Abstract
Alcohol and nicotine are often co-used and data from human and animals studies have demonstrated that common genes underlie responses to these two drugs. Recently, the genes that code for the subunits of the nicotinic acetylcholine receptors have been implicated as a common genetic mediator for alcohol and nicotine responses. The mammalian genes that code for the α6 and β3 subunits of the nicotinic acetylcholine receptor (Chrna6 and Chrnb3, respectively) are located adjacent to each other on human and mouse chromosome 8. These subunits have gained attention as potential regulators of drug behaviors because of their expression in the striatum where they have been shown to modulate dopamine release. Human genetic studies have shown that variation in these genes is associated with alcohol phenotypes. In the current experiments, mice lacking the Chrna6 or Chrnb3 gene were tested for three ethanol behaviors: choice ethanol consumption, ataxia, and sedation. Wildtype (WT), heterozygous (HET), and knockout (KO) mice of each strain went through a standard 2-bottle choice drinking paradigm, the balance beam, and the Loss of Righting Reflex (LORR) paradigm. No genotypic effects on any of the 3 behavioral tasks were observed in Chrnb3 animals. While the Chrna6 gene did not significantly influence ethanol consumption (g/kg) or ataxia, mice lacking the α6 subunit took significantly longer to recover their righting reflex than WT animals. These data provide evidence that receptors containing this subunit modulate the sedative effects of ethanol. Further work examining other models of ethanol consumption and behavioral responses to ethanol is needed to fully characterize the role of these receptor subunits in modulating ethanol responses.
Keywords: Ethanol, Nicotinic Acetylcholine Receptors, Ataxia, Sedation, Consumption
Introduction
Alcohol and nicotine are widely used and frequently co-abused substances. Alcoholics are more likely to be dependent on nicotine, and tobacco dependence is correlated with increased severity of alcohol dependence (Daeppen et al., 2000; Falk et al., 2006; John et al., 2003; Marks et al., 1997). In addition, several behavioral genetics studies have provided strong evidence that shared genes contribute to this co-morbidity (Kendler et al., 2007; Madden and Heath, 2002; Swan et al., 1997; True et al., 1999). It is widely accepted that nicotine exerts its pharmacological effects through activation of nicotinic acetylcholine receptors and several studies have provided evidence these receptors may also be targets for alcohol (Bhutada et al., 2010a; Bhutada et al., 2010b; Blomqvist et al., 1996; Blomqvist et al., 1992; Ericson et al., 2000; Larsson et al., 2002; Le et al., 2000.
Nicotinic acetylcholine receptors are ligand gated ion channels formed of five subunits centered around a membrane spanning pore. In the central nervous system there are 2 varieties of these receptors: homomeric and hetermeric. The primary homomeric receptor is the α7 receptor. Heteromeric receptors are comprised of a combination of α and β subunits. Acetylcholine receptors are located on both pre- and post-synaptic terminals and when depolarized allow the influx of Na+ or Ca2+ ion (Arias et al., 2006; Jensen et al., 2005; McGehee, 1999; Romanelli and Gualtieri, 2003).
The α6 and β3 subunits of the nicotinic acetylcholine receptors have relatively limited expression in the brain, but they are both expressed in the ventral tegmental area and substantia nigra (Grady et al., 2007). Recently, the α6 and β3 nicotinic acetylcholine receptors subunits have been found to have elevated mRNA expression in the ventral tegmental area following an acute injection of ethanol (Hendrickson et al., 2010), suggesting that acetylcholine receptors containing these subunits may modulate the response to ethanol. Receptors containing the α6 and β3 subunits have also been implicated in dopamine release. In particular, five combinations of nicotinic acetylcholine receptors have been localized to mouse dopaminergic terminals in the striatum. Of these, three contain either the α6, β3 or both receptor subunits (α4α6β2β3, α6β2β3, α6β2) and have been shown to influence agonist stimulated dopamine release when measured with a synaptosome preparation (Grady et al., 2007). Moreover, blockade of these receptors have been shown to modulate elevations in dopamine levels observed in the nucleus accumbens following systemic administration of alcohol (Larsson et al., 2004). Thus these subunits form functional receptors in a prime location to alter the behavioral effects of drugs (including ethanol).
Due to the expression of these receptor subunits in the mesolimbic dopamine system, human genetic studies have focused on genes that encode the α6 (CHRNA6) and β3 (CHRNB3) subunits. The genes encoding these subunits are located contiguous to each other on human and mouse chromosome 8. Accumulating evidence from human genetics studies has provided strong support that variation in these receptor subunits influences nicotine behaviors (Ehringer et al., 2010; Greenbaum et al., 2006; Hoft et al., 2009b; Saccone et al., 2007; Thorgeirsson et al., 2010; Zeiger et al., 2008). This includes a meta-analysis of over 31,000 individuals in which data for number of cigarettes smoked per day was assessed. In this meta-analysis, rs6474412, a SNP located 5′ to the start of CHRNB3, was one of the top 3 variants associated with this phenotype (Thorgeirsson et al., 2010). Given that genetic correlations have been observed among alcohol and nicotine behaviors in both human and animal models (Bergstrom et al., 2003; de Fiebre et al., 1990; de Fiebre et al., 1987; de Fiebre et al., 1991; de Fiebre et al., 2002; Swan et al., 1997; True et al., 1999; Tsuang et al., 2001), the genes encoding the α6 and β3 subunits have also been examined for associations with alcohol behaviors. In a recent study, three SNPs in CHRNA6 and one SNP in CHRNB3 were associated with alcohol consumption in a nationally representative sample (Hoft et al., 2009a). Furthermore, a CHRNA6 haplotype has been associated with heavy alcohol use in an independent sample (Landgren et al., 2009). These data provide evidence that genes that code for the α6 and β3 nicotinic acetylcholine receptors subunits may modulate alcohol behaviors.
To date, relatively little is known regarding the involvement of the α6 or β3 subunit in modulating ethanol behaviors in animal models. To our knowledge only one study has examined the expression of the Chrna6 gene (but not Chrnb3 gene) in lines of mice that were selectively bred for high (FAST) and low (SLOW) ethanol-induced locomotor stimulation (Crabbe et al., 1999; Phillips et al., 1991; Phillips et al., 2002; Shen et al., 1995). The FAST and SLOW mice have been bred for this phenotype for over 35 generations, and these animals display a markedly different response to ethanol within the first 5 minutes after administration, thus naïve mice were examined. Mice that show minimal locomotor stimulation (SLOW) following an acute injection of ethanol had greater expression of Chrna6 mRNA compared to mice that exhibited a robust stimulant response (FAST; Kamens and Phillips, 2008). Together with the aforementioned data that an acute injection of ethanol has been shown to increase expression of the Chrna6 and Chrnb3 genes in activated ventral tegmental neurons (Hendrickson et al., 2010), these data provide evidence that these receptor subunits may be important for modulating ethanol responses.
To determine more fully the role of Chrna6 and Chrnb3 in ethanol behaviors we tested mice lacking these receptor subunits for a range of ethanol behaviors. Mice were tested for voluntary ethanol consumption using a standard two-bottle choice paradigm, ethanol-induced ataxia using the balance beam, and ethanol-induced sedation using the Loss of Righting Reflex (LORR) paradigm. Finally, mice were tested for ethanol metabolism to rule out this as a confounding factor to any behavioral differences observed. We hypothesized that mice lacking the Chrna6 and Chrnb3 gene would display altered responses to ethanol because of their prominent location on dopaminergic nerve terminals.
