scholar]
- 47.Berg M, Hilibi H, Dimroth P. The acyl carrier protein of malonate decarboxylase of Malonomas rubra contains 2′-(5″-phosphoribosyl)-3′-dephosphocoenzyme A as a prosthetic group. Biochemistry. 1996;35:4689–4696. doi: 10.1021/bi952873p. [DOI] [PubMed] [Google Scholar]
- 48.Bott M, Dimroth P. Klebisella pneumoniae genes for citrate lyase and citrate lyase ligase: localization, sequening, and expression. Mol Microbiol. 1994;14:347–356. doi: 10.1111/j.1365-2958.1994.tb01295.x. [DOI] [PubMed] [Google Scholar]
- 49.Debabov DV, Heaton MP, Zhang Q, Stewart K, Lambalot RH, Neuhaus FC. The D-alanyl carrier protein in Lactobacillus casei: cloning, sequencing, and expression of dltC. J Bacteriol. 1996;178:3869–3876. doi: 10.1128/jb.178.13.3869-3876.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koglin A, Mofid MR, Lohr F, Schafer B, Rogov VV, Blum M-M, Mittag T, Marahiel MA, Bernhard F, Dotsch V. Conformational switches modulate protein interactions in peptide antibiotic synthetases. Science. 2006;312:273. doi: 10.1126/science.1122928. [DOI] [PubMed] [Google Scholar]
- 51.Eyal E, Yang L-W, Bahar I. Anisotropic network model: systematic evaluation and a new web interface. Bioinformatics. 2006;22:2619–2627. doi: 10.1093/bioinformatics/btl448. Available at: http://ignmtest.ccbb.pitt.edu/cgi-bin/anm/anm1.cgi. [DOI] [PubMed] [Google Scholar]
- 52.Colizzi F, Recanatini M, Cavalli A. Mechanical features of Plasmodium falciparum acyl carrier protein in the delivery of substrates. J Chem Inf Model. 2008;48:2289–2293. doi: 10.1021/ci800297v. [DOI] [PubMed] [Google Scholar]
- 53.UniProt Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. Available at: http://www.uniprot.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. Available at: http://blast.ncbi.nlm.nih.gov/Blast.cgi/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.National Center for Biotechnology Information (NCBI) The nr Database. Available at ftp.ncbi.nih.gov. Last accessed: October, 2011.
- 56.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. Available at: http://www. drive5.com/muscle/index.htm/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. Available at: http://www.mega software.net. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Desper R, Gascuel O. Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. J Comp Biol. 2002;9:687–705. doi: 10.1089/106652702761034136. [DOI] [PubMed] [Google Scholar]
- 59.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 60.Shatsky M, Nussinov R, Wolfson HJ. A method for simultaneous alignment of multiple protein structures. Proteins. 2004;56:143–156. doi: 10.1002/prot.10628. Available at: http://bioin fo3d.cs.tau.ac.il/MultiProt/ [DOI] [PubMed] [Google Scholar]
- 61.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69:108–117. doi: 10.1002/prot.21702. Available at: http://zhanglab.ccmb.med.umich.edu/I-TASSER/ [DOI] [PubMed] [Google Scholar]
- 63.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. Available at: http://zhanglab.ccmb.med.umich.edu/I-TASSER/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 65.Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvary I, Wong KF, Paesani F, Vanicek J, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, YE X, Wang J, Hseih M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Sagui C, Babin V, Luchko T, Gusarov S, Kovalchenko A, Kollman PA. 2010. AMBER11. University of California, San Francisco. Available at: http://ambermd.org/
- 66.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. Available at: http://iupred.enzim.hu/ [DOI] [PubMed] [Google Scholar]
- 67.Wu S, Zhang Y. LOMETS: a local meta-threading-server for protein structure predictions. Nucl Acids Res. 2007;35:3375–3382. doi: 10.1093/nar/gkm251. Available at: http://zhanglab.ccmb. med.umich.edu/LOMETS/ [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