Materials and Methods
Animals
All mice tested in these experiments were produced by breeder pairs at the Institute for Behavioral Genetics animal facility. Mice deficient in either Chrna6 (Champtiaux et al., 2002) or Chrnb3 (Cui et al., 2003) were previously produced using homologous recombination technology. Briefly, Chrna6 were developed by the deletion of exons 1 and 2 of the Chrna6 gene in embryonic cells from the 129Sv/Pas strain of mice and bred onto a CD-1 background (Champtiaux et al., 2002). Null mutant Chrnb3 mice were created by the interruption of exon 5 of the Chrnb3 gene in 129Svj embryonic stem cells and were injected into a C57BL/J blastocysts (Cui et al., 2003). Null mutant mice used in these experiments had been backcrossed to the C57BL/6 strain for at least 10 generations prior to testing. Male and female wild-type (WT), heterozygous (HET), and knockout (KO) animals were produced by HET breeder pairs to allow for the testing of littermates in this study. Mice were housed 2-4 per cage in standard mouse cages with ad libitum water and rodent chow. The lighting in the animal colony was maintained on a 12-hour light/dark cycle with lights on at 0700 hours. All testing was approved by the University of Colorado's Institutional Animal Care and Use Committee.
Drugs
Ethyl alcohol (200 proof; Pharmco, Brookfield, CT, USA) was the ethanol source for all experiments. For drinking solutions, it was diluted in tap water to the appropriate concentration while for injections it was diluted in physiological saline (0.9% NaCl; 20% v/v). Injection volumes were adjusted for the individual body weight of the animal to achieve the desired dose. All injections were given intraperitoneal.
Two-bottle choice ethanol consumption
Ethanol consumption was measured in a standard 2-bottle free choice paradigm (Kamens et al., 2010a; Kamens et al., 2006). Eighty-three naïve Chrna6 and 55 Chrnb3 mice were tested. Mice were singly housed in a standard mouse cage and presented with two 25-ml graduated cylinders fitted with drinking spouts. Mice were initially given 2 water bottles to allow time to habituate (2 days) to the test environment. On Day 1 of the experiment, a tube containing 3% ethanol replaced one of the water tubes. Every 2 days the side of the cage the ethanol was presented on was switched. Every 4th day the concentration of ethanol changed (3, 7, 10 and 20%). Two empty cages located on the same rack as the test animals provided a measure of evaporation/leakage which was used to correct individual drinking values. For each ethanol concentration, the average consumption on days 2 and 4 was used for the analysis because these data represent stable consumption (Phillips et al., 1994). For each concentration three dependent variables were obtained: ethanol consumption (g/kg), ethanol preference (ml of ethanol/total ml fluid) and total volume consumed (ml).
Balance Beam
To test for effects on ethanol-induced ataxia we used the balance beam, based on a published procedure (Crabbe et al., 2003; Kamens et al., 2010b; Linsenbardt et al., 2009). Briefly, 51 naïve Chrna6 and 57 naive Chrnb3 mice were used. The balance beam consisted of a white, 104.1 cm long by 1.9 cm wide, PVC board. It was elevated 54.6 cm above the floor by supports located on each end of the beam. Mice were trained to run the length of the beam prior to testing. To complete the training, each mouse had to traverse the beam twice, which all mice did easily. The mouse was then made to cross the beam a third time and the number of foot slips the mouse made was counted as the baseline ataxia measurement. At least 1 hr later each animal was given an injection of ethanol (1.5 g/kg, a dose based on prior literature (Kamens et al., 2010b; Linsenbardt et al., 2009)) and was placed into a holding cage. Ten min after the ethanol injection, the mouse was placed back on the balance beam and the number of hind foot missteps was counted by an experimenter unaware of the animal's genotype. During the training and test sessions, if an animal stopped crossing the balance beam, its tail was gently pressed to encourage movement. On the occasion that an animal fell from the balance beam, it was replaced on the beam at the location from which it fell and allowed to finish crossing the beam (Crabbe et al., 2003).
LORR
The sedative-hypnotic effects of ethanol were measured using LORR, based on standard published methods (Crabbe et al., 2006; McClearn and Kitahama, 1981). Sixty experimentally naïve Chrna6 and 51 Chrnb3 mice were tested. Each mouse was challenged with ethanol (4.1 g/kg), and placed into a holding cage until it appeared intoxicated. It was then placed on its back in a plexiglass V-shaped trough. If the mouse remained on its back for at least 30 seconds it was determined to have lost its righting reflex. Nine (three Chrna6 and six Chrnb3) mice did not reach this criteria within 3 minutes and these animals were excluded from further testing because they likely received a misplaced injection (Ponomarev and Crabbe, 2002). Mice were observed until they righted themselves, defined as turning over onto all four paws. The mouse was returned to its back and a mouse was deemed to have regained its righting reflex when it was able to right itself 3 times in 1 minute. Once the animal was deemed to have regained its righting reflex, a 10-μl blood sample was taken from the retro-orbital sinus for measurement of blood ethanol concentrations (BEC) using a modified enzymatic assay (Ehringer et al., 2009; Smolen et al., 1986).
Ethanol Metabolism
To determine if either the Chrna6 or Chrnb3 gene influenced ethanol metabolism, a standard ethanol metabolism protocol was used (Ehringer et al., 2009). Forty-eight Chrna6 and 54 Chrnb3 mice were tested. Briefly, mice were given a single challenge injection of 3 g/kg ethanol and placed into individual holding cages. At 10, 30, 60, 120 and 180 min after the injection a 10-μl blood sample was taken from the retro-orbital sinus. BEC were measured as described previously (Ehringer et al., 2009; Smolen et al., 1986).
Statistical Analysis
Primary dependent variables examined were ethanol consumption, ethanol preference, total volume consumed, footslips, duration of LORR, and BEC. Data from the ethanol consumption and metabolism studies were analyzed using repeated measures analysis of variance (ANOVA). Data from the balance beam and LORR studies were analyzed with a factorial ANOVA. Significant interactions were subsequently analyzed with ANOVA that included fewer factors and Tukey's HSD for post hoc comparisons. Sex, genotype, ethanol concentration and time were possible independent factors. α < 0.05 was considered significant.
Results
Ethanol Consumption
Preference ratios were as high as 80%, reflective of the high-drinking C57BL/J background strain for each knock-out. No differences in ethanol consumption were observed in either Chrna6 or Chrnb3 mice (Fig 1 and 2). When Chrna6 mice were analyzed for their choice ethanol consumption there was a significant main effect of sex and a significant sex X concentration interaction, thus males and females were analyzed independently. A significant main effect of concentration (F3,120 =72.1, p<0.001) on ethanol consumption was detected when female Chrna6 mice were examined, but no other significant main effects or interactions were observed. Mice drank significantly more ethanol when increasing concentrations of ethanol were available (all p-values < 0.01; Fig 1a). Preference for the ethanol containing solution also increased as the concentration of ethanol increased as evident by a significant main effect of concentration (F3, 120=59.5, p<0.001), but no other factors significantly altered ethanol preference. Ethanol preference followed an inverted U-shaped pattern. Preference increased between 3 and 7% ethanol, leveled off between 7 and 10% ethanol, and decreased at 20% ethanol (Fig 1c). There was a significant difference in ethanol preference between all concentrations of ethanol (all p-values < 0.001) except between 7 and 10% ethanol. When the total fluid consumed among female Chrna6 animals was examined there were no significant main effects or interactions (Fig 1e).
Fig. 1.
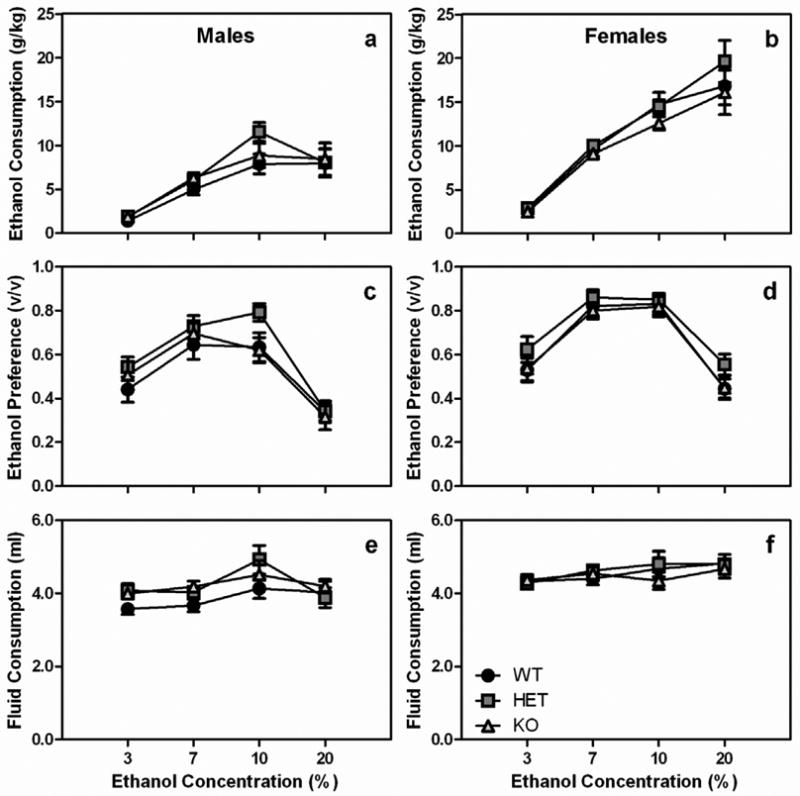
The Chrna6 gene does not modulate ethanol consumption in male or female mice. Data (mean ± SEM) represent ethanol consumption of (a) male and (b) female Chrna6 mice, ethanol preference of (c) male and (d) female Chrna6 mice, and total fluid consumption of (e) male and (f) female Chrna6 mice.
Fig. 2.
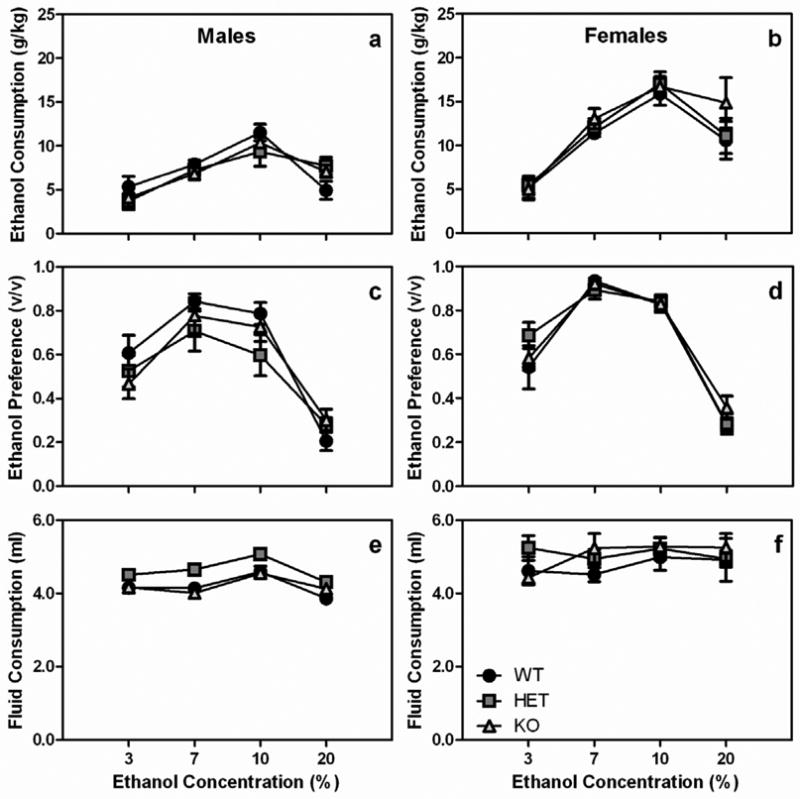
The Chrnb3 gene does not modulate ethanol consumption in male or female mice. Data (mean ± SEM) represent ethanol consumption of (a) male and (b) female Chrnb3 mice, ethanol preference of (c) male and (d) female Chrnb3 mice, and total fluid consumption of (e) male and (f) female Chrnb3 mice.
When male Chrna6 mice were examined for g/kg ethanol consumption, similar to female Chrna6 mice, there was a significant main effect of concentration (F3, 111=34.4, p<0.001), but no other significant main effects or interactions. Ethanol consumption increased with increasing concentrations of ethanol up to 10% ethanol, but leveled off at 20% ethanol (Fig 1b). Pairwise, there were differences in ethanol consumption between all concentrations of ethanol (all p-values < 0.05) except 10 and 20% ethanol. When preference was examined, there was a significant main effect of concentration (F3, 111=49.2, p<0.001), but no other significant main effects or interactions. Preference for ethanol increased between 3 and 7% ethanol (p<0.01), leveled off between 7 and 10% ethanol, then decreased at 20% ethanol (p<0.01; Fig 1d). Similar to ethanol consumption and preference, there was a main effect of concentration on total fluid consumption (F3, 111=5.2, p<0.01). Although the difference was small, male Chrna6 mice drank significantly more fluid when 10% ethanol was available compared to when 3 or 7% ethanol was available (all p-values <0.05; Fig 1f).
Similar to results with Chrna6 mice, when data from Chrnb3 mice were examined, there were no genotypic effects on ethanol consumption (Fig 2). Data were initially analyzed with a 3-way ANOVA with sex, genotype and concentration as factors. Due to a significant main effect of sex (F1, 49=48.2, p<0.001) and sex X concentration interaction (F3, 147=5.6, p<0.01) on ethanol consumption, data were further analyzed within each sex independently. When female Chrnb3 mice were analyzed for their ethanol consumption, there was a significant main effect of concentration (F3, 75=28.8, p<0.001). Ethanol consumption was significantly different between all concentrations of ethanol except between 7 and 20% ethanol (all p-values < 0.01; Fig 2a). In general, ethanol consumption increased as the concentration of ethanol increased up to 20% ethanol at which it started to decrease. When female Chrnb3 mice were examined for ethanol preference, there was a significant main effect of concentration (F3, 75=125.8, p<0.001) but no other significant effects. Preference for ethanol exhibited an inverted U-shaped pattern. Preference increased between 3 and 7% ethanol (p<0.01), was similar between 7 and 10%, and decreased at 20% ethanol (p<0.01; Fig 2c). No significant effects were observed when total fluid consumption was analyzed (Fig 2e).
Male Chrnb3 mice showed a similar pattern of results as female Chrnb3 mice. There was a significant main effect of ethanol concentration on ethanol consumption (F3, 72=25.1, p<0.001), ethanol preference (F3, 72=62.6, p<0.001), and total fluid consumption (F3, 72=5.3, p<0.01), but no other significant effects were observed. Both ethanol consumption and ethanol preference exhibited an inverted-U dose response function. For intake, ethanol consumption increased with increasing concentrations of ethanol up to 10% ethanol then decreased at 20% ethanol (Fig 2b). Pairwise, there were significant differences in ethanol intake at all concentrations except between 7 and 20% ethanol (all p-values <0.05). A similar pattern was observed for ethanol preference. There were significant differences in ethanol preference between all concentrations of ethanol except between 7 and 10% ethanol (all p-values <0.05; Fig 2d). There was a very small, but statistically significant, difference in total fluid consumption between 10 and 20% ethanol concentrations. Total fluid consumption was lower when 20% ethanol was available compared to when the 10% ethanol concentration was available (Fig 2f).
Balance Beam
Neither the Chrna6 nor Chrnb3 gene influenced ethanol-induced ataxia as measured on the balance beam (Fig 3). There were no statistically significant effects on baseline footslips when either Chrna6 or Chrnb3 mice were tested (data not shown). Thus to simplify the data we corrected ethanol footslips by the number of baseline footslips each animal made. This difference score was subjected to a 2-way ANOVA with sex and genotype as independent variables. There were no significant main effects or interactions when either the Chrna6 or Chrnb3 strains were examined.
Fig. 3.
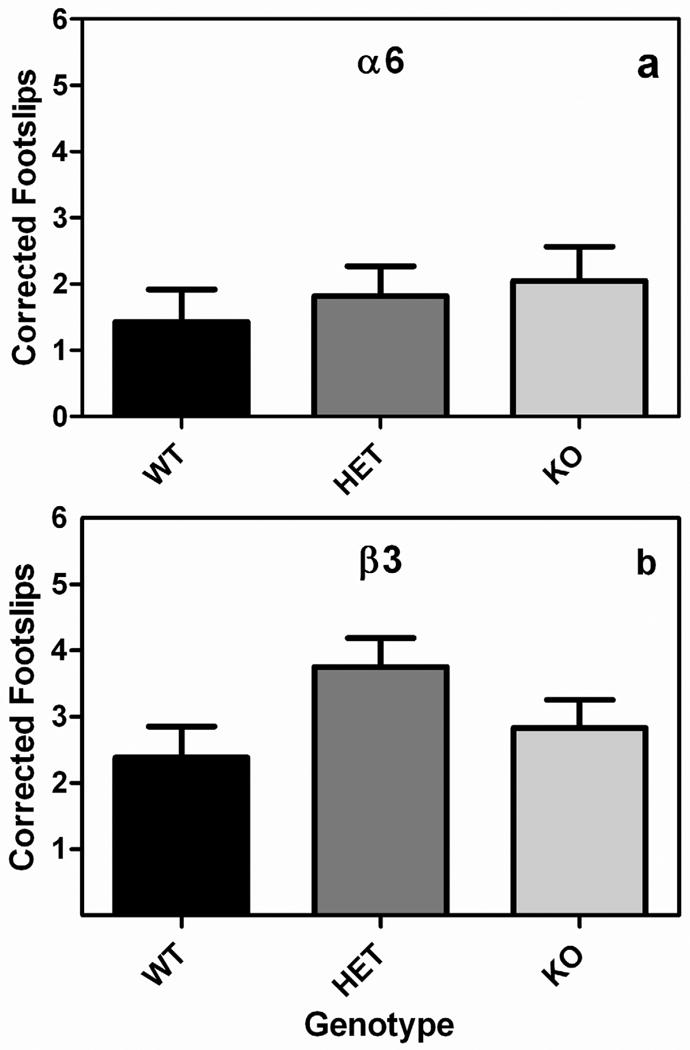
Neither the Chrna6 nor Chrnb3 gene influences ethanol-induced ataxia. Data (mean ± SEM) represent ethanol-induced ataxia when corrected for baseline ataxia in (a) Chrna6 and (b) Chrnb3 mice.
LORR
The Chrna6, but not Chrnb3, gene influenced the sedative-hypnotic effects of ethanol (Fig 4). LORR data were analyzed with a 2-way ANOVA with sex and genotype as independent factors in the Chrna6 and Chrnb3 strains. When Chrna6 mice were examined there was a significant main effect of genotype (F2, 51=3.6, p<0.05), but no other significant main effects or interactions. Mice lacking the Chrna6 gene were unable to right themselves on average 10 minutes longer than WT animals. When mice regained their righting reflex there were no significant differences in BEC (Chrna6 WT 361 ± 11, Chrna6 HET 346 ± 11, Chrna6 KO 331 ± 12). In contrast to the Chrna6 animals, there were no significant main effects or interactions when the Chrnb3 mice were examined. Nor were there any significant differences in the BEC when the animals recovered (Chrnb3 WT 366 ± 7, Chrnb3 HET 368 ± 7, Chrnb3 KO 349 ± 6).
Fig. 4.
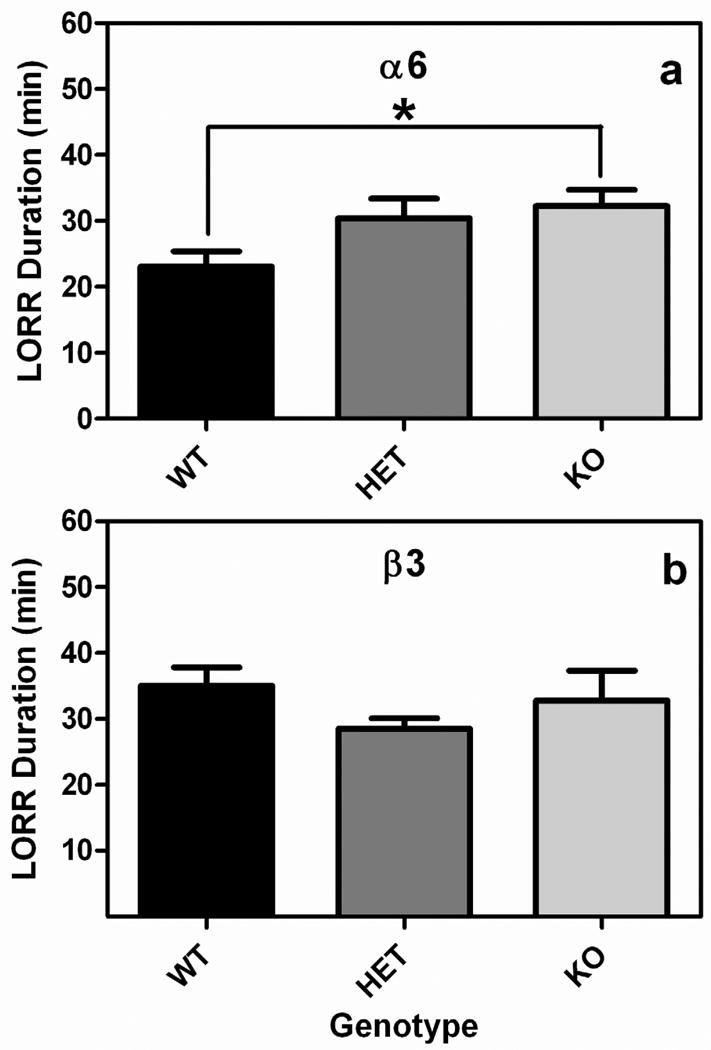
The sedative-hypnotic effects of alcohol are modulated by the α6, but not β3, subunit of the nicotinic acetylcholine receptor. Data (mean ± SEM) represent ethanol-induced LORR in (a) Chrna6 and (b) Chrnb3 mice. *; p < 0.05
Metabolism
Ethanol metabolism was not affected by either the Chrna6 or Chrnb3 genes (Fig 5). When BEC was examined over a 3 hour period following an acute injection of 3 g/kg ethanol, in both the Chrna6 and Chrnb3 strains, there was a significant main effect of time (F4, 168=279.5, p<0.001, F4, 192=273.1, p<0.001, respectively), but no other significant main effects or interactions. These data suggest that BEC decreases over time following an acute injection of ethanol, but that the genotype of the animal does not influence the rate of metabolism.
Fig. 5.
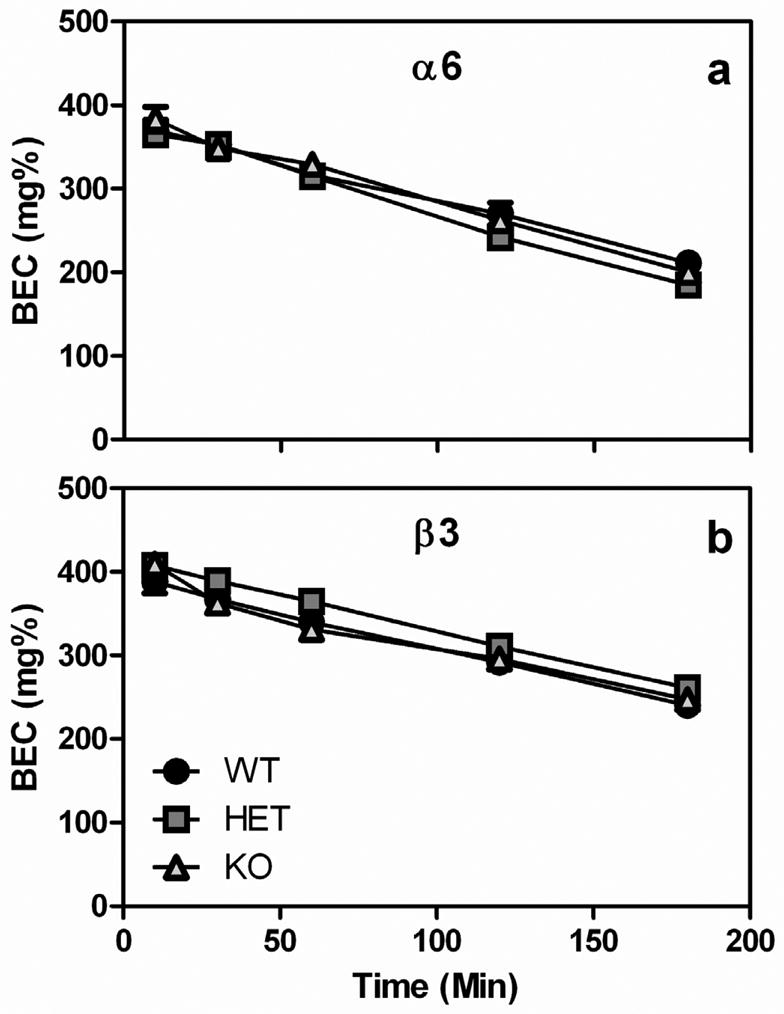
Metabolism of an acute injection of ethanol (3 g/kg) is not influence by either the Chrna6 or Chrnb3 gene. Data (mean ± SEM) represent blood ethanol concentrations (BEC) in (a) Chrna6 and (b) Chrnb3 mice.
Discussion
We set out to determine if nicotinic acetylcholine receptors, particularly those containing a α6 or β3 subunit, were involved in ethanol behaviors in mice. We examined 3 different behavioral responses: 2-bottle choice ethanol consumption, ataxia as measured on the balance beam, and the sedative-hypnotic effects of ethanol. The α6 subunit had a significant effect on the sedative effects of ethanol. Mice that lacked the α6 subunit took significantly longer to regain their righting reflex than did WT animals. This behavioral difference could not be explained by a difference in ethanol metabolism between the genotypes. Although the α6 subunit modulated the sedative effects of ethanol it did not alter any other behavior measured. Similarly, the β3 subunit did not alter any behavioral response tested, nor did it influence ethanol metabolism.
Our inability to detect a significant effect of the Chrna6 or Chrnb3 gene on ethanol consumption suggests that nicotinic acetylcholine receptors containing these subunits may not be involved in this effect of ethanol. Studies in rats and mice have implicated these subunits in the neurochemical response to ethanol. For example, in mice α-contoxin-MII, but not the α-contoxin-PIA-analogue, has been shown to attenuate the elevations in dopamine observed in the nucleus accumbens following peripheral administration of ethanol (Jerlhag et al., 2006; Larsson et al., 2004) implicating α3β2 or β3 containing receptors in this response. Moreover in rats α-contoxin-MII has been shown to decrease ethanol intake in a limited access paradigm (Larsson et al., 2004) as well as modulate the conditioned reinforcing properties of ethanol-associated cues (Lof et al., 2007). Interestingly, in a limited access ethanol consumption paradigm in adult male C57BL/6 mice α-contoxin-MII decreased ethanol consumption compared to baseline conditions, but did not significantly reduce consumption compared to the control group that received the vehicle injection (Larsson et al., 2004). Thus, more work will be needed to fully examine this system. For example, it is possible that these receptors modulate ethanol consumption in rats, but not in mice.
Given that mecamylamine, a non-specific nicotinic acetylcholine receptor antagonist, has been shown to alter ethanol consumption in rats and mice (Blomqvist et al., 1996; Ericson et al., 2000; Farook et al., 2009; Hendrickson et al., 2009; Le et al., 2000), it is likely that nicotinic receptors modulate this behavior in both species. However, the current study suggests receptor subtypes containing subunits other than α6 and β3 are likely to be involved in mediating effects of mecamylamine on ethanol consumption in mice. The α7 subunit has been shown to modulate ethanol intake in a 2-bottle choice paradigm similar to the one used in the current study (Kamens et al., 2010a). Moreover, mice that overexpress the α5, α3, and β4 subunits of the nicotinic acetylcholine receptor have reduced ethanol consumption in a 24-hour ethanol preference paradigm similar to the one reported here (Gallego et al., In press). This finding is consistent with work done in rats that showing that a partial agonist of α3β4 receptors reduces ethanol intake (Chatterjee et al., 2011). The α4 nicotinic acetylcholine receptor subunit has been implicated in alcohol drinking using the Drinking in the Dark model (Hendrickson et al., 2010), although these results were obtained after a saline injection and thus should be interpreted with caution. The acetylcholine receptors that influence alcohol consumption in the Drinking in the Dark model may be different than those in the 24-hour choice paradigm. The current data do not rule out the possibility that acetylcholine receptors containing the α6 and β3 subunits may modulate ethanol consumption when measured with the binge-like Drinking in the Dark model or through intravenous self-administration. Similar to how there may be species differences, there may be differences in acetylcholine receptors that modulate ethanol consumption in different behavioral paradigms.
Female mice consumed more ethanol than male mice when both the α6 and β3 null mutant mice were examined. Sex differences in ethanol consumption are widely reported in the literature, with female mice consistently consuming more ethanol than male mice (e.g., Finn et al., 2004; Kamens et al., 2010a). Important for the interpretation of the current findings, sex did not interact with genotype in either study. Thus while female mice consume more ethanol than male mice, the α6 and β3 nicotinic acetylcholine receptor subunits do not modulate this response.
We were unable to detect a significant effect of either Chrna6 or Chrnb3 on ethanol-induced ataxia when measured by the balance beam test. Nicotinic acetylcholine receptors have been implicated in ethanol's ataxic effects in two recent studies. In the first, injection of an α4β2 selective agonist, RJR-2403, directly into the cerebellum decreased ethanol-induced ataxia (Taslim et al., 2008). Furthermore, varenicline, an α4β2 partial agonist, increased ethanol-induced ataxia measured both by the balance beam and dowel test (Kamens et al., 2010b). These data support the role of α4β2 nicotinic acetylcholine receptors in this effect of ethanol, and when combined with the current data do not support a role of nicotinic receptors containing either an α6 or β3 subunit. Although we found no evidence for a role of Chrna6 or Chrnb3 in ethanol-induced ataxia it is important to note that we only used one behavioral measure of this trait. There are many different measures of ethanol-induced ataxia and it is known that a no single measure fully captures all aspects of this complex behavior (Crabbe et al., 2005). It is possible we may have observed different results if we had chosen a different task to measure ataxia.
We were able to detect a significant effect of Chrna6 on ethanol-induced LORR. In contrast Chrnb3 had no effect. The difference in duration of LORR in Chrna6 animals was independent of a difference in ethanol metabolism, because no significant differences were observed between the Chrna6 genotypes when ethanol metabolism was studied over a 3 hour time course. We saw no differences in BECs at the time the animals regained their righting reflex, even though we observed a significant behavioral difference. This is likely due to the relatively little difference in behavioral response (10 min) not allowing us to observe statistically significant differences in BECs. The α7 nicotinic acetylcholine receptor subunit has also been implicated in this ethanol behavior. Similar to the results obtained in the α6 KO mice, mice lacking the α7 subunit took longer to right themselves in the LORR paradigm compared to WT animals (Bowers et al., 2005). These data suggest that at least 2 different subtypes of nicotinic acetylcholine receptors modulate this behavioral trait, since α6 and α7 subunits are found in separate receptor subtypes (Gotti et al., 2006).
While we found no evidence for a role of Chrnb3 in ethanol behaviors, this is limited to the behaviors we examined. Mecamylamine has been shown to decrease ethanol-stimulated activity in a variety of mouse strains (Blomqvist et al., 1992; Kamens et al., 2009; Kamens and Phillips, 2008; Larsson et al., 2002). A number of groups have used pharmacologic approaches and genetic animal models to try to elucidate which acetylcholine receptor subtypes are modulating this effect. The cholinergic antagonists, dihydro-β-erythroidine (α4β2-specific) and methyllycaconitine (α7-specific) were found to have no effect on ethanol-induced stimulation (Larsson et al., 2002). Additional experiments with specific conotoxins provided further elucidation of which receptor subtypes were involved. α-conotoxin MII (α3β2-, β3-, and α6-specific), but not α-conotoxin PIA-analogue (α6-specific), attenuated ethanol-induced locomotor stimulation (Jerlhag et al., 2006; Larsson et al., 2004). These data provide evidence that α3β2- or β3-containing nicotinic acetylcholine receptors are involved in this response. α3 KO mice demonstrate an altered ethanol stimulant response (Kamens et al., 2009), providing evidence that receptors containing this subunit may modulate this behavior. However, it is possible that β3-containing receptors may also influence this response. Unfortunately, our β3 KO exists on a C57BL/6 background that shows a minimal stimulate response to ethanol (Demarest et al., 1999; Randall et al., 1975).
These results complement the human genetics studies, providing support for the role of CHRNA6 in alcohol behaviors. Consistent with what we observed in animal models, human genetic association studies have implicated CHRNA6 in alcohol behaviors with CHRNB3 being involved to a lesser extent (Hoft et al., 2009a; Landgren et al., 2009). Identifying the subunits and receptor subtypes involved in mediating specific alcohol and nicotine behaviors is an area of active research (recently reviewed by Tuesta et al., 2011). The data presented here contribute to this body of work and support the hypothesis that behavioral responses to alcohol and nicotine are likely to be differentially modulated by specific acetylcholine receptor subunits.
Acknowledgments
We would like to thank Michael Marks and Sharon Grady for helpful comments on the manuscript. These studies were performed with support from AA017889 (MAE), DA026901 (MAE), AA019447 (HMK), and DA015663 (Michael Marks).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arias HR, Bhumireddy P, Bouzat C. Molecular mechanisms and binding site locations for noncompetitive antagonists of nicotinic acetylcholine receptors. Int J Biochem Cell Biol. 2006;38:1254–1276. doi: 10.1016/j.biocel.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Bergstrom HC, Palmer AA, Wood RD, Burkhart-Kasch S, McKinnon CS, Phillips TJ. Reverse selection for differential response to the locomotor stimulant effects of ethanol provides evidence for pleiotropic genetic influence on locomotor response to other drugs of abuse. Alcohol Clin Exp Res. 2003;27:1535–1547. doi: 10.1097/01.ALC.0000091226.18969.B9. [DOI] [PubMed] [Google Scholar]
- Bhutada PS, Mundhada YR, Bansod KU, Dixit PV, Umathe SN, Mundhada DR. Inhibitory influence of mecamylamine on the development and the expression of ethanol-induced locomotor sensitization in mice. Pharmacol Biochem Behav. 2010a;96:266–273. doi: 10.1016/j.pbb.2010.05.015. [DOI] [PubMed] [Google Scholar]
- Bhutada PS, Mundhada YR, Bansod KU, Umathe SN, Kahale VP, Dixit PV, Mundhada DR. Inhibitory influence of mecamylamine on ethanol withdrawal-induced symptoms in C57BL/6J mice. Behav Pharmacol. 2010b;21:90–95. doi: 10.1097/FBP.0b013e328337be54. [DOI] [PubMed] [Google Scholar]
- Blomqvist O, Ericson M, Johnson DH, Engel JA, Soderpalm B. Voluntary ethanol intake in the rat: effects of nicotinic acetylcholine receptor blockade or subchronic nicotine treatment. Eur J Pharmacol. 1996;314:257–267. doi: 10.1016/s0014-2999(96)00583-3. [DOI] [PubMed] [Google Scholar]
- Blomqvist O, Soderpalm B, Engel JA. Ethanol-induced locomotor activity: involvement of central nicotinic acetylcholine receptors? Brain Res Bull. 1992;29:173–178. doi: 10.1016/0361-9230(92)90023-q. [DOI] [PubMed] [Google Scholar]
- Bowers BJ, McClure-Begley TD, Keller JJ, Paylor R, Collins AC, Wehner JM. Deletion of the alpha7 nicotinic receptor subunit gene results in increased sensitivity to several behavioral effects produced by alcohol. Alcohol Clin Exp Res. 2005;29:295–302. doi: 10.1097/01.alc.0000156116.40817.a2. [DOI] [PubMed] [Google Scholar]
- Champtiaux N, Han ZY, Bessis A, Rossi FM, Zoli M, Marubio L, McIntosh JM, Changeux JP. Distribution and pharmacology of alpha 6-containing nicotinic acetylcholine receptors analyzed with mutant mice. J Neurosci. 2002;22:1208–1217. doi: 10.1523/JNEUROSCI.22-04-01208.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Steensland P, Simms JA, Holgate J, Coe JW, Hurst RS, Shaffer CL, Lowe J, Rollema H, Bartlett SE. Partial agonists of the alpha3beta4* neuronal nicotinic acetylcholine receptor reduce ethanol consumption and seeking in rats. Neuropsychopharmacology. 2011;36:603–615. doi: 10.1038/npp.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Metten P, Cameron AJ, Wahlsten D. An analysis of the genetics of alcohol intoxication in inbred mice. Neurosci Biobehav Rev. 2005;28:785–802. doi: 10.1016/j.neubiorev.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Metten P, Ponomarev I, Prescott CA, Wahlsten D. Effects of genetic and procedural variation on measurement of alcohol sensitivity in mouse inbred strains. Behav Genet. 2006;36:536–552. doi: 10.1007/s10519-006-9067-6. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Metten P, Yu CH, Schlumbohm JP, Cameron AJ, Wahlsten D. Genotypic differences in ethanol sensitivity in two tests of motor incoordination. J Appl Physiol. 2003;95:1338–1351. doi: 10.1152/japplphysiol.00132.2003. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Wahlsten D, Dudek BC. Genetics of mouse behavior: interactions with laboratory environment. Science. 1999;284:1670–1672. doi: 10.1126/science.284.5420.1670. [DOI] [PubMed] [Google Scholar]
- Cui C, Booker TK, Allen RS, Grady SR, Whiteaker P, Marks MJ, Salminen O, Tritto T, Butt CM, Allen WR, et al. The beta3 nicotinic receptor subunit: a component of alpha-conotoxin MII-binding nicotinic acetylcholine receptors that modulate dopamine release and related behaviors. J Neurosci. 2003;23:11045–11053. doi: 10.1523/JNEUROSCI.23-35-11045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daeppen JB, Smith TL, Danko GP, Gordon L, Landi NA, Nurnberger JI, Jr, Bucholz KK, Raimo E, Schuckit MA. Clinical correlates of cigarette smoking and nicotine dependence in alcohol-dependent men and women. The Collaborative Study Group on the Genetics of Alcoholism Alcohol Alcohol. 2000;35:171–175. doi: 10.1093/alcalc/35.2.171. [DOI] [PubMed] [Google Scholar]
- de Fiebre CM, Marks MJ, Collins AC. Ethanol-nicotine interactions in long-sleep and short-sleep mice. Alcohol. 1990;7:249–257. doi: 10.1016/0741-8329(90)90014-4. [DOI] [PubMed] [Google Scholar]
- de Fiebre CM, Medhurst LJ, Collins AC. Nicotine response and nicotinic receptors in long-sleep and short-sleep mice. Alcohol. 1987;4:493–501. doi: 10.1016/0741-8329(87)90092-9. [DOI] [PubMed] [Google Scholar]
- de Fiebre CM, Romm E, Collins JT, Draski LJ, Deitrich RA, Collins AC. Responses to cholinergic agonists of rats selectively bred for differential sensitivity to ethanol. Alcohol Clin Exp Res. 1991;15:270–276. doi: 10.1111/j.1530-0277.1991.tb01868.x. [DOI] [PubMed] [Google Scholar]
- de Fiebre NC, Dawson R, Jr, de Fiebre CM. The selectively bred high alcohol sensitivity (HAS) and low alcohol sensitivity (LAS) rats differ in sensitivity to nicotine. Alcohol Clin Exp Res. 2002;26:765–772. [PubMed] [Google Scholar]
- Demarest K, McCaughran J, Jr, Mahjubi E, Cipp L, Hitzemann R. Identification of an acute ethanol response quantitative trait locus on mouse chromosome 2. J Neurosci. 1999;19:549–561. doi: 10.1523/JNEUROSCI.19-02-00549.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehringer MA, Hoft NR, Zunhammer M. Reduced alcohol consumption in mice with access to a running wheel. Alcohol. 2009;43:443–452. doi: 10.1016/j.alcohol.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Ehringer MA, McQueen MB, Hoft NR, Saccone NL, Stitzel JA, Wang JC, Bierut LJ. Association of CHRN genes with “dizziness” to tobacco. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:600–609. doi: 10.1002/ajmg.b.31027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson M, Engel JA, Soderpalm B. Peripheral involvement in nicotine-induced enhancement of ethanol intake. Alcohol. 2000;21:37–47. doi: 10.1016/s0741-8329(99)00099-3. [DOI] [PubMed] [Google Scholar]
- Falk DE, Yi HY, Hiller-Sturmhofel S. An epidemiologic analysis of co-occurring alcohol and tobacco use and disorders: findings from the National Epidemiologic Survey on Alcohol and Related Conditions. Alcohol Res Health. 2006;29:162–171. [PMC free article] [PubMed] [Google Scholar]
- Farook JM, Lewis B, Gaddis JG, Littleton JM, Barron S. Effects of mecamylamine on alcohol consumption and preference in male C57BL/6J mice. Pharmacology. 2009;83:379–384. doi: 10.1159/000219488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn DA, Sinnott RS, Ford MM, Long SL, Tanchuck MA, Phillips TJ. Sex differences in the effect of ethanol injection and consumption on brain allopregnanolone levels in C57BL/6 mice. Neuroscience. 2004;123:813–819. doi: 10.1016/j.neuroscience.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Gallego X, Ruiz J, Valverde O, Crabbe JC, Dierssen M. Transgenic over expression of nicotinic receptor alpha 5, alpha 3, and beta 4 subunit genes reduces ethanol intake in mice. Alcohol. doi: 10.1016/j.alcohol.2011.11.005. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotti C, Zoli M, Clementi F. Brain nicotinic acetylcholine receptors: native subtypes and their relevance. Trends Pharmacol Sci. 2006;27:482–491. doi: 10.1016/j.tips.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Grady SR, Salminen O, Laverty DC, Whiteaker P, McIntosh JM, Collins AC, Marks MJ. The subtypes of nicotinic acetylcholine receptors on dopaminergic terminals of mouse striatum. Biochem Pharmacol. 2007;74:1235–1246. doi: 10.1016/j.bcp.2007.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum L, Kanyas K, Karni O, Merbl Y, Olender T, Horowitz A, Yakir A, Lancet D, Ben-Asher E, Lerer B. Why do young women smoke? I Direct and interactive effects of environment, psychological characteristics and nicotinic cholinergic receptor genes. Mol Psychiatry. 2006;11:312–322. 223. doi: 10.1038/sj.mp.4001774. [DOI] [PubMed] [Google Scholar]
- Hendrickson LM, Zhao-Shea R, Pang X, Gardner PD, Tapper AR. Activation of alpha4* nAChRs is necessary and sufficient for varenicline-induced reduction of alcohol consumption. J Neurosci. 2010;30:10169–10176. doi: 10.1523/JNEUROSCI.2601-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson LM, Zhao-Shea R, Tapper AR. Modulation of ethanol drinking-in-the-dark by mecamylamine and nicotinic acetylcholine receptor agonists in C57BL/6J mice. Psychopharmacology (Berl) 2009;204:563–572. doi: 10.1007/s00213-009-1488-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoft NR, Corley RP, McQueen MB, Huizinga D, Menard S, Ehringer MA. SNPs in CHRNA6 and CHRNB3 are associated with alcohol consumption in a nationally representative sample. Genes Brain Behav. 2009a;8:631–637. doi: 10.1111/j.1601-183X.2009.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoft NR, Corley RP, McQueen MB, Schlaepfer IR, Huizinga D, Ehringer MA. Genetic association of the CHRNA6 and CHRNB3 genes with tobacco dependence in a nationally representative sample. Neuropsychopharmacology. 2009b;34:698–706. doi: 10.1038/npp.2008.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen AA, Frolund B, Liljefors T, Krogsgaard-Larsen P. Neuronal nicotinic acetylcholine receptors: structural revelations, target identifications, and therapeutic inspirations. J Med Chem. 2005;48:4705–4745. doi: 10.1021/jm040219e. [DOI] [PubMed] [Google Scholar]
- Jerlhag E, Grotli M, Luthman K, Svensson L, Engel JA. Role Of The Subunit Composition Of Central Nicotinic Acetylcholine Receptors For The Stimulatory And Dopamine-Enhancing Effects Of Ethanol. Alcohol Alcohol. 2006;41:486–493. doi: 10.1093/alcalc/agl049. [DOI] [PubMed] [Google Scholar]
- John U, Meyer C, Rumpf HJ, Schumann A, Thyrian JR, Hapke U. Strength of the relationship between tobacco smoking, nicotine dependence and the severity of alcohol dependence syndrome criteria in a population-based sample. Alcohol Alcohol. 2003;38:606–612. doi: 10.1093/alcalc/agg122. [DOI] [PubMed] [Google Scholar]
- Kamens HM, Andersen J, Picciotto MR. Modulation of ethanol consumption by genetic and pharmacological manipulation of nicotinic acetylcholine receptors in mice. Psychopharmacology (Berl) 2010a;208:613–626. doi: 10.1007/s00213-009-1759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamens HM, Andersen J, Picciotto MR. The nicotinic acetylcholine receptor partial agonist varenicline increases the ataxic and sedative-hypnotic effects of acute ethanol administration in C57BL/6J mice. Alcohol Clin Exp Res. 2010b;34:2053–2060. doi: 10.1111/j.1530-0277.2010.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamens HM, Burkhart-Kasch S, McKinnon CS, Li N, Reed C, Phillips TJ. Ethanol-related traits in mice selectively bred for differential sensitivity to methamphetamine-induced activation. Behav Neurosci. 2006;120:1356–1366. doi: 10.1037/0735-7044.120.6.1356. [DOI] [PubMed] [Google Scholar]
- Kamens HM, McKinnon CS, Li N, Helms ML, Belknap JK, Phillips TJ. The alpha 3 subunit gene of the nicotinic acetylcholine receptor is a candidate gene for ethanol stimulation. Genes Brain Behav. 2009;8:600–609. doi: 10.1111/j.1601-183X.2008.00444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamens HM, Phillips TJ. A role for neuronal nicotinic acetylcholine receptors in ethanol-induced stimulation, but not cocaine- or methamphetamine-induced stimulation. Psychopharmacology (Berl) 2008;196:377–387. doi: 10.1007/s00213-007-0969-7. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Myers J, Prescott CA. Specificity of genetic and environmental risk factors for symptoms of cannabis, cocaine, alcohol, caffeine, and nicotine dependence. Arch Gen Psychiatry. 2007;64:1313–1320. doi: 10.1001/archpsyc.64.11.1313. [DOI] [PubMed] [Google Scholar]
- Landgren S, Engel JA, Andersson ME, Gonzalez-Quintela A, Campos J, Nilsson S, Zetterberg H, Blennow K, Jerlhag E. Association of nAChR gene haplotypes with heavy alcohol use and body mass. Brain Res. 2009;1305(Suppl):S72–79. doi: 10.1016/j.brainres.2009.08.026. [DOI] [PubMed] [Google Scholar]
- Larsson A, Jerlhag E, Svensson L, Soderpalm B, Engel JA. Is an alpha-conotoxin MII-sensitive mechanism involved in the neurochemical, stimulatory, and rewarding effects of ethanol? Alcohol. 2004;34:239–250. doi: 10.1016/j.alcohol.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Larsson A, Svensson L, Soderpalm B, Engel JA. Role of different nicotinic acetylcholine receptors in mediating behavioral and neurochemical effects of ethanol in mice. Alcohol. 2002;28:157–167. doi: 10.1016/s0741-8329(02)00244-6. [DOI] [PubMed] [Google Scholar]
- Le AD, Corrigall WA, Harding JW, Juzytsch W, Li TK. Involvement of nicotinic receptors in alcohol self-administration. Alcohol Clin Exp Res. 2000;24:155–163. doi: 10.1111/j.1530-0277.2000.tb04585.x. [DOI] [PubMed] [Google Scholar]
- Linsenbardt DN, Moore EM, Gross CD, Goldfarb KJ, Blackman LC, Boehm SL., 2nd Sensitivity and tolerance to the hypnotic and ataxic effects of ethanol in adolescent and adult C57BL/6J and DBA/2J mice. Alcohol Clin Exp Res. 2009;33:464–476. doi: 10.1111/j.1530-0277.2008.00857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lof E, Olausson P, deBejczy A, Stomberg R, McIntosh JM, Taylor JR, Soderpalm B. Nicotinic acetylcholine receptors in the ventral tegmental area mediate the dopamine activating and reinforcing properties of ethanol cues. Psychopharmacology (Berl) 2007;195:333–343. doi: 10.1007/s00213-007-0899-4. [DOI] [PubMed] [Google Scholar]
- Madden PA, Heath AC. Shared genetic vulnerability in alcohol and cigarette use and dependence. Alcohol Clin Exp Res. 2002;26:1919–1921. doi: 10.1097/01.ALC.0000040960.15151.30. [DOI] [PubMed] [Google Scholar]
- Marks JL, Hill EM, Pomerleau CS, Mudd SA, Blow FC. Nicotine dependence and withdrawal in alcoholic and nonalcoholic ever-smokers. J Subst Abuse Treat. 1997;14:521–527. doi: 10.1016/s0740-5472(97)00049-4. [DOI] [PubMed] [Google Scholar]
- McClearn GE, Kakihana R. Selective breeding for ethanol sensitivity: short-sleep and long-sleep mice. In: McClearn GE, Deitrich RA, Erwin VG, editors. Development of Animal Models as Pharmacogenetic Tools. Rockville, MD: USDHHS/NIAAA; 1981. pp. 147–159. [Google Scholar]
- McGehee DS. Molecular diversity of neuronal nicotinic acetylcholine receptors. Ann N Y Acad Sci. 1999;868:565–577. doi: 10.1111/j.1749-6632.1999.tb11330.x. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Burkhart-Kasch S, Terdal ES, Crabbe JC. Response to selection for ethanol-induced locomotor activation: genetic analyses and selection response characterization. Psychopharmacology (Berl) 1991;103:557–566. doi: 10.1007/BF02244259. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Crabbe JC, Metten P, Belknap JK. Localization of genes affecting alcohol drinking in mice. Alcohol Clin Exp Res. 1994;18:931–941. doi: 10.1111/j.1530-0277.1994.tb00062.x. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Shen EH, McKinnon CS, Burkhart-Kasch S, Lessov CN, Palmer AA. Forward, relaxed, and reverse selection for reduced and enhanced sensitivity to ethanol's locomotor stimulant effects in mice. Alcohol Clin Exp Res. 2002;26:593–602. [PubMed] [Google Scholar]
- Ponomarev I, Crabbe JC. A novel method to assess initial sensitivity and acute functional tolerance to hypnotic effects of ethanol. J Pharmacol Exp Ther. 2002;302:257–263. doi: 10.1124/jpet.302.1.257. [DOI] [PubMed] [Google Scholar]
- Randall CL, Carpenter JA, Lester D, Friedman HJ. Ethanol-induced mouse strain differences in locomotor activity. Pharmacol Biochem Behav. 1975;3:533–535. doi: 10.1016/0091-3057(75)90069-6. [DOI] [PubMed] [Google Scholar]
- Romanelli MN, Gualtieri F. Cholinergic nicotinic receptors: competitive ligands, allosteric modulators, and their potential applications. Med Res Rev. 2003;23:393–426. doi: 10.1002/med.10037. [DOI] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau O, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen EH, Harland RD, Crabbe JC, Phillips TJ. Bidirectional selective breeding for ethanol effects on locomotor activity: characterization of FAST and SLOW mice through selection generation 35. Alcohol Clin Exp Res. 1995;19:1234–1245. doi: 10.1111/j.1530-0277.1995.tb01606.x. [DOI] [PubMed] [Google Scholar]
- Smolen A, Marks MJ, Smolen TN, Collins AC. Dose and route of administration alter the relative elimination of ethanol by long-sleep and short-sleep mice. Alcohol Clin Exp Res. 1986;10:198–204. doi: 10.1111/j.1530-0277.1986.tb05071.x. [DOI] [PubMed] [Google Scholar]
- Swan GE, Carmelli D, Cardon LR. Heavy consumption of cigarettes, alcohol and coffee in male twins. J Stud Alcohol. 1997;58:182–190. doi: 10.15288/jsa.1997.58.182. [DOI] [PubMed] [Google Scholar]
- Taslim N, Al-Rejaie S, Saeed Dar M. Attenuation of ethanol-induced ataxia by alpha(4)beta(2) nicotinic acetylcholine receptor subtype in mouse cerebellum: a functional interaction. Neuroscience. 2008;157:204–213. doi: 10.1016/j.neuroscience.2008.08.046. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True WR, Xian H, Scherrer JF, Madden PA, Bucholz KK, Heath AC, Eisen SA, Lyons MJ, Goldberg J, Tsuang M. Common genetic vulnerability for nicotine and alcohol dependence in men. Arch Gen Psychiatry. 1999;56:655–661. doi: 10.1001/archpsyc.56.7.655. [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Bar JL, Harley RM, Lyons MJ. The Harvard Twin Study of Substance Abuse: what we have learned. Harv Rev Psychiatry. 2001;9:267–279. [PubMed] [Google Scholar]
- Tuesta LM, Fowler CD, Kenny PJ. Recent advances in understanding nicotinic receptor signaling mechanisms that regulate drug self-administration behavior. Biochem Pharmacol. 2011;82:984–995. doi: 10.1016/j.bcp.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiger JS, Haberstick BC, Schlaepfer I, Collins AC, Corley RP, Crowley TJ, Hewitt JK, Hopfer CJ, Lessem J, McQueen MB, et al. The neuronal nicotinic receptor subunit genes (CHRNA6 and CHRNB3) are associated with subjective responses to tobacco. Hum Mol Genet. 2008;17:724–734. doi: 10.1093/hmg/ddm344. [DOI] [PubMed] [Google Scholar]


